Abstract
The Kv2.1 voltage-gated K+ channel is widely expressed throughout mammalian brain where it contributes to dynamic activity-dependent regulation of intrinsic neuronal excitability. Here we show that somatic plasma membrane Kv2.1 clusters are juxtaposed to clusters of intracellular ryanodine receptor (RyR) Ca2+-release channels in mouse brain neurons, most prominently in medium spiny neurons (MSNs) of the striatum. Electron microscopy-immunogold labeling shows that in MSNs, plasma membrane Kv2.1 clusters are adjacent to subsurface cisternae, placing Kv2.1 in close proximity to sites of RyR-mediated Ca2+ release. Immunofluorescence labeling in transgenic mice expressing GFP in specific MSN populations reveals the most prominent juxtaposed Kv2.1-RyR clusters in indirect pathway MSNs. Kv2.1 in both direct and indirect pathway MSNs exhibits markedly lower levels of labeling with phosphospecific antibodies directed against the S453, S563, and S603 phosphorylation site compared to levels observed in neocortical neurons, although labeling for Kv2.1 phosphorylation at S563 was significantly lower in indirect pathway MSNs compared to those in the direct pathway. Finally, acute stimulation of RyRs in heterologous cells causes a rapid hyperpolarizing shift in the voltage-dependence of activation of Kv2.1, typical of Ca2+/calcineurin-dependent Kv2.1 dephosphorylation. Together, these studies reveal that striatal MSNs are distinct in their expression of clustered Kv2.1 at plasma membrane sites juxtaposed to intracellular RyRs, as well as in Kv2.1 phosphorylation state. Differences in Kv2.1 expression and phosphorylation between MSNs in direct and indirect pathways provide a cell- and circuit-specific mechanism for coupling intracellular Ca2+ release to phosphorylation-dependent regulation of Kv2.1 to dynamically impact intrinsic excitability.
Keywords: ion channel, localization, intracellular Ca2+ release, striatum, immunohistochemistry
INTRODUCTION
The wide variety of voltage-gated K+ (Kv) channels plays distinct and diverse roles in regulating intrinsic excitability of mammalian neurons (Vacher et al., 2008). The delayed rectifier Kv2.1 channel is present in large clusters on the somata and proximal dendrites of most mammalian brain neurons (Trimmer, 1991; Scannevin et al., 1996; Du et al., 1998; Misonou et al., 2005b; Vacher et al., 2008). In cultured hippocampal neurons, Kv2.1 comprises up to 80% of the delayed rectifier current (Murakoshi and Trimmer, 1999), and establishes the firing frequency and peak width of action potentials (Du et al., 2000). Under periods of increased neuronal excitation, Kv2.1 undergoes extensive dephosphorylation resulting in a hyperpolarizing shift of the voltage dependence of activation (Murakoshi et al., 1997; Misonou et al., 2006; Misonou et al., 2008; Mulholland et al., 2008). More than 30 phosphorylation sites have been identified within the Kv2.1 C-terminus (Murakoshi et al., 1997; Park et al., 2006; Cerda and Trimmer, 2010; Baek et al., 2011; Trimmer and Misonou, 2014), a number of which regulate either Kv2.1 clustering (Lim et al., 2000) or the voltage-dependence of Kv2.1 activation (Murakoshi et al., 1997; Park et al., 2006; Mulholland et al., 2008). A subset of these sites exhibit bidirectional activity-dependent changes in phosphorylation state (Misonou et al., 2006), as a result of activity-dependent regulation via specific neuronal protein kinases and phosphatases (Misonou et al., 2005a; Misonou et al., 2006; Misonou et al., 2008; Mulholland et al., 2008; Cerda and Trimmer, 2011; Ikematsu et al., 2011). The major protein phosphatase known to modulate Kv2.1 activity is the Ca2+- and calmodulin-dependent protein phosphatase calcineurin/PP2B (Misonou et al., 2005a; Misonou et al., 2006).
A common mechanism to regulate ion channel activity is through specific association with modulatory proteins (Dai et al., 2009). For example, Ca2+-activated K+ channels colocalize with voltage-dependent Ca2+ channels (Berkefeld et al., 2006), IP3-stimulated intracellular Ca2+-release channels (Kaufmann et al., 2009) and/or Ca2+-regulated protein kinases (Bildl et al., 2004) to confer robust regulation of the plasma membrane K+ channel by intracellular Ca2+. In cultured hippocampal neurons, somatic plasma membrane Kv2.1 clusters are found juxtaposed to clusters of intracellular ryanodine receptor (RyR) Ca2+-release channels (Antonucci et al., 2001; Misonou et al., 2005b), and Kv2.1 clusters on the axon initial segment are found near RyR-rich cisternal organelles (King et al., 2014). RyRs are found in a variety of neurons where they mediate diverse local signaling events, including modulation of synaptic plasticity in the spine apparatus (Holbro et al., 2009) and neurotransmitter release from presynaptic terminals (Emptage et al., 2001). Importantly, in neuronal somata, intracellular Ca2+ release leads to activation of protein kinases and phosphatases (Berridge, 1998), and RyRs interact directly with these enzymes, including the key Kv2.1 modifying enzyme calcineurin/PP2B (Marx et al., 2001). Associations between somatic Kv2.1 and macromolecular complexes containing proteins that may regulate its dynamic, phosphorylation-dependent regulation in mammalian brain have not been addressed. Here we use multiple label immunofluorescence and light microscopy, and immunogold electron microscopy, to gain insights into the nature of Kv2.1-RyR localization in mammalian brain. We also studied the impact of RyR stimulation on the function of Kv2.1 expressed in heterologous cells. Finally, using Kv2.1-specific phosphospecific antibodies in multiple label immunofluorescence labeling of brain sections, we assayed the phosphorylation state of Kv2.1 in specific neuronal populations. These studies reveal a novel cell- and circuit-specific association of Kv2.1 and RyRs, and distinct Kv2.1 phosphorylation state, in striatal output neurons that may yield circuit-specific modulation of Kv2.1 and neuronal excitability.
MATERIALS AND METHODS
Animals used in this study
All animal use procedures were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals described by the National Institutes of Health, and were approved by the UC Davis Institutional Animal Care and Use Committee. Mice were maintained under standard light-dark cycles and allowed to feed and drink ad libitum. Kv2.1-KO mice (RRID:IMSR_MGI:3806050) were previously described (Jacobson et al., 2007; Speca et al., 2014), and were generated from breeding of Kv2.1+/− mice that had been backcrossed on the C57/BL6J background (RRID:IMSR_JAX:000664), such that all experiments with Kv2.1-KO mice used wild-type littermates as controls. The Tg(Drd1a-EGFP)×60Gsat/Mmmh (RRID:IMSR_MMRRC:000297) and Tg(Drd2-EGFP)S118Gsat/Mmnc (RRID:IMSR_MMRRC:000230) mice were obtained from the Mutant Mouse Regional Resource Centers (MMRRC) supported by the NIH. All mice used were adult females, over 10 weeks old.
Generation of novel native and recombinant monoclonal antibodies
A hybridoma producing the L61/14 (IgG1) monoclonal antibody (mAb) was generated from a mouse immunized with the pS603 phosphopeptide used previously (Misonou et al., 2006; Park et al., 2006) to generate rabbit polyclonal antibodies. A hybridoma producing the L86A/37 (IgG2b) mAb was generated from a mouse immunized with a recombinant fragment corresponding to human AMIGO-1 amino acids 395-493 (Peltola et al., 2011). Candidate L61 and K86A mAbs were screened and validated by standard procedures including ELISA, and immunoblots against brain samples from Kv2.1-KO and AMIGO-KO mouse brain samples, respectively. The L61/14 mAb was also validated by immunohistochemistry against brain samples from wild-type mice subjected to CO2 inhalation to induce dephosphorylation of Kv2.1 (Misonou et al., 2005a), as detailed in the Results section.
For generation of recombinant mAb K89/34 (IgG2a), total RNA was extracted from K89/34 hybridoma cells (RNAqueous Extraction Kit, Life Technologies) and cDNA transcripts synthesized (SuperScript III First-Strand cDNA Synthesis Kit; Life Technologies) using the total RNA extract as template. Other procedures were as previously described (Crosnier et al., 2010) including cloning into the immunoglobulin expression plasmid (generously provided by Dr. Gavin Wright), with the exception that the variable light chain amplification product was digested with BciVI (Fermentas) to eliminate aberrant SP2/0 transcript (Juste et al., 2006), and the heavy chain constant region in the expression plasmid was modified to produce an IgG2a mAb. Recombinant mAb plasmids were transfected into COS-1 cells using Lipofectamine 2000 (Life Technologies), and recombinant mAbs present in the media of transiently transfected cell cultures were assayed by multiple label immunofluorescence against heterologous cells expressing Kv2.1 using native K89/34 (IgG1) as a control, employing subclass-specific secondary antibodies (Manning et al., 2012) to identify plasmids encoding functional recombinant K89/34 mAb (IgG2a). Recombinant K89/34 was produced from cultures of transiently transfected COS-1 cells.
Antibody characterization
Table 1 contains a list of all antibodies used in this study. We refer the reader to the JCN antibody database (http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1096-9861/homepage/jcn_antibody_database.htm) for details and links to further information on the antibodies used in this study.
Table 1. Antibodies used in this study.
Details of the polyclonal (pAb) and monoclonal (mAb) antibodies used in this study.
|
Antigen and
Antibody Name |
Immunogen | Manufacturer Information |
Concentra
tion used |
|---|---|---|---|
| Kv2.1 (KC) | Synthetic peptide, rat Kv2.1 aa 837- 853, accession NP_037318, HMLPGGGAHGSTRDQSI |
In House (Trimmer Lab), affinity purified rabbit polyclonal antibody, RRID:AB_2315767 |
1:100 Affinity Purified |
| Kv2.1 (K89/34) | Synthetic peptide, rat Kv2.1 aa 837- 853, accession NP_037318, HMLPGGGAHGSTRDQSI |
In House (Trimmer Lab), recombinant mouse monoclonal antibody expressed in COS cells, IgG2a, RRID: AB_2315768 |
1:2 Tissue Culture Supernatant |
| Kv2.1 (K89/34) | Synthetic peptide, rat Kv2.1 aa 837- 853, accession NP_037318, HMLPGGGAHGSTRDQSI |
NeuroMab (Davis, CA), Clone# K89/34, Cat# 73-014, mouse monoclonal antibody, IgG1 RRID: AB_10672253 |
10 μg/mL Purified |
| pS603-Kv2.1 (L61/14) |
Synthetic phospho-peptide at S603, rat Kv2.1 aa 595-616, accession NP_037318, PEATRFSH[pS]PLASLSSKAGSST |
In House (Trimmer Lab), purified mouse monoclonal antibody, IgG1, RRID:AB_2315769 |
10 μg/mL Purified |
| pS453-Kv2.1 | Synthetic peptide, rat Kv2.1 aa 447- 467, accession NP_037318, KDAFAR[pS]IEMMDIVVEKNGES |
In House (Trimmer Lab), affinity purified rabbit polyclonal antibody, RRID: AB_2315784 |
1:100 Serum |
| pS563-Kv2.1 | Synthetic peptide, rat Kv2.1 aa 554- 574, accession NP_037318, EELEMSSMP[pS]PVAPLPARTEG |
In House (Trimmer Lab), affinity purified rabbit polyclonal antibody, RRID: AB_2315785 |
1:100 Serum |
| AMIGO-1 (L86A/37) |
Fusion protein, human AMIGO-1 aa 395-493, accession AA048948.1, PCRCWCRGVEKPSSHQGDSLSS SMLSTTPNHDPMAGGDKDDGFDR RVAFLEPAGPGQGQNGKLKPGNT LPVPEATGKGQRRMSDPESVSSV FSDTPIVV |
NeuroMab (Davis, CA), Clone# L86A/37, Cat# 73-329, mouse monoclonal antibody, IgG2b, RRID:AB_2315765 |
1:5 Tissue Culture Supernatant |
| Ryanodine Receptor (34C) |
Partially purified chicken pectoral muscle ryanodine receptor |
Pierce (Rockford, IL), Clone 34C, Cat# MA3925, purified mouse monoclonal antibody, IgG1,RRID:AB_2254138 |
1 μg/mL Purified |
| GFP | Purified GFP | Invitrogen (Grand Island, NY),Cat # A11122, rabbit polyclonal antibody, RRID:AB_221569 |
5 μg/mL Purified |
| GFP | Fusion protein amino acids 1-238 (full-length) of jellyfish green fluorescent protein (also known as GFP; accession number P42212) |
NeuroMab (Davis, CA), Clone# N86/8, Cat# 73-131, mouse monoclonal antibody, IgG2A, RRID: AB_2313651 |
10 μg/mL Purified |
The anti- ryanodine receptor mouse monoclonal antibody 34C (Thermo Pierce Cat# MA3-925 RRID:AB_2254138) was raised against partially purified chicken pectoral muscle ryanodine receptor. All three isoforms of RyR are detected by this antibody (manufacturer’s specifications).
The rabbit polyclonal antibody against GFP (Life Technologies Cat# A11122 RRID:AB_221569) was raised against GFP directly isolated from the jellyfish Aequorea victoria. The IgG fraction was purified from the rabbit serum by ion-exchange. Specificity was tested by ELISA (manufacturer’s specifications) as well as by immunohistochemistry in our experimental controls. The mouse monoclonal antibody against GFP (NeuroMab Cat#73-131 RRID:AB_2313651) was validated by immunocytochemistry of transfected COS cells expressing GFP (manufacturer’s specifications), as well as by immunohistochemistry in our experimental controls. Cross-reactivity to CFP and YFP was confirmed by immunoblot against transfected COS cell lysates expressing MHC-GFP, MHC-CFP and H3-3-YFP (manufacturer’s specifications).
The anti-Kv2.1 rabbit polyclonal antibody KC (Trimmer, 1991) (Trimmer Lab RRID:AB_2315767) was previously validated was validated by immunoblot against wild-type and Kv2.1-KO mouse brain samples (Misonou et al., 2006). The anti-Kv2.1 K89/34 monoclonal antibody (NeuroMab Cat# 73-014 RRID:AB_10672253) was validated by immunoblot against wild type and Kv2.1-KO mouse brain samples.
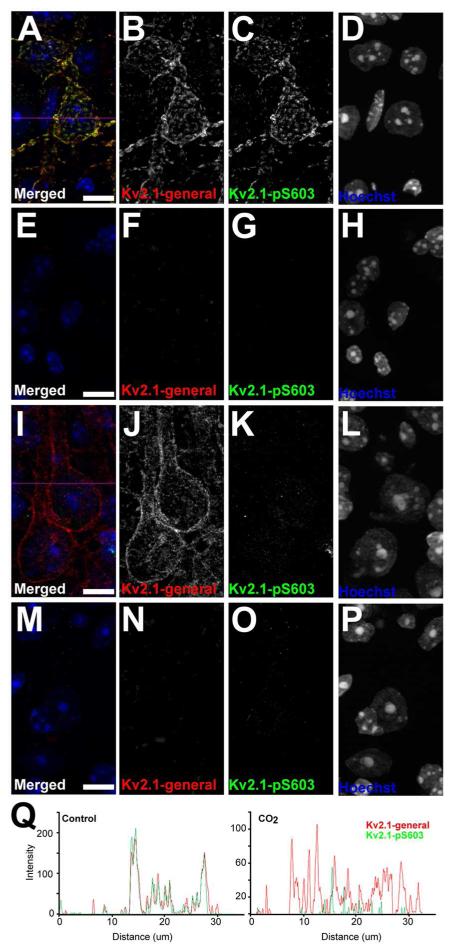
Anti-Kv2.1 K89/34 recombinant mouse mAb (Trimmer Lab RRID:AB_2315768) was validated by immunohistochemistry against wild type and Kv2.1-KO mouse brain sections, as detailed in the Results section. Phosphospecific anti-pS603-Kv2.1 L61/14 mouse mAb (Trimmer Lab RRID:AB_2315769) was validated by ELISA for phosphorylation specificity using identical synthetic peptides (rat Kv2.1 a.a. 595-616) with or without S603 phosphorylation. As detailed in the Results section, L61/14 was further validated by immunohistochemistry against wild-type and Kv2.1-KO mouse brain sections prepared from control mice, and from mice subjected to CO2 inhalation, which results in dephosphorylation of pS603 (Misonou et al., 2005a; Misonou et al., 2006). Brain sections prepared from wild-type mice under control conditions exhibit similar labeling for anti-Kv2.1 recombinant K89/34 mAb and phosphospecific anti-Kv2.1 pS603 L61/14 mAb, and that is eliminated in samples from Kv2.1-KO mice. Under hypoxic conditions, clustered labeling for anti-Kv2.1 recombinant K89/34 mAb becomes diffuse, while labeling for the phosphospecific anti-Kv2.1 pS603 L61/14 mAb is eliminated, as detailed in the Results section.
Rabbit polyclonal phosphospecific antibodies against Kv2.1 phosphorylated at S453 (Trimmer Lab RRID:AB_2315785) and S563 (Trimmer Lab RRID:AB_2315784) were validated for phosphospecificity by immunoblotting against phosphosite mutants expressed in HEK293 cells, and by immunoprecipitation reactions against rat bran membrane fractions without or with alkaline phosphatase digestion (Park et al., 2006). The S563 antibody was also validated by immunoblot against wild type and Kv2.1-KO brain samples (Misonou et al., 2006). Additional validation for phosphospecificity was performed by ELISA against phospho- and dephosphopeptides (not shown), and for molecular specificity by immunohistochemistry against wild-type and Kv2.1-KO mouse brain sections, as detailed in the Results section.
NeuroMab L86A/37 anti-AMIGO-1 mouse mAb (NeuroMab Cat# 73-329 RRID:AB_2315765) was validated by immunoblot against AMIGO-1-KO brain samples (manufacturer’s specifications). L86A/37 does not detect other isoforms of AMIGO in immunoblots using cell lysates from heterologous cells expressing either AMIGO-1, -2 or -3 (manufacturer’s specifications).
Multiple label immunofluorescence labeling
Adult female mice were anesthetized with 50 mg/kg pentobarbital and transcardially perfused with ≈10 mL PBS (150 mM NaCl, 10 mM Na-phosphate buffer, pH7.4) followed by ≈30 mL ice-cold 4% formaldehyde (freshly prepared from paraformaldehyde) in 0.1M Na-phosphate buffer (PB), pH7.4. Brains were removed and cryoprotected with 10% sucrose/0.1 M PB, pH 7.4 overnight, followed by a subsequent cryoprotection with 30% sucrose/0.1 M PB. Brains were frozen at −80°C prior to sagittal sectioning on a freezing stage microtome at 30 μm per section. All resultant labeling steps were performed on free-floating sections. Sections were blocked in a vehicle solution containing 10% goat serum and 0.3% Triton X-100 in PB, pH 7.4 for 1 hour at 4°C. Sections were then labeled overnight at 4°C with primary antibodies (Table 1) diluted in vehicle. Following three 10 min washes in vehicle, sections were incubated with Alexa conjugated, mouse IgG subclass-specific (Manning et al., 2012) or anti-rabbit IgG secondary antibodies (secondary antibodies from Invitrogen) at 1–2000 for 1 hour. Following three 10 min washes in vehicle, sections were mounted and dried onto gelatin coated slides, and labeled with 0.05% Sudan Black (Schnell et al., 1999) (EM Sciences) in 70% ethanol for 5 min, extensively washed in water and mounted with Prolong Gold (Invitrogen). All staining reported is representative of at least 3 staining sets from different animals, except in the case of EGFP-D1 and EGFP-D2 animals where the staining was repeated 3 times, but from the same animal. Low magnification images were acquired on Zeiss Axio Observer Z1 microscope using a 10×/0.5 NA Fluar objective and an AxioCam MRm digital camera, and reconstructed as tiled mosaics using Axiovision 4.0 (Carl Zeiss Microimaging RRID:SciRes_000111). High magnification images were acquired on a Zeiss Axio Imager M2 microscope using a 20×/0.8 NA plan-Apochromat objective, or a 100×/1.40 NA plan-Apochromat oil immersion objective, and an AxioCam MRm digital camera, and optical sections were acquired using an ApoTome 2 structured illumination system (Carl Zeiss Microimaging). Images were processed for brightness and contrast only in Axiovision (Carl Zeiss Microimaging) and Photoshop (Adobe Systems) software. High-resolution images were acquired with Zeiss Elyra system (SR-SIM) using a 63×/1.4 NA plan-Apochromat oil immersion objective and Z-stacks were reconstructed using Zen software (Carl Zeiss Microimaging).
Immunofluorescence intensity quantification
Three sets of mouse brain sections from EGFP-D1 or EGFP-D2 transgenic mice were multiple immunofluorescence labeled for Kv2.1 and RyR, for Kv2.1 and Kv2.1 phosphorylated at pS453, p563, or pS603, and for AMIGO-1. Single optical sections were acquired from the striatum at 20× using an epifluorescence microscope equipped with an ApoTome. Brain sections were chosen based on evenness of Kv2.1 and marker staining to exclude any sections that had a poor antibody penetration from tissue folding during the staining process. Regions of interest (ROIs) are from single brain sections that contained ~200 clearly labeled somata. Fluorescence intensities from individual somata were measured from single optical sections using Photoshop. Cells were grouped as being either positive or negative for the EGFP marker, and signal intensities of RyR or AMIGO-1 were normalized to the signal intensity for Kv2.1 in each cell. Statistical analysis was done using non-parametric tests (Mann-Whitney) compared by a two-sided z-test. A z value of p<0.01 was considered to be statistically significant. Immunofluorescence intensity line plots were generated using the ImageJ software (NIH, RRID:nif-0000-30467). Line sections were chosen for regions that contained edges with clear Kv2.1 clusters. Single lines were sampled as indicated in the figures. The plot profile function in ImageJ was used to measure the fluorescence along the sample line. Plot profiles were graphed in Excel (Microsoft).
Immunogold labeling and electron microscopy
All mouse handling and sample preparation for electron microscopy was done at the National Institute for Physiological Sciences, Okazaki, Japan and were conducted under regulatory guidelines of the institution. Adult mice were anesthetized with pentobarbital and transcardially perfused with 100 mL of ice cold 4% formaldehyde (freshly prepared from paraformaldehyde) in 15% saturated picric acid/0.05% glutaraldehyde in 0.1 M PB, pH 7.4. Brains were removed and sectioned to 50 μm using a vibratome. Sections were then blocked with 10% goat serum/0.025% Triton X-100 in 50 mM TBS (150 mM NaCl/50 mM Tris HCl pH 7.4), at 4 °C for 1 hour. Sections were labeled free floating and probed with primary antibodies diluted in vehicle (2% goat serum/50 mM TBS), for 48 hours. Sections were washed with 50 mM TBS 3 times for 20 min each and then incubated with 1.4 μm gold-conjugated secondary antibodies (Nanoprobes) diluted in vehicle for 12 hours at 4°C. Sections were then washed at room temperature 3 times for 10 min each with 50 mM TBS, followed by 25 mM PBS (150 mM NaCl/25 mM phosphate buffered pH 7.4). Sections were then postfixed with 1% glutaraldehyde/25 mM PBS for 10 min, washed with water 3 times for 5 min each. Silver enhancement was performed using HQ silver (Nanoprobes) EM intensification kit. After washing with water 3 times for 5 min each and with 0.1 M PB 2 times for 10 min each, osmification was performed by treatment with 1% OsO4/PB for 40 min. After washing sections with water 3 times for 10 min each, counterstaining was done using 1% uranyl acetate for 35 min. Sections were then dehydrated with 50%, 70%, 90%, 95%, and 100% ethanol, followed by 100% propylenoxide. Dehydrated sections were infiltrated with Durcupan resin overnight and cured on glass slides at 60°C overnight. Regions of interest were then excised and re-embedded in Durcupan blocks to be sectioned at 70 μm using a microtome (Leica). Ultra thin sections were mounted on single slot copper grids using pioloform support film and imaged using a CCD camera connected to a Philips EM208S transmission electron microscope.
Electrophysiology
HEK293 cells were cultured in modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin and 1% non-essential amino acids under 5% CO2 atmosphere. HEK293 cells were transiently co-transfected with 50 ng Kv2.1/RBG4 (Shi et al., 1994) and either 0.5 μg RBG4 plasmid (Lee et al., 1991), full length rabbit RyR1 cDNA in the pCIneo plasmid (Nakai et al., 1998), full length mouse RyR2 in the pcDNA3 plasmid (Zhao et al., 1999) or rabbit RyR3 in the pcDNA3 plasmid (RyR plasmids generously provided by Dr. Paul D. Allen). For each transfection, GFP (0.5 μg) was co-transfected as a transfected cell marker. Transfections were performed on 70% confluent HEK293 cultures according to standard Lipofectamine 2000 (Invitrogen) protocol. Cells were trypsinized 16 to 24 h after transfection, replated and used for electrophysiological analysis within 5 hours after replating.
Whole cell currents were recorded at room temperature (22 – 23 °C) using an Axopatch-200B amplifier (Axon Instruments). Current recordings were low-pass filtered and sampled at 1-10 kHz using a Digidata 1200A data-acquisition system (Axon Instruments). The pClamp10 software (Axon Instruments) was used to control command voltages and data storage. Patch pipettes were pulled with a laser puller P2000 (Sutter Instruments) from 1.2 mm borosilicate glass (World Precision Instruments) and heat polished. An extracellular solution containing (in mM), 145 NaCl, 4 KCl, 1.8 CaCl2, 1 MgCl2, 10 HEPES, 10 glucose and adjusted to pH 7.35 with NaOH was used to superfuse the cells continuously. The patch pipettes were filled with an intracellular solution containing (in mM), 110 KCl, 5 K4BAPTA, 5 K2ATP, 1 MgCl2 and 10 HEPES with the pH adjusted to 7.2 using KOH. Junction potentials between the extracellular and intracellular solution were zeroed with the filled pipette in the bath solution. To ensure that voltage errors did not exceed 5 mV, cells were excluded from analysis if the series resistance exceeded 3 MV after compensation.
Ionomycin and ryanodine treatments were performed by incubating the HEK293 cells for 10 min in 1 μM ionomycin (Invitrogen) or 1 μM ryanodine (Tocris Bioscience) dissolved in Hank’s balanced salt solution with Ca2+/Mg2+ and without Ca2+/Mg2+, respectively. The ionomycin and ryanodine solutions were applied to the recording chamber via bath perfusion.
Voltage protocols are given in the figures and/or figure legends. The voltage dependence of activation or inactivation was fitted with a Boltzmann equation according to, y = 1/(1 + exp[−(V − V1/2)/k]) in which V represents the voltage applied, V1/2 the voltage at which 50% of the channels are activated or inactivated and k the slope factor. Results are presented as mean ± S.E. Student’s t test or Mann-Whitney U Rank Sum test were used for statistical analysis with p values < 0.05 considered to be statistically significant.
RESULTS
Clusters of Kv2.1 are juxtaposed to clustered RyR in specific mouse brain neurons
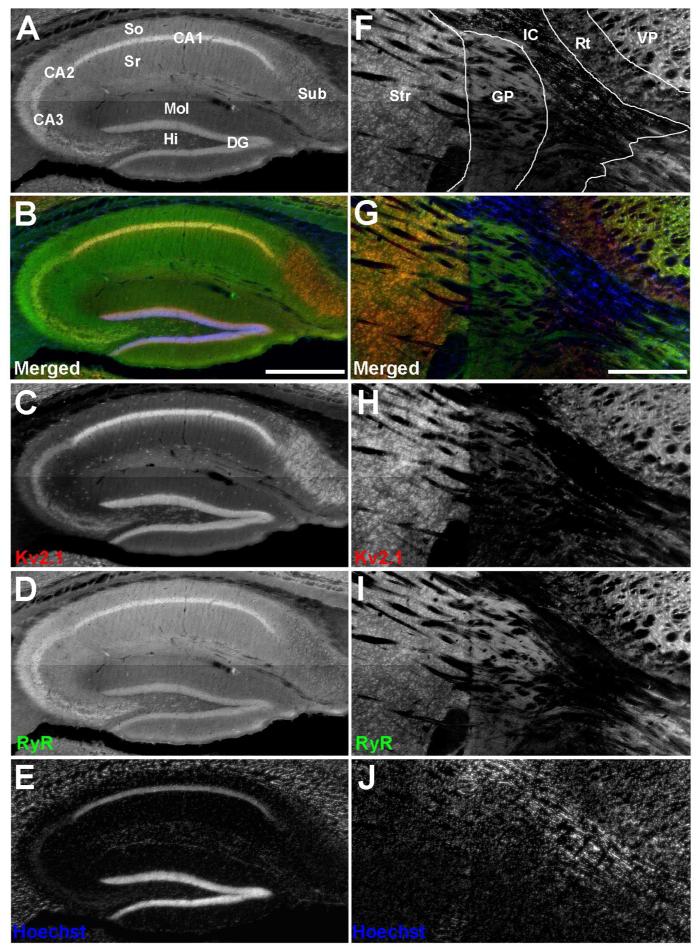
Previous studies demonstrated that in cultured rat hippocampal neurons, plasma membrane Kv2.1 clusters overlay clusters of RyRs (Antonucci et al., 2001; Misonou et al., 2005b). To determine whether these proteins exhibit overlapping localization in brain neurons in situ, we performed multiple label immunofluorescence labeling for Kv2.1 and RyR on mouse brain sections. We refer the reader to the JCN antibody database (http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1096-9861/homepage/jcn_antibody_database.htm) for details and links to further information on the antibodies used in this study. We found that labeling for Kv2.1 and RyR overlaps in certain classes of hippocampal neurons. For example (Fig. 1A and 1B), extensive overlap is observed in stratum pyramidale of CA1 and CA3 (yielding an overall yellow tint in CA1 and CA3 in Fig. 1B), but to a lesser extent in other regions, for example subiculum, which exhibits high levels of labeling for Kv2.1 but lower levels of labeling for RyR (yielding an overall red tint in subiculum in Fig. 1B). We found that RyR labeling extends into the neuropil of stratum radiatum and stratum oriens of the CA1 and CA3 regions in the absence of Kv2.1 labeling (Fig. 1A and 1B). Somatic Kv2.1 and RyR labeling is also present in dentate granule cells, and in a subset of Kv2.1-positive interneurons within the hilus of the dentate gyrus. However, not all cells within the hippocampus exhibit somatic RyR expression, such as interneurons within stratum radiatum and stratum oriens of CA1, and within the molecular layer of the dentate gyrus (Fig. 1A and 1B). We next determined whether the juxtaposed clustering of Kv2.1 and RyR seen in certain classes of hippocampal neurons is present in other regions of the mouse brain. In the basal ganglia and thalamus, we found that labeling for Kv2.1 and RyR overlaps in striatum, in the reticular and ventral posterior nuclei of the thalamus, but not in globus pallidus (Fig. 1F and 1G).
Figure 1.
Distinct patterns of Kv2.1 and RyR coexpression in mouse brain. Mouse brain sections were double immunofluorescence labeled for Kv2.1 (red) and RyR (green), and nuclei were labeled with Hoechst 33258 (blue). Images were acquired with an epifluorescence microscope using an automated stage for mosaic images of brain regions. A-E: Hippocampus. F-J: Striatum and adjacent brain regions. A: Grayscale version of the color image shown in panel B labeled for anatomical regions. B: Merged image of triple immunofluorescence labeling. C: Labeling for Kv2.1. D: Labeling for RyR. E: Labeling for nuclei with Hoechst. F: Grayscale version of the color image shown in panel G labeled for anatomical regions. G: Merged image of triple immunofluorescence labeling. H: Labeling for Kv2.1. I: Labeling for RyR. J: Labeling for nuclei with Hoechst. Labels on panel A: Sub: Subiculum; So: Stratum oriens; Sr: Stratum radiatum; CA1-CA3: respective pyramidal cell layers; Mol: Dentate gyrus molecular layer; HI: Dentate gyrus hilus; DG: Dentate gyrus granule cell layer. Labels on panel F: Str: Striatum; Gp, Globus pallidus; IC, Internal capsule; RT, Reticular nucleus (Thalamic); VP, Ventral posterior nucleus (Thalamic). Scale bar in panel B is 500 μm and is for panels A-E. Scale bar in panel G is 500 μm and is for panels F-J.
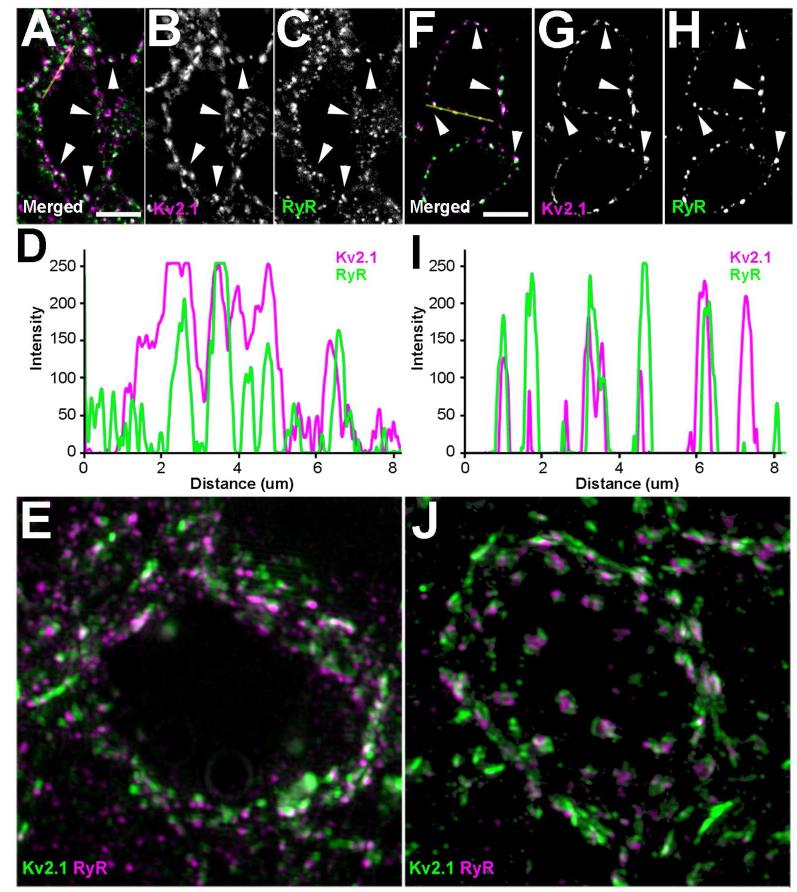
Higher magnification images of CA1 pyramidal cell bodies reveal that the overlap in labeling seen as a yellow tint in Fig. 1B is actually from juxtaposed clusters of plasma membrane Kv2.1 and intracellular clusters of RyRs (Fig. 2A-C). Line plots of fluorescent intensities shows that the Kv2.1 and RyR clusters are different in size but are localized in the same regions (Fig. 2D). Super-resolution, structured illumination microscopy of CA1 pyramidal neuron somata supports the juxtaposed localization of Kv2.1 and RyR (Fig. 2E). There is high density labeling of both molecules, but consistent with what was previously found in cultured rat hippocampal neurons (Antonucci et al., 2001; Misonou et al., 2005b), Kv2.1 clusters in mouse hippocampal neurons are not always found juxtaposed to RyR clusters, and intracellular RyR clusters in general are smaller than plasma membrane Kv2.1 clusters. At higher magnification, the overlap of Kv2.1 and RyR labeling in striatum (Fig. 1G) is seen to be in striatal medium spiny neurons (MSNs), where Kv2.1 and RyR labeling is present as juxtaposed clusters (Fig. 2F-H), similar to that seen in somata of CA1 pyramidal neurons. However, the line plots of MSN fluorescent intensities show highly congruent clusters of Kv2.1 and RyR labeling (Fig. 2I). Super-resolution imaging of MSNs (Fig. 2J) demonstrates the juxtaposed clustering of Kv2.1 and RyRs. As expected from their respective membrane compartments, RyR clusters in the intracellular membranes tend to be below/interior to the plasma membrane Kv2.1 clusters, as particularly evident in a rotated animation of three-dimensional reconstructed images of MSNs (Supplementary Fig. 1).
Figure 2.
Kv2.1 clusters are juxtaposed to clusters of RyR in the mouse brain. Mouse brain sections were double immunofluorescence labeled for Kv2.1 (magenta) and RyR (green). Images in panels A-C and F-H were acquired using an epifluorescence microscope equipped with an ApoTome. Yellow lines in A and F represent line traces of fluorescence intensities depicted in D and I respectively. Line traces were generated using the plot profile function in ImageJ for each channel. High-resolution images in panels E and J were acquired with Zeiss Elyra system (SR-SIM) and Z-stacks were reconstructed using Zen software. A-C: Single optical section of CA1 pyramidal neuron. A: Merged image of double immunofluorescence labeling. B: Labeling for Kv2.1. C: Labeling for RyR. E: CA1 pyramidal neuron imaged using SR-SIM. F-H: Single optical section of a striatal MSN neuron. F: Merged image of double immunofluorescence labeling. G: Labeling for Kv2.1. H: Labeling for RyR. H: Striatal MSNs imaged using SR-SIM. White arrowheads indicate prominent examples of juxtaposed clusters of Kv2.1 and RyR. Scale bar in panel A is 5 μm and is for panels A-C. Scale bar in panels E, J are 1 μm. Scale bar in panel F is 5 μm and is for panels E-G.
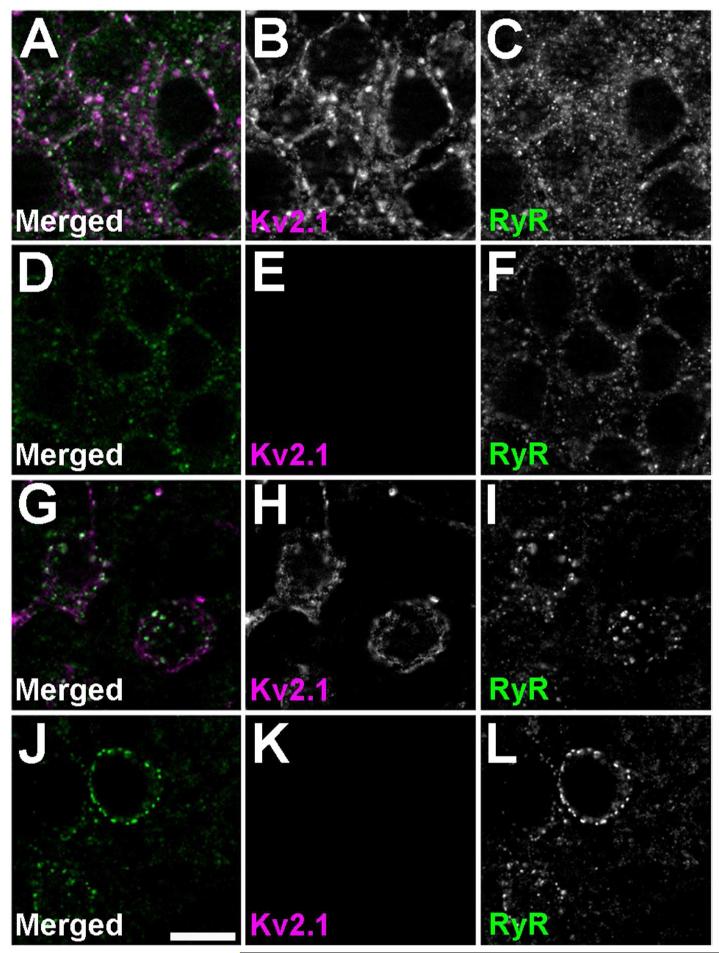
To determine whether the presence of Kv2.1 clusters was instructive to the generation and maintenance of the cell-type specific clustering of RyRs, we examined localization of RyRs in Kv2.1-KO mice (Jacobson et al., 2007; Speca et al., 2014). We found no striking differences in RyR clustering or expression throughout the brains of Kv2.1-KO mice relative to that seen in their WT littermates. Fig. 3 shows labeling for RyRs in CA1 pyramidal neurons of the hippocampus (Fig. 3A-F) and striatal MSNs (Fig. 3G-L) in wild type (Fig. 3A-C, G-I) and Kv2.1-KO (Fig. 3D-F, J-L) mice. This suggests that the clustered expression of Kv2.1 is not required for the cell type-specific somatic clustering of RyRs.
Figure 3.
RyR expression and localization is not grossly altered in brains of Kv2.1-KO mice. Kv2.1 labeling is shown in magenta, and RyR labeling in green. Images were acquired using an epifluorescence microscope equipped with an ApoTome. Images of single optical sections from mouse brain sections prepared from A-C, G-I: Wild type; and D-F, J-L: Kv2.1-KO mice. A-F: CA1 pyramidal neurons; G-L: striatal MSNs. A, D, G, J: Merged images. B, E, H, K: Kv2.1 labeling. C, F, I, L: RyR labeling. Scale bar in panel J is 10 μm and is for all panels.
Kv2.1 and RyR clusters are found juxtaposed in multiple regions of the brain
The extensive co-clustering of Kv2.1 and RyR in CA1 pyramidal neurons and striatal medium spiny neurons led us to question whether other brain regions and neuronal cell types exhibited similar patterns of clustering. To aid in region identification, we performed multiplex immunofluorescence labeling for Kv2.1 and RyR in serial coronal sections with various interneuron markers including calretinin, calbindin and parvalbumin (data not shown). Regions that were identified with confidence and contained the striatum from more than three brain sections were scored for expression level of Kv2.1 and for somatic clustering with RyR. The striatum was used as a reference for relative expression of Kv2.1 so that each brain section had an internal control. Brain regions containing few cells expressing somatically clustered RyR were scored as negative. We found that although Kv2.1 is widely expressed in multiple brain regions, there are striking differences in expression levels per cell between brain regions, as seen above for hippocampus (Fig. 1B) and for striatum, globus pallidus and thalamic nuclei (Fig. 1G). Table 2 summarizes the regional expression profiles for Kv2.1, and for somatically clustered RyRs. We found that numerous brain regions exhibit synaptic/neuropil labeling for RyR, while certain regions, especially striatum and thalamus, exhibit prominent clustered somatic RyR. In all regions analyzed, when somatic clusters of RyRs were present they were found juxtaposed to Kv2.1 clusters. Within the thalamus, neurons within the anterior nucleus group, mediodorsal nucleus, ventral nuclear complex and posterior nucleus exhibit somatic juxtaposed clustering of Kv2.1 and RyR through the majority of the subdivisions. Regions that did not exhibit predominant somatically clustered RyR were the neocortex, habenula, globus pallidus, and a few thalamic nuclei including the lateral nuclear group and reticular nucleus. Not all neurons in a given brain region have distinctive somatic RyR clustering patterns, for example the amygdala and thalamic nuclei, including the intralaminar nuclear group, midline nuclear group, and geniculate nucleus, have very clear divisions of cell populations that do and do not express somatically clustered RyR. In summary, diverse neurons in distinct brain regions have somatically clustered RyRs, which are juxtaposed to clusters of Kv2.1.
Table 2. Regional expression of Kv2.1 juxtaposed clustering with RyR.
Multiple label immunofluorescence was performed on serial coronal brain sections for Kv2.1 and RyR. Sections that included the striatum were scored for levels of Kv2.1 per cell, with the highest levels being scored as four:plus signs (++++), equal to the intensities found in striatal MSNs. Regions were also noted for having a majority of the cells exhibiting somatic clustering of RyR. Levels of RyR were not indicated due to heterogeneity within structures.
| Brain Region |
Kv2.1
expression |
Somatic
Clustered RYR |
|---|---|---|
| Cortex | +++++ | |
| Hippocampus | ||
| Pyramidal neurons | ++++ | Yes |
| Dentate Gyrus | ++++ | Yes |
| Subiculum | ++++ | |
| Habenular complex | ||
| Medial habenular nucleus (MHb) | +++ | |
| Lateral habenular nucleus (LHb) | ++ | |
| Amygdala | ||
| Lateral amygdaloid nucleus | ||
| Dorsolateral part (LaDL) | ++++ | |
| Ventrolateral part (LaVL) | ++++ | |
| Central amygdaloid nucleus | ||
| capsular part (CeC) | ++ | Yes |
| lateral division (CeL) | ++ | Yes |
| Striatum | ||
| Caudate putamen (Cpu) | ++++ | Yes |
| Globus pallidus (GP) | + | |
| Thalamus: Anterior nuclear group | ||
| Anteroventral nucleus (AV) | +++ | Yes |
| Anteromedial nucleus (AM) | ++ | Yes |
| Thalamus: Mediodorsal nucleus (MD) | ||
| Intermediate level | ||
| Medial part (MDM) | ++ | Yes |
| Central part (MDC) | ++ | Yes |
| Lateral part (MDL) | +++ | Yes |
| Thalamus: Ventral nuclear complex | ||
| Ventromedial nucleus (VM) | +++ | Yes |
| Ventrolateral nucleus (VL) | ++ | Yes |
| Ventral posterolateral nucleus (VPL) | ++ | Yes |
| Ventral posteromedial nucleus (VPM) | ++ | Yes |
| Thalamus: Lateral nuclear group | ||
| Laterodorsal nucleus (LD) | ||
| Dorsomedial part (LDDM) | ++++ | |
| Ventrolateral part (LDVL) | +++ | |
| Lateroposterior nucleus (LP) | ||
| Laterorostral part (LPLR) | +++ | |
| Mediorostral part (LPMR) | +++ | |
| Thalamus: Posterior nucleus (Po) | +++ | Yes |
| Thalamus: Intralaminar nuclear group | ||
| Central medial nucleus (CM) | +++ | Yes |
| Centrolateral nucleus (CL) | ++ | |
| Parafascicular nucleus (PF) | +++ | |
| Thalamus: Midline nuclear group | ||
| Paraventricular nucleus (PV) | + | |
| Ineranteromedial nucleus (IAM) | ++ | |
| Intermediodorsal nucleus (IMD) | ++ | Yes |
| Thalamus: Reticular nucleus (Rt) | ++ | |
| Thalamus: Geniculate Nucleus | ||
| Pregeniculate nucleus | ||
| Magnocellular part (PGMC) | ++ | |
| Parvicellular part (PGPC) | +++ | |
| Lateral geniculate nucleus | ||
| Dorsal part (DLG) | +++ | Yes |
| Anterior pretectal nucleus | ||
| Dorsal part (APTD) | +++ | |
| Sub-thalamus: Zona incerta (ZI) | ||
| Dorsal part (ZID) | ++ | |
| Ventral (ZIV) | ++ | |
| Subthalamic nucleus (STh) | +++ | Yes |
| Parasubthalamic nucleus | ++ |
Kv2.1 clusters are found near subsurface cisternae in striatal and thalamic neurons
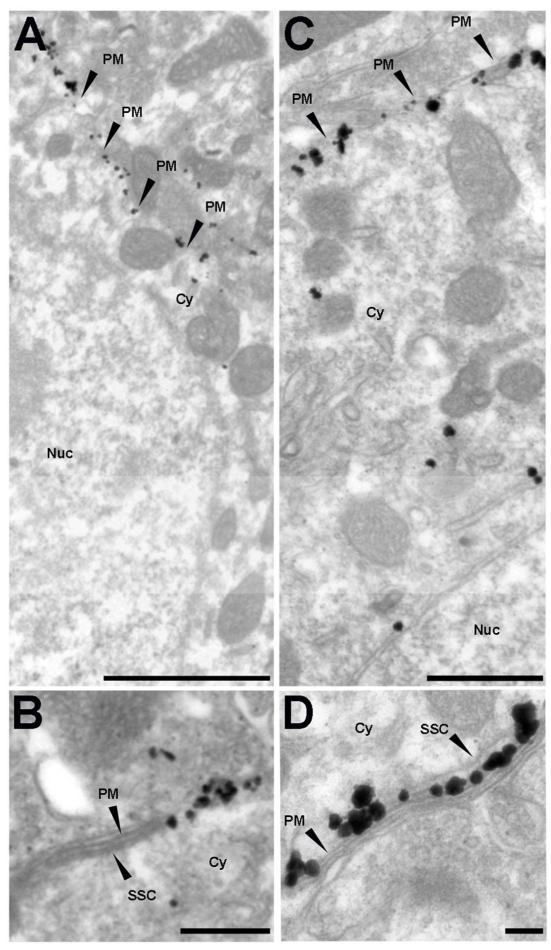
Previous immunogold electron microscopic studies of rat CA1 pyramidal neurons revealed that plasma membrane Kv2.1 clusters are adjacent to contact sites of glial cell processes, and to the open end of stacks of the endoplasmic reticulum/subsurface cisternae (SSC) (Du et al., 1998). Kv2.1 in plasma membrane domains adjacent to SSC was also observed in immunoperoxidase electron microscopy labeling of C-terminal synapses on spinal motor neurons (Muennich and Fyffe, 2004). We used a pre-embedding immunogold labeling technique (Parajuli et al., 2012) on mouse brain sections to examine the ultrastructural localization of Kv2.1 in areas of mouse brain (hippocampal CA1 pyramidal neurons, relay neurons of the LGN, and MSNs of the striatum) that exhibit clustered RyRs and Kv2.1 (Table 2). We included the CA1 region in our study to ensure that our technique provided labeling in mouse that was consistent with what was previously reported in rat (Du et al., 1998).
We found Kv2.1 immunogold particles in mouse MSNs clustered on the plasma membrane of the soma (Fig. 4A) and dendrites (data not shown). These somatic Kv2.1 clusters in MSNs are found on the edges of SSC (Fig. 4B). A similar localization pattern was observed for LGN thalamic neurons, in which Kv2.1 is found highly clustered on the plasma membrane (Fig. 4C) at sites associated with SSC (Fig. 4D). We also found Kv2.1 immunogold labeling in mouse hippocampal CA1 pyramidal neurons similar to what was previously reported in rat (Du et al., 1998), with clustering of plasma membrane Kv2.1 gold particles adjacent to the open ends of SSC on both the soma and proximal dendrites (data not shown). Kv2.1 immunogold particles were rarely observed in dendritic spines, and were not in myelinated axons or synapses (data not shown). These electron microscopy studies show that somatic Kv2.1 clusters labeled with immunogold particles are in close proximity to SSC in the same classes of neurons that by multiple label immunofluorescence light microscopy exhibit juxtaposed somatic Kv2.1 and RyR clusters.
Figure 4.
Kv2.1 clusters on the plasma membrane near subsurface cisternae in striatal and thalamic neurons. Ultrastructural localization of Kv2.1 in A, B: striatal and C, D: thalamic neurons, as revealed by pre-embedding immunogold labeling method. B and D depict Kv2.1 clusters adjacent to sub-surface cisternae in striatal and thalamic neurons respectively. Black arrowheads point to specific structures. SSC: subsurface cisternae; PM: plasma membrane; Cy: cytosol; Nuc: nucleus. Scale bars in A: 1 μm; B, D: 0.1 μm; and C: 0.5 μm.
Kv2.1 clusters are juxtaposed to higher levels of RyR in MSNs of the indirect pathway
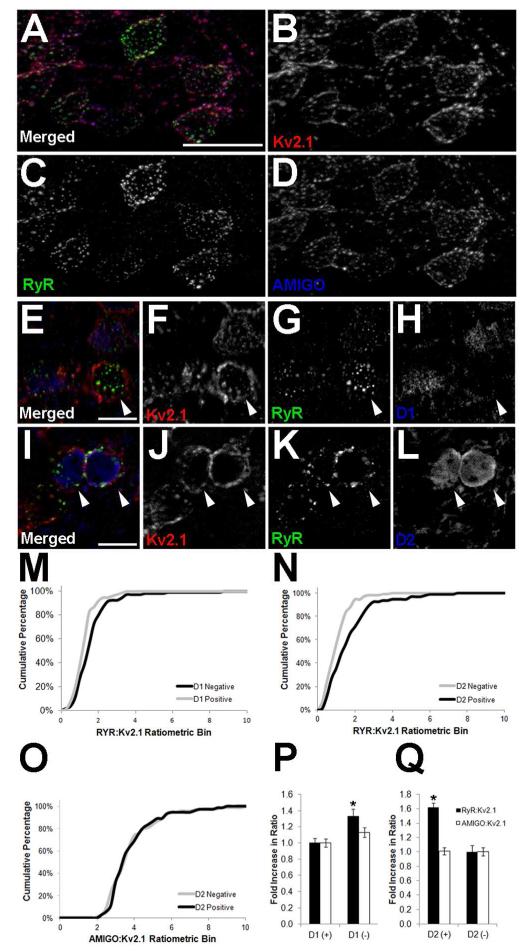
Somatic Kv2.1 clusters are juxtaposed to RyR clusters in striatal MSNs (Fig. 1G). In brain sections labeled Kv2.1 and its auxiliary subunit AMIGO-1, we found MSNs across the striatum exhibit similar labeling for these two Kv channel subunits (Fig. 5A). However, labeling for RyRs exhibited clear heterogeneity (Fig. 5A). The GABAergic MSNs are roughly divided into two equal populations corresponding to the striatonigral (direct) pathway neurons that express D1 dopamine receptors, and the striatopallidal (indirect) pathway neurons that express D2 dopamine receptors (Kreitzer, 2009; Gerfen and Surmeier, 2011). The expression of these receptors is mutually exclusive and can be used to differentiate the basal ganglia circuitry by immunolabeling, or by using BAC transgenic mice that express EGFP under control of the D1 or D2 dopamine receptor promoters (Gong et al., 2003; Gerfen and Surmeier, 2011). To determine whether the heterogeneity in RyR labeling across MSNs corresponded to differences between neurons of the direct and indirect pathways, we measured fluorescence intensities of immunolabeled Kv2.1, AMIGO-1 and RyR in sections prepared from the dopamine receptor-EGFP BAC transgenic mice. As expected from the uniform appearance of Kv2.1 immunolabeling, we found no significant difference (p ≤ 0.01) in the Kv2.1 fluorescence intensity in direct (D1+, D2−) versus indirect (D1−, D2+) pathway MSNs in the striatum of both the EGFP-D1 (p = 0.0251; n = 205) and EGFP-D2 (p = 0.6312; n = 172) mice. We next examined the level of AMIGO-1 expression and found that there is also no significant difference between direct and indirect pathway neurons in the striatum of both the EGFP-D1 (p = 0.1336; n = 205) and EGFP-D2 (p = 0.6527; n = 172) mice. However, we found that the level of RyR immunolabeling is significantly higher in indirect pathway MSNs in the striatum of both the EGFP-D1 (Fig. 5B, p = 0.0014; n = 180) and EGFP-D2 (Fig. 5C, p = 0.0001; n = 200) mice. The RyR:Kv2.1 ratios were binned and plotted as cumulative frequencies, and higher RyR:Kv2.1 ratios were found in MSNs of the indirect pathway in both the EGFP-D1 (Fig. 5M) and EGFP-D2 (Fig. 5N) mice. This distinction is not observed for the AMIGO-1:Kv2.1 ratio (Fig. 5O, showing EGFP-D2 striatal measurements only). RyR:Kv2.1 fluorescence ratios are higher in MSNs within the indirect pathway in both the EGFP-D1 (D1- neurons, Fig. 5P) and EGFP-D2 (D2+ neurons, Fig. 5Q) mouse lines. However, the AMIGO-1:Kv2.1 ratio is the same across these MSN populations (Fig. 5P and 5Q). These data demonstrate that D2+/D1− MSNs of the indirect pathway express elevated levels of RyR expression relative to that of Kv2.1 and its auxiliary subunit AMIGO-1.
Figure 5.
Indirect pathway striatal MSNs have higher levels of RyR labeling. Mouse brain sections were multiple immunofluorescence labeled for Kv2.1 (red) and RyR (green), and single optical sections were acquired using an epifluorescence microscope equipped with an ApoTome.. A-D: Wild type, labeling for AMIGO-1 shown in blue. In panels E-L, GFP labeling in shown in blue for E-H: EGFP-D1 transgenic mice, or I-L: EGFP-D2 transgenic mice. Scale bar in panel A is 20 μm, and is for panels A-D. Scale bars in panels E and I are 10 μm and are for panels E-H and I-L, respectively. White arrowheads indicate D1−/D2+ (indirect pathway) MSNs. Panels M-Q: Fluorescence intensities were measured from single optical sections taken at 20× using Adobe Photoshop. Cumulative percentages in panels M and N show the RyR:Kv2.1 ratios in the EGFP-D1 and EGFP-D2 mouse striatum, respectively. Panel O shows the AMIGO-1:Kv2.1 ratio in D2+ and D2− MSNs in the EGFP-D2 mouse striatum. Black lines represent D2+/D1− (indirect pathway) MSNs and gray lines represent D2−/D1+ (direct pathway) MSNs. P, Q: Average ratios RyR:Kv2.1 (black bars) and AMIGO-1:Kv2.1 (white bars) ratios from the striatum of P: EGFP-D1, and Q: EGFP-D2 mice. D1+ cell values were used to normalize for corresponding D2 measurements in each independent sample. Data are mean ± SEM (n>150 per group). *p<0.01 versus D2 measurements.
Distinct patterns of Kv2.1 phosphorylation in striatal MSN populations
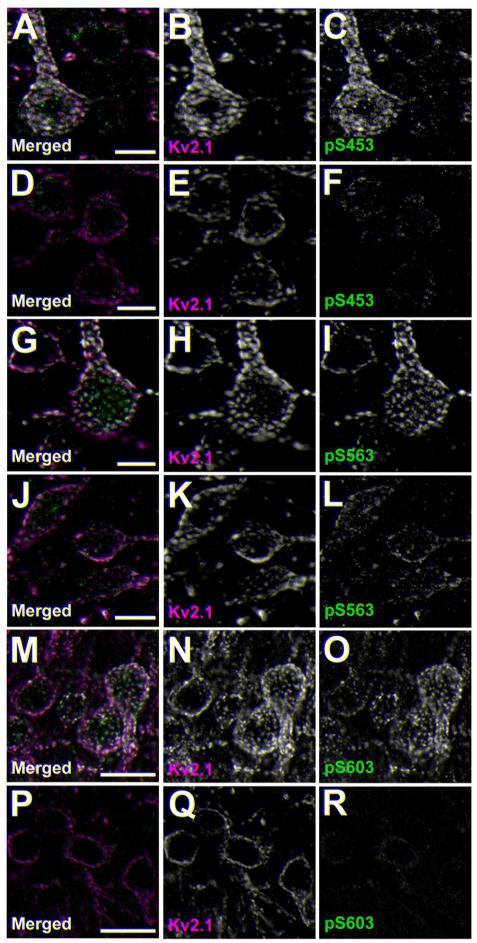
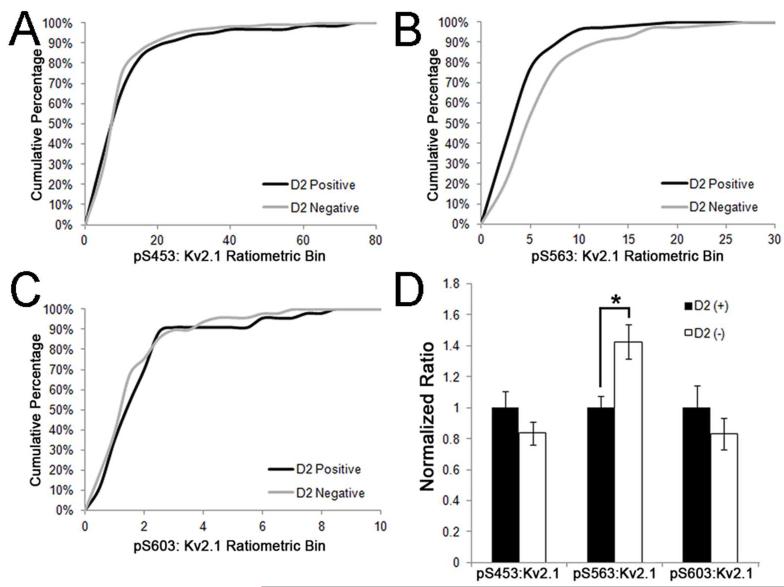
Previous studies using phosphospecific antibodies directed against specific phosphorylation sites on the regulatory C-terminus of Kv2.1 showed high levels of labeling in neocortical neurons in rat brain slices (Misonou et al., 2006). However, differences in phosphorylation between different populations of neurons in mammalian brain have not been examined. We examined Kv2.1 phosphorylation state in mouse brain neurons that express different RyR:Kv2.1 ratios, as detailed above. We used phosphospecific antibodies (Misonou et al., 2006; Park et al., 2006) against Kv2.1 phosphorylated at the S453, S563, and S603 phosphorylation sites (i.e., pS453, pS563 and pS603, respectively) as markers for Kv2.1 phosphorylation state. We found that Kv2.1 is highly phosphorylated at pS453 (Fig. 6A), pS563 (Fig. 6G) and pS603 (Fig. 6M) in neocortical neurons, where juxtaposed clustering of Kv2.1 with RyR is not observed (Table 2). However, Kv2.1 in striatal MSNs had markedly lower levels of labeling for all three phosphorylation sites-pS453 (Fig. 6D), pS5633 (Fig. 6J) and pS603 (Fig. 6P). We next examined the levels of phosphorylation at these sites in direct and indirect pathway MSNs using the EGFP-D2 mouse line. We found that, in addition to the differences in the ratio of RyR:Kv2.1 expression between neurons in these pathways described above, the pS563:Kv2.1 labeling ratio was also distinct, in being significantly higher (p=0.00168, n=218; Fig. 7B, D) in the D2− (i.e., D1+) MSNs compared to the D2+ MSNs. No significant difference in the pS453:Kv2.1 (p=0.69654, n=229; Fig. 7A, D) or pS603-Kv2.1 (p=0.28462, n=92; Fig. 7C, D) labeling ratios was observed between D2− (i.e., D1+) and D2+ neurons. Importantly, the phosphospecific and general anti-Kv2.1 antibodies used in these labeling experiments were validated for specificity by immunohistochemistry against samples from mice in which Kv2.1 phosphorylation was acutely reduced by brief hypoxia, and/or samples from Kv2.1 KO mice (Figures 8-10). In conclusion, Kv2.1 expressed in MSNs, which have high overall levels of RyR:Kv2.1 colocalization/juxtaposition, is maintained at a lower overall level of steady-state phosphorylation than Kv2.1 in neocortical neurons, which have little or no colocalized Kv2.1 and RyRs. Within stratum, MSNs of the direct pathway, which have a lower Kv2.1:RyR ratio than do MSNs of the direct pathway, have higher levels of phosphorylation at the pS563 site, compared to direct pathway MSNs.
Figure 6.
Kv2.1 phosphorylation at pS453 and pS563 and pS603 is reduced in striatum relative to cortex. Mouse brain sections obtained from an EGFP-D2 mouse were immunofluorescence labeled for Kv2.1 (magenta) and with phosphospecific antibodies specific for Kv2.1 phosphorylated at pS453 A-F, pS563 G-L or pS603 M-R (green). Images were acquired using an epifluorescence microscope equipped with an ApoTome. Images are reconstructed Z-stacks from optical sections taken at the same exposure. A-C, G-I and M-O: cortex. D-F, J-L and P-R: striatum. Scale bars are 10 μm for panels A-L, and 20 μm panels M-O.
Figure 7.
Indirect pathway striatal MSNs have lower levels of pS563 labeling. Mouse brain sections were multiple immunofluorescence labeled for Kv2.1 and pS603, pS453 and pS563, and single optical sections acquired using an epifluorescence microscope equipped with an ApoTome. Fluorescence intensities were measured from single optical sections taken at 20x using Adobe Photoshop. Cumulative percentages in panels A, B and C show the pS453:Kv2.1, pS563:Kv2.1 and pS603:Kv2.1 ratios in the EGFP-D2 mouse striatum, respectively. Black lines represent D2+/D1− (indirect pathway) MSNs and gray lines represent D2−/D1+ (direct pathway) MSNs. D: Ratios of phosphorylation to total Kv2.1 from the striatum of EGFP-D2 mice: D2+ (black bars) and D2− (white bars). D2+ cell values were used to normalize for corresponding D2− measurements in each independent sample. Data are mean ± SEM (n>90 per group). *p<0.01 versus D2+ measurements.
Figure 8.
Kv2.1-pS603 validation in Kv2.1-KO mice and CO2 induced dephosphorylation mouse models. Mouse brain sections from wild type mice, A-D and I-L, and Kv2.1-KO mice, E-H and M-P, were double immunofluorescence labeled for general Kv2.1 with the K89/34 mAb (red), and Kv2.1-pS603 (green), and nuclei were labeled using Hoechst (blue). Images of single optical sections of cortical neurons were acquired at equal exposures using an epifluorescence microscope equipped with an ApoTome. Kv2.1 dephosphorylation was induced using CO2 in panels I-L and M-P. Scale bars are 10 μm. Magenta lines in A and D represent line traces of fluorescence intensities depicted in the left and right graphs in panel Q. Q. Line traces were generated using the plot profile function in ImageJ for each channel.
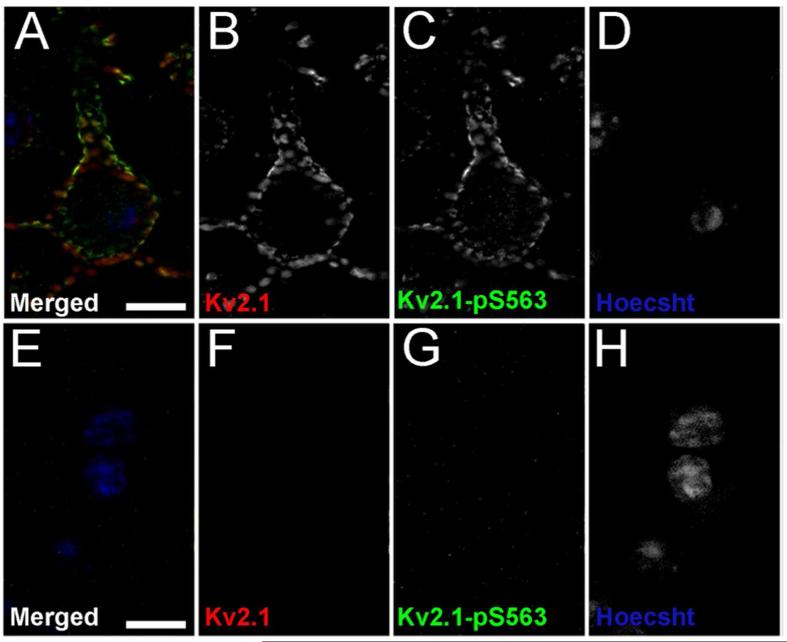
Figure 10. Kv2.1-pS563 validation in Kv2.1-KO mice.
Mouse brain sections from wild type mice, A-D and Kv2.1-KO mice, E-H were double immunofluorescence labeled for general Kv2.1 with the K89/34 mAb (red), and Kv2.1-pS563 (green), and nuclei were labeled using Hoechst (blue). Images of single optical sections of cortical neurons were acquired at equal exposures using an epifluorescence microscope equipped with an ApoTome. Scale bars are 10 μm.
Stimulation of RyRs in HEK293 cells leads to modulation of Kv2.1 function
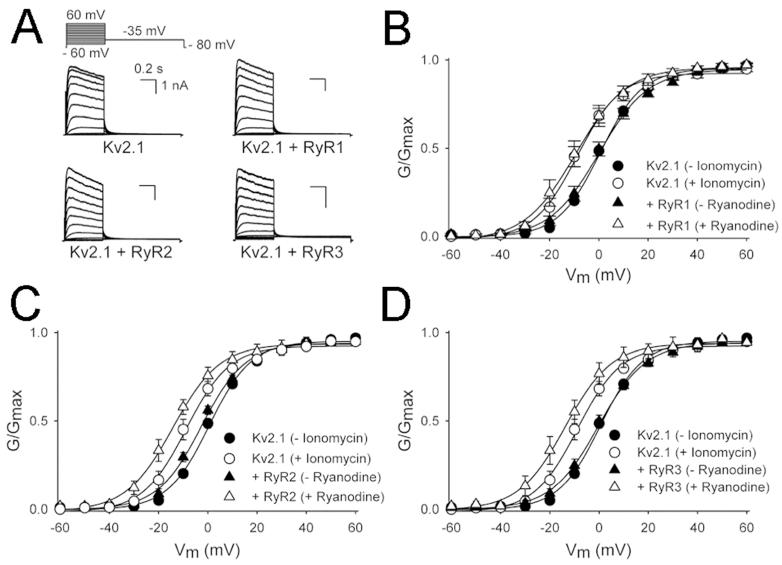
The data presented above reveal a spatial relationship between Kv2.1 and RyR labeling, and Kv2.1 phosphorylation, that differs between different classes of mouse brain neurons. To determine whether Kv2.1 and RyRs are functionally coupled, we coexpressed Kv2.1 with the three RyR subtypes in HEK293 cells, and used the whole cell patch clamp technique to record Kv2.1 currents. Typical recordings of Kv2.1 current when expressed alone and with co-expression with the three RyR subtypes (RyR1, RyR2 and RyR3) are shown in Fig. 11A. Co-expression of Kv2.1 with all three RyRs did not change the voltage dependence of inactivation of Kv2.1 (Table 3). However, co-expression of Kv2.1 with RyR2 shifted the voltage dependence of activation of Kv2.1 in the hyperpolarized direction (Fig. 11C and Table 3). Furthermore, triggering RyR-mediated Ca2+ release by treating the cells with 1 μM ryanodine for 10 min resulted in a further shift of the voltage dependence of activation of Kv2.1 to hyperpolarizing potentials in cells expressing each the three RyR subtypes (Fig. 11B-D and Table 3). Stimulation of RyR in cells overexpressing Kv2.1 alone exhibited a slight hyperpolarizing shift, but this is likely due to endogenously expressed RyRs. The dramatic leftward shift observed in cells overexpressing both Kv2.1 and RyR isoforms was similar to the shift observed upon treatment with ionomycin (1 μM in media that contained external Ca2+) that served as a positive control for Ca2+ dependent dephosphorylation of Kv2.1 in HEK293 cells (Mohapatra and Trimmer, 2006; Park et al., 2006) and neurons (Mohapatra et al., 2009). Together these data show that all coexpression of all three RyR isoforms leads to enhanced Kv2.1 modulation.
Figure 11.
Stimulation of RyR causes a hyperpolarizing shift in the voltage dependence of activation of Kv2.1. A: Typical current recordings of Kv2.1 alone and upon co-expression with the ryanodine receptor 1 (RyR1), ryanodine receptor 2 (RyR2) and ryanodine receptor 3 (RyR3). The voltage protocol is given on top. B-D: Conductance-voltage relationship of voltage-dependence of activation of Kv2.1 currents. Circles in all panels correspond to data from cells expressing Kv2.1 alone, before (black) and after (open) ionomycin treatment. Triangles in all panels correspond to data from cells expressing Kv2.1 with an RyR isoform (B: RyR1; C: RyR2; D: RyR3), before (black) and after (open) ryanodine treatment. The voltage-dependence of activation is derived from plotting the normalized tail current amplitudes at −35 mV as a function of the prepulse potential (ranging from 60 mV to −60 mV in 10-mV steps). Solid lines represent the Boltzmann fit. Note that the voltage dependence of activation is significantly shifted in the hyperpolarized direction upon co-expression of Kv2.1 with RyR2 even without ryanodine treatment.
Table 3. Biophysical properties of Kv2.1 in response to RyR expression and stimulation.
The midpoints of activation and inactivation (V1/2) and slope factors (k) were obtained from a single Boltzmann fit. Values are mean ± SE and n represents the number of experiments. Values in bold font are statistically significant (p < 0.05) compared with the corresponding Kv2.1 properties before treatment with 1 NM ionomycin (IM) or 1 NM ryanodine (Ry).
| Activation | Inactivation | |||||
|---|---|---|---|---|---|---|
| V1/2 | k | n | V1/2 | k | n | |
| Kv2.1 | 1.06 ± 0.55 | 9.35 ± 0.47 | 16 | −27.17 ± 1.43 | 6.05 ± 0.30 | 7 |
| + IM | −9.03 ± 2.00 | 7.95 ± 1.00 | 6 | −25.58 ± 1.22 | 5.58 ± 0.67 | 5 |
| + Ry | −2.36 ± 4.39 | 11.27 ± 0.81 | 5 | −28.34 ± 1.65 | 5.27 ± 0.24 | 5 |
| Kv2.1 + RyR1 | 0.00 ± 1.75 | 10.10 ± 0.68 | 15 | −27.98 ± 2.94 | 5.80 ± 0.25 | 6 |
| + Ry | −10.84 ± 3.04 | 9.94 ± 0.71 | 6 | −30.47 ± 3.71 | 5.40 ± 0.19 | 6 |
| Kv2.1 + RyR2 | −3.09 ± 0.94 | 8.55 ± 0.42 | 21 | −30.31 ± 2.64 | 5.37 ± 0.14 | 12 |
| + Ry | −15.25 ± 2.43 | 9.18 ± 1.09 | 6 | −32.25 ± 3.07 | 5.31 ± 0.16 | 6 |
| Kv2.1 + RyR3 | −0.89 ± 1.76 | 8.85 ± 0.68 | 20 | −24.96 ± 2.56 | 5.67 ± 0.43 | 5 |
| + Ry | −14.43 ± 2.82 | 8.85 ± 0.68 | 6 | −33.72 ± 2.79 | 5.63 ± 0.28 | 4 |
DISCUSSION
The major finding from this study is the novel region- and cell-specific relationship between the localization of the plasma membrane Kv2.1 channel and intracellular RyR Ca2+ release channels. Kv2.1 acts a homeostatic regulator of neuronal activity (Misonou et al., 2004) through bidirectional activity-dependent regulation of phosphorylation at multiple sites, including pS453, pS563 and pS603 (Misonou et al., 2006; Park et al., 2006). Here we show by multiple label immunofluorescence microscopy that Kv2.1 clusters are found juxtaposed to RyR clusters in neurons in specific brain regions, and that this is especially prominent in striatal MSNs. We show using immunogold electron microscopy that plasma membrane Kv2.1 clusters are adjacent to SSC in these neurons. We also show that Kv2.1 has low steady-state levels of phosphorylation at the pS453 pS563 and pS603 phosphorylation sites in striatal MSNs, which have extensive Kv2.1:RyR juxtaposed clusters. Conversely, neocortical neurons have low levels of juxtaposed Kv2.1:RyR, and higher levels of Kv2.1 phosphorylation at the pS453 pS563 and pS603 phosphorylation sites. Within striatum, we found that Kv2.1 clusters are found juxtaposed to higher levels of clustered RyR, and have lower levels of phosphorylation at pS563, in indirect pathway MSNs, compared to direct pathway MSNs. Finally, experiments in heterologous cells show that expression and stimulation of RyR is coupled to changes in Kv2.1 gating similar to those seen upon Kv2.1 dephosphorylation. The cell type specificity of the expression patterns of Kv2.1 relative to RyRs, and differences in phosphorylation state, are expected to influence Kv2.1 function and the dynamic regulation of intrinsic excitability in these distinct neuronal populations, including MSNs in the direct and indirect output pathways of the striatum.
The mechanism that underlies the juxtaposed clustering of plasma membrane Kv2.1 and intracellular RyR is unknown. Kv2.1 clusters are present in neurons in brain regions that do not have somatically clustered RyR (e.g., neocortex), while clustering of RyRs is not disrupted Kv2.1-KO mice. This suggests that Kv2.1 and RyR form clusters independent of one another. There exist numerous examples of plasma membrane and intracellular membrane ion channels that exist in very specific relationships. These include those in which there is a direct physical association between the plasma membrane and intracellular membrane channels (e.g., Ca2+ channels and RyRs, respectively, at the junctional triad of skeletal muscle), and those where a direct interaction has not been shown (e.g., large conductance Ca2+-activated BK channels and IP3 receptors, respectively, in cerebellar Purkinje neurons) (Kaufmann et al., 2009), although functional coupling between IP3-mediated Ca2+ release and BK channel gating has been demonstrated in cerebellar Purkinje neurons (Khodakhah and Ogden, 1993). Co-immunoprecipitation experiments have not provided consistent data supporting a physical association between Kv2.1 and RyR. However, Kv2.1 and RyRs are found juxtaposed in many brain neurons, and RyR coexpression and activation leads to modulation of Kv2.1 gating in heterologous cells. This suggests that functional coupling between Kv2.1 and RyRs, via RyR-mediated Ca2+ release and calcineurin/PP2B dephosphorylation of Kv2.1, could occur in these cells. There are multiple triggers for RyR activation and intracellular Ca2+ release in neurons, including Ca2+ (Verkhratsky and Shmigol, 1996), membrane potential (Kim et al., 2007) and RyR nitrosylation (Kakizawa et al., 2012). Future studies dissecting these RyR activation pathways and which, if any, impact phosphorylation and gating of neuronal Kv2.1 will better define the signaling pathways regulating this important determinant of neuronal excitability. Emerging evidence shows that RyRs play a role in regulating intrinsic excitability of neurons (Cheong et al., 2011), but the mechanism linking RyR function to ion channel function and membrane excitability is unknown. A recent study showed that RyRs impact the firing of thalamic neurons, and that caffeine stimulation of RyRs resulted in less frequent spiking (Cheong et al., 2011). It is interesting to speculate that the juxtaposed clusters of Kv2.1 and RyR in thalamic neurons provide a mechanism whereby RyR activation leads to altered Kv2.1 phosphorylation and gating, contributing to suppression of thalamic neuron excitability.
The most striking example of juxtaposed clustering of somatic Kv2.1 and RyRs is in striatal MSNs. When comparing MSNs to neocortical neurons, there emerges a consistent relationship between the extent of colocalized/juxtaposed RyR and Kv2.1, and the level of Kv2.1 phosphorylation. Kv2.1 in neocortical neurons has low levels of RyR found juxtaposed to somatodendritic Kv2.1, and high levels of labeling with phosphospecific anti-Kv2.1 antibodies. Kv2.1 in MSNs is found juxtaposed to RyRs, and has an overall lower levels of phosphospecific antibody labeling. Moreover, within the population of MSNs, we observed that the D1+/D2− direct pathway MSNs have a relatively low Kv2.1:RyR ratio, and a relatively high pS563:Kv2.1 phosphorylation ratio, compared to D2+/D1− indirect pathway MSNs (high Kv2.1:RyR ratio; low pS563:Kv2.1 phosphorylation ratio). That phosphorylation at the pS563 site appears to be maintained at higher levels in direct pathway MSNs than phosphorylation at either the pS453 or pS603 sites suggest distinct regulation of phosphorylation at these sites, as has been shown previously in hippocampal neurons in dissociated culture (Misonou et al., 2006; Ikematsu et al., 2011). It is intriguing to note that direct pathway (D1+) MSNs have lower intrinsic excitability compared to D2+ MSNs of the indirect pathway (Cepeda et al., 2008; Gertler et al., 2008; Planert et al., 2013); reviewed in (Gerfen and Surmeier, 2011), which may contribute to the increased vulnerability of indirect pathway MSNs in neurodegenerative disorders such as Huntington’s Disease (Han et al., 2010). Stimulation by DA has opposite effects on the excitability of direct and indirect MSNs, counteracting these constitutive differences by D1 receptor-mediated increases in excitability of direct pathway neurons, and D2 receptor-mediated decreases in excitability of indirect pathway neurons (Planert et al., 2013). A number of plasma membrane ion channels including Cav channels (Hernandez-Lopez et al., 2000) and inwardly rectifying Kir2 K+ channels (Shen et al., 2007; Gertler et al., 2008) have been identified as targets of these dopamine pathways, although this does not preclude the involvement of other ion channels such as Kv2.1. One of the major effects of D1 receptor activation in direct pathway neurons is elevated cAMP and activation of PKA, leading to elevated phosphorylation of target proteins (e.g., DARPP-32), and increased excitability, while D2 receptor activation in indirect pathway MSNs leads to increased intracellular Ca2+, leading to activation of calcineurin/PP2B, and decreased excitability (Gerfen and Surmeier, 2011). The distinct responses of direct and indirect pathway MSNs to DA may be consistent with a role for Kv2.1 in modulation of MSN excitability, in that calcineurin-mediated dephosphorylation of Kv2.1 enhances its activity (Misonou et al., 2004; Misonou et al., 2006), leading to suppression of neuronal excitability (Misonou et al., 2005a; Mohapatra et al., 2009). This could be consistent with the effects of DA to reduce excitability of D2+ indirect pathway MSNs. Conversely, DA-triggered increases in phosphorylation of Kv2.1 in D1+ direct pathway MSNs would be expected to suppress Kv2.1 function and enhance excitability. A specific role for Kv2.1 in DA-mediated regulation of MSN excitability has not been described, although the reduced expression of Kv2.1 in an animal model of Huntington’s Disease has been proposed to contribute to increased neuronal excitability and susceptibility to excitotoxic damage (Ariano et al., 2005).
That RyR expression and stimulation leads to changes in coexpressed Kv2.1 activity in heterologous cells presumably reflects a pathway acting through calcineurin/PP2B activation, as has been previously shown for the effects of Ca2+ ionophore and muscarinic stimulation (Mohapatra and Trimmer, 2006). However, the protein phosphatases PP1, PP2A and calcineurin/PP2B also interact with RyRs (Marx et al., 2001) and could potentially contribute to RyR-mediated dephosphorylation of Kv2.1. Although both PP1 (Misonou et al., 2004; Misonou et al., 2005b; Cerda and Trimmer, 2011) and calcineurin/PP2B (Misonou et al., 2004; Misonou et al., 2005a; Mohapatra and Trimmer, 2006) regulate Kv2.1 phosphorylation, PP1 appears to be more important for constitutive maintenance of Kv2.1 phosphorylation state in a dynamic equilibrium, while calcineurin/PP2B is the key mediator of the dephosphorylation triggered by stimuli that lead to increased intracellular Ca2+ (Misonou et al., 2004). Future studies will determine whether specific protein kinases and phosphatases associate with these unique Kv2.1:RyR assemblies in MSNs, and their role on regulating Kv2.1 in response to stimulation by dopaminergic, muscarinic and other modulatory signaling pathways crucial to regulating the physiology of MSNs.
Supplementary Material
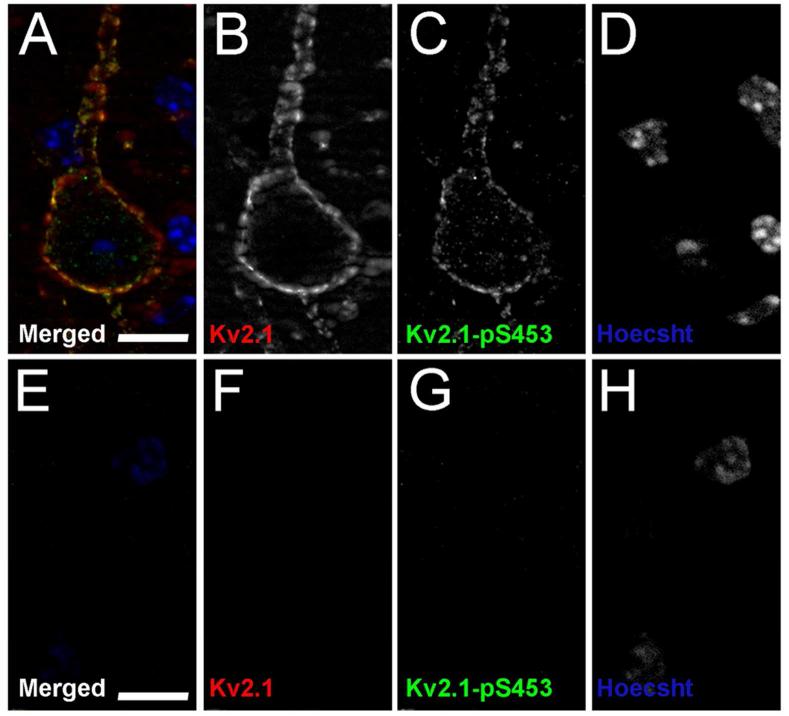
Figure 9. Kv2.1-pS453 validation in Kv2.1-KO mice.
Mouse brain sections from wild type mice, A-D and Kv2.1-KO mice, E-H were double immunofluorescence labeled for general Kv2.1 with the K89/34 mAb (red), and Kv2.1-pS453 (green), and nuclei were labeled using Hoechst (blue). Images of single optical sections of cortical neurons were acquired at equal exposures using an epifluorescence microscope equipped with an ApoTome. Scale bars are 10 μm.
OTHER ACKNOWLEDGMENTS
We are grateful to Dr. David Speca and Dr. Karl Murray for their intellectual and technical contributions, and to Thinh Chau, Ashleigh Evans and Lauren Guy for technical assistance with monoclonal antibody generation and characterization. We thank Bryant Chhun (Carl Zeiss Microscopy) for assistance with super-resolution imaging, Dr. Gavin Wright of the Wellcome Trust Sanger Institute for providing the immunoglobulin expression vector, and Dr. Paul D. Allen of the University of California Davis for the RyR expression plasmids.
Support and Grant Information: This work was funded by National Institutes of Health (NIH) research grant R01 NS42225 to J. S. Trimmer. D. Mandikian was supported by National Institutes of Health (NIH) institutional training grant T32 GM7377. E. Bocksteins was supported by a postdoctoral fellowship of the Research Foundation-Flanders (FWO).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no competing financial interests.
ROLE OF AUTHORS
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: DM, OC and JST. Acquisition of data: DM, EB, LP and HB. Analysis and interpretation of data: DM, EB, LP and HB. Drafting of the manuscript: DM and JST. Critical revision of the manuscript for important intellectual content: EB, LP, RS and JST. Statistical analysis: DM and EB. Obtained funding: EB and JST. Administrative, technical, and material support: JST. Study supervision: RS and JST.
Literature Cited
- Antonucci DE, Lim ST, Vassanelli S, Trimmer JS. Dynamic localization and clustering of dendritic Kv2.1 voltage-dependent potassium channels in developing hippocampal neurons. Neuroscience. 2001;108(1):69–81. doi: 10.1016/s0306-4522(01)00476-6. [DOI] [PubMed] [Google Scholar]
- Ariano MA, Cepeda C, Calvert CR, Flores-Hernandez J, Hernandez-Echeagaray E, Klapstein GJ, Chandler SH, Aronin N, DiFiglia M, Levine MS. Striatal potassium channel dysfunction in Huntington’s disease transgenic mice. J Neurophysiol. 2005;93(5):2565–2574. doi: 10.1152/jn.00791.2004. [DOI] [PubMed] [Google Scholar]
- Baek JH, Cerda O, Trimmer JS. Mass spectrometry-based phosphoproteomics reveals multisite phosphorylation on mammalian brain voltage-gated sodium and potassium channels. Semin Cell Dev Biol. 2011;22(2):153–159. doi: 10.1016/j.semcdb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkefeld H, Sailer CA, Bildl W, Rohde V, Thumfart JO, Eble S, Klugbauer N, Reisinger E, Bischofberger J, Oliver D, Knaus HG, Schulte U, Fakler B. BKCa-Cav channel complexes mediate rapid and localized Ca2+-activated K+ signaling. Science. 2006;314(5799):615–620. doi: 10.1126/science.1132915. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Neuronal calcium signaling. Neuron. 1998;21(1):13–26. doi: 10.1016/s0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- Bildl W, Strassmaier T, Thurm H, Andersen J, Eble S, Oliver D, Knipper M, Mann M, Schulte U, Adelman JP, Fakler B. Protein kinase CK2 is coassembled with small conductance Ca(2+)-activated K+ channels and regulates channel gating. Neuron. 2004;43(6):847–858. doi: 10.1016/j.neuron.2004.08.033. [DOI] [PubMed] [Google Scholar]
- Cepeda C, Andre VM, Yamazaki I, Wu N, Kleiman-Weiner M, Levine MS. Differential electrophysiological properties of dopamine D1 and D2 receptor-containing striatal medium-sized spiny neurons. Eur J Neurosci. 2008;27(3):671–682. doi: 10.1111/j.1460-9568.2008.06038.x. [DOI] [PubMed] [Google Scholar]
- Cerda O, Trimmer JS. Analysis and functional implications of phosphorylation of neuronal voltage-gated potassium channels. Neurosci Lett. 2010;486(2):60–67. doi: 10.1016/j.neulet.2010.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda O, Trimmer JS. Activity-dependent phosphorylation of neuronal Kv2.1 potassium channels by CDK5. J Biol Chem. 2011;286(33):28738–28748. doi: 10.1074/jbc.M111.251942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong E, Kim C, Choi BJ, Sun M, Shin HS. Thalamic ryanodine receptors are involved in controlling the tonic firing of thalamocortical neurons and inflammatory pain signal processing. J Neurosci. 2011;31(4):1213–1218. doi: 10.1523/JNEUROSCI.3203-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Staudt N, Wright GJ. A rapid and scalable method for selecting recombinant mouse monoclonal antibodies. BMC Biology. 2010:8–76. doi: 10.1186/1741-7007-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Hall DD, Hell JW. Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol Rev. 2009;89(2):411–452. doi: 10.1152/physrev.00029.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Haak LL, Phillips-Tansey E, Russell JT, McBain CJ. Frequency-dependent regulation of rat hippocampal somato-dendritic excitability by the K+ channel subunit Kv2.1. J Physiol. 2000;522(Pt 1):19–31. doi: 10.1111/j.1469-7793.2000.t01-2-00019.xm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tao-Cheng JH, Zerfas P, McBain CJ. The K+ channel, Kv2.1, is apposed to astrocytic processes and is associated with inhibitory postsynaptic membranes in hippocampal and cortical principal neurons and inhibitory interneurons. Neuroscience. 1998;84(1):37–48. doi: 10.1016/s0306-4522(97)00519-8. [DOI] [PubMed] [Google Scholar]
- Emptage NJ, Reid CA, Fine A. Calcium stores in hippocampal synaptic boutons mediate short-term plasticity, store-operated Ca2+ entry, and spontaneous transmitter release. Neuron. 2001;29(1):197–208. doi: 10.1016/s0896-6273(01)00190-8. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Surmeier DJ. Modulation of striatal projection systems by dopamine. Annu Rev Neurosci. 2011;34:441–466. doi: 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler TS, Chan CS, Surmeier DJ. Dichotomous anatomical properties of adult striatal medium spiny neurons. J Neurosci. 2008;28(43):10814–10824. doi: 10.1523/JNEUROSCI.2660-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Han I, You Y, Kordower JH, Brady ST, Morfini GA. Differential vulnerability of neurons in Huntington’s disease:the role of cell type-specific features. J Neurochem. 2010;113(5):1073–1091. doi: 10.1111/j.1471-4159.2010.06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Lopez S, Tkatch T, Perez-Garci E, Galarraga E, Bargas J, Hamm H, Surmeier DJ. D2 dopamine receptors in striatal medium spiny neurons reduce L-type Ca2+ currents and excitability via a novel PLC[beta]1-IP3-calcineurin-signaling cascade. J Neurosci. 2000;20(24):8987–8995. doi: 10.1523/JNEUROSCI.20-24-08987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro N, Grunditz A, Oertner TG. Differential distribution of endoplasmic reticulum controls metabotropic signaling and plasticity at hippocampal synapses. Proc Natl Acad Sci U S A. 2009;106(35):15055–15060. doi: 10.1073/pnas.0905110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N, Dallas ML, Ross FA, Lewis RW, Rafferty JN, David JA, Suman R, Peers C, Hardie DG, Evans AM. Phosphorylation of the voltage-gated potassium channel Kv2.1 by AMP-activated protein kinase regulates membrane excitability. Proc Natl Acad Sci U S A. 2011;108(44):18132–18137. doi: 10.1073/pnas.1106201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson DA, Kuznetsov A, Lopez JP, Kash S, Ammala CE, Philipson LH. Kv2.1 ablation alters glucose-induced islet electrical activity, enhancing insulin secretion. Cell Metab. 2007;6(3):229–235. doi: 10.1016/j.cmet.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juste M, Muzard J, Billiald P. Cloning of the antibody kappa light chain V-gene from murine hybridomas by bypassing the aberrant MOPC21-derived transcript. Anal Biochem. 2006;349(1):159–161. doi: 10.1016/j.ab.2005.10.046. [DOI] [PubMed] [Google Scholar]
- Kakizawa S, Yamazawa T, Chen Y, Ito A, Murayama T, Oyamada H, Kurebayashi N, Sato O, Watanabe M, Mori N, Oguchi K, Sakurai T, Takeshima H, Saito N, Iino M. Nitric oxide-induced calcium release via ryanodine receptors regulates neuronal function. EMBO J. 2012;31(2):417–428. doi: 10.1038/emboj.2011.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WA, Ferraguti F, Fukazawa Y, Kasugai Y, Shigemoto R, Laake P, Sexton JA, Ruth P, Wietzorrek G, Knaus HG, Storm JF, Ottersen OP. Large-conductance calcium-activated potassium channels in purkinje cell plasma membranes are clustered at sites of hypolemmal microdomains. J Comp Neurol. 2009;515(2):215–230. doi: 10.1002/cne.22066. [DOI] [PubMed] [Google Scholar]
- Khodakhah K, Ogden D. Functional heterogeneity of calcium release by inositol trisphosphate in single Purkinje neurones, cultured cerebellar astrocytes, and peripheral tissues. Proc Natl Acad Sci U S A. 1993;90(11):4976–4980. doi: 10.1073/pnas.90.11.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ridgway LD, Zou S, Chiu YH, Dryer SE. Alternatively spliced C-terminal domains regulate the surface expression of large conductance calcium-activated potassium channels. Neuroscience. 2007;146(4):1652–1661. doi: 10.1016/j.neuroscience.2007.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AN, Manning CF, Trimmer JS. A unique ion channel clustering domain on the axon initial segment of mammalian neurons. J Comp Neurol. 2014;522(11):2594–2608. doi: 10.1002/cne.23551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer AC. Physiology and pharmacology of striatal neurons. Annu Rev Neurosci. 2009;32:127–147. doi: 10.1146/annurev.neuro.051508.135422. [DOI] [PubMed] [Google Scholar]
- Lee BS, Gunn RB, Kopito RR. Functional differences among nonerythroid anion exchangers expressed in a transfected human cell line. J Biol Chem. 1991;266(18):11448–11454. [PubMed] [Google Scholar]
- Lim ST, Antonucci DE, Scannevin RH, Trimmer JS. A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron. 2000;25(2):385–397. doi: 10.1016/s0896-6273(00)80902-2. [DOI] [PubMed] [Google Scholar]
- Manning CF, Bundros AM, Trimmer JS. Benefits and pitfalls of secondary antibodies: why choosing the right secondary is of primary importance. PLoS One. 2012;7(6):e38313. doi: 10.1371/journal.pone.0038313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx SO, Reiken S, Hisamatsu Y, Gaburjakova M, Gaburjakova J, Yang YM, Rosemblit N, Marks AR. Phosphorylation-dependent regulation of ryanodine receptors: a novel role for leucine/isoleucine zippers. J Cell Biol. 2001;153(4):699–708. doi: 10.1083/jcb.153.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Menegola M, Mohapatra DP, Guy LK, Park KS, Trimmer JS. Bidirectional activity-dependent regulation of neuronal ion channel phosphorylation. J Neurosci. 2006;26(52):13505–13514. doi: 10.1523/JNEUROSCI.3970-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Menegola M, Trimmer JS. Calcium- and metabolic state-dependent modulation of the voltage-dependent Kv2.1 channel regulates neuronal excitability in response to ischemia. J Neurosci. 2005a;25(48):11184–11193. doi: 10.1523/JNEUROSCI.3370-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Park EW, Leung V, Zhen D, Misonou K, Anderson AE, Trimmer JS. Regulation of ion channel localization and phosphorylation by neuronal activity. Nat Neurosci. 2004;7(7):711–718. doi: 10.1038/nn1260. [DOI] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Trimmer JS. Kv2.1:a voltage-gated K+ channel critical to dynamic control of neuronal excitability. Neurotoxicology. 2005b;26(5):743–752. doi: 10.1016/j.neuro.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Misonou H, Thompson SM, Cai X. Dynamic regulation of the Kv2.1 voltage-gated potassium channel during brain ischemia through neuroglial interaction. J Neurosci. 2008;28(34):8529–8538. doi: 10.1523/JNEUROSCI.1417-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra DP, Misonou H, Pan SJ, Held JE, Surmeier DJ, Trimmer JS. Regulation of intrinsic excitability in hippocampal neurons by activity-dependent modulation of the KV2.1 potassium channel. Channels (Austin) 2009;3(1):46–56. doi: 10.4161/chan.3.1.7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra DP, Trimmer JS. The Kv2.1 C terminus can autonomously transfer Kv2.1-like phosphorylation-dependent localization, voltage-dependent gating, and muscarinic modulation to diverse Kv channels. J Neurosci. 2006;26(2):685–695. doi: 10.1523/JNEUROSCI.4620-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muennich EA, Fyffe RE. Focal aggregation of voltage-gated, Kv2.1 subunit-containing, potassium channels at synaptic sites in rat spinal motoneurones. J Physiol. 2004;554(Pt 3):673–685. doi: 10.1113/jphysiol.2003.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Carpenter-Hyland EP, Hearing MC, Becker HC, Woodward JJ, Chandler LJ. Glutamate transporters regulate extrasynaptic NMDA receptor modulation of Kv2.1 potassium channels. J Neurosci. 2008;28(35):8801–8809. doi: 10.1523/JNEUROSCI.2405-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Shi G, Scannevin RH, Trimmer JS. Phosphorylation of the Kv2.1 K+ channel alters voltage-dependent activation. Mol Pharmacol. 1997;52(5):821–828. doi: 10.1124/mol.52.5.821. [DOI] [PubMed] [Google Scholar]
- Murakoshi H, Trimmer JS. Identification of the Kv2.1 K+ channel as a major component of the delayed rectifier K+ current in rat hippocampal neurons. J Neurosci. 1999;19(5):1728–1735. doi: 10.1523/JNEUROSCI.19-05-01728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Sekiguchi N, Rando TA, Allen PD, Beam KG. Two regions of the ryanodine receptor involved in coupling with L-type Ca2+ channels. J Biol Chem. 1998;273(22):13403–13406. doi: 10.1074/jbc.273.22.13403. [DOI] [PubMed] [Google Scholar]
- Parajuli LK, Nakajima C, Kulik A, Matsui K, Schneider T, Shigemoto R, Fukazawa Y. Quantitative regional and ultrastructural localization of the Ca(v)2.3 subunit of R-type calcium channel in mouse brain. J Neurosci. 2012;32(39):13555–13567. doi: 10.1523/JNEUROSCI.1142-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Mohapatra DP, Misonou H, Trimmer JS. Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science. 2006;313(5789):976–979. doi: 10.1126/science.1124254. [DOI] [PubMed] [Google Scholar]
- Peltola MA, Kuja-Panula J, Lauri SE, Taira T, Rauvala H. AMIGO is an auxiliary subunit of the Kv2.1 potassium channel. EMBO reports. 2011;12(12):1293–1299. doi: 10.1038/embor.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planert H, Berger TK, Silberberg G. Membrane properties of striatal direct and indirect pathway neurons in mouse and rat slices and their modulation by dopamine. PLoS One. 2013;8(3):e57054. doi: 10.1371/journal.pone.0057054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannevin RH, Murakoshi H, Rhodes KJ, Trimmer JS. Identification of a cytoplasmic domain important in the polarized expression and clustering of the Kv2.1 K+ channel. J Cell Biol. 1996;135(6 Pt 1):1619–1632. doi: 10.1083/jcb.135.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell SA, Staines WA, Wessendorf MW. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem. 1999;47(6):719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
- Shen W, Tian X, Day M, Ulrich S, Tkatch T, Nathanson NM, Surmeier DJ. Cholinergic modulation of Kir2 channels selectively elevates dendritic excitability in striatopallidal neurons. Nat Neurosci. 2007;10(11):1458–1466. doi: 10.1038/nn1972. [DOI] [PubMed] [Google Scholar]
- Shi G, Kleinklaus AK, Marrion NV, Trimmer JS. Properties of Kv2.1 K+ channels expressed in transfected mammalian cells. J Biol Chem. 1994;269(37):23204–23211. [PubMed] [Google Scholar]
- Speca DJ, Ogata G, Mandikian D, Bishop HI, Wiler SW, Eum K, Wenzel HJ, Doisy ET, Matt L, Campi KL, Golub MS, Nerbonne JM, Hell JW, Trainor BC, Sack JT, Schwartzkroin PA, Trimmer JS. Deletion of the Kv2.1 delayed rectifier potassium channel leads to neuronal and behavioral hyperexcitability. Genes Brain Behav. 2014;13(4):394–408. doi: 10.1111/gbb.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS. Immunological identification and characterization of a delayed rectifier K+ channel polypeptide in rat brain. Proc Natl Acad Sci U S A. 1991;88(23):10764–10768. doi: 10.1073/pnas.88.23.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS, Misonou H. Phosphorylation of voltage-gated ion channels. In: Zheng J, Trudeau MC, editors. Handbook of Ion Channels. CRC Press; Boca Raton, FL: 2014. [Google Scholar]
- Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88(4):1407–1447. doi: 10.1152/physrev.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky A, Shmigol A. Calcium-induced calcium release in neurones. Cell Calcium. 1996;19(1):1–14. doi: 10.1016/s0143-4160(96)90009-3. [DOI] [PubMed] [Google Scholar]
- Zhao M, Li P, Li X, Zhang L, Winkfein RJ, Chen SR. Molecular identification of the ryanodine receptor pore-forming segment. J Biol Chem. 1999;274(37):25971–25974. doi: 10.1074/jbc.274.37.25971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.