Abstract
The ANR1 MADS-box gene in Arabidopsis is a key gene involved in regulating lateral root development in response to the external nitrate supply. There are five ANR1-like genes in Oryza sativa, OsMADS23, OsMADS25, OsMADS27, OsMADS57 and OsMADS61, all of which belong to the AGL17 clade. Here we have investigated the responsiveness of these genes to fluctuations in nitrogen (N), phosphorus (P) and sulfur (S) mineral nutrient supply. The MADS-box genes have been shown to have a range of responses to the nutrient supply. The expression of OsMADS61 was transiently induced by N deprivation but was not affected by re-supply with various N sources. The expression of OsMADS25 and OsMADS27 was induced by re-supplying with NO3 − and NH4NO3, but downregulated by NH4 +. The expression of OsMADS57 was significantly downregulated by N starvation and upregulated by 3 h NO3 − re-supply. OsMADS23 was the only gene that showed no response to either N starvation nor NO3 − re-supply. OsMADS57 was the only gene not regulated by P fluctuation whereas the expression of OsMADS23, OsMADS25 and OsMADS27 was downregulated by P starvation and P re-supply. In contrast, all five ANR1-related genes were significantly upregulated by S starvation. Our results also indicated that there were interactions among nitrate, sulphate and phosphate transporters in rice.
Introduction
Nitrogen, phosphorus and sulfur are three major macronutrients essential for plant growth and development [1]. Nitrogen (N) deficiency is a vital factor limiting agricultural quality and productivity. In aerobic soil conditions, nitrate is the major source of nitrogen for many higher plant species [2]–[4]. Nitrate acts not only as a nutrient but also as a signal to regulate gene expression, energy transfer, protein activation, metabolic and physiological activities, and plant growth and development [5]–[8]. Phosphorus (P) is a component of many key biomolecules and participates in various enzymatic reactions and metabolic pathways [9]. Due to low availability of phosphate (Pi) in soil, it acts as the second most important limiting macronutrients for plant growth and development [10]–[12]. Plants can modify their root architecture to improve phosphate acquisition in phosphate deficient soil. Many studies have demonstrated that P deficiency affects root development, including root hair elongation, reduced primary root length and increased number and length of lateral roots in Arabidopsis thaliana [13]–[19]. Sulfur (S) is also an essential macronutrients, and its deficiency adversely affects plant growth and development as well as the quality of crops [20], [21].
Plants can modify their root architecture to forage for sources of N that are distributed unevenly in soil [6]. One important kind of foraging response involves increased proliferation of lateral roots within soil patches enriched in certain nutrients, such as NH4 + and NO3 − [22], [23]. There are signaling mechanisms in roots to measure the levels of intrinsic and extrinsic nutritional factors so that they can modify their growth and development [24], [25]. In Arabidopsis, localized nitrate treatment stimulated a localized increase in lateral root (LR) numbers and elongation. The stimulation of LR elongation was found to be the result of a signaling effect of external NO3 − ion itself rather than downstream metabolites [26], [27]. An important breakthrough in understanding NO3 − in stimulating LR growth was the identification of the ANR1 gene, which is a vital component in regulatory signaling pathway [26]. ANR1 is a member of the plant MADS-box family of transcription factors, which covers more than 100 members in Arabidopsis [28]–[30]. In addition to their roles in regulation of reproductive development, the MADS-box transcription factors are also widely expressed in vegetative tissues [28], [31]. Recent research has demonstrated that at least 50 MADS-box genes are expressed in roots of Arabidopsis, with ANR1 as the only member so far to have a known function in lateral root development in Arabidopsis [32], [33]. Previous reports revealed that ANR1 is a positive regulator of lateral root growth and is not present in the primary root tip [34]. Initial studies using Arabidopsis root cultures had demonstrated that ANR1 gene was NO3 – inducible [26]. Subsequent results obtained from hydroponically culture experiments illustrated that the expression of ANR1 was induced by N deprivation and rapidly downregulated when NO3 – or other N source was re-supplied [29]. OsMADS23, OsMADS25, OsMADS27, OsMADS57 and OsMADS61 are five ANR1-like homologs in rice and their functions have not been well understood [35], [36]. Recent work from Meng et al. (2013) has suggested that OsMADS27 could play a key role in the response to cold and salt stress [37]. To gain further insight into the possible regulatory functions of ANR1-like genes in roots, we have investigated their root expression patterns in response to N, P, and S fluctuation using quantitative real-time PCR (qPCR).
Materials and Methods
Plant Materials
Rice seeds (Oryza sativa L. cv. Nipponbare) were used for all experiments.
Hydroponic culture
Rice seeds were surface-sterilized by treatment with 70% ethanol for 1 min and 10% sodium hypochlorite for 20 min, followed by five rinses with sterile distilled water. Seeds were germinated in the dark by placing into the incubator at 28°C for 2 days (d). Uniform seedlings were selected and transferred to black plastic buckets containing 4 L nutrient solution, where the growth conditions were 30/28°C day/night temperature with a 14/10 h light/dark at a relative humidity of 65–70%. The complete nutrient solution contained: 1.44 mM NH4NO3, 0.32 mM NaH2PO4, 0.5 mM K2SO4, 1 mM CaCl2·2H2O, 1.6 mM MgSO4·7H2O, 50 µM Fe-EDTA, 15 µM H3BO3, 9 µM MnCl2·4H2O, 0.12 µM CuSO4·5H2O, 0.12 µM ZnSO4·7H2O, 40.5 µM citric acid and, 0.39 µM Na2MoO4·2H2O [38]. 1 M HCl or NaOH was added to adjust pH to 5.5 and the nutrient solution was replaced every 2 d.
Nitrogen treatments
For the analysis of gene expression in response to nitrate, rice seedlings were grown in liquid culture for 14 d with 1.44 mM NH4NO3 as the N source. The solution was changed every 2 d. After 10 d, the medium was changed to 2.88 mM KNO3 as the sole N source for 4 d. Then, the seedlings were deprived of N for 3 d before being re-supplied with KNO3 or KCl to a final concentration of 2.88 mM. The control plants were grown in continuous 2.88 mM KNO3 as the sole nitrogen source. The plants from different treatments were harvested at 4 h, 6 h and 8 h after re-supply separately. The roots were frozen in liquid N2 and stored at −80°C for later analysis.
For analyses of gene expression in response to different N sources, the plants were grown in complete nutrient solution with 2.88 mM KNO3 as the sole nitrogen source for 14 d [39]. Then, the plants were starved for N for 3 d before being transferred to fresh nutrient solution with the same concentration of different N sources. Roots were harvested 3 h later. For the control treatment, the plants were continuously supplied with 2.88 mM KNO3 as the only N source. All the nutrient experiments were repeated at least twice with similar results.
P and S treatments
To initiate different P and S treatments, two-week-old seedlings grown in complete nutrient solution as described above. For the P and S starvation treatments, the complete nutrient solution was replaced with nutrient solution lacking P or S for 3 d with PO4 − or SO4 2− being replaced by chloride. For the control plants, the seedlings grown in continuous complete nutrient solution were transferred to fresh complete nutrient solution at the same time. For re-supply, the appropriate nutrient, 0.32 mM PO4 − or 2.1 mM SO4 2− was added in the light period for 3 h before the roots were harvested and frozen in liquid N2 and stored at −80°C for gene expression analyses [38].
RNA extraction and qPCR
Primers for OsMADS23, OsMADS25, OsMADS27, OsMADS57 and OsMADS61 for qPCR (see Table 1) were designed using Primer Premier 5. Total RNA was extracted using the RNAiso Plus reagent (Takara) according to the manufacturer’s instructions. The first-strand cDNA was synthesized using PrimeScript RT reagent Kit with gDNA Eraser (Takara). The qPCR was performed in 96-well plates using SYBR Premix Ex Taq II (Takara) according to the manufacturer’s instructions. The qPCR was conducted in the following cycling conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Melt curve analysis was used to confirm the absence of non-specific amplification products. Relative expression levels were calculated by subtracting the threshold cycle (Ct) values for OsActin (Os03g0718100) from those of the target gene (to give △Ct) and then calculating 2–△Ct as we described before [40]–[42]. RT-PCR experiments were performed with three biological replicates with the representative being shown.
Table 1. List of primers used for Real-time PCR.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
| OsMADS23 | TCTTCTCCAGCACCAGCCGTCT | TGCTGCCTCCTGTTGCCAAAGC |
| OsMADS25 | CCAGCTCAAGCATGAAATCAA | AAAGTTGCCTGTTGTTGTGGTGT |
| OsMADS27 | GAAGCGGAGGAACGGGATCTTCAA | TGCCATACCGATCTATAACTGACT |
| OsMADS57 | ACGAGCAGGCAGGTGACGTT | ACTCATAGAGCCTGCCGGTGCT |
| OsMADS61 | GGGAGGGGCAAGATAGTGAT | TGGTGCTGGCATACTCGTAG |
| OsActin | CTTCATAGGAATGGAAGCTGCGGGT | CGACCACCTTGATCTTCATGCTGCT |
Statistics
The results were analyzed for variance by IBM SPSS Statistics 20. Student’s t-test was calculated at the probability at either at 1% (P<0.01 with significant level **) or 5% (P<0.05 with significant level *) as described before [43], [44].
Results
Five ANR1-like genes have different expression patterns in response to nitrate
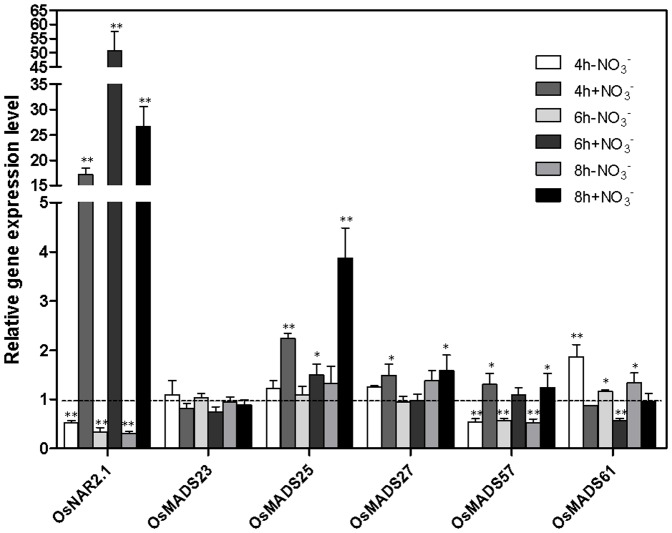
The Arabidopsis ANR1 gene was identified as a key gene controlling lateral root growth through NO3 − signaling [26]. To understand whether these five ANR1 homologous genes in rice could play similar roles to ANR1 in Arabidopsis, we begin by investigating their expression patterns and levels in response to nitrate. Rice seedlings were grown in complete nutrient solution for 14 d with 2.88 mM KNO3 as the sole N source They were then deprived of N for 3 d before being re-supplied with 2.88 mM KNO3 (or 2.88 mM KCl as control). The rice nitrate transporter gene OsNAR2.1, which is known to be nitrate-inducible [45], was used as a positive control. As shown in Figure 1, the expression of OsNAR2.1 was very strongly upregulated by nitrate re-supply as expected. Each of the five ANR1-related genes was found to have a different expression pattern in response to nitrate starvation and re-supply (Figure 1). Similar to ANR1 gene in Arabidopsis, the expression of OsMADS61 was significantly induced by nitrate starvation at 4 h, 6 h and 8 h and significantly suppressed by nitrate re-supply at 6 h in comparison to the seedlings continuously supplied with nitrate. In contrast, the expression of OsMADS25 was not significantly affected by nitrate starvation but was induced by nitrate re-supply at 4 h, 6 h and 8 h in comparison to the continuous nitrate treatment. Similarly, the expression of OsMADS27 was not significantly affected by nitrate starvation but was induced by nitrate re-supply at 4 h and 8 h in comparison to the continuous nitrate treatment. The expression of OsMADS57 was downregulated by nitrate starvation at 4 h, 6 h and 8 h and was upregulated by nitrate re-supply at 4 h in comparison to the continuous nitrate treatment (Figure 1). OsMADS23 was the only gene to show no significant response to either nitrate starvation or nitrate re-supply (Figure 1).
Figure 1. Effect of N deprivation and nitrate resupply on the expression of five ANR1 related genes in rice roots.
Ten-day old rice seedlings grown in complete nutrient solutions were transferred to modified nutrient solutions during which 2.88 mM KNO3 was the sole nitrogen source for 4 days. Transcript abundance was assayed by qPCR and was expressed relative to the abundance in roots of plants of the same age grown under continuous N (CK). The OsNAR2.1 gene, a known nitrate-regulated gene, was included for comparison. Treatments: CK: continuous KNO3; −N4h: starved of N for 3 d and resupplied with KCl for 4 h; +N4h: resupplied with KNO3 for 4 h; −N6h: starved of N for 3 d and resupplied with KCl for 6 h; +N6h: resupplied with KNO3 for 6 h. The mRNA of OsActin was used as the reference. A Student’s t-test was calculated at the probability of either 5% (*, p<0.05) or (**, P<0.01).
Five ANR1-like genes respond differently to different N sources
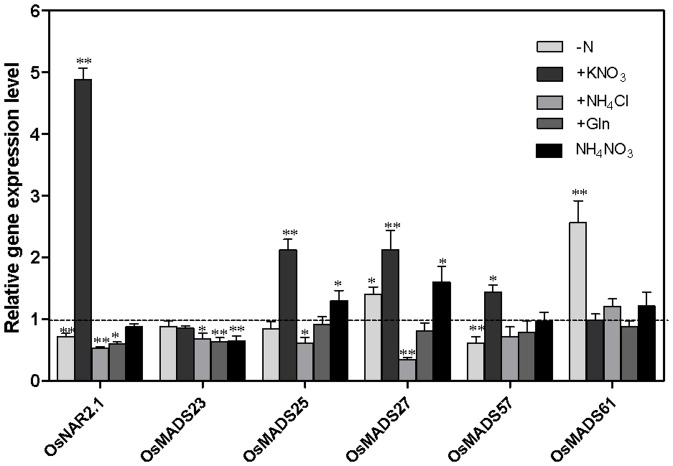
To investigate whether these five ANR1 homologous genes in rice were also regulated by the other N sources, we further investigated their expression patterns in response to different N sources. The plants were grown in the complete nutrient solution for 10 d before changing media with 2.88 mM KNO3 as the sole nitrogen source for 4 days. Then, the plants were starved for N nutrient for 3 d followed by re-supply with the same concentrations of different N sources. As shown in Figure 2, the expression of rice transporter OsNAR2.1 was down–regulated in the roots of N-starved rice plants and then rapidly upregulated when supplied with 2.88 mM KNO3 for 3 h which was consistent with a previous report [39]. OsMADS61 and OsMADS57 were the only genes that were affected by nitrate not by any other N sources, whereas OsMADS23 was the only gene regulated by various different N sources but not by nitrate. In contrast, OsMADS25 and OsMADS27 were upregulated by both nitrate and ammonium nitrate but downregulated by ammonium chloride.
Figure 2. Effect of different N sources on the expression of five ANR1-related genes in rice roots.
Rice seedlings were grown in liquid culture for 14 days with 2.88 mM KNO3 as the sole nitrogen source and then N starved for 3 d. CK, continuous N; −N, starved for 3 d; +KNO3, resupplied with 2.88 mM nitrate; +NH4Cl resupplied with 2.88 mM NH4 +; and +Gln, resupplied with 2.88 mM glutamine; +NH4NO3, resupplied with 1.44 mM NH4NO3. The value of related genes were normalized to its CK control respectively. The mRNA of OsActin was used as the reference. Error bars represent SE. LSD values were calculated at the probability of either 5% (*, p<0.05) or (**, P<0.01).
Effect of P and S deprivation and re-supply on the expressions of five ANR1-like genes in rice roots
To analyze whether ANR1-like homologous genes are involved in the regulation of gene expression in response to other nutritional stresses, we investigated the effect of P and S deprivation and re-supplementation. As shown in Figure 3, in contrast to the phosphate transporter OsIPS1, OsMADS23, OsMADS25 and OsMADS27 were all downregulated by P starvation and P re-supply whereas OsMADS57 and OsMADS61 were not significantly affected. For the S treatment, OsMADS23, OsMADS25, OsMADS27 and OsMADS57 were all upregulated by S starvation but not by S re-supply whereas OsMADS61 was upregulated by both S starvation and re-supply in comparison to the continuous S nutrient treatment.
Figure 3. Effect of deprivation and re-supply of phosphate (P) and sulfate (S) on the expression of five ANR1-related genes in rice.
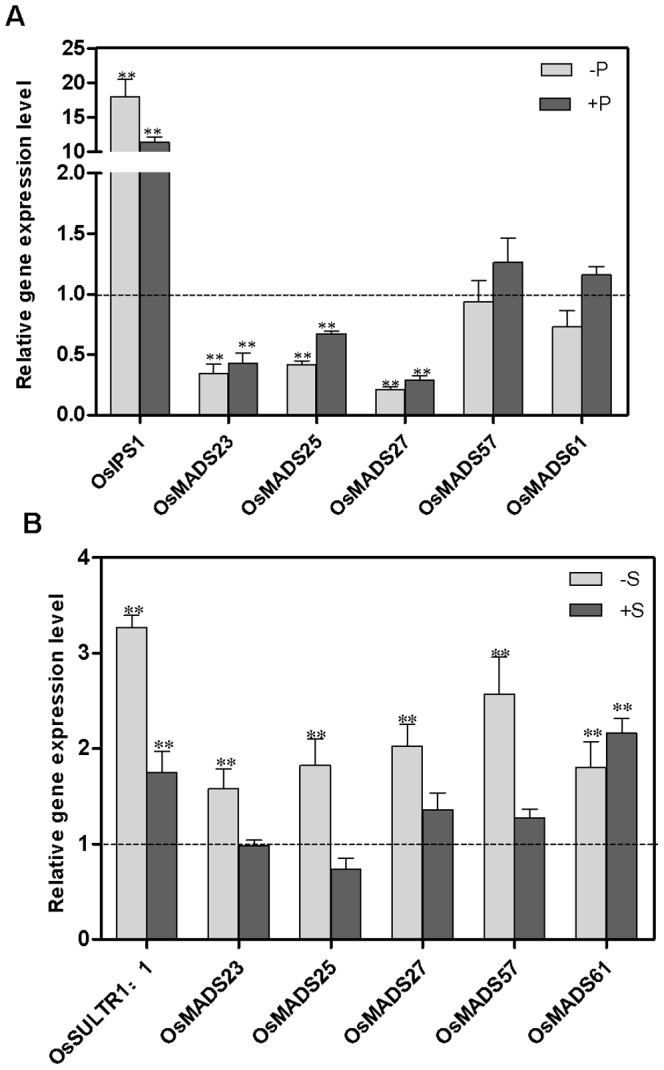
Two-week old rice seedlings grown hydroponically in complete nutrient solution were deprived of P or S or were maintained on complete nutrient supply for 3 d. In the light period on the day of the experiment, one set of the P-starved and S-starved plants were re-supplied with 0.32 mM H2PO4 − and 2.1 mM SO4 2− respectively. Roots were harvested 3 h later from controls: continuous nutrient supply (CK); P-deprived (−P); P-resupply (+P); S-deprived (−S); S-resupply (+S). Total RNA was extracted from roots and qPCR reactions were performed in triplicate for each RNA sample. The mRNA of OsActin was used as the reference. The value of related genes were normalized to its CK control respectively. A Student’s t-test was calculated at the probability of either 5% (*, p<0.05) or (**, P<0.01).
Effect of N-deprivation and re-supply on the expressions of OsNRT2.1, OsNAR2.1, OsIPS1 and OsSULTR1;1 in rice roots
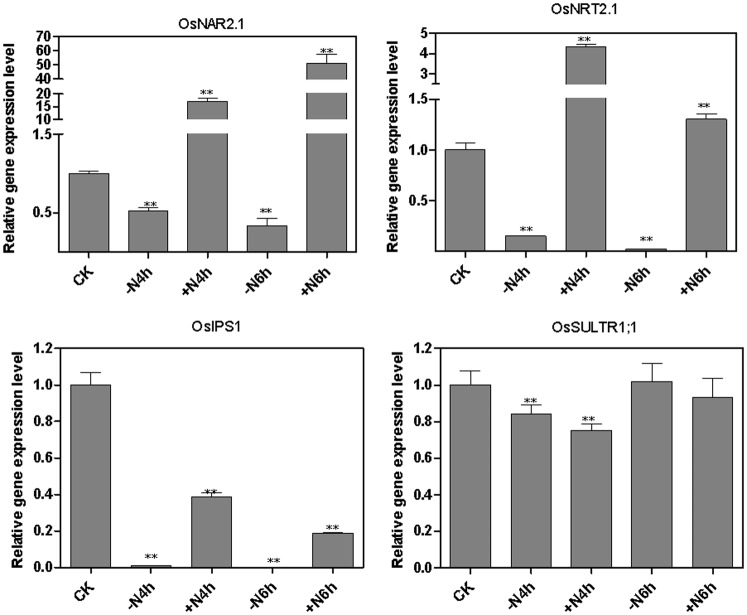
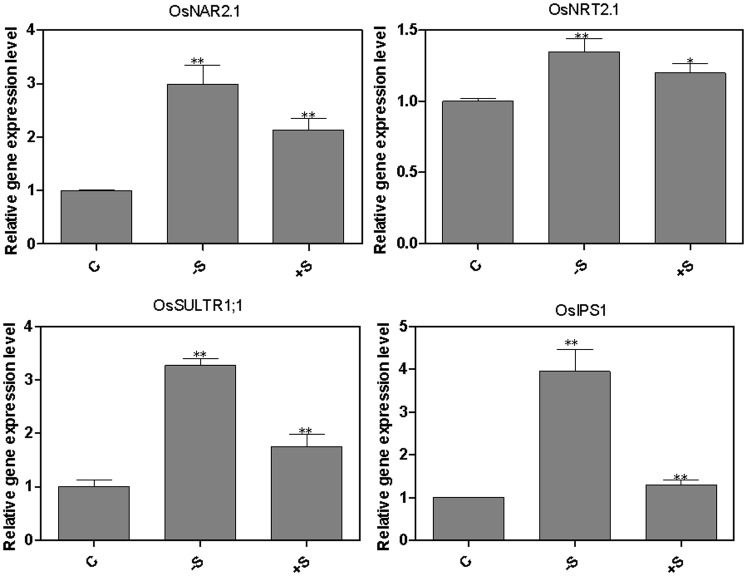
We have investigated whether there is crosstalk between the N, P and S regulatory pathways in the regulation of expression of the the OsNRT2.1, OsNAR2.1, OsIPS1 and OsSULTR1;1 genes. We first investigated whether the expression of phosphate transporter OsIPS1 and sulphate transporter OsSULTR1;1 was regulated in response in nitrate starvation and nitrate re-supply. The OsNRT2.1 and OsNAR2.1 genes, which encode components of the high affinity nitrate transport system (HATS) in rice, were used as the positive controls OsNRT2.1 and OsNAR2.1, were previously shown to be upregulated by nitrate and suppressed by NH4 +. As expected, OsNRT2.1 and OsNAR2.1 were downregulated by nitrate starvation and rapidly upregulated by nitrate re-supply (Figure 4), confirming the results of previous studies [45], [46]. However, the expression of phosphate transporter OsIPS1 was significantly downregulated by both nitrate starvation and nitrate re-supply at both 4 h and 6 h time points, whereas OsSULTR1;1 was only significantly downregulated by both nitrate starvation and re-supply at 4 h and not at 6 h.
Figure 4. Effect of N-deprivation and re-supply on expression of OsNRT2.1, OsNAR2.1, OsIPS1 and OsSULTR1;1 in rice roots.
Rice seedlings were grown hydroponically in a growth cabinet. Nitrogen treatments were as described in Fig. 1. CK: continuous KNO3; −N4h: starved of N for 3 d and resupplied with KCl for 4 h; +N4h: resupplied with KNO3 for 4 h; −N6h: starved of N for 3 d and resupplied with KCl for 6 h; +N6h: resupplied with KNO3 for 6 h. Total RNA was extracted from roots and qPCR reactions were performed in triplicate for each RNA sample. The mRNA of OsActin was used as the reference. A Student’s t-test was calculated at the probability of either 5% (*, p<0.05) or (**, P<0.01).
Effect of P, S deprivation and re-supply on the expression of OsNRT2.1, OsNAR2.1, OsIPS1 and OsSULTR1;1 in rice roots
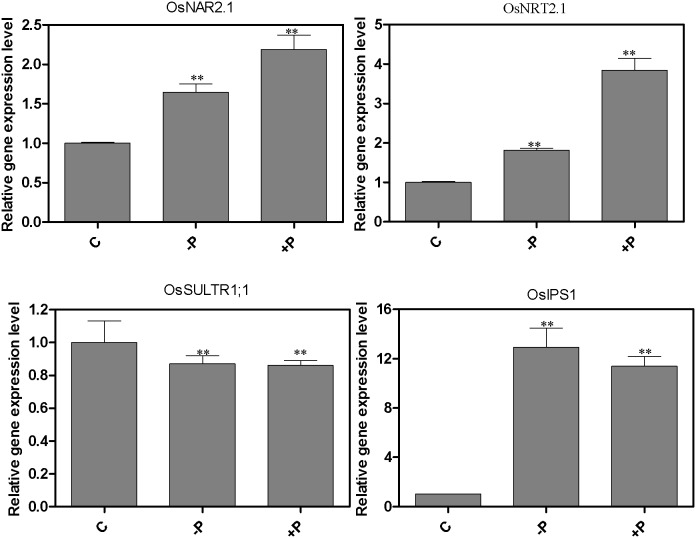
In this experiment two-week old rice seedlings grown in complete nutrient solution were deprived of phosphate or sulfate for 3 d and re-supplied with phosphate or sulfate for 3 h. OsIPS1 was used as P control gene and OsSULTR1;1 was used as S control gene. As shown in Figure 5, the expression of OsIPS1 was notably upregulated by P deprivation and downregulated by P re-supply, which was consistent with previous study [14]. Surprisingly, the gene expression patterns of OsNRT2.1 and OsNAR2.1 is very similar to the expression pattern of OsIPS1, which were upregulated by both starvation and P re-supply in comparison to the continuous P treatment (Figure 5). However, the expression of OsSULTR1;1 was significantly downregulated by P starvation and P re-supply in comparison to the continuously P supply treatment. For the S fluctuation treatment, as shown in Figure 6, the mRNA level of OsSULTR1;1 was significantly increased by sulfate starvation, which was consistent with previous study [47]. The expression patterns of OsNRT2.1, OsNAR2.1 and OsIPS1 were the same as OsSULTR1;1, being upregulated by both S starvation and S re-supply in comparison to the continuous S treatment (Figure 6).
Figure 5. Effect of P-deprivation and re-supply on expression of OsNRT2.1, OsNAR2.1, OsIPS1 and OsSULTR1;1 in rice roots.
Rice seedlings were grown hydroponically in a growth cabinet. Phosphorous treatments were as described in Fig. 3. C: continuous complete nutrient supply; −P: starved of P; +P: resupplied with H2PO4 − for 3 h. Total RNA was extracted from roots and qPCR reactions were performed in triplicate for each RNA sample. The mRNA of OsActin was used as the reference. A Student’s t-test was calculated at the probability of either 5% (*, p<0.05) or (**, P<0.01).
Figure 6. Effect of S-deprivation and re-supply on expression of OsNRT2.1, OsNAR2.1, OsIPS1 and OsSULTR1;1 in rice roots.
Rice seedlings were grown hydroponically in a growth cabinet. Sulfur treatments were as described in Fig. 3. C: continuous complete nutrient supply; −S: starved of S; +S: resupplied with SO4 2− for 3 h. Total RNA was extracted from roots and qPCR reactions were performed in triplicate for each RNA sample. The mRNA of OsActin was used as the reference. A Student’s t-test was calculated at the probability of either 5% (*, p<0.05) or (**, P<0.01).
Discussion
Five ANR1-like genes have different expression patterns in response to nitrogen
It was previously demonstrated that expression of ANR1 in roots of hydroponically grown Arabidopsis plants was induced by N deprivation and rapidly downregulated by N re-supply [29]. This pattern of N responsiveness differs from the NO3 − inducibility of ANR1 as previously obtained in Arabidopsis root cultures [26]. In the present study we have shown that the five ANR1-related genes in rice have diverse expression patterns in response to N starvation and N re-supply. The expression of OsMADS61 was only significantly induced by N starvation and significantly down regulated by N re-supply, which is very similar to the expression pattern of ANR1 in Arabidopsis. In contrast, the expression of OsMADS57 was significantly downregulated by N starvation and significantly upregulated by nitrate re-supply. Furthermore, OsMADS25 and OsMADS27 were only significantly upregulated by nitrate re-supply not by N starvation. These results were partly consistent with the results from [48], in which the rice seedlings were grown in half-strength liquid Murashige and Skoog medium without N and re-supplied with 3 mM KNO3. They found that OsMADS23 and OsMADS61 were not significantly affected by N fluctuation and that OsMADS25, OsMADS27, OsMADS57 were all significantly upregulated by nitrate re-supply [48]. They therefore suggested that the AGL17-like related genes in rice had specific functions differing from those of their A. thaliana homologs. However, our finding found that the expression pattern of OsMADS61 is very similar to that of ANR1 pattern in Arabidopsis.
It has recently been shown that microRNA444 (miR444), which targets three OsMADS-box genes (OsMADS23, OsMADS27, OsMADS57) in rice, has multiple roles in the NO3 – signaling pathway [49]. In this study, OsMADS23, OsMADS27 and OsMADS57 were downregulated under conditions of N-deprivation and were unaffected following 0.5, 1, or 2 h of 5 mM KNO3 supplementation [49]. The different sampling times may account for the partial differences between their results and ours.
The phenotype of transgenic lines overexpressing miR444 provided evidence that OsMADS23, OsMADS27 and OsMADS57 could have a role in regulating the lateral response to localised nitrate in rice [49], as ANR1 does in Arabidopsis [26]. In addition, there was evidence that this group of genes is involved in controlling the root architecture in response to P starvation [49]. However, it has been noted that miR444 has additional target genes (non-MADS box) in rice and there could be other unknown genes involved in the root developmental responses to the nutrient supply [50].
The five ANR1-like homologous gene in rice are regulated by P and S fluctuations
Previous results from Arabidopsis had demonstrated that expression of ANR1 was not regulated by fluctuations in P and S supplies and that and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) was the only type-II MADS-box gene that responded to phosphate and sulfate deprivation and re-supplementation [29], [41]. A later study found that AGL12, AGL18 and AGL19 were downregulated by P and S re-supply [41]. In this study, the OsMADS23, OsMADS25 and OsMADS27 genes were downregulated by both P starvation and P re-supply whereas expression of OsMADS57 and OsMADS61 was not significantly regulated by P fluctuations, which was consistent with what was observed for ANR1 in Arabidopsis [29]. These results were partly consistent with the results from [49], in which phosphate starvation reduced the mRNA levels of two miR444 targets (OsMADS27 and OsMADS57). In our result, the expression level of OsMADS23 and OsMADS27 were downregulated by P starvation, however in [49], the mRNA abundance of OsMADS27 and OsMADS57 were downregulated by phosphate starvation. We used Yoshida [38] rice nutrient solution and Yan chose 1/2 MS culture solution [49] and the time course of phosphate-deprivation was different, which may account for the different gene expression patterns. However, for the S treatment, the expression of all five ANR1-related genes in rice was modulated by S starvation, in contrast to what was seen with ANR1 in Arabidopsis [29], suggesting that these rice genes have specific functions differing from their Arabidopsis homologs. Furthermore, the result of the expression of OsMADS25 regulated by phosphate deprivation was consistent with previous report by [51]. Like SOC1 in Arabidopsis, rice OsMADS25, OsMADS27 and OsMADS57 in roots were responsive to nitrate, phosphate and sulfate fluctuations, which suggest that these three genes may be involved in a general stress response pathway to these three macronutrients.
Crosstalk among OsNRT2.1, OsNAR2.1, OsIPS1 and OsSULTR1;1
As already discussed, depriving seedlings of one mineral nutrient may lead to the disruption of the metabolism of other nutrients [29], [52], [53]. For example, molybdenum deficiency had positive impacts on genes involved in nitrate and sulfate assimilation and phosphate transport [54]. Our results show that OsNRT2.1 and OsNAR2.1 were regulated by both P and S starvation and re-supply, which are partly consistent with previous research in which the gene expression level of AtNRT2.1 in Arabidopsis was found to be upregulated by P and S re-supply [44]. Our results also indicated that OsIPS1 was sensitive to N and S fluctuation and that the expression of OsSULTR1;1 was downregulated by phosphate re-supply, indicating that there is crosstalk between the signaling pathways regulating expression of nitrate, phosphate and sulfate transporters. This is similar to the finding that some nitrate-inducible genes are involved in sulfate metabolism [51], [55]. These results suggest that there is a complex regulatory network among these three macronutrients and their transporters. Further work using mutants or other genomic approach needs to be done to identify the mechanisms of the crosstalk among nitrate, phosphate and sulfate transporters.
Acknowledgments
We thank Prof. Hao Yu from the National University of Singapore for critical reading of this manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
The research was supported by the International Scientific and Technological Cooperation Project of the Ministry of Science and Technology of China (grant number 2010DFA34430), International Scientific and Technological Cooperation Project of Science and Technology Department of Zhejiang Province (grant number 2013C34G2010017), National Natural Science Foundation of China (Grant No. 31370215; 31228002), and Zhejiang Provincial Natural Science Foundation of China (Grant No. Z3110004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rouached H, Stefanovic A, Secco D, Bulak Arpat A, Gout E, et al. (2011) Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis . Plant J 65: 557–570. [DOI] [PubMed] [Google Scholar]
- 2. Robertson GP, Vitousek PM (2009) Nitrogen in agriculture: balancing the cost of an essential resource. Annu Rev Environ Resour 34: 97–125. [Google Scholar]
- 3. Fan X, Shen Q, Ma Z, Zhu H, Yin X, et al. (2005) A comparison of nitrate transport in four different rice (Oryza sativa L.) cultivars. Sci China C Life Sci 48: 897–911. [PubMed] [Google Scholar]
- 4. Forde BG, Clarkson DT (1999) Nitrate and ammonium nutrition of plants: physiological and molecular perspectives. Adv Bot Res 30: 1–90. [Google Scholar]
- 5.Crawford NM, Forde BG (2002) Molecular and developmental biology of inorganic nitrogen nutrition. The Arabidopsis Book. American Society of Plant Biologists. e0011. [DOI] [PMC free article] [PubMed]
- 6. Vidal EA, Gutiérrez RA (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis . Curr Opin Plant Biol 11: 521–529. [DOI] [PubMed] [Google Scholar]
- 7. Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, et al. (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182. [DOI] [PubMed] [Google Scholar]
- 8. Marschner H (1988) Mineral nutrition of higher plants. Plant Cell Environ 11: 147–148. [Google Scholar]
- 9. Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116: 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raghothama KG (1999) Phospate acquisition. Annu Rev Plant Phys 50: 665–693. [DOI] [PubMed] [Google Scholar]
- 11.Aziz T, Sabir M, Farooq M, Maqsood MA, Ahmad H, et al. (2014) Phosphorus deficiency in plants: responses, adaptive mechanisms, and signaling. In: K. R Hakeem, R. U Rehman and I Tahir, editors. Plant signaling: Understanding the molecular crosstalk. India: Springer. 133–148. [Google Scholar]
- 12. Chiou T-J, Lin S-I (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206. [DOI] [PubMed] [Google Scholar]
- 13. Bates TR, Lynch JP (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19: 529–538. [Google Scholar]
- 14. Hou XL, Wu P, Jiao FC, Jia QJ, Chen HM, et al. (2005) Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ 28: 353–364. [Google Scholar]
- 15. Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, et al. (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana . Plant Cell Physiol 46: 174–184. [DOI] [PubMed] [Google Scholar]
- 16. Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, et al. (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796. [DOI] [PubMed] [Google Scholar]
- 17. Ticconi CA, Delatorre CA, Abel S (2001) Attenuation of phosphate starvation responses by phosphite in Arabidopsis . Plant Physiol 127: 963–972. [PMC free article] [PubMed] [Google Scholar]
- 18.Wu P, Xu G, Lian X (2013) Nitrogen and phosphorus uptake and utilization. In: Q Zhang and R. A Wing, editors. Genetics and Genomics of Rice. New York: Springer. 217–226.
- 19. Dai X, Wang Y, Yang A, Zhang W-H (2012) OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses androot architecture in rice. Plant Physiol 159: 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, et al. (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134: 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tabe LM, Droux M (2002) Limits to sulfur accumulation in transgenic lupin seeds expressing a foreign sulfur-rich protein. Plant Physiol 128: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson D (1994) Tansley review No. 73. the responses of plants to non-uniform supplies of nutrients. New Phytol 127: 635–674. [DOI] [PubMed] [Google Scholar]
- 23. Wang X, Wu P, Xia M, Wu Z, Chen Q, et al. (2002) Identification of genes enriched in rice roots of the local nitrate treatment and their expression patterns in split-root treatment. Gene 297: 93–102. [DOI] [PubMed] [Google Scholar]
- 24. Walch-Liu P, Liu L-H, Remans T, Tester M, Forde BG (2006) Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana . Plant Cell Physiol 47: 1045–1057. [DOI] [PubMed] [Google Scholar]
- 25. Walch-Liu P, Ivanov Ii, Filleur S, Gan Y, Remans T, et al. (2006) Nitrogen regulation of root branching. Ann Bot-london 97: 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409. [DOI] [PubMed] [Google Scholar]
- 27. Linkohr BI, Williamson LC, Fitter AH, Leyser HMO (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis . Plant J 29: 751–760. [DOI] [PubMed] [Google Scholar]
- 28. Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-Box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana . Planta 222: 730–742. [DOI] [PubMed] [Google Scholar]
- 30. Gan Y, Bernreiter A, Filleur S, Abram B, Forde BG (2012) Overexpressing the ANR1 MADS-Box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol 53: 1003–1016. [DOI] [PubMed] [Google Scholar]
- 31. Messenguy F, Dubois E (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316: 1–21. [DOI] [PubMed] [Google Scholar]
- 32. Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, et al. (2000) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J 24: 457–466. [DOI] [PubMed] [Google Scholar]
- 33. Burgeff C, Liljegren S, Tapia-López R, Yanofsky M, Alvarez-Buylla E (2002) MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214: 365–372. [DOI] [PubMed] [Google Scholar]
- 34. Filleur S, Walch-Liu P, Gan Y, Forde B (2005) Nitrate and glutamate sensing by plant roots. Biochem Soc Trans 33: 283–286. [DOI] [PubMed] [Google Scholar]
- 35. Lee S, Kim J, Son J-S, Nam J, Jeong D-H, et al. (2003) Systematic reverse genetic screening of T-DNA tagged genes in rice for functional genomic analyses: MADS-box genes as a test case. Plant Cell Physiol 44: 1403–1411. [DOI] [PubMed] [Google Scholar]
- 36. Arora R, Agarwal P, Ray S, Singh A, Singh V, et al. (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meng Y, Shao C, Wang H, Ma X, Chen M (2013) Construction of gene regulatory networks mediated by vegetative and reproductive stage-specific small RNAs in rice (Oryza sativa). New Phytol 197: 441–453. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida S, Forno D, Cock J, Gomez K (1976) Routine procedure for growing rice plants in culture solution. In: S Yoshida, D Forno, J Cock and K Gomez, editors. Laboratory manual for physiological studies of rice. Philippines: The International Rice Research Institut. 61–66.
- 39. Cai C, Wang J-Y, Zhu Y-G, Shen Q-R, Li B, et al. (2008) Gene structure and expression of the high-affinity nitrate transport system in rice roots. J Integr Plant Biol 50: 443–451. [DOI] [PubMed] [Google Scholar]
- 40. Zhou Z, An L, Sun L, Zhu S, Xi W, et al. (2011) Zinc Finger Protein5 Is required for the control of trichome initiation by acting upstream of Zinc Finger Protein8 in Arabidopsis . Plant Physiol 157: 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gan Y-b, Zhou Z-j, An L-j, Bao S-j, Forde BG (2011) A comparison between northern blotting and quantitative real-time PCR as a means of detecting the nutritional regulation of genes expressed in roots of Arabidopsis thaliana . Agric Sci China 10: 335–342. [Google Scholar]
- 42. An L, Zhou Z, Sun L, Yan A, Xi W, et al. (2012) A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis . Plant J 72: 474–490. [DOI] [PubMed] [Google Scholar]
- 43. Gan Y, Zhou Z, An L, Bao S, Liu Q, et al. (2010) The effects of fluctuations in the nutrient supply on the expression of ANR1and 11 other MADS box genes in shoots and roots of Arabidopsis thaliana . Botany 88: 1023–1031. [Google Scholar]
- 44. Bao S, An L, Su S, Zhou Z, Gan Y (2011) Expression patterns of nitrate, phosphate, and sulfate transporters in Arabidopsis roots exposed to different nutritional regimes. Botany 89: 647–653. [Google Scholar]
- 45. Feng H, Yan M, Fan X, Li B, Shen Q, et al. (2011) Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot 62: 2319–2332. [DOI] [PubMed] [Google Scholar]
- 46. Araki R, Hasegawa H (2006) Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breeding Sci 56: 295–302. [Google Scholar]
- 47. Kumar S, Asif MH, Chakrabarty D, Tripathi RD, Trivedi PK (2011) Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funct Integr Genomics 11: 259–273. [DOI] [PubMed] [Google Scholar]
- 48. Puig J, Meynard D, Khong GN, Pauluzzi G, Guiderdoni E, et al. (2013) Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr Patterns 13: 160–170. [DOI] [PubMed] [Google Scholar]
- 49. Yan Y, Wang H, Hamera S, Chen X, Fang R (2014) miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J 78: 44–55. [DOI] [PubMed] [Google Scholar]
- 50. Forde BG (2014) Glutamate signalling in roots. J Exp Bot 65: 779–787. [DOI] [PubMed] [Google Scholar]
- 51. Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate,iron, and sulfate metabolism. Plant Physiol 132: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Prosser IM, Purves JV, Saker LR, Clarkson DT (2001) Rapid disruption of nitrogen metabolism and nitrate transport in spinach plants deprived of sulphate. J Exp Bot 52: 113–121. [PubMed] [Google Scholar]
- 53. Takehisa H, Sato Y, Antonio B, Nagamura Y (2013) Global transcriptome profile of rice root in response to essential macronutrient deficiency. Plant Signal Behav 8: e24409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ide Y, Kusano M, Oikawa A, Fukushima A, Tomatsu H, et al. (2011) Effects of molybdenum deficiency and defects in molybdate transporter MOT1 on transcript accumulation and nitrogen/sulphur metabolism in Arabidopsis thaliana . J Exp Bot 62: 1483–1497. [DOI] [PubMed] [Google Scholar]
- 55. Leustek T, Martin MN, Bick J-A, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Phys 51: 141–165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.