Background: ZO-1 overexpression inhibits DbpA/ZONAB overactivation, suggesting that ZO-1 sequesters DbpA at junctions.
Results: Simultaneous depletion of ZO-1, ZO-2, and ZO-3 is required to decrease DbpA localization at junctions.
Conclusion: The junctional localization of DbpA is regulated redundantly by ZO proteins.
Significance: Clarifying how junctional transcription factors are regulated is essential to understand epithelial proliferation.
Keywords: Junction, Nucleus, Proliferation, Scaffold Protein, Transcription Factor
Abstract
The localization and activities of DbpA/ZONAB and YAP transcription factors are in part regulated by the density-dependent assembly of epithelial junctions. DbpA activity and cell proliferation are inhibited by exogenous overexpression of the tight junction (TJ) protein ZO-1, leading to a model whereby ZO-1 acts by sequestering DbpA at the TJ. However, mammary epithelial cells and mouse tissues knock-out for ZO-1 do not show increased proliferation, as predicted by this model. To address this discrepancy, we examined the localization and activity of DbpA and YAP in Madin-Darby canine kidney cells depleted either of ZO-1, or one of the related proteins ZO-2 and ZO-3 (ZO proteins), or all three together. Depletion of only one ZO protein had no effect on DbpA localization and activity, whereas depletion of ZO-1 and ZO-2, which is associated with reduced ZO-3 expression, resulted in increased DbpA localization in the cytoplasm. Only depletion of ZO-2 reduced the nuclear import of YAP. Mammary epithelial (Eph4) cells KO for ZO-1 showed junctional DbpA, demonstrating that ZO-1 is not required to sequester DbpA at junctions. However, further depletion of ZO-2 in Eph4 ZO-1KO cells, which do not express ZO-3, caused decreased junctional localization and expression of DbpA, which were rescued by the proteasome inhibitor MG132. In vitro binding assays showed that full-length ZO-1 does not interact with DbpA. These results show that ZO-2 is implicated in regulating the nuclear shuttling of YAP, whereas ZO proteins redundantly control the junctional retention and stability of DbpA, without affecting its shuttling to the nucleus.
Introduction
The apicolateral junctional complex of vertebrate epithelial cells comprises tight junctions (TJ)4 and adherens junctions, and plays fundamental roles in the establishment of tissue barriers and in the maintenance of tissue integrity. In addition, proteins of TJ and adherens junctions have been implicated in the regulation of signaling, gene expression, and cell proliferation, through several different mechanisms (reviewed in Refs. 1–4). For example, the adherens junctions proteins E-cadherin and α-catenin, and the TJ proteins angiomotin and ZO-2 were shown to modulate the nucleo-cytoplasmic shuttling of transcription factor YAP, a downstream target of the Hippo signaling pathway (5–10). The TJ proteins occludin, cingulin, JAM-A, and ZO-3 are implicated in the regulation of cell proliferation by modulating signaling pathways involving Raf, RhoA, Akt, and cyclin-D1, respectively (11–14).
ZO-1 (zonula occludens-1) (15), together with ZO-2 and ZO-3, forms the ZO family of TJ proteins. ZO proteins comprise three PDZ domains, which are involved in protein-protein interactions and scaffolding of membrane proteins, central SH3 and GUK domains, and a C-terminal domain, which is involved in anchoring to the actin cytoskeleton (16–19). ZO-1 overexpression counteracts the effects of overexpression of the transcription factor DbpA (DNA-binding Protein A, also called Mouse Y-Box protein 3, or ZONAB) on gene expression and cell proliferation, and DbpA localization shifts from nuclear to junctional when sparse cells become confluent (20, 21). In addition, fragments of ZO-1, comprising either the SH3, or the PDZ3 plus the SH3 domains, were reported to interact in vitro with DbpA (20). Based on these findings, a model was proposed, whereby ZO-1 functions as an inhibitor of cell proliferation, by directly interacting with and sequestering DbpA at junctions, in a cell density-dependent manner (20, 21). Since in confluent cells DbpA and ZO-1 are localized at junctions, this model predicts that when ZO-1 is depleted, there should be activation of DbpA, through its decreased junctional localization. However, whether the localization of DbpA in MDCK cells is affected by depletion of either ZO-1 or other ZO proteins has not been determined. In contrast, evidence from knock-out studies is at variance with the model proposed by Balda and Matter (20), because mammary epithelial cells and mouse tissues lacking ZO-1 do not show altered growth curves, or increased cell proliferation (16, 22, 23).
To address these discrepancies, and clarify the role of ZO proteins in the control of DbpA localization, DbpA-dependent gene expression, and cell proliferation, we examined different clonal MDCK lines depleted of ZO-1, ZO-2, or ZO-3, or ZO-1 and ZO-2, and clonal lines of Eph4 cells, either WT or KO for ZO-1. Our results show that (i) the junctional localization of DbpA is not affected by ZO-1 knockdown or knock-out, in MDCK and Eph4 cells, respectively; (ii) depletion and/or KO of all three ZO proteins is required to observe any effect on DbpA localization and expression; (iii) full-length ZO-1 does not interact with DbpA, and (iv) only ZO-2 regulates the nuclear shuttling of YAP.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used: ZO-1 (Zymed Laboratories Inc./Invitrogen, 61-7300 and 33-9100), occludin (Invitrogen, 71-1500), cingulin (Invitrogen, 36-440, rabbit-C532 (24)), DbpA (Invitrogen, 40-2800), YAP (25), HA (Invitrogen, 32-6700), β-tubulin (Zymed Laboratories Inc., 32-2600), ZO-2 (Zymed Laboratories Inc., 71-1400 and 37-4700), ZO-3 (Zymed Laboratories Inc., 36-4100), GEF-H1 (B4/7, Abcam), symplekin (Transduction Laboratories, 605-259-1550). Secondary antibodies for immunofluorescence and immunoblotting were from Jackson ImmunoResearch Laboratories and Zymed Laboratories Inc., respectively.
Cell Culture, Transfection, and siRNA
Wild-type MDCK-II (Madin-Darby Canine Kidney) cells, ZO-1, ZO-2, and ZO-1/ZO-2-depleted cells were previously described (19, 26, 27). To generate ZO-3-depleted MDCK clones, a short hairpin shRNA to target endogenous canine ZO-3 (target sequence: 5′-GCAGTCAGATCTTCATCAA-3′) was cloned into the BglII/HindIII sites of the pTER vector, and used to transfect MDCK tet-off cells, using Lipofectamine 2000 (Invitrogen). Cells were selected in medium containing 0.6 mg/ml of zeocin, and clones were isolated by cloning rings and cell sorting (12). To re-express endogenous ZO-3 (rescue cells), cells were incubated in medium containing 40 μg/ml of doxycycline, which activates the Tet-repressor and inhibits transcription of the shRNA. MDCK, HEK293T, and Eph4 WT (a kind gift of E. Reichmann, University of Zurich) and ZO-1 KO cells (a kind gift of S. Tsukita, Osaka University (22)) were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma) containing 10% fetal bovine serum (FBS), 1× minimal essential medium non-essential amino acids (Invitrogen). For immunofluorescence experiments on sparse versus confluent cells, cells were seeded in 24-well plates at a density of either 62,500 cells/cm2, and grown for 4 days (confluent), or at a density of 5,000 cells/cm2, and grown for 24 h (sparse). Caco-2 cells were cultured once in DMEM (Sigma) containing 20% FBS, 1× minimal essential medium non-essential amino acids, and penicillin-streptomycin. For transient depletion using siRNA, cells were transfected with Lipofectamine RNAiMAX (Invitrogen) 24 h after plating, according to the manufacturer's instructions. The following siRNA for ZO-1 were used for MDCK and Caco-2 cells, respectively: cZO-1, forward, 5′-CCTCTGGAATGCATCATGA, reverse, 5′-TCATGATGCATTCCAGAGG; hZO-1, forward, 5′-CTGATCAAGAACTAGATGA, reverse, 5′-TCATCTAGTTCTTGATCAG. siRNA negative control was obtained from Sigma (number SIC001). For mouse ZO-2 (Eph4 cells) we used: forward, 5′-CTCCTATCACGAAGCTTAT and reverse, 5′-ATAAGCTTCGTGATAGGAG. Cells were analyzed by Western blotting and immunofluorescence microscopy 48–72 h after transfection. BrdU cell proliferation assays were carried out as described previously (12).
Treatment with Proteasome Inhibitor
60 h after transfection with siRNAs, cells were treated with the proteasome inhibitor MG132 (25 μm, C-2211, Sigma), followed 8 h later by immunofluorescence or cell lysis.
Immunofluorescence Microscopy
For immunofluorescence, cells were plated on collagen-coated glass coverslips (0.01% collagen, Sigma number C9791). Conventional fixation for junctional proteins was with methanol (10 min at −20 °C), and incubations with antibodies were for 30 min at 37 °C. For detection of DbpA, cells were incubated in actin stabilization buffer (100 mm KCl, 3 mm MgCl2, 1 mm CaCl2, 200 mm sucrose, 10 mm Hepes, pH 7.1) containing 0.1% Triton X-100 for 1 min, and fixed in ice-cold methanol for 7 min at −20 °C, followed by ice-cold acetone for 30 s at room temperature. Nonspecific sites were blocked in blocking buffer (PBS containing either 0.5% BSA, 10 mm glycine or 5% BSA and 0.3% gelatin) for 30 min. Primary antibodies were diluted in blocking buffer and incubated overnight at 4 °C. For the detection of YAP, cells were fixed and permeabilized with PBS containing 3% paraformaldehyde, and 0.1% Triton X-100 (3 min at room temperature), and post-fixed with 3% paraformaldehyde for 20 min at room temperature, prior to rinsing with PBS and incubation with primary antibody (overnight at 4 °C). After staining, coverslips were mounted with Vectashield (Reactolab), and observed either with a Zeiss Axiovert S100 fluorescence microscope, or a Zeiss LSM700 confocal microscope, equipped with ×63 objective. For MDCK single KD lines, the images of one clonal line are representative of all available clones for each KD. For semiquantitative analysis of YAP localization, a maximum of 10 optical fields were analyzed for each sample (total of at least 500 cells, as determined by staining with DAPI). The number of cells showing an exclusively cytoplasmic or nuclear localization of YAP, or both localizations, was counted using ImageJ software, and data were expressed as percentage of cells showing a specific subcellular localization (10). Data were from three separate experiments, for a total of at least 250 or 500 cells for sparse or confluent conditions, respectively.
Plasmids
HA-tagged DbpA was obtained by amplifying the full-length canine DbpA sequence from a bacterial expression construct (a gift from K. Matter, University College London (20)), using primers coding for the HA sequence, and cloning into the BamHI and EcoRI sites of pcDNA 3.1A(+). The CFP-CGN-HA plasmid was generated by cloning the full-length canine CGN (28) into the BamHI and XhoI sites of pcDNA 3.1A(+), and the CFP-HA plasmid was obtained by the excision of CGN from this plasmid, using EcoRI/NotI digestion. Myc-tagged ZO-1 was described previously (19).
Immunoprecipitation and Immunoblotting
Total lysates were obtained in RIPA buffer (150 mm NaCl, 40 mm Tris-HCl, pH 7.6, 2 mm EDTA, 10% glycerol, 1% Triton X-100, 0.5% sodium deoxycholate, 0.2% SDS, 5 mg/ml of antipain, 5 mg/ml of leupeptin, 5 mg/ml of pepstatin, 1 mm PMSF), followed by sonication (3 s at 33% amplitude with a Brandon digital sonifier). Protein loadings were normalized by immunoblotting with anti-tubulin antibody. For co-immunoprecipitation (coIP) experiments, HEK293T cells were transiently transfected using Lipofectamine 2000 (Invitrogen) and the following constructs: HA-tagged CFP, Myc-tagged ZO-1, HA-tagged CGN, and HA-tagged DbpA. Lysates were prepared 48 h after transfection by washing cells twice in ice-cold PBS, followed by lysis in coIP buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 1 mm EDTA, 5 mg/ml of antipain, 5 mg/ml of leupeptin, 5 mg/ml of pepstatin, 1 mm PMSF) for 15 min at 4 °C. Lysates were sonicated and centrifuged for 15 min at 13,000 × g. Mouse antibodies (4 μl of anti-HA) were conjugated to 20 μl of pre-washed G-protein Dynabeads (Invitrogen) (1 h and 30 min at 4 °C) and the beads were incubated with lysates (overnight at 4 °C). Beads were washed in coIP buffer, and proteins were eluted by boiling in SDS sample buffer, and analyzed by SDS-PAGE and immunoblotting with the indicated rabbit antibodies. After incubation with HRP-labeled secondary antibodies, the reaction was developed using the SuperSignal WestPico chemiluminescence kit (Pierce). To carry out coIPs of endogenous proteins from MDCK lysates using anti-ZO-1 cross-linked antibodies, Sepharose CL-4B beads (Invitrogen, 10-101042) were incubated with rat anti-ZO-1 McAb R40-76 (in PBS + 1 mg/ml of BSA), washed, and incubated with the cross-linker dimyristoylphosphate (in PBS containing 0.1 m triethanolamine) for 30 min at room temperature. The beads were washed (PBS containing 0.2 m triethanolamine), and the incubation with dimyristoylphosphate and wash was repeated 2 times. Then beads were quenched with 50 mm ethanolamine in PBS, followed by washing once with PBS. Un-cross-linked antibody was removed by washing twice with 1 m glycine, pH 3, followed by washing in coIP buffer. The beads were used for coIP using the same protocol described before. Soluble and insoluble MDCK lysate fractions were prepared as described in Ref. 30.
Recombinant Protein Expression and Glutathione S-Transferase (GST) Pulldown Assays
Constructs coding either GST, or GST in-frame with either the head domain of cingulin (residues 1–406), or the ZPSG (residues (417–806) or ZS1 (SH3 domain, residues 509–597) constructs of recombinant ZO-1 were previously described (31, 32). Purification of GST fusion proteins and GST pulldowns with full-length ZO-1 expressed in baculovirus-infected insect cells were described previously (33, 34). For the titration curve of interaction between GST-ZSPG and HA-ZONAB we used increasing amounts of HEK293T cells lysate (0, 50, 100, 200, 300, and 400 μl).
Quantitative RT-PCR
Total RNA was isolated using the RNeasy mini kit (Qiagen), retrotranscribed using iScript cDNA synthesis kit (Bio-Rad), and analyzed by SYBR Green-based PCR, using a Roche Light Cycler 480 (software version 1.5, 384 wells). For each PCR, three different couples of oligonucleotide primers were tested on serial dilutions of MDCK cDNA of known concentrations (10, 2.5, 0.625, 0.156, and 0.039 ng/μl), to generate a standard curve (Absolute Quantification analysis software), where the concentration of standard samples is plotted against the crossing point(s). Crossing point values from unknown samples were obtained by interpolation with the standard curve. The slope of the standard curve calculated by the software describes the kinetics of the PCR amplification and is referred to as the “efficiency” of the amplification reaction. The highest quality PCRs run at an efficiency of 2 (the number of the target molecules doubles with every PCR cycle). The primer pairs that exhibited an efficiency >1.95 and that showed the best fit with the points of the standard curve were selected for qRT-PCR analysis and were: HPRT (internal standard), forward, 5′-TGG ACA GGA CTG AGC GGC-3′ and reverse, 5′-TGA GCA CAC AGA GGG CTA CG-3′; ZO-1 forward, 5′-ATG TCA CTG ACA GAT GCA AAG ACT TT-3′ and reverse, 5′-GAC ATT CAA TAG GGT AGC CCG T-3′; ZO-2, forward, 5′-GCG ACG GTT CTT TCT AGG GA and reverse 5′-TCC CCT TGA GGA AAT GGG AG; ZO-3 forward, 5]prime]-CAT CCA GGA GGG AGA TCA GA-3′ and reverse, 5′-TGT GTG CGG ATG TAG AAG GA; CGN, forward, 5′-CTG AAG TAG CTT CCC CAG G-3′ and reverse, 5′ TGT TGA TGA GTG AGT CCA CTG-3′; CGNL1, forward, 5′-CTC AAG GAC CTG GAA TAC GAG C-3′ and reverse, 5′-TCC GAG AGC AAA TCC GAG TT-3′; DbpA, forward, 5′-AGG AGG GAG TCC CAG AGG G-3′ and reverse, 5′-TGG GCG GTA AGT TGG ATT TC-3′; YAP, forward, 5′-CAC AGC ATG TTC GAG CTC AT and reverse, 5′-CAG GCA GAG GTA CAT CGT CA; Cyclin D1, forward 5′-GCT TAA GGC GGA GGA GAC TT-3′ and reverse, 5′-TTC ATT TCC AAC CCA CCC TC-3′; PCNA, forward, 5′-GCC TTC TGG AGA ATT TGC AC-3′ and reverse, 5′-AGA GCG GAG TGG CTT TTG TA-3′; ErBb2, forward, 5′-GCG GAT CCT GAA AGA GAC AG-3′ and reverse, 5′-GTT GGC TTT GGG AGA TGT GT-3′; CTGF, forward, 5′-CCT GGT CCA GAC CAC AGA GT-3′ and reverse, 5′-CAC GCT GGT ACA ACC AGA AA-3′. For each gene, 10 ng/μl of cDNA were used for qRT-PCR, and the values were analyzed by the Relative Analysis Quantification software, which expresses results as a normalized ratio, based on the comparison of two ratios: 1) ratio between the concentration of the target gene of interest and the concentration of the internal reference gene (HPRT) in the unknown sample, and 2) the same ratio in a “calibrator” sample (positive control sample, with a stable target/reference ratio, which is a function of PCR efficiency and of crossing points for each gene).
Statistical Analysis
All experiments were carried out at least in triplicate, and histogram values show mean ± S.D. Values were considered statistically significant (*) when p < 0.05 between experiments (Student's t tests). For immunoblots and immunofluorescence data, one representative example of 2–3 independent experiments is shown. Quantification of optical density of bands after immunoblotting was carried out using ImageJ.
RESULTS
Simultaneous but Not Individual Depletion of ZO-1, ZO-2, or ZO-3 Is Required to Decrease the Junctional Localization of DbpA in Confluent MDCK Monolayers
To study the role of ZO proteins in regulating the localization and activity of transcription factors DbpA and YAP, we used WT MDCK cells, and distinct clonal lines of MDCK cells stably expressing shRNA, and rescue clones: three clones depleted of ZO-1 (clones ZO-1KD1,2,3), a rescue ZO-1 clone (ZO1-R) (26), one clone depleted of ZO-2 (ZO-2KD) (26), one clone depleted of ZO-3 (ZO-3KD), a rescue ZO-3 clone (ZO-3R) (this study), and three MDCK clones with a double depletion of ZO-1 and ZO-2 (dKD-1, dKD-2, and dKD-3) (19).
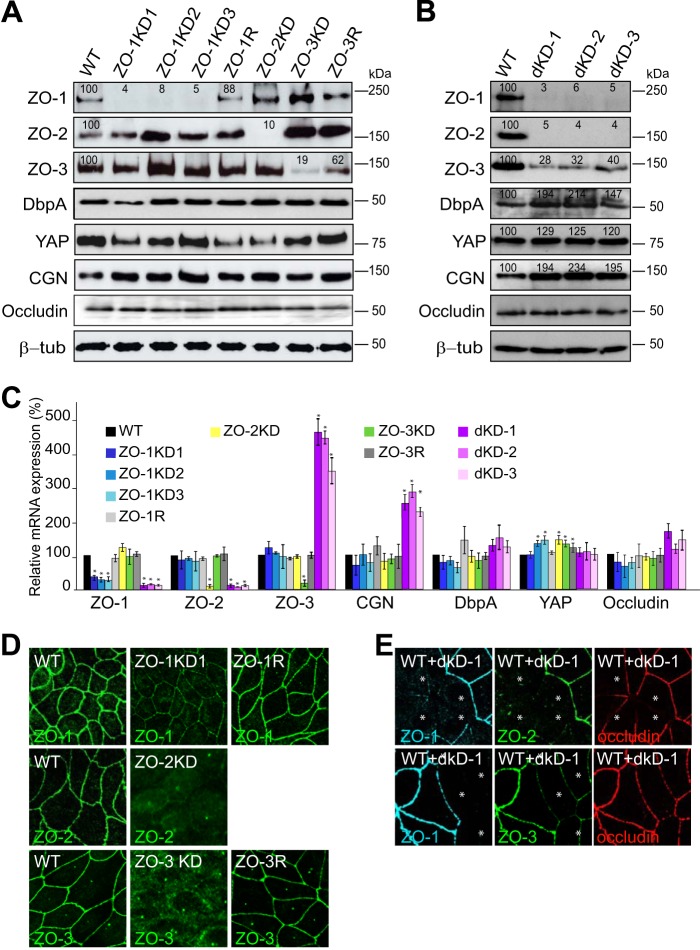
First, we examined the expression of each of the three ZO proteins, of transcription factors DbpA and YAP, and cingulin and occludin (controls), in the clonal MDCK cell lines, by immunoblotting, qRT-PCR, and immunofluorescence (Fig. 1). Immunoblotting and qRT-PCR analyses showed that the levels of ZO-1, ZO-2, and ZO-3 protein and mRNA were decreased by at least 80% in the knockdown lines, compared with WT, whereas in the rescued clones they returned to between 60 and 90% of wild-type levels (Fig. 1, A and B). As previously noted, the expression of ZO-3 was significantly decreased (to 20–40% of WT levels) in cells depleted of both ZO-1 and ZO-2 (Fig. 1B) (19), which effectively renders these cells triple KD for ZO proteins. The mRNA levels of ZO-3 and cingulin were also significantly increased in the double KD lines (Fig. 1C), suggesting that despite increased ZO-3 mRNA levels, the ZO-3 protein is either not translated efficiently, or is less stable. No significant changes in the expression of cingulin, occludin, DbpA, and YAP were detected in the single knockdown or rescue clones, either by immunoblotting or qRT-PCR (Fig. 1, A and C). However, in the double KD MDCK lines we observed a roughly 2- and 1.5-fold increase in the levels of cingulin and DbpA protein, respectively, by immunoblotting (Fig. 1B). The decrease in ZO protein levels was accompanied by a strong decrease in immunofluorescent junctional labeling for ZO-1, ZO-2, or ZO-3 in the respective knockdown clones, and recovered junctional signal in the rescued clones (Fig. 1, D and E). The junctional labeling for occludin was similar in wild-type, single knockdown, and rescued cells (Fig. 1E and data not shown), suggesting that knockdown of ZO proteins individually in MDCK cells does not prevent the formation of structurally normal TJ (26). However, in MDCK cells depleted of both ZO-1 and ZO-2 there was a decreased accumulation of occludin at junctions (Fig. 1E) (19).
FIGURE 1.
Characterization of clonal MDCK lines depleted of either ZO-1, ZO-2, ZO-3, or both ZO-1 and ZO-2. Immunoblotting (A and B), qRT-PCR (C), and immunofluorescence (D and E) analyses of clonal cell lines depleted of either of ZO-1 or ZO-2 or ZO-3 (A, C, and D), or of both ZO-1 and ZO-2 (B, C, and E). KD indicates the depleted line and R the rescued line. In A and B the numbers in the immunoblots indicate the relative levels of the proteins signal, as detected by densitometry, taking the signal of WT cells as 100%. The numbers on the right indicate migration of molecular mass markers, in kDa. β-Tubulin was used to normalize protein loadings. In C, for each gene, relative mRNA levels were normalized to that of WT cells (100%). Values represent the mean ± S.D. of three independent RNA preparations. *, p < 0.05. For immunofluorescence (D and E), junctions of confluent cells were labeled with anti-occludin antibodies (not shown for panel D), and ZO-1, ZO-2, or ZO-3, as indicated in each panel.
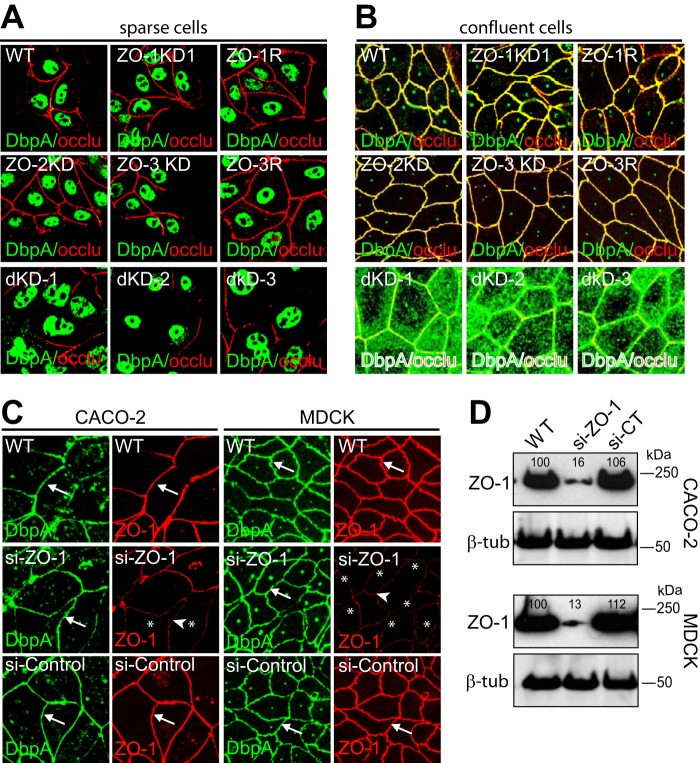
If ZO-1 inhibits cell proliferation by sequestering DbpA in confluent epithelial cells, as previously proposed (20), then depletion of ZO-1 should result in decreased junctional localization and increased nuclear localization of DbpA in confluent cells. To test this hypothesis, we examined the immunofluorescent localization of DbpA in either sparse or confluent monolayers of either wild-type, or single or double knockdown MDCK clonal lines. In sparsely grown cultures, cells formed islands of 5–15 cells, and DbpA was detected mostly in a nuclear localization, as previously observed (20), in all clonal lines: WT, ZO-1KD and rescue, ZO-2KD, ZO-3KD, and ZO-3R, and ZO-1/ZO-2 double KD (Fig. 2A). In confluent cultures DbpA displayed an exclusively junctional localization in WT cells and in all the single KD and rescue clones (Fig. 2B). However, in the double KD MDCK clones DbpA was detected not only at junctions, but also diffusely in the cytoplasm, whereas no nuclear labeling was detected (Fig. 2B). These observations demonstrate that single depletion of any one of the three ZO proteins does not affect the junctional localization of DbpA, but that depletion of ZO-1 and ZO-2, together with reduced levels of ZO-3, can lead to a partial redistribution of junctional DbpA to the cytoplasm, but not to the nucleus.
FIGURE 2.
The localization of DbpA in MDCK cells is not affected by depletion of ZO-1, ZO-2, or ZO-3 alone, but is affected by double depletion of ZO-1 and ZO-2 together. A and B, immunofluorescent localization of DbpA (green) and occludin (red, occlu) in sparse (A) and confluent (B) wild-type MDCKII cells (WT), and in clonal lines depleted of either ZO-1 (ZO-1KD1/2/3), ZO-2 (ZO-2KD), or ZO-3 (ZO-3KD), or both ZO-1 and ZO-2 (dKD-1/2/3). Rescue clones for ZO-1 (ZO-1R) and ZO-3 (ZO-3R) are also shown. Merge images, with occludin in red and DbpA in green, are shown. The occludin localization was not affected in single KD clones, but was decreased in double KD clones. C, localization of DbpA (green) and ZO-1 (red) in confluent MDCK and Caco-2 cells, either WT, or after transfection with either ZO-1-specific siRNA or control siRNA. Arrows and arrowheads indicate normal or reduced junctional staining, respectively. D, immunoblotting analysis of ZO-1 in lysates of WT Caco-2 and MDCK cells after transfection with either siRNA against ZO-1 or siRNA control. The numbers below the blots indicate relative intensity, as determined by densitometry, taking WT lysates as 100%.
To rule out the possibility that the results on ZO-1 could be dependent on cell type- or clone-dependent variations, we transiently depleted ZO-1 from either Caco-2 or MDCK cells, using a siRNA approach (Fig. 2, C and D). Immunoblot analysis showed a significant decrease in ZO-1 protein levels (Fig. 2D), and immunofluorescence analysis showed that DbpA displayed a junctional localization in cells depleted of ZO-1, indistinguishable from WT cells, and cells treated with control siRNA (Fig. 2C). Taken together, these data demonstrate that the junctional localization of DbpA in confluent epithelial cells is regulated redundantly, and is not affected by depletion of only ZO-1.
The Effects of Depletion of ZO Proteins on the Expression of DbpA Target Genes and Cell Proliferation in MDCK Cells
Previous studies reported that overexpression of the SH3 domain of ZO-1 in MDCK cell results in the up-regulation of expression of cyclin D1 and PCNA, and down-regulation of ErbB2 expression, through DbpA inhibition (35). If DbpA is inhibited by ZO-1, a prediction would be that ZO-1 depletion should lead to DbpA activation, followed by decreased ErbB2 expression, and increased expression of cyclin D1 and PCNA. To test this hypothesis, we examined the mRNA levels for these genes in synchronized populations of proliferating cells, using single or double KD MDCK cell lines. We observed no significant increase in the expression of either cyclin D1 or PCNA in different clonal lines depleted of ZO-1, ZO-2, or ZO-3. In fact, the mRNA levels of cyclin D1 were significantly decreased in two of the three ZO-1KD clones examined, the mRNA levels for PCNA were decreased in one of the three ZO-1KD clones (Fig. 3A). Furthermore, the mRNA levels for ErbB2 were decreased in only one of three ZO-1KD clones (Fig. 3A). In contrast, the ZO-1 rescue clone showed increased mRNA levels for PCNA and ErbB2, although these increases were not statistically significant. In ZO-2- and ZO-3-depleted cells we observed no significant changes (Fig. 3A). Two of the three different clonal lines depleted of both ZO-1 and ZO-2 showed significantly increased mRNA levels of PCNA (by about 50%), and decreased cyclin D1 (by about 40%). In addition, there was a roughly 4-fold increase in the expression of ErbB2 in the double KD MDCK lines. Taken together, these results indicate that changes in the expression of the DbpA target genes are not consistent with an increased nuclear activity of DbpA in cells depleted of ZO proteins.
FIGURE 3.
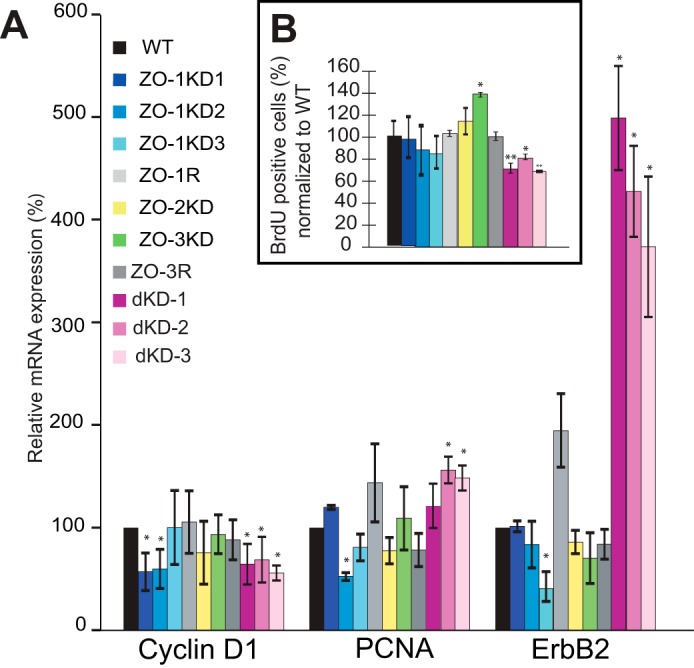
Effect of ZO protein depletion on DbpA target genes and cell proliferation. A, relative mRNA levels of DbpA target genes (cyclin-D1, PCNA, and ErbB2), as determined by qRT-PCR, in MDCK cells WT, ZO-1KD (clones ZO-1KD1, ZO-1KD2, and ZO-1KD3), ZO-2KD, ZO-3KD, and rescue, and double KD clones (dKD-1/2/3). B, percentage of cells positive for BrdU incorporation in proliferating cultures of MDCK cells WT, ZO-1KD (clones ZO-1KD1, ZO-1KD2, ZO-1KD3), ZO-2KD, ZO-3KD, and rescue clones, and double KD clones (dKD-1/2/3). The number of BrdU positive in wild-type cultures was taken as 100%.
Next, we investigated the role of ZO-1, ZO-2, and ZO-3 in cell proliferation, by performing BrdU proliferation assays on the different knockdown and rescue clonal lines. The BrdU assay measures the number of cells in S phase, by detecting nuclei that incorporate BrdU in synchronized cultures. The three distinct ZO-1KD clonal lines showed a small, but not significant decrease in the number of dividing cells, compared with wild-type cells (Fig. 3B). In ZO-2KD cells we observed an increase in the number of cells in S phase, but it was not statistically significant (Fig. 3B). In contrast, there was an increase in the number of proliferating cells in ZO-3KD cells, and this was statistically significant, and was reverted in the ZO-3R cells (Fig. 3B). This was in agreement with the previously established role of ZO-3 in the regulation of cell proliferation (14). In double KD cells we observed a small, but statistically significant decrease of the number of cells in S-phase (Fig. 3B), suggesting decreased proliferation. Taken together, these results show that only depletion of ZO-3, but not either ZO-1 or ZO-2 results in increased cell proliferation in MDCK cells, whereas depletion of ZO-1 and ZO-2 together in MDCK cells results in decreased proliferation.
Knockdown of ZO-2 in ZO-1KO Eph4 Cells Results in Loss of DbpA from Junctions
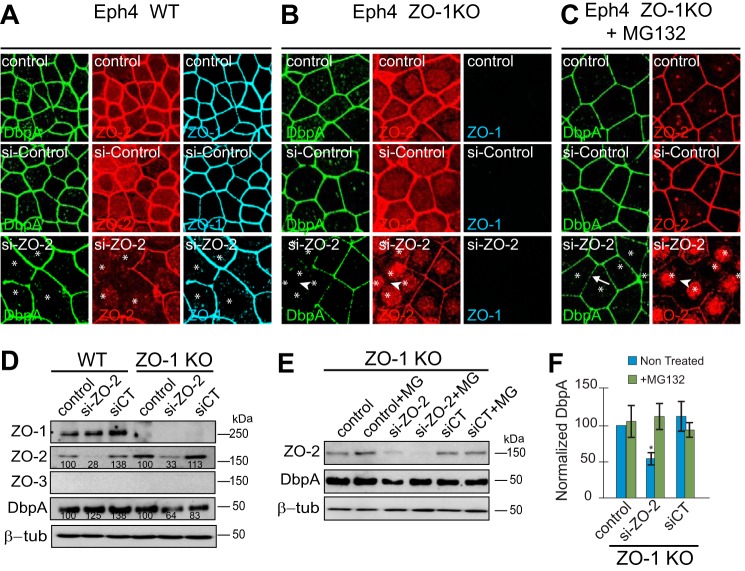
To examine an alternative model of depletion/KO of ZO proteins, we examined expression and localization of DbpA in mouse mammary epithelial (Eph4) cells (Fig. 4). These cells lack ZO-3, and upon knock-out of ZO-1 they display apparently normal TJ, and loss of junctional cingulin (22). Further depletion of ZO-2 in the background of ZO-1KO Eph4 cells results in loss of TJ barrier and claudin organization, whereas epithelial polarity and adherens junctions are maintained (16). In confluent WT Eph4 cells, and upon depletion of ZO-2 from WT cells, DbpA was localized at junctions, with no detectable labeling in either nucleus or cytoplasm (Fig. 4A). In confluent ZO-1KO Eph4 cells treated with control siRNA, DbpA was also normally localized at junctions (Fig. 4B), demonstrating that ZO-1 is not required to sequester DbpA at junctions. However, depletion of ZO-2 in the background of ZO-1KO cells resulted both in a 40% decrease in the levels of DbpA, as detected by immunoblotting (Fig. 4E), and in the loss of junctional staining for DbpA, with no increased staining in the nucleus and cytoplasm (Fig. 4B). Treatment of these cells with the proteasome inhibitor MG132 resulted in a rescue both of the junctional immunofluorescent localization of DbpA (Fig. 4C) and DbpA protein levels, as determined by immunoblotting (Fig. 4, D-F). The effect of ZO-1KO/ZO-2KD on DbpA target gene expression and BrdU incorporation could not be evaluated, because the 5-day protocols could not be overlapped with the 3-day siRNA treatments.
FIGURE 4.
Combined KO/depletion of ZO-1 and ZO-2 in Eph4 cells results in decreased junctional localization and stability of DbpA. A–C, immunofluorescent localization of DbpA (green), ZO-2 (red), and ZO-1 (blue) in confluent Eph4 cells, WT (A), ZO-1KO (B), and ZO-1 KO treated with the proteasome inhibitor MG132 (C). In each panel, the top panels series shows untreated cells, the middle panels show cells treated for 3 days with control siRNA, and the bottom panels show cells treated with ZO-2-specific siRNA. D and E, immunoblotting analysis of ZO-2 and DbpA (ZO-1 and ZO-3 are shown as controls in A) in tubulin-normalized lysates of WT and ZO-1KO Eph4 cells, either untreated (control), or treated with control siRNA (siCT), or ZO-2-specific siRNA (si-ZO-2), in the absence or presence (+MG) of the proteasome inhibitor MG132. F, histogram showing the quantification of DbpA protein levels, as determined by densitometry, taking WT lysates as 100%, in ZO-1KO cells treated as shown in E (E is a representative gel blot of three experiments).
Full-length ZO-1 Does Not Interact with DbpA
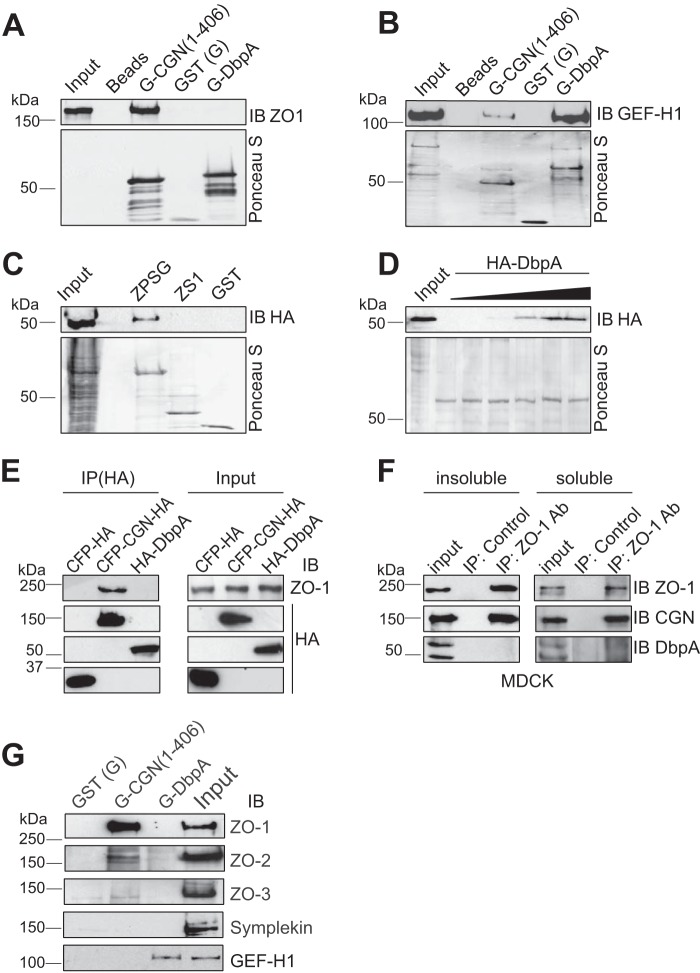
Previous studies showed an interaction in vitro of DbpA with recombinant fragments of ZO-1, comprising either SH3 or the PDZ3-SH3 domains (20, 35). However, the interaction of full-length DbpA with full-length ZO-1 has not been reported. To explore this question, we used a pulldown assay, where a bacterially expressed protein encoding full-length DbpA fused to GST was incubated with full-length ZO-1, expressed in baculovirus-infected insect cells. As a positive control, we used the bacterially expressed cingulin head domain, which interacts with high affinity with full-length ZO-1 (33), and full-length recombinant GEF-H1, which interacts with DbpA (36). As negative controls, we used either GST-coated beads, or uncoated beads. Immunoblot analysis using anti-ZO-1 antibodies showed that ZO-1 interacts with the cingulin head construct, but is not detectable in pulldowns of full-length DbpA, or in negative control pulldowns (Fig. 5A). In contrast, bacterially expressed DbpA was competent to interact in vitro with full-length recombinant GEF-H1 (Fig. 5B).
FIGURE 5.
Interaction of DbpA with ZO proteins. A and B, immunoblot (IB) analysis of GST pulldowns of either full-length recombinant ZO-1 (A) or GEF-H1 (B) produced in baculovirus-infected insect cells (input) using either Sepharose beads (Beads, negative control), or beads linked to GST (G) fused to the head region of cingulin (G-CGN(1–406)), or GST alone (GST), or GST fused to full-length DbpA (G-DbpA). The bottom panels in each image show Ponceau-S staining of the recombinant proteins used as baits in the pulldown assay. C, immunoblot analysis of GST pulldowns from lysates of HEK293 cells expressing HA-tagged DbpA, using as bait GST fused to the PDZ3/SH3/GUK domain of ZO-1 (ZPSG), GST fused to the SH3 domain (ZS1) or GST alone. D, saturation curve for the binding between ZPSG and HA-DbpA, using a fixed amount of bait, and increasing amounts of DbpA prey. E and F, immunoblot analysis (using either anti-ZO-1 antibodies or anti-HA antibodies) of: E, HA immunoprecipitates (IP) from lysates of HEK293 cells expressing myc-tagged-ZO-1 together with either HA-tagged CFP (negative control) or HA-tagged cingulin (positive control) of HA-tagged DbpA, and F, cytoskeleton-insoluble and -soluble fractions from MDCK cells (endogenous proteins), immunoprecipitated with either control or anti-ZO-1 antibodies. G, immunoblot analysis, using antibodies against ZO-1, ZO-2, ZO-3, symplekin, and GEF-H1, of MDCK lysates after pulldown using beads incubated with GST fused to the CGN head domain (G-CGN(1–406)), GST alone (G), or GST-DbpA (G-DbpA). Numbers on the left indicate the migration of pre-stained markers of known size (kDa).
Next, we asked whether bacterially expressed fragments of ZO-1 could interact with DbpA. Pull-down experiments showed that DbpA interacts with a GST fusion protein comprising PDZ3, SH3, and GUK domains of ZO-1 (ZPSG, residues 417–806), in agreement with previous observations (20), but not with a fusion protein comprising only the SH3 domain of ZO-1 (ZS1, residues 509–597) (Fig. 5C). The interaction between the ZPSG recombinant fragment of ZO-1 showed saturable binding (Fig. 5D). In summary, only a bacterially expressed fragment of ZO-1, but not full-length ZO-1, was found to interact with DbpA in vitro.
To examine whether ZO-1 and DbpA can form a complex, we expressed exogenous constructs of HA-tagged DbpA, and myc-tagged ZO-1 in HEK293 cells. As negative and positive controls, we used HA-tagged CFP and HA-tagged cingulin, respectively. ZO-1 was detected in cingulin, but not in DbpA or control immunoprecipitates (Fig. 5E). We also carried out immunoprecipitation of endogenous ZO-1 from MDCK soluble and insoluble fractionated lysates, and we detected cingulin, but not DbpA in ZO-1 immunoprecipitates (Fig. 5F), indicating that ZO-1 can be detected in a complex with cingulin, but not with DbpA. Next, we asked whether DbpA can interact with any of the ZO proteins, or with symplekin or GEF-H1, by carrying out a GST pulldown assay from lysates of MDCK cells, using bacterially expressed DbpA as a bait. As a positive control bait we used the head domain of cingulin. Immunoblot analysis showed that ZO-1, ZO-2, and ZO-3 are not detected in pulldowns using DbpA, but they are detected in pulldowns of cingulin (33) (Fig. 5G). Furthermore, the endogenous GEF-H1 in MDCK lysates was used as a positive control, and was detected in pulldowns using DbpA as a bait (Fig. 5G). This suggested that the failure of recombinant DbpA to interact with ZO proteins in MDCK lysates was not due to improper folding.
ZO-2, but Not ZO-1 and ZO-3, Regulate YAP Localization and Activity
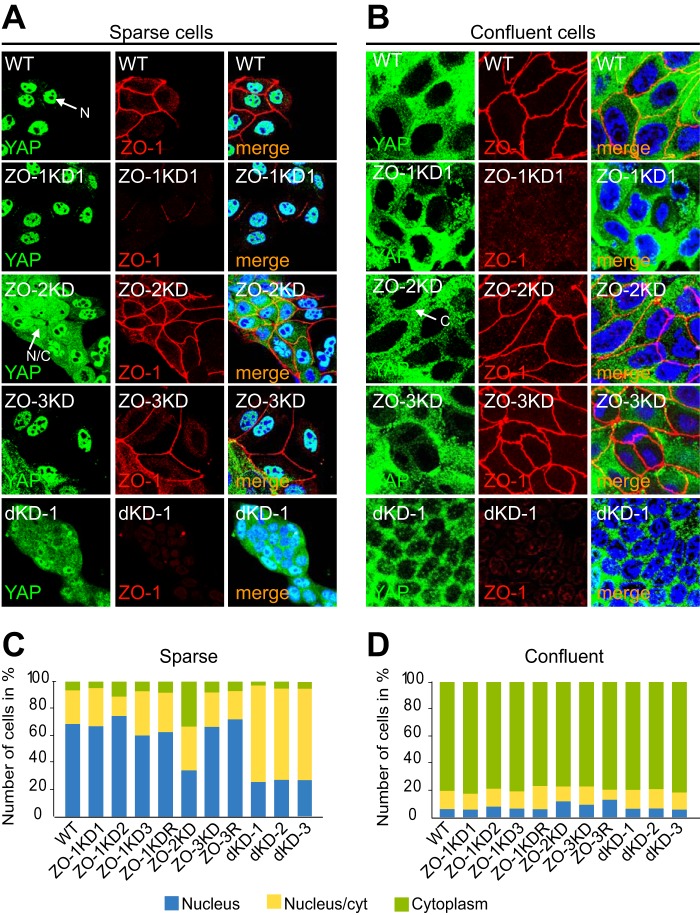
To explore alternative, DbpA-independent mechanisms through which ZO proteins may regulate cell proliferation, we studied the effects of depletion of ZO-1, or ZO-2, or ZO-3 on localization of the transcription factor YAP, and the expression of its downstream target, CTGF. YAP and TAZ, as components of the Hippo signaling pathway, control organ size and cell growth in vertebrates and invertebrates (37). In sparsely growing cells YAP is localized in the nucleus, whereas it migrates to the cytoplasm in contact-inhibited cells (38). Importantly, YAP has been shown to interact directly with ZO-2 (7, 39). The nucleo-cytoplasmic shuttling of YAP is regulated by different junctional proteins, including ZO-2 (3, 5–10). Cultures of the clonal KD and rescue MDCK lines were immunofluorescently labeled with antibodies against YAP, and the localization of YAP was scored as either exclusively nuclear, or nuclear and cytoplasmic, or exclusively cytoplasmic. In sparsely grown cultures the localization of YAP was exclusively nuclear in over 50% of the cells, with less than 10% of the cells displaying an exclusively cytoplasmic staining in WT, and all clonal lines ZO-1KD and ZO-3KD and rescue cells (Fig. 6, A and C, and data not shown). In contrast, in sparsely grown cells depleted of ZO-2 only, or of both ZO-1 and ZO-2, 30% of the cells showed an exclusively nuclear localization of YAP, and 30% showed an exclusively cytoplasmic localization (Fig. 6, A and C). This is in agreement with previous observations, indicating that ZO-2 is required to facilitate shuttling of YAP to the nucleus (7). Unexpectedly, upon depletion of both ZO-1 and ZO-2, we observed a reduced accumulation of YAP in the cytoplasm, with respect to depletion of ZO-2 alone, indicating that in these cells the reduced levels of ZO-2 do not correlate with reduced nuclear shuttling. In confluent monolayers, the behavior of the single and double KD and rescue lines was indistinguishable from wild-type, because in all cases only around 20% of the cells showed nuclear staining for YAP, with typically around 80% of the cells showing exclusively cytoplasmic staining (Fig. 6, B and D, and data not shown).
FIGURE 6.
ZO-2 but not ZO-1 or ZO-3 is required for the nuclear localization of YAP in MDCK cells. A and B, immunofluorescent localization of Yap (green) and ZO-1 (red) in sparse (A) and confluent (B) MDCK WT cells, and cells depleted of ZO-1 (ZO-1KD1), ZO-2 (ZO-2KD), or ZO-3 (ZO-3KD). Merge images (merge) show nuclei labeled in blue by DAPI. C and D, composite histograms showing the percentage of cells with exclusively nuclear (blue), exclusively cytoplasmic (green), or mixed nuclear-cytoplasmic (yellow) localization (see legend) in the different clonal and rescue lines, cultured at either sparse (C) or confluent (D) density. N, N/C, and C in immunofluorescent panels indicate an example of nuclear, nuclear/cytoplasmic, and cytoplasmic localization, respectively.
Next, we measured the levels of mRNA for CTGF, a target gene for nuclear YAP (40) in sparse and confluent cultures, by qRT-PCR. In sparsely grown wild-type cultures, the mRNA levels of CTGF were high, and they decreased significantly when cells were confluent (Fig. 7). Clonal lines of MDCK cells depleted of either ZO-1 or ZO-3, and the respective rescue clones behaved very similarly to wild-type cells (Fig. 7). In contrast, the mRNA levels of CTGF were low in sparsely grown cultures of ZO-2KD cells, and similar to confluent cultures of the same cells (Fig. 7), consistent with the notion that ZO-2 depletion prevents YAP shuttling to the nucleus. However, in double KD cells mRNA levels for CTGF were much higher, and sparse cells showed 2–3-fold higher mRNA levels of CTGF mRNA than confluent cells. Taken together, these data indicate that ZO-1 and ZO-3 do not affect the nucleo-cytoplasmic shuttling and the transcriptional activity of YAP, whereas ZO-2 is required to promote the nuclear translocation and activity of YAP in sparse cells.
FIGURE 7.
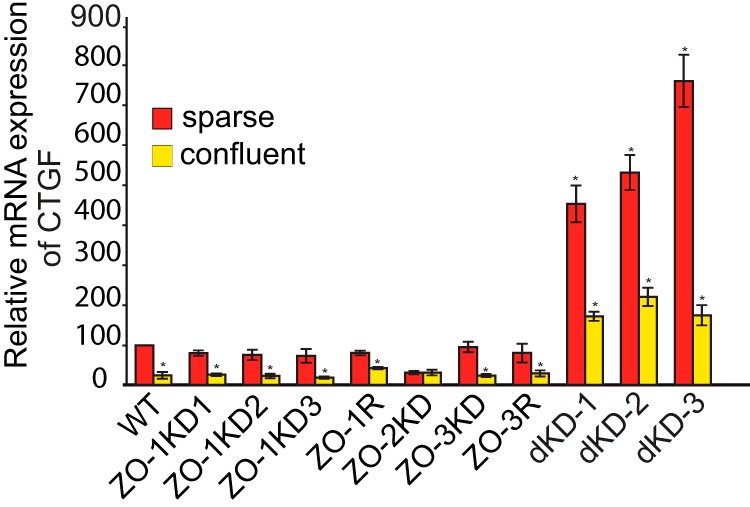
Effect of ZO protein depletion on the expression of the YAP target gene CTGF in MDCK cells. Relative levels of mRNA expression of CTGF in sparse (red) and confluent (yellow) MDCK WT cells, ZO-1KD clones (ZO-1KD1, ZO-1KD2, ZO-1KD3), ZO-2KD cells, ZO-3KD, and ZO-3 rescue clone, and double KD clones (dKD-1/2/3), as determined by qRT-PCR analysis. Note that CTGF activity in sparse cells is lowered following ZO-2KD, but not following double KD of ZO-1 and ZO-2.
DISCUSSION
Understanding the mechanisms through which junctional proteins control epithelial proliferation and gene expression is essential to clarify the molecular basis of physiological and pathological growth. Cell proliferation in vertebrate epithelial cells is regulated by multiple and complex signaling pathways, and the cross-talk between junction-associated transcription factors and junctional proteins is emerging as a novel and important pathway (2–4). Here we provide evidence from two different models of cultured epithelial cells that the localization of the TJ-associated transcription factor DbpA is redundantly regulated by all three ZO proteins, and not exclusively by ZO-1, as previously proposed (20, 21, 35). Furthermore, we show that full-length ZO-1 and DbpA do not interact together in vitro, and that among ZO proteins, only ZO-2 is required for YAP nuclear import.
The results reported here confirm the finding that DbpA shuttles between junction and nuclei in a cell density-dependent manner, linking TJ formation to the control of cell proliferation (20, 21, 35). However, they are not in agreement with a previous model (2, 4, 41, 42) that ZO-1 alone controls cell proliferation and gene expression by junctional sequestration of DbpA. Indeed, the localization of DbpA in cells either depleted of ZO-1, or knock-out for ZO-1 was not described in previous studies (20, 22, 23, 26, 35), and our observations that these cells do not display increased expression of DbpA target genes or cell proliferation are in agreement with data from ZO-1 knock-out cells and tissues, which do not show any effect of ZO-1 knock-out on cell proliferation (22, 23, 26). It is also interesting that the unique fly homologue of ZO-1 (polychaetoid) does not genetically interact with the unique fly DbpA homologue (Yps) (43). The concept that ZO-1 controls DbpA was based on overexpression studies, and we speculate that exogenous ZO-1 overexpression in epithelial cells interferes with DbpA localization and function by its indirect action on junctional complex proteins and additional DbpA partners, such as the cell division kinase CDK4, the RNA processing factor symplekin, and the RhoA activator GEF-H1 (21, 35, 36, 44), rather than directly on DbpA. Future studies should clarify the molecular mechanisms of action of overexpressed ZO-1.
We examined the localization and activity of DbpA in different cell types depleted individually not only of ZO-1, but also of MAGUK-ZO family members ZO-2 and ZO-3. We could not find significant effects on the localization and activity of DbpA in MDCK cells depleted of only one ZO protein. However, when we used MDCK cells depleted of both ZO-1 and ZO-2, which also display reduced ZO-3 expression, we observed increased cytoplasmic labeling for DbpA, indicating that the efficient junctional retention of DbpA requires a threshold level of expression of all three ZO proteins. We found no correlation between ZO protein depletion and either activation/repression of DbpA target genes or cell proliferation, in agreement with the lack of nuclear accumulation of DbpA in cells depleted of ZO proteins. This indicates that RhoA activation (36), rather than the ZO protein complex, is critical for regulation of nuclear DbpA activity. Interestingly, double KD MDCK cells showed a 4-fold increase in ZO-3 mRNA, despite a 70% decrease in ZO-3 protein levels. Because ZO proteins exist at least in part as hetero-oligomers (45), we speculate that this may be due to the decreased stability of ZO-3 protein, due to the reduced levels of ZO-1 and ZO-2, which elicits a compensatory increase in mRNA. This hypothesis must be tested by future studies, and we cannot rule out that increased ZO-3 mRNA may be due to alternative transcriptional or post-transcriptional mechanisms.
By using Eph4 KO cells for ZO-1, we were able to confirm that ZO-1 is not required for junctional sequestration of DbpA, and ruled out the possibility that the residual ZO-1 in ZO-1KD MDCK cells might have been sufficient to recruit DbpA to junctions. Furthermore, because ZO-2 depletion in the absence of ZO-1 and ZO-3 in Eph4 cells results in the loss of junctional DbpA and decreased DbpA levels, we conclude that ZO-1 and ZO-2 redundantly control not only the junctional retention of DbpA, as they do in double KD MDCK cells, but also the stability of DbpA. In contrast to what observed in Eph4 cells, we did not observe a decrease in total levels of DbpA in the double KD MDCK cells, and we speculate that a stronger depletion of each ZO protein, or even better, a KO, might be required to observe a decrease in the expression of DbpA in this context.
Whether the decreased levels of DbpA in Eph4 ZO-1KO/ZO-2KD cells is due to a reduced interaction between DbpA and ZO-1/ZO-2 at the TJ remains to be clarified. Our experiments confirm the interaction of DbpA with a recombinant fragment of ZO-1, which was observed previously (20, 35), but show that full-length ZO-1 is unable to interact with DbpA in vitro. This suggests that ZO-1 and DbpA either do not interact directly in vivo, or their interaction is transient and/or regulated. For example, the conformation of the PDZ3/SH3 domains within the full-length recombinant ZO-1 may be affected by neighboring domains, and/or by post-translational modifications. Crystallographic analysis shows that the SH3 domain of ZO-1 forms an interdependent structural unit with the GUK domain (46), suggesting that the SH3 domain may not be available for interaction with DbpA within the full-length molecule. Another possibility is that interaction of ZO proteins with DbpA is regulated by mechanical forces, which stretch ZO protein conformation in the cytoplasmic plaque of the junction. Because DbpA could not in our hands be co-immunoprecipitated with ZO-1, it appears that the DbpA-ZO-1 interaction, if it occurs in vivo, is not stable. Similarly, a direct interaction with either ZO-2 or ZO-3 is not supported by our pulldown experiments. Therefore, the precise mechanism by which ZO proteins stabilize the junctional localization of DbpA remains to be clarified, and further studies should address this issue. The observation that the proteasome inhibitor MG132 rescues the junctional localization and expression levels of DbpA suggests that cytoplasmic DbpA is unstable, and is rapidly degraded in ZO-1KO/ZO-2KD cells. However, when its degradation is inhibited, DbpA can be retained at junctions, even with no ZO-1/ZO-3, and with reduced levels of ZO-2.
To investigate alternative mechanisms through which ZO proteins may interact with signaling proteins that regulate gene expression and cell proliferation, we examined the potential functional interactions of ZO-1, and also ZO-2 and ZO-3 with the YAP signaling pathway, using a depletion approach that was not used in previous studies (7). We could not find an effect of either ZO-1 or ZO-3 depletion on the subcellular localization of YAP, or on transcription of the YAP target gene, suggesting that ZO-1 and ZO-3 do not control YAP function. However, we cannot rule out that interaction of the PDZ1 domain of ZO-1 with the YAP orthologue TAZ, which has been identified in previous studies (29), may be physiologically relevant. Indeed, polychaetoid, the fly ZO-1 homologue, regulates wing shape through genetic interaction with Expanded, a regulator of Hippo signaling in flies (43). In contrast, when cells had reduced levels of ZO-2, they failed to efficiently translocate YAP to the nucleus, in agreement with previous studies showing that mutated constructs of ZO-2 that lacked the nuclear localization signal resulted in accumulation of YAP in the cytoplasm, rather than the nucleus (7). It is unclear why in double KD MDCK cells the reduced levels of ZO-2 do not correlate with the reduced activity of CTGF in sparse growing conditions. The localization of TAZ was similar in single ZO-2KD and double ZO-1/ZO-2 KD cells (data not shown), suggesting that the very high CTGF levels in double KD cells is not due to an increase in the nuclear localization of YAP/TAZ, but rather an increase in their intrinsic activity, possibly related to the increased cytoskeletal tension in double KD cells (19), or to activation of additional factors or signaling pathways.
In summary, our study establishes that the subcellular localization and activity of DbpA are not regulated by ZO-1 alone, but instead the three ZO proteins (ZO-1, ZO-2, and ZO-3) redundantly control the junctional retention and stability of DbpA. Furthermore, we show that depletion of ZO-2 alone inhibits the shuttling of YAP to the nucleus of MDCK cells. These results improve and refine currently held models about the roles of ZO proteins in the control of gene expression and cell proliferation.
Acknowledgments
We are very grateful to Prof. Sachiko Tsukita for the generous gift of Eph4 ZO-1 KO cells, and we thank all other colleagues cited in the text for kind gift of reagents.
This work was supported in part by Swiss National Fund Grants 31003A103637 and 31003A116763, Swiss Cancer League Grant KFS-2813-08-2011 (to S. C.), and the Section of Biology of the Faculty of Sciences of the University of Geneva.
- TJ
- tight junction
- CTGF
- connective tissue growth factor
- DbpA
- DNA-binding protein A
- GEF-H1
- Rho/Rac guanine nucleotide exchange factor (GEF) H1
- HPRT
- hypoxanthine-guanine phosphoribosyltransferase
- MDCK
- Madin-Darby canine kidney
- PDZ
- Psd95-Dlg1-ZO-1 domain
- qRT
- quantitative RT
- SH3
- Src homology 3
- YAP
- Yes-associated protein
- Yps
- Yipsilon schachtel
- ZO
- zonula occludens
- coIP
- co-immunoprecipitation
- PCNA
- proliferating cell nuclear antigen
- KD
- knock-down
- KO
- knockout.
REFERENCES
- 1. Citi S., Pulimeno P., Paschoud S. (2012) Cingulin, paracingulin and PLEKHA7: signalling and cytoskeletal adaptors at the apical junctional complex. Ann. N.Y. Acad. Sci. 1257, 125–132 [DOI] [PubMed] [Google Scholar]
- 2. McCrea P. D., Gu D., Balda M. S. (2009) Junctional music that the nucleus hears: cell-cell contact signaling and the modulation of gene activity. Cold Spring Harbor Perspect. Biol. 1, a002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spadaro D., Tapia R., Pulimeno P., Citi S. (2012) The control of gene expression and cell proliferation by the apical junctional complex. Essays Biochem. 53, 83–93 [DOI] [PubMed] [Google Scholar]
- 4. Balda M. S., Matter K. (2009) Tight junctions and the regulation of gene expression. Biochim. Biophys. Acta 1788, 761–767 [DOI] [PubMed] [Google Scholar]
- 5. Silvis M. R., Kreger B. T., Lien W. H., Klezovitch O., Rudakova G. M., Camargo F. D., Lantz D. M., Seykora J. T., Vasioukhin V. (2011) α-Catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal 4, ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J. R., Zhou D., Kreger B. T., Vasioukhin V., Avruch J., Brummelkamp T. R., Camargo F. D. (2011) Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144, 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oka T., Remue E., Meerschaert K., Vanloo B., Boucherie C., Gfeller D., Bader G. D., Sidhu S. S., Vandekerckhove J., Gettemans J., Sudol M. (2010) Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem. J. 432, 461–472 [DOI] [PubMed] [Google Scholar]
- 8. Zhao B., Li L., Lu Q., Wang L. H., Liu C. Y., Lei Q., Guan K. L. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang W., Huang J., Chen J. (2011) Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 286, 4364–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim N. G., Koh E., Chen X., Gumbiner B. M. (2011) E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. U.S.A. 108, 11930–11935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li D., Mrsny R. J. (2000) Oncogenic Raf-1 disrupts epithelial tight junctions via down-regulation of occludin. J. Cell Biol. 148, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guillemot L., Citi S. (2006) Cingulin regulates claudin-2 expression and cell proliferation through the small GTPase RhoA. Mol. Biol. Cell 17, 3569–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nava P., Capaldo C. T., Koch S., Kolegraff K., Rankin C. R., Farkas A. E., Feasel M. E., Li L., Addis C., Parkos C. A., Nusrat A. (2011) JAM-A regulates epithelial proliferation through Akt/α-catenin signalling. EMBO Rep. 12, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Capaldo C. T., Koch S., Kwon M., Laur O., Parkos C. A., Nusrat A. (2011) Tight function zonula occludens-3 regulates cyclin D1-dependent cell proliferation. Mol. Biol. Cell 22, 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stevenson B. R., Siliciano J. D., Mooseker M. S., Goodenough D. A. (1986) Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 103, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M., Tsukita S. (2006) ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126, 741–754 [DOI] [PubMed] [Google Scholar]
- 17. Itoh M., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S. (1999) Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 147, 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fanning A. S., Jameson B. J., Jesaitis L. A., Anderson J. M. (1998) The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 273, 29745–29753 [DOI] [PubMed] [Google Scholar]
- 19. Fanning A. S., Van Itallie C. M., Anderson J. M. (2012) Zonula occludens (ZO)-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell 23, 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Balda M. S., Matter K. (2000) The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 19, 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Balda M. S., Garrett M. D., Matter K. (2003) The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 160, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Umeda K., Matsui T., Nakayama M., Furuse K., Sasaki H., Furuse M., Tsukita S. (2004) Establishment and characterization of cultured epithelial cells lacking expression of ZO-1. J. Biol. Chem. 279, 44785–44794 [DOI] [PubMed] [Google Scholar]
- 23. Katsuno T., Umeda K., Matsui T., Hata M., Tamura A., Itoh M., Takeuchi K., Fujimori T., Nabeshima Y., Noda T., Tsukita S., Tsukita S. (2008) Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol. Biol. Cell 19, 2465–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cardellini P., Davanzo G., Citi S. (1996) Tight junctions in early amphibian development: detection of junctional cingulin from the 2-cell stage and its localization at the boundary of distinct membrane domains in dividing blastomeres in low calcium. Dev. Dyn. 207, 104–113 [DOI] [PubMed] [Google Scholar]
- 25. Oka T., Mazack V., Sudol M. (2008) Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). J. Biol. Chem. 283, 27534–27546 [DOI] [PubMed] [Google Scholar]
- 26. Van Itallie C. M., Fanning A. S., Bridges A., Anderson J. M. (2009) ZO-1 stabilizes the tight junction solute barrier through coupling to the perijunctional cytoskeleton. Mol. Biol. Cell 20, 3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodgers L. S., Beam M. T., Anderson J. M., Fanning A. S. (2013) Epithelial barrier assembly requires coordinated activity of multiple domains of the tight junction protein ZO-1. J. Cell Sci. 126, 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paschoud S., Citi S. (2008) Inducible overexpression of cingulin in stably transfected MDCK cells does not affect tight junction organization and gene expression. Mol. Membr. Biol. 25, 1–13 [DOI] [PubMed] [Google Scholar]
- 29. Remue E., Meerschaert K., Oka T., Boucherie C., Vandekerckhove J., Sudol M., Gettemans J. (2010) TAZ interacts with zonula occludens-1 and -2 proteins in a PDZ-1 dependent manner. FEBS Lett. 584, 4175–4180 [DOI] [PubMed] [Google Scholar]
- 30. Guillemot L., Paschoud S., Jond L., Foglia A., Citi S. (2008) Paracingulin regulates the activity of Rac1 and RhoA GTPases by recruiting Tiam1 and GEF-H1 to epithelial junctions. Mol. Biol. Cell 19, 4442–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. D'Atri F., Nadalutti F., Citi S. (2002) Evidence for a functional interaction between cingulin and ZO-1 in cultured cells. J. Biol. Chem. 277, 27757–27764 [DOI] [PubMed] [Google Scholar]
- 32. Fanning A. S., Little B. P., Rahner C., Utepbergenov D., Walther Z., Anderson J. M. (2007) The unique-5 and -6 motifs of ZO-1 regulate tight junction strand localization and scaffolding properties. Mol. Biol. Cell 18, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cordenonsi M., D'Atri F., Hammar E., Parry D. A., Kendrick-Jones J., Shore D., Citi S. (1999) Cingulin contains globular and coiled-coil domains and interacts with ZO-1, ZO-2, ZO-3, and myosin. J. Cell Biol. 147, 1569–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Citi S., D'Atri F., Cordenonsi M., Cardellini P. (2001) Tight junction protein expression in early Xenopus development and protein interaction studies. in Cell-Cell Interactions (Fleming T. P., ed) 2nd Ed., pp. 153–176, IRL Press, Oxford [Google Scholar]
- 35. Sourisseau T., Georgiadis A., Tsapara A., Ali R. R., Pestell R., Matter K., Balda M. S. (2006) Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol. Cell. Biol. 26, 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nie M., Aijaz S., Leefa Chong San I. V., Balda M. S., Matter K. (2009) The Y-box factor ZONAB/DbpA associates with GEF-H1/Lfc and mediates Rho-stimulated transcription. EMBO Rep. 10, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu F. X., Guan K. L. (2013) The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oka T., Schmitt A. P., Sudol M. (2012) Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene 31, 128–134 [DOI] [PubMed] [Google Scholar]
- 40. Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 [DOI] [PubMed] [Google Scholar]
- 41. Matter K., Balda M. S. (2007) Epithelial tight junctions, gene expression and nucleo-junctional interplay. J. Cell Sci. 120, 1505–1511 [DOI] [PubMed] [Google Scholar]
- 42. Bauer H., Zweimueller-Mayer J., Steinbacher P., Lametschwandtner A., Bauer H. C. (2010) The dual role of zonula occludens (ZO) proteins. J. Biomed. Biotechnol. 2010, 402593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Djiane A., Shimizu H., Wilkin M., Mazleyrat S., Jennings M. D., Avis J., Bray S., Baron M. (2011) Su(dx) E3 ubiquitin ligase-dependent and -independent functions of polychaetoid, the Drosophila ZO-1 homologue. J. Cell Biol. 192, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kavanagh E., Buchert M., Tsapara A., Choquet A., Balda M. S., Hollande F., Matter K. (2006) Functional interaction between the ZO-1-interacting transcription factor ZONAB/DbpA and the RNA processing factor symplekin. J. Cell Sci. 119, 5098–5105 [DOI] [PubMed] [Google Scholar]
- 45. Wittchen E. S., Haskins J., Stevenson B. R. (1999) Protein interactions at the tight junction. Actin has multiple binding partners, and ZO-1 forms independent complexes with ZO-2 and ZO-3. J. Biol. Chem. 274, 35179–35185 [DOI] [PubMed] [Google Scholar]
- 46. Lye M. F., Fanning A. S., Su Y., Anderson J. M., Lavie A. (2010) Insights into regulated ligand binding sites from the structure of ZO-1 Src homology 3-guanylate kinase module. J. Biol. Chem. 285, 13907–13917 [DOI] [PMC free article] [PubMed] [Google Scholar]