Abstract
Innate immune sensors of intracellular nucleic acids must be regulated to prevent inappropriate activation by endogenous DNA and RNA. The exonuclease Trex1 regulates the DNA sensing pathway by metabolizing potential DNA ligands that trigger it. However, an analogous mechanism for regulating the RIG-I-like receptors (RLRs) that detect RNA remains unknown. We show that the SKIV2L RNA exosome potently limits the activation of RLRs. We find that the unfolded protein response (UPR), which generates endogenous RLR ligands through IRE-1 endonuclease cleavage of cellular RNAs, triggers type I interferon (IFN) production in SKIV2L-depleted cells. Humans with SKIV2L deficiency have a type I IFN signature in their peripheral blood. Our findings reveal a mechanism for intracellular metabolism of immunostimulatory RNA, with implications for specific autoimmune disorders.
INTRODUCTION
Antiviral immunity is initiated within virus-infected cells by innate immune sensors of viral nucleic acids1. These sensors must detect viral RNA or DNA among a vast excess of cellular RNA and DNA, which presents a challenge of self/non-self discrimination and a risk of inappropriate immune responses to self nucleic acids. Specificity of these innate antiviral sensors is primarily accomplished by the detection of unique structural features that distinguish viral nucleic acids. For example, the RIG-I RNA helicase binds to 5′ triphosphate RNA that is present in the genomes of many classes of RNA viruses but scarce within host cells2. Similarly, the MDA5 RNA helicase is activated by long, double-stranded viral RNA that is not routinely formed in host cells3. For intracellular DNA sensing by the interferon stimulatory DNA (ISD) pathway, the mechanisms of self/non-self discrimination are less clear because the known DNA sensors are activated in a sequence-independent fashion by nearly any double-stranded DNA4–6. Indeed, recently published crystal structures of several important DNA receptors reveal that most molecular contacts with immunostimulatory DNA are made with the sugar-phosphate backbone and not the specific bases7–9.
Recently, intracellular nucleic acid metabolism was identified as an essential mechanism for limiting the activation of the ISD pathway. We identified the 3′ repair exonuclease 1 (Trex1) in a biochemical screen for ISD-binding proteins10. Loss-of-function mutations in the human TREX1 gene cause Aicardi-Goutières syndrome (AGS), a severe type I interferon (IFN)-associated autoimmune disease11. Using Trex1-deficient mice as a model of AGS, we defined Trex1 as an essential negative regulator of the ISD pathway10,12. Moreover, we found that the reverse-transcribed cDNAs of endogenous retroelements accumulate within Trex1-deficient cells and that Trex1 is a potent anti-retroviral enzyme10. These studies provide a framework for understanding the pathogenic mechanisms of AGS and related diseases, and reveal an important source of endogenous intracellular nucleic acids that can trigger innate immune sensors of DNA if they fail to be properly metabolized.
Based on these findings, we wondered whether an analogous mechanism exists to metabolize intracellular RNA for regulation of the RIG-I-like receptors (RLRs), and whether there is a source of relevant endogenous immunostimulatory RNAs that could trigger the RLRs upon their accumulation. We show here that the cytosolic 3′-to-5′ RNA exosome, defined by the SKIV2L RNA helicase, is an important negative regulator of the RLR-mediated antiviral response. We identify the RNA cleavage products of the inositol-requiring enzyme 1 (IRE-1) endonuclease as immunostimulatory RNAs that activate the RLR pathway in SKIV2L-depleted cells upon activation of the unfolded protein response (UPR). We show that SKIV2L-deficient humans have a type I IFN signature in their peripheral blood cells. These findings reveal a mechanism that may contribute to IFN-associated autoimmune diseases.
RESULTS
SKIV2L limits the RLR antiviral response
We began our exploration of potential negative regulators of the RNA-activated antiviral response by considering the ubiquitous pathways for RNA degradation that mediate turnover of mRNAs and elimination of incompletely spliced RNA transcripts. These pathways are often initiated by endonucleolytic cleavage of the RNA, followed by degradation of the two resulting products by distinct enzyme complexes13. The XRN1 exonuclease metabolizes RNA in the 5′-to-3′ direction, and the RNA exosome degrades RNA in the 3′-to-5′ direction13. The RNA exosome is a multi-protein complex composed of several core factors associated with key accessory proteins that determine its subcellular localization and RNA substrate specificity14. The RNA exosome responsible for RNA turnover in the cytoplasm of human cells is formed by the RNA helicase SKIV2L together with additional subunits that are well-characterized in yeast (SKI3, SKI8, ref. 15) but remain poorly defined in humans. Intriguingly, a previous study identified SKIV2L as a potential susceptibility gene for the autoimmune disorder systemic lupus erythematosus (SLE) in humans16. We established stable, robust lentiviral knockdown of SKIV2L and XRN1 in primary mouse bone marrow-derived macrophages (BMDMs), confirming depletion of these proteins by immunoblot (Fig. 1a). We stimulated the knockdown cells by transfection of a pure triphosphate RNA ligand for RIG-I17, and measured IFN-β mRNA transcription as the primary and most direct cytokine readout of RLR pathway activation. We found that SKIV2L-depleted cells exhibited a substantially enhanced IFN-β response to RIG-I ligands as compared to cells with scramble shRNA, and that XRN1 knockdown had no effect on IFN-β production (Fig. 1b). SKIV2L-depleted cells also had an enhanced IFN-β response to transfection of poly(I:C), which activates both RIG-I and MDA5 (Fig. 1c). The abundance of mRNAs encoding IFNα4, the inflammatory chemokine CXCL10 and interleukin 6 (IL-6), were similarly enhanced in SKIV2L-depleted cells but not XRN-1-depleted cells after RIG-I ligand stimulation (Fig. 1d–f), demonstrating broad enhancement of the RLR-activated antiviral response conferred by SKIV2L knockdown.
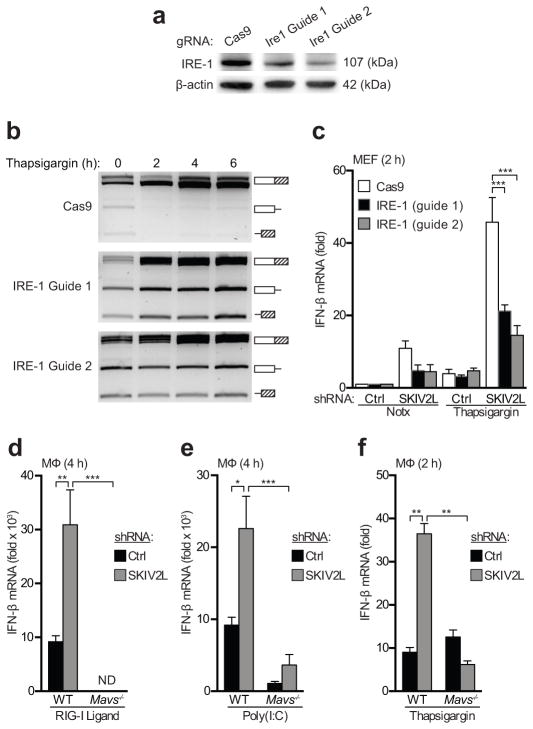
Figure 1. SKIV2L is a negative regulator of the RIG-I-like receptor-mediated antiviral response.
(a) SKIV2L and XRN1 were stably knocked down in primary macrophages, and the respective protein levels were evaluated by immunoblotting.
(b,c) Primary mouse macrophages, stably transduced as indicated with shRNA knockdown constructs, were treated with an HCV RIG-I ligand (b) or Poly (I:C) (c) and assessed for IFN-β production by quantitative RT-PCR.
(d–f) RIG-I ligand-stimulated macrophages with the indicated knockdown constructs were evaluated by quantitative RT-PCR for expression of IFNa4 (d), CXCL10 (e) and IL-6 (f).
(g,h) Macrophages were stimulated with DMXAA to activate STING directly, and with LPS to activate TLR4. *P < 0.05; **P < 0.001; ***P < 0.0005; ****P < 0.0001 (2-way ANOVA, b–f). Data are representative of at least three independent experiments with biological triplicates (mean + s.d., a–h).
The enhancement of the antiviral response in SKIV2L-depleted cells could be a result of increased mRNA stability conferred by loss of a key participant in the regulation of mRNA turnover. To control for the specificity of SKIV2L depletion, we stimulated cells with two non-nucleic acid ligands that each potently induce the type I IFN response. The chemotherapeutic agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA) binds to and activates the STING adapter protein18,19, and lipopolysaccharide (LPS) triggers IFN-β expression through the TLR4-Trif pathway20. We found that SKIV2L-depleted cells did not exhibit enhanced responses to these non-RNA ligands (Fig. 1g,h). These data suggest that SKIV2L is a specific negative regulator of the RNA-activated RLR response.
The UPR triggers IFN production in SKIV2L-depleted cells
We considered the potential existence of endogenous immunostimulatory RNAs that could trigger the RLRs upon accumulation within cells. We hypothesized that the UPR might provide a source of such RNAs. The UPR is a stress response that is activated when the burden of newly synthesized polypeptides in the endoplasmic reticulum (ER) exceeds its protein folding capacity21. UPR activation induces the expression of genes that encode ER protein chaperones, ER membrane biosynthetic enzymes, and the ER-associated protein degradation machinery, thus restoring homeostasis by increasing the size and functional capacity of the ER22. One key component of the UPR is the ER-resident, transmembrane kinase/endoribonuclease IRE-1 (refs. 23,24). The ER luminal domain of IRE-1 is activated directly by misfolded proteins25,26, leading to the activation of its cytosolic RNA endonuclease domain. The primary role or IRE-1 in UPR induction is to catalyze the removal of a small intron from the mRNA encoding the XBP1 transcription factor by precisely cleaving the XBP1 mRNA at each end of the intron27,28. The resulting exons are then spliced by tRNA ligase29, and this newly spliced mRNA encodes functional XBP1 protein, which migrates to the nucleus and induces the expression of dozens of UPR-inducible genes21. In addition to its role in XBP1 mRNA splicing, IRE-1 in metazoan organisms also mediates the destructive cleavage of many ER-localized mRNAs30,31. This regulated IRE-1-dependent RNA decay (RIDD) transiently reduces the burden of newly synthesized proteins entering the ER, protecting it from further proteotoxic stress30. IRE-1 endonuclease cleavage generates RNAs with a 3′ cyclic phosphate moiety, and these RNAs are efficiently cleared by the SKI2 RNA exosome30. Interestingly, RNAs containing 3′ cyclic phosphates are capable of activating the RLRs32,33. Moreover, a recent study found that UPR activation by cholera toxin triggers an IRE-1-dependent, RIG-I-mediated inflammatory response34. Based on these findings, we reasoned that activation of the UPR in SKIV2L-depleted cells might result in an accumulation of endogenous IRE-1 RNA cleavage products that could elicit an ectopic type I IFN response.
We knocked down SKIV2L or XRN1 in immortalized mouse embryonic fibroblasts and then stimulated these cells with thapsigargin, a chemical inhibitor of the sarco/endoplasmic reticulum calcium ATPase (SERCA) that depletes calcium stores from the ER and triggers a potent UPR. Upon induction of the UPR, we found that SKIV2L-depleted cells activated significant IFN-β production that we did not observe in either XRN1-depleted cells or cells transduced with scramble shRNA (Fig. 2a). We observed a similar induction of IFN-β in SKIV2L-depleted primary BMDMs treated with thapsigargin, but again not in control or XRN1-depleted macrophages (Fig. 2b). To extend these findings, we treated BMDMs with tunicamycin, a nucleoside antibiotic that inhibits N-linked glycosylation and induces the UPR by causing the accumulation of unglycosylated proteins in the ER21. We treated macrophages with thapsigargin or tunicamycin and monitored the kinetics of UPR activation by tracking the IRE-1-dependent splicing of XBP1 mRNA35. We found that thapsigargin-induced XBP1 mRNA splicing was nearly complete at two hours post treatment and returned to baseline by 24 hours (Fig. 2c). In contrast, tunicamycin-induced XBP1 mRNA splicing occurred with delayed kinetics, commencing at 6 hours and continuing through 24 hours (Fig. 2c). Interestingly, both thapsigargin and tunicamycin activated IFN-β production in SKIV2L-depleted cells with kinetics that precisely mirrored activation of IRE-1 (Fig. 2d,e). Thus, two mechanistically distinct activators of the UPR trigger a substantial antiviral response in SKIV2L-depleted cells.
Figure 2. The UPR activates an antiviral response in SKIV2L-depleted cells.
(a) SV40 Large T antigen-immortalized mouse embryonic fibroblasts were transduced with the indicated knockdown constructs and treated with thapsigargin (TG). Ifnb mRNA induction was evaluated using quantitative RT-PCR at the indicated time points. Data are representative of 3 experiments.
(b) Primary mouse macrophages were transduced and treated as in (a).
(c) Endonuclease activity of IRE-1 is responsible for removal of an intron in XBP-1 mRNA, as well as destructive cleavage of ER-localized RNAs. The kinetics of UPR activation were assessed using a splicing assay for XBP-1 mRNA. The illustration shows the PstI digest site in the intron of unspliced XBP-1 mRNA used to distinguish spliced (S) and unspliced (U1, U2) cDNAs.
(d–e) Primary mouse macrophages were treated with thapsigargin (TG; e) or tunicamycin (TM; f) treatment. *P < 0.05; **P < 0.0001 (2-way ANOVA, b–e). Data are representative of at least three independent experiments with biological triplicates (mean + s.d., b–e).
We next tested whether the enhanced UPR-induced IFN response in SKIV2L-depleted cells required IRE-1 activity. To do this, we used the CRISPR/Cas9 genome editing approach36,37 to target the endogenous Ern1 gene that encodes IRE-1α. We developed a construct similar to one described in a recent report38 that enables co-expression of a guide RNA and the Cas9 endonuclease from a single lentiviral vector. We targeted Ern1 coding exons in immortalized MEFs using two different guide RNAs, and we found near complete modification of the target sites in these cells several days after selection for transduced cells (Supplementary Fig. 1). The targeted cells had reduced expression of IRE-1α protein and were severely compromised in UPR-induced XBP1 mRNA splicing compared to control cells transduced with Cas9 alone (Fig. 3a,b), demonstrating extensive loss of IRE-1α function in the targeted cells. We next knocked down SKIV2L in the Cas9 control cells and in the Ern1-targeted cells and stimulated them with thapsigargin to induce the UPR. SKIV2L depletion in the control cells resulted in an enhanced UPR-induced IFN response, however this enhancement was significantly blunted in both lines of Ern1-targeted cells (Fig. 3c).
Figure 3. SKIV2L specifically regulates the MAVS-dependent antiviral response.
(a) Immortalized MEFs were transduced with lenti-CRISPR constructs encoding CAS9 alone or two different guide RNAs targeting Ern1. IRE-1α protein was measured by immunoblot.
(b) CAS9 control and IRE-1α-targeted cells were treated with thapsigargin to induce the UPR, and IRE-1- mediated XBP-1 mRNA splicing was quantitated as in Fig. 2d.
(c) Control and IRE-1-targeted cells were treated with thapsigargin, and IFN-β mRNA was measured by quantitative RT-PCR at the indicated time points. Data are representative of one experiment with biological triplicates testing two independent guide RNAs (mean + s.d., a–c).
(d–f) Bone marrow macrophages of the indicated genotypes were transduced with a control or SKIV2L shRNA, treated with RIG-I ligand (d), Poly I:C (e), or thapsigargin (f), and harvested for analysis at the indicated time points. *P < 0.001; **P < 0.0005; ***P < 0.0001; (2-way ANOVA, c–f). Data are representative of at least three independent experiments with biological triplicates (mean + s.d., d–f).
To formally establish a role for RLR signaling in the enhanced antiviral response in SKIV2L-depleted cells, we compared IFN-β induction in wild-type BMDMs to MAVS-deficient BMDMs that lack RIG-I- and MDA5-induced type I IFN production39. As expected, the IFN response to RIG-I ligand transfection in wild-type cells, as well as the enhanced IFN response in SKIV2L-depleted cells, was entirely MAVS-dependent (Fig. 3d). Similarly, the increased response to poly(I:C) transfection in SKIV2L-depleted cells required MAVS, with the residual IFN induction in MAVS-deficient cells likely mediated by the TLR3-Trif pathway (Fig. 3e;20). Importantly, we found that the induction of IFN during UPR activation in SKIV2L-depleted cells was also dependent on MAVS (Fig. 3f). Together, these data reveal that SKIV2L is an important negative regulator of the RLR response to exogenous RNA ligands. Moreover, we identify the UPR as a source of endogenous RNA ligands that trigger an ectopic antiviral response in SKIV2L-depleted cells through an IRE-1α– and MAVS-dependent pathway.
SKIV2L-deficient humans have a type I IFN signature
We next explored a role for SKIV2L in regulating the type I IFN response in humans. Loss-of-function mutations in the human SKIV2L gene cause tricohepatoenteric syndrome (THES; OMIM 222470; ref. 40). THES is an extremely rare disease characterized by growth retardation, facial dysmorphism, immunodeficiency, liver and intestine abnormalities and intractable diarrhea that requires parenteral nutrition41. These severe symptoms likely reflect the broad role for the cytosolic RNA exosome in maintaining RNA homeostasis within cells, as discussed below. In addition to the mutations in SKIV2L that cause THES, loss-of function mutations in TTC37, the human ortholog of the yeast SKI3 RNA exosome component, also cause the same disease42,43.
We obtained peripheral blood samples from two of the four living THES patients with biallelic SKIV2L mutations, as well as from three THES patients with TTC37 mutations and three controls heterozygous for THES mutations (Table 1). We confirmed complete loss of SKIV2L protein in extracts derived from a lymphoblastoid cell line derived from a THES patient with biallelic SKIV2L mutations (Fig. 4a), providing further support for the notion that these SKIV2L mutations are indeed loss-of-function. We measured the relative abundance of six IFN-stimulated genes (ISGs) by quantitative RT-PCR using cDNA prepared from fresh blood samples: IFI27, IFI44L, IFIT1, ISG15, RSAD2, and SIGLEC1. To establish the range of ISG expression in a well-characterized, IFN-mediated human autoimmune disorder and for comparison with our THES samples, we reproduced the recently published ISG profiles of blood samples from 82 AGS patients and 29 healthy controls, adding three additional controls of individuals heterozygous for THES mutations (Fig. 4b;44). Remarkably, we found that both of the SKIV2L-deficient THES patients had a type I IFN signature that was even more robust than the mean ISG scores in AGS patients (Fig. 4b,c). In contrast, we found no evidence for an IFN signature in the three THES patients with TTC37 mutations (Fig. 4b,c). Our findings reveal a strong IFN signature in SKIV2L-deficient THES patients, establishing an important role for SKIV2L in the negative regulation of the IFN-mediated antiviral response in humans. However, the lack of IFN signature in TTC37-deficient THES patients raised the possibility that SKIV2L and TTC37 contribute differently to this negative regulation.
Table 1.
Details of ancestry and mutations in THES peripheral blood samples
| Status | Gene Mutated | Mutation 1 | Mutation 2 | Protein | Gender | Age at Harvest | Ancestry | Consanguinity |
|---|---|---|---|---|---|---|---|---|
| Control | SKIV2L | c.1635insA | - | p.Gly546Argfs*35 | Male | 49 | North Africa | - |
| Control | SKIV2L | c.1635insA | - | p.Gly546Argfs*35 | Female | 43 | North Africa | - |
| Control | TTC37 | c.3960C>A | - | p.(Tyr1320*) | Male | 50 | North Africa | - |
| THES | SKIV2L | c.3561_3581del | c.3561_3581del | p.(Ser1189_Leu1195del) | Male | 1 | Kuwait | Yes |
| THES | SKIV2L | c.848G>A | c.1022T>G | p.(Trp283*), p.Val341Gly | Female | 4 | France | No |
| THES | TTC37 | c.3960C>A | c.3960C>A | p.(Tyr1320*) | Male | 10 | France | Yes |
| THES | TTC37 | c.2808G>A | c.2808G>A | p.Trp936* | Male | 5 | Pakistan | Yes |
| THES | TTC37 | c.2779-2G>A | c.2779-2G>A | p.Glu974Glyfs*19 | Male | 9 | Pakistan | Yes |
Figure 4. SKIV2L-deficient humans have a robust type I IFN signature.
(a) SKIV2L protein was evaluated by immunoblot in control and THES (c.1635insA/c.1635insA) patient lymphoblastoid cell lines.
(b) Quantitative RT-PCR measuring six human interferon-stimulated genes (ISGs) in cDNA prepared from whole blood. For each gene, we present values from 29 AGS control patients (black closed circles) together with 3 controls with heterozygous THES mutations (red filled squares), 82 AGS patients (black open triangles), 2 THES patients with SKIV2L mutations (red filled triangles), and 3 THES patients with TTC37 mutations (blue open circles). Horizontal bars represent the median relative quantification (RQ) value for each ISG probe in each group, normalized to HPRT and 18S rRNA expression within each sample. Note that the data from AGS controls and AGS patients were recently published44 and are reproduced here for comparison to the THES controls and THES patients.
(c) Interferon scores were calculated using the median fold change in RQ value for all of the six interferon-stimulated genes in each individual as described44. Horizontal bars show the mean interferon scores in patients and controls. (Note: Cannot provide P value, as there are only 2 SKIV2L THES patient samples)
Differential roles of SKIV2L and TTC37 RLR regulation
Based on the presence of an IFN signature in SKIV2L-deficient THES patients and its absence in TTC37-deficient patients with the same disease, we wondered whether SKIV2L has a unique function in RLR regulation that is distinct from its role in the classical cytosolic RNA exosome defined by both SKIV2L and TTC37. We therefore established stable and efficient lentiviral knockdown of TTC37 in primary murine macrophages (Fig. 5a). We found that SKIV2L depletion markedly enhanced the IFN response to transfected RLR ligands. In contrast, TTC37-depleted macrophages exhibited an IFN response that was similar to control cells (Fig. 5b). Similarly, SKIV2L knockdown enhanced the IFN response to thapsigargin-induced UPR activation, but TTC37 knockdown had no effect (Fig. 5c). Together with the THES patient ISG data presented in Fig. 4, these findings reveal that SKIV2L plays a unique role in the regulation of RLR responses that is distinct from its role in the SKIV2L-TTC37 cytosolic RNA exosome.
Figure 5. Differential role of SKIV2L and TTC37 in RLR regulation.
(a) Immunoblots of primary BMDM lysates showing TTC37 protein expression in knockdown and control cells. Data are representative of two independent experiments.
(b,c) Cells were transduced with the indicated lentiviral shRNA constructs and treated with thapsigarin or RIG-I ligands and assessed for IFN-β production by quantitative RT-PCR. RIG-I ligand-treated samples were harvested at 4 h and thapsigargin-treated samples were harvested at 2 h. *P < 0.01; **P < 0.005; ***P < 0.0001; (one-way ANOVA, b–c). Data are representative of two independent experiments with biological triplicates (mean + s.d., b–c).
DISCUSSION
Our data extend the paradigm of negative regulation of cell-intrinsic innate antiviral responses that was first described for the DNA sensing pathway10 by identifying a key negative regulator of the RLRs that detect RNA. We find that the cytosolic RNA exosome, defined by the SKIV2L RNA helicase, is important for limiting the activation of RLRs. We identify the UPR as a cellular stress response that generates endogenous RLR ligands, and we show that cells undergoing a UPR trigger an aberrant IFN-mediated antiviral response if these endogenous RNAs fail to be metabolized. We show that humans with SKIV2L deficiency have a strong type I IFN signature in their peripheral blood, indicative of a chronic antiviral response that may be relevant to human autoimmune disorders. Finally, we present evidence in human patients and in mouse cells that the role of SKIV2L in RLR regulation is independent of another exosome protein, TTC37. Taken together, these findings reveal a new mechanism of RLR regulation and emphasize the importance of intracellular nucleic acid metabolism for preventing aberrant innate immune responses.
We found that SKIV2L-depleted cells, but not XRN1-depleted cells, had enhanced IFN-β response to RLR ligands. Perhaps more relevant to autoimmune disease, we identify the RNA degradation products of IRE-1 that are generated during the UPR as a source of endogenous immunostimulatory RNAs that trigger a MAVS-dependent IFN response in SKIV2L-depleted cells, but not in wild-type cells. Importantly, this finding reveals a mechanism by which a “sterile” stress response could be misinterpreted as a viral infection if cells fail to metabolize the RNA degradation products that are formed as a result of the stress. The UPR-activated RLR response in SKIV2L-depleted cells was robust and significant, but much less potent than the response to transfected triphosphate RNAs. This likely reflects the number of RIG-I ligands introduced by transfection: one microgram of transfected RIG-I ligand contains approximately 1013 molecules, which would amount to ~10 million molecules delivered per cell in our experimental system. In contrast, IRE-1-mediated cleavage of cellular mRNAs would generate far fewer endogenous RIG-I ligands. Moreover, it is also likely that 3′ cyclic phosphate RNAs are less efficient activators of RLRs than 5′ triphosphate RNAs32,34. Despite the relatively weaker IFN response triggered by the UPR, we propose that such an IFN response in a chronic setting would be sufficient to drive substantial pathology.
Our findings suggest a new potential mechanism for the pathogenesis of certain autoimmune diseases that affect highly secretory cells that routinely undergo ER stress, including Sjögren’s syndrome (which affects salivary and lacrimal glands) and Type I diabetes (which affects insulin-secreting β cells of the exocrine pancreas). We propose a simple model in which regular, episodic activation of the UPR would generate endogenous RIG-I ligands. In individuals with deficiencies in their ability to metabolize these RNAs, this UPR would trigger a chronic, inappropriate antiviral response, leading to immune activation and eventual destruction of the cells over time. Importantly, our model emphasizes the role of the secretory cells themselves as active participants in the autoimmune process. By activating the cell-intrinsic antiviral response, these cells become the targets of an adaptive immune response, similar to the manner in which virus-infected cells are targeted for elimination by cytotoxic T cells.
We show that human patients with SKIV2L deficiency have a potent IFN signature in their peripheral blood cells, thus providing important evidence for the relevance of this regulatory mechanism to human disease. Our findings raise two important questions. First, does this IFN signature contribute to the pathogenesis of THES? Based on the lack of IFN signature in TTC37-deficient THES patients, we propose that most of the symptoms of this severe disease are the consequence of loss of the ‘housekeeping’ function of the cytosolic RNA exosome in general RNA turnover, rather than the aberrant IFN response that is found only in SKIV2L-deficient patients. Second, given the strong IFN signature, why do SKIV2L-deficient patients not develop autoimmunity? Interestingly, some THES patients also exhibit immunodeficiency and require supplemental immunoglobulins41, suggesting an essential role for the RNA exosome in lymphocyte function. Indeed, we found that the survival of our SKIV2L-knockdown cells was compromised after stimulation, precluding a number of experiments that we attempted to perform, including RNA virus infections (data not shown). Taken together, we suggest that defective lymphocyte function in THES patients prevents overt autoimmune disease, despite the potent IFN signature evident in their peripheral blood cells. However, the immunological aspects of THES have thus far been incompletely defined, in large part because of the extreme rarity of the disease and the severity of the other symptoms. It will be informative to further explore potential autoimmune phenotypes in THES and their differential manifestation in patients with SKIV2L mutations and TTC37 mutations, in light of the findings presented above.
We present evidence that SKIV2L contributes to RLR regulation, but the other defined component of the cytosolic RNA exosome, TTC37, does not. The subcellular localization and RNA substrate specificity of the RNA exosome is determined by its associated cofactors, such that the exosome associated with SKIV2L and TTC37 is primarily involved in cytosolic RNA decay14. Moreover, the RNA exonucleases that also associate with the core exosome components can each mediate degradation of unique subsets of RNAs within cells14. We propose that the role for SKIV2L uncovered here is distinct from its involvement in the conventional cytosolic RNA exosome, and it will be interesting to determine whether there are proteins that partner with SKIV2L that are uniquely involved in RLR regulation. Such proteins may serve to target SKIV2L specifically to immunostimulatory RNAs.
In summary, we identify a regulatory mechanism in which metabolism of intracellular RNA ligands limits activation of the RIG-I-like receptors. We propose that deficient function of this regulatory mechanism would predispose to autoimmune diseases, particularly those that affect highly secretory cells. Identification of other components of this pathway may provide insight into the pathogenesis of certain IFN-associated autoimmune disorders.
METHODS
Mice and cells
All mice used were on an inbred C57BL/6 background, and C57BL/6 mice were purchased from the Jackson Laboratories. Mavs−/− mice (provided by M. Gale, Jr., University of Washington, Seattle, WA) were generated on a C57BL/6 background as previously described12. BMDMs were harvested from leg bones and cultured in complete RPMI media supplemented with MCSF for 8 days. WT controls for all experiments were age-matched C57BL/6 mice. Mice used were 4–20 weeks old and littermates were used when available. All mice were maintained in accordance with guidelines of the University of Washington Institutional Animal Care and Use Committee. HEK293T cells were obtained from the ATCC. SV40 Large T antigen-immortalized fibroblasts were generated by retroviral transduction of primary MEFs. All cell lines tested negative for mycoplasma contamination.
Cell treatments and analysis
RIG-I ligand was transcribed and purified in vitro as previously described using the T7 Megashortscript kit (Ambion)17. PU/UC A template sequence is available in Supplementary Table 1. BMDMs were plated at a density of 0.7 x 106 in 12-well plates for transfection. For transfections, 5 μg poly(I:C), or 1 μg RIG-I ligand were complexed with Lipofectamine 2000 (Life Technologies) at a ratio of 1 μg nucleic acid to 1 μl lipid in a final volume of 1 ml. Final concentrations of 30 μg/ml DMXAA (Sigma-Aldrich), 10 ng/ml LPS (Sigma Aldrich), 500 nM Thapsigargin (Life Technologies), and 2.5 μg/ml Tunicamycin (Santa Cruz Biotechnology) were added directly to the culture media. Treatment times are indicated in figures. For quantitative RT-PCR of IFN-β mRNA, cells were harvested into RNA-Bee (Teltest). RNA was treated with DNase (Ambion) and primed with Oligo(dT), then reverse transcribed with Superscript III (Life Technologies). cDNA was used for PCR with EVA Green reagents (Bio-Rad Laboratories) on a Bio-Rad CFX96 Real-Time System. The abundance of each cytokine mRNA was normalized to HPRT expression and compared with untreated cells transduced with the same shRNA to calculate the relative induction. Primers are available in Supplementary Table 1.
Lentiviral shRNA knockdown and CRISPR
SKIV2L, TTC37 and control shRNA constructs were designed and cloned into the pLKO.1 vector. XRN1 shRNA was designed and cloned into the retroviral MSCV-LMP vector. SKIV2L was targeted by the sequence (sense): 5′-CGCATCATGGAGTCTGTGAAT-3′. TTC37 was targeted by a combination of two plasmids: (sense): 5′-AGAAGATTATGTGCCTGCCTT-3′ and (sense): 5′-TTCAGAATTCCGCTTCAGCTT-3′. XRN1 was targeted by the sequence (sense): 5′-GAGTAGCTTCTAGAGATAA-3′. The control shRNA targeted eGFP: 5′-CAACAAGATGAAGAGCACCAA-3′. Knockdowns were validated by immunoblotting of whole-cell extracts with mouse monoclonal antibodies according to standard techniques. Antibodies were: SKIV2L (Proteintech Group, Inc. catalog number 11462-1-AP), TTC37 (Abcam, catalog number ab122421), XRN1 (Santa Cruz Biotechnology, Catalog #sc-98459), and actin (Sigma, clone AC-74).
For SKIV2L, XRN1 and TTC37 knockdowns in BMDMs, VSV-G-pseudotyped lentivirus was produced by transfecting 2.5 x 106 HEK293T cells in 10-cm plates with 10 μg of shRNA knockdown construct, 9 μg psPAX-2, a lentiviral packaging plasmid with a puromycin resistance gene, and 1 μg pVSV-G envelope plasmid for 48 h. 4 x 106 BMDMs were transduced with HEK293T viral supernatants on days 3 and 4 after harvest, selected with 5 μg/ml puromycin (Life Technologies) for 3 d, and then plated at equal numbers for treatments (described above).
For CRISPR-Cas9 targeting of Ern1, we developed a lentivirus vector similar to the one recently reported by Zhang and colleagues38 in which a U6 promoter-driven guide RNA and a MND promoter-driven Cas9-T2A-blasticidin resistance cassette were constitutively expressed from a single, self-inactivating lentivirus upon integration into the host cell genome. Immortalized MEFs were transduced and selected as described above, and CRISPR targeting of the Ern1 locus was evaluated by restriction fragment length polymorphism (RFLP) using a restriction site that overlapped the CRISPR targeting site (IRE-1 guide 1: BsajI, IRE-1 guide 2: MlucI). Products were run on a 3% MetaPhor agarose gel (Lonza, Supplementary Figure 1). The sequences of the guide RNA target sequences are: IRE-1α gRNA #1 (sense): 5′-GCTTGTTGTTTGTCTCGACCC-3′; IRE-1α gRNA #2 (sense): 5′-GGGGAGGCCTGAACCAATTC-3′.
Splicing Assay
RT-PCR Xbp1 splicing assays were performed as described previously35. cDNA was obtained as described above. Product was amplified using sense primer mXBP1.3S (sense): 5′-AAACAGAGTAGCAGCGCAGACTGC-3′, and antisense primer mXBP1.2AS (antisense): 5′-GGATCTCTAAAACTAGAGGCTTGGTG-3′. PCR products were then digested with PstI and run on a 3% MetaPhor agarose gel (Lonza).
Human Interferon Signature Scores
Human peripheral blood samples were obtained with informed, written consent, processed, and analyzed for the expression of six human interferon-stimulated genes as described previously 44. AGS sample collection was approved by the Leeds (East) Research Ethics Committee (reference number 10/H1307/132). THES patient sample collection was approved by the South Birmingham Research Ethics Committee, and by the French ministry of Health, authorization number: AC-2011-1312.
Statistical Analysis
RT-qPCR data were analyzed in Graphpad Prism software using a 2-way ANOVA. Significant differences were considered at P values of < 0.05. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Supplementary Material
Acknowledgments
We are grateful to M. Gale, Jr (University of Washington) for providing Mavs−/− mice, and to members of the Stetson lab for helpful discussions. Supported by the National Institute of Allergy and Infectious Disease (AI084914; D.B.S.), the European Union (FP7/2007–2013) grant agreement number 241779 to D.B.S. (NIMBL: http://www.NIMBL.eu), the Lupus Research Institute (D.B.S.), and the Cancer Research Institute (E.E.G). D.B.S. is a scholar of the Rita Allen Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
D.B.S. and S.C.E. designed the study; S.C.E. performed all of the experiments in murine cells; G.I.R. and Y.J.C. performed the ISG analysis of human cells and helped write the manuscript; E.E.G. developed the Lenti-CRISPR system; A.F., C.B. and J.L.H. obtained peripheral blood samples from THES patients and controls, provided intellectual input and important insight into human THES, and helped write the manuscript; D.B.S. and S.C.E. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 2.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Xiao TS, Fitzgerald KA. The cGAS-STING Pathway for DNA Sensing. Mol Cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin T, et al. Structures of the HIN domain: DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao JC, et al. Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure. 2011;19:418–429. doi: 10.1016/j.str.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Civril F, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow YJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 12.Gall A, et al. Autoimmunity Initiates in Nonhematopoietic Cells and Progresses via Lymphocytes in an Interferon-Dependent Autoimmune Disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 14.Schneider C, Tollervey D. Threading the barrel of the RNA exosome. Trends Biochem Sci. 2013;38:485–493. doi: 10.1016/j.tibs.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernando MM, et al. Identification of two independent risk factors for lupus within the MHC in United Kingdom families. PLoS Genet. 2007;3:e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts ZJ, et al. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J Exp Med. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conlon J, et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto M, et al. Role of Adaptor TRIF in the MyD88-Independent Toll-Like Receptor Signaling Pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 21.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 22.Travers KJ, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 23.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 24.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 25.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 28.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 29.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 30.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 31.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Cho JA, et al. The unfolded protein response element IRE1alpha senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe. 2013;13:558–569. doi: 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Han D, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Q, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Fabre A, et al. SKIV2L mutations cause syndromic diarrhea, or trichohepatoenteric syndrome. Am J Hum Genet. 2012;90:689–692. doi: 10.1016/j.ajhg.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabre A, Martinez-Vinson C, Goulet O, Badens C. Syndromic diarrhea/Tricho-hepato-enteric syndrome. Orphanet J Rare Dis. 2013 Jan 9;8:5. doi: 10.1186/1750-1172-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartley JL, et al. Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy) Gastroenterology. 2010;138:2388–2398. doi: 10.1053/j.gastro.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabre A, et al. Novel mutations in TTC37 associated with tricho-hepato-enteric syndrome. Hum Mutat. 2011;32:277–281. doi: 10.1002/humu.21420. [DOI] [PubMed] [Google Scholar]
- 44.Rice GI, et al. Assessment of interferon-related biomarkers in Aicardi-Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurology. 2013;12:1159–1169. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.