Abstract
Background
Defining the RNA transcriptome in Alzheimer’s Disease (AD) will help understand disease mechanisms and provide biomarkers. Though the AD blood transcriptome has been studied, effects of white matter hyperintensities (WMH) were not considered. This study investigated the AD blood transcriptome and accounted for WMH.
Methods
RNA from whole blood was processed on whole-genome microarrays.
Results
293 probe sets were differentially expressed in AD versus controls, 5 of which were significant for WMH status. The 288 AD-specific probe-sets classified subjects with 87.5% sensitivity and 90.5% specificity. They represented 188 genes of which 29 have been reported in prior AD blood and 89 in AD brain studies. Regulated blood genes included MMP9, MME (Neprilysin), TGFβ1, CA4, OCLN, ATM, TGM3, IGFR2, NOV, RNF213, BMX, LRRN1, CAMK2G, INSR, CTSD, SORCS1, SORL1 and TANC2.
Conclusions
RNA expression is altered in AD blood irrespective of WMH status. Some genes are shared with AD brain.
Keywords: Alzheimer’s disease, RNA, white matter hyperintensities, blood, brain
1. Introduction
The diagnosis of Alzheimer’s disease (AD) is based mainly on clinical criteria. Though research guidelines recommend confirmation using PET imaging or CSF biomarkers, high cost and low patient compliance make introducing these in the typical clinical setting challenging. Peripheral blood is accessible, and provides information on systemic factors involved in disease. It has been suggested that blood-based biomarkers could be useful for evaluating diagnosis, pathogenesis and progression of AD 1–5.
Clinical dementia phenotypes, however, are complex syndromes manifested by various diseases potentially leading to both false positive and negative associations. For example, cerebrovascular disease (CVD) and white matter hyperintensities (WMH) commonly co-occur with AD, and contribute to the dementia syndrome. In fact, mixed AD and CVD pathologies may be the most common presentation of the AD phenotype in community populations. Further understanding of the genetic underpinning of AD, therefore, should account for CVD and WMH. This approach has been used to understand the effects of SORL1 on dementia risk6.
White matter hyperintensities (WMH) are commonly found on brain MRI T2-weighted and fluid-attenuated inversion recovery images and are associated with advancing age, vascular risk factors, stroke risk, incident cognitive impairment and incident dementia 7. Neuropathological and amyloid PET imaging studies show that WMH are not correlated with AD pathology, but associated with cerebral arteriosclerosis, supporting the relationship between WMH and CVD 7. Our previous study suggested that WMH found in normal aging and in AD patients are associated with distinctive RNA expression profiles in peripheral blood that are not secondary to AD and may be related in part to oxidative stress 8.
RNA expression in blood of patients with AD has been assessed in several recent independent studies 1–4, 9, 10. However, none has considered the possible effects of WMH on RNA expression in blood as a confounder for AD gene profiles 8. The aim of this study was to examine RNA expression in blood of AD patients with and without WMH as compared to controls. We determined that 188 AD genes were not affected by WMH, and of these 29 had been previously reported in AD blood and 89 had previously been reported in AD brain.
2. Methods
Subjects (n=17 AD (9 WMH+), and n=21 non-AD controls (11 WMH+)) were recruited from the Alzheimer’s disease (AD) Center at University of California at Davis (UCD). The UCD institutional review board approved this study.
AD diagnosis was made according to the NINCDS and Communication Disorders and Stroke/AD and Related Disorders Association (NINDS-ADRDA) criteria. AD with and without WMH, as well as controls (cognitively normal subjects with and without WMH) were included in the present study 8.
Whole blood was collected from each subject and mRNA hybridized on Affymetrix Human U133 Plus 2.0 Arrays. A multivariate ANCOVA was performed on AD status with adjustments for potential confounders, including batch, sex, age, heart disease, hyperlipidemia, education and presence of WMH. Differentially expressed genes (DEGs) were identified based on significance of p<0.005 and absolute fold-change (|FC|) expression >1.2. The results of the gene sets enrichment analysis (GSEA) and functional enrichment analysis were filtered for FDR p-value < 0.05. Prediction analysis was performed as described 8.
Microarray datasets for AD blood transcriptome studies were downloaded from Gene Expression Omnibus (GEO). The major datasets re-analyzed in this study included GSE6613 10 and GSE4229 4 from GEO. In addition, DEG lists were retrieved directly from published papers 1–3. We also conducted a re-analysis of our independent blood microarray study in a Chinese cohort 5. For the re-analysis, DEGs were identified using the method of RankProd (|FC|>1.5, FDR<0.05). Detailed methods are presented in Supplementary Material 1.
3. Results
3.1 Demographics
Demographic information for AD and control subjects with and without WMH is provided in Table 1 and Supplementary Table 1A. A closely matched proportion of the subjects in the AD and non-AD (control) groups had coexisting WMH. To evaluate the influence of each possible confounder on the RNA expression profile, we conducted a one-way ANOVA on the demographic information (Table 1). There were no significant differences in age, gender, race, years of education, history of hyperlipidemia, or heart disease between those with and without AD. There was a significant difference in history of diabetes (Table 1).
3.2 Distinctive RNA expression profile in the blood for AD
ANCOVA was performed for AD versus non-AD to control for expression changes caused by potential confounders, including batch, gender, age, heart disease, hyperlipidemia, diabetes and WMH, since they are known risk factors for developing AD/WMH, were different between the groups and/or because could affect gene expression. The ANCOVA showed 293 differentially expressed probe sets between the AD and non-AD subjects. Among them, 5 genes were also regulated by WMH 8. To avoid confounding effects of WMH, these 5 genes were removed from the gene list, which resulted in a DEG list of 288 probe sets (188 annotated genes) exclusively regulated by AD (Supplementary Table 1B). These 288 probe sets were used for AD classification analyses described below.
3.3 Discrimination of AD vs controls regardless of the WMH status
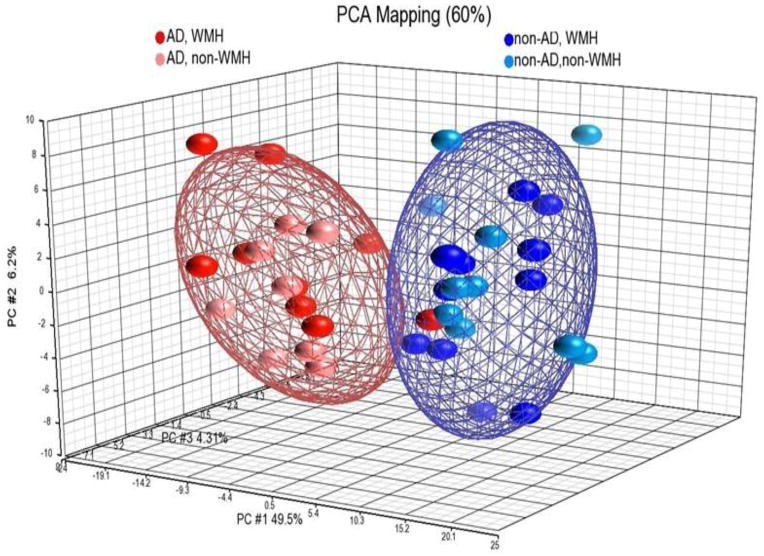
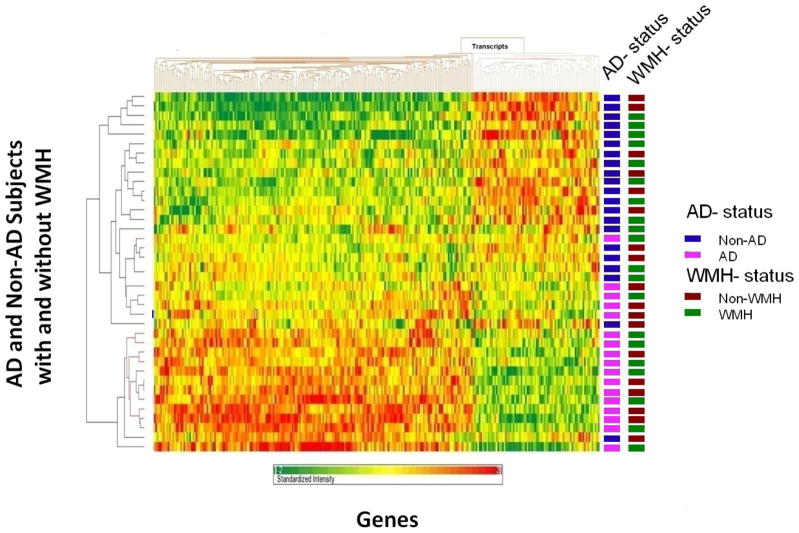
Three different approaches were used to determine how well the 288 probe sets could differentiate AD vs non-AD subjects. We conducted a Principal Components Analysis (PCA) on all the subjects regardless of the coexisting WMH using the 288 probe sets. AD subjects were separated from non-AD subjects, the AD with and without WMH forming a separate centroid from non-AD with and without WMH (Figure 1). The three principal components accounted for 60% of the total variance. To further evaluate the 288 probe set classifier, a non-supervised cluster analysis was performed (Figure 2). The AD and non-AD subjects generally cluster separately, and do not cluster based upon WMH status (Figure 2). Prediction Analysis of Microarray (PAM) was also performed using 10-fold leave-one-out cross-validation in the derivation cohort, and showed sensitivity for AD diagnosis of 87.5% and specificity of 90.5% (Supplementary Figure 1).
Figure 1.
Principal Components Analysis (PCA) on the 288 probe sets that were differentially expressed between AD and control subjects (non-AD). AD subjects with WMH (AD, WMH – red dots) and AD without WMH (AD, non-WMH, pink dots) are included within the same centroids, and are separated from the centroids for control non-AD with WMH (non-AD, WMH – blue dots) and control non-AD without WMH (non-AD, non-WMH, light blue dots). The PCA accounts for 60% of the variance.
Figure 2.
Cluster analysis for the 288 probe sets that separated AD from control (Non-AD). Note that subjects with WMH (WMH) and without WMH (Non-WMH) do not cluster separately. Subjects are on the Y axis, and genes/probe sets are on the X axis. Dark red represents a 2 fold increase in expression and dark green a 2 fold decrease in expression compared to controls.
3.4 Genes with most significantly altered gene expression levels
Among the 288 DEGs between AD and controls, the largest FCs were for TGM3 (transglutaminase 3, FC=2.18), NOV (nephroblastoma overexpressed, FC=1.96), BMX (BMX non-receptor tyrosine kinase, FC=1.93), CA4 (carbonic anhydrase IV, FC=1.88), MMP9 (matrix metallopeptidase 9, FC=1.82), LRRN1 (leucine rich repeat neuronal 1, FC=1.77), INSC (inscrutable homolog, FC=1.74), and OCLN (occludin, FC=−1.74).
3.5 Gene functions/ pathways
Functional enrichment analysis of the 188 genes showed up-regulation of membrane-bound vesicle pathways, endocytosis, cytoskeleton, actomyosin, apoptosis, and protein kinase cascades; and down-regulation of transcription (Supplementary Table 1C). Canonical pathway analysis using the Ingenuity database showed cell cycle, nervous system signaling, hypoxia - angiogenesis, inflammatory signaling and cancer signaling were over represented (Supplementary Table 1D).
3.6 Comparison with blood and brain transcriptome from other AD studies
The comparative analysis of AD transcriptome was based on datasets from five published studies and our own independent microarray experiment on a Chinese cohort 5. We also conducted another analysis of the data from this study using the method of rank product and a stringent threshold of |FC|>1.5 (FDR<0.05). In total, we found 44 genes regulated in AD blood in this study that had been either reported in previous studies of AD blood, or were confirmed using our separate analytical method (Supplementary Table 1E). Of these 44 genes, 29 have been reported to be regulated in blood of AD patients in previous studies (Supplementary Table 1E) 1–5, 9, 10. In addition, of these 29 genes regulated in blood in this and previous studies, 15 have been reported to be regulated in AD brain (Table 2).
Among the 15 genes, most were regulated similarly in blood and brain (e.g. TGFB1, NDE1 and HIST1H4C), while others showed discordant regulation in blood and brain (e.g. ATP6V0D10) - that is being up regulated in one tissue and down regulated in the other. In addition, of the 15 genes regulated in AD blood and brain, MME, MMP9, CAMK2G and TGFB1 were identified as AlzGene (reported in previous genetic studies).
3.7 Cellular Functions of the 44 overlapping genes
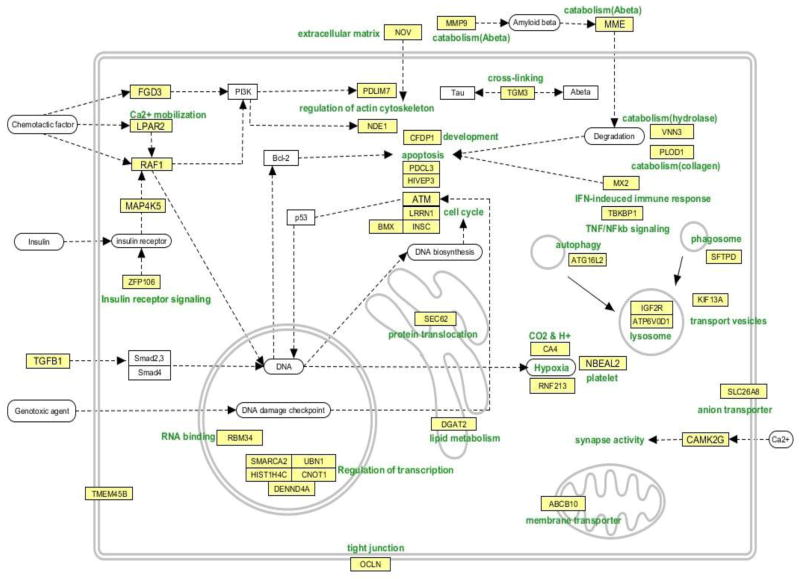
In order to summarize the 44 genes that have been replicated from previous AD studies and/or in from both our analytical approaches, we manually constructed a pathway map based on information from Entrez Gene (Figure 3). Genes involved in survival/death signaling were mainly up-regulated. ZFP106 and NOV participate in insulin signaling, MAP4K5 in insulin and TNF receptor signaling, and TGFB1 is upstream of the TGFB signaling pathway. TBKBP1 participates in TNF/NFkb signaling, ATM responds to DNA damage, LRRN1 is involved in cell proliferation, and INSC is involved in cell division and nervous system development. HIVEP3 is involved in apoptosis, PDCL3 modulates caspase activation, and RAF1 is a MAP3K kinase involved in apoptosis, and regulation of actin cytoskeleton. PDLIM7, FGD3 and NDE1 regulate the cytoskeleton (Figure 3).
Figure 3.
Cellular functions for the 44 genes listed in Supplementary Table 1D. These genes represent those that were regulated in AD blood in this study (ANCOVA, p <0.005, |FC| > 1.2) and which were reported to be regulated in other AD blood studies, or were shown to be regulated in this study also using the rank product method and a |FC| threshold of > 1.5.
Several genes related to immune response and catabolism were up-regulated. MX2 is up-regulated by interferon-alpha, SFTPD is involved in phagosome function, ATG16L2 participates in autophagy, ATP6V0D1 is a lysosomal proton transporter, KIF13A is involved in endosome to lysosome transport and IGF2R is involved insulin response and in trafficking of lysosomal enzymes. MME interacts with and degrades amyloid beta, MMP9 degrades extracellular matrix at the blood brain barrier and is also involved in Aβ degradation. TGM3 has cross-linking activity which may be involved in the aggregation of Aβ and tau (Figure 3).
Many of the down-regulated genes were linked to transcriptional regulation. SMARCA2 is involved in transcriptional activation, CNOT1 is involved in RNA degradation, DENND4A regulates the transcription factor c-myc, UBN1 is involved in transcriptional repression, and RBM34 is a RNA binding protein. Sec62 is involved in post-translational protein translocation and ABCB10 is a mitochondrion-associated membrane transporter. OCLN is part of vascular endothelial cell tight junctions (Figure 3).
3.8 Genes with altered expression in AD blood and in AD brain studies
In our previous studies we conducted comprehensive analyses of the AD brain transcriptome 11, 12. For the present analyses we included all of the published studies on AD brain, as well as genes recorded in the AlzGene database for GWAS findings in AD. When comparing our DEG list of 288 probe sets (188 genes) with these datasets, we found that 89 genes regulated in blood in this study were altered in brain transcriptome and/or identified as AlzGene (Supplementary Table 1F). This is a high percentage considering that the 288 genes/probes represented only 188 unique genes with clear annotation. Among the 89, some genes of interest were IL6R, IL17R, ANKRD13D, SORCS1, PFDN2, PREP, HES1, CRISPLD2, MYH11, SYMPK, and several genes were identified as AlzGenes, including: SORCS1, SORL1, INSR, CTSD and TANC2
4. Discussion
This is the first study to identify differences of RNA expression in blood of Alzheimer’s disease (AD) patients compared to controls that considers the possible confounding effects of WMH. Though there are only a few overlapping genes between the two entities, it is important to consider co-morbid conditions when searching for AD biomarkers. There were several remarkable findings. First, of the 188 genes regulated in AD blood in this study, 89 have been reported to be altered RNA expression in AD brain or have been confirmed to have increased risk for clinical AD based on genetic studies. Second, of the 188 genes regulated in AD blood in this study 44 genes were reported in previous AD blood studies or were replicated using an independent analytical method; and, 15 of these were previously reported to have altered expression in AD brain. Of the genes regulated in AD blood a number have been implicated in AD pathogenesis including MMP9, MME (Neprilysin) and Transforming growth factor 1, and several have been reported in previous genetic association studies including: MMP9, MME, TGFβ1, CAMK2G, SORCS1, SORL1, INSR, CTSD and TANC2.
Significance of regulated RNAs in AD blood
The majority of AD biomarker studies have examined proteins from CSF as a direct expression of brain pathology. Studies of peripheral blood have been more difficult to replicate. In this study RNA from whole blood was examined, an important difference from most previous studies. The RNA is almost all intracellular, and thus represents expression of RNA in blood leukocytes and immature platelets and red blood cells. Thus, the data reported represents changes mostly in immune cells in AD patients, and supports previous reports of changes in the immune system in AD. The following discussion will focus mainly on a few molecules that have been reported in previous AD studies.
MMP9 (Metalloproteinase 9) participates in NGF-induced α-secretase cleavage of AβPP. Thus, it contributes to the shift of AβPP processing towards the non-amyloidogenic pathway precluding the formation of neurotoxic Aβ peptides. The apolipoprotein E4 fragment affects matrix metalloproteinase 9 in brain cell lines and estrogen activates MMP9 to increase beta amyloid degradation. MMP9 not only degrades Aβ but also affects cytoskeleton organization, and also plays a key role in opening the blood brain barrier following cerebral ischemia and other acute injuries to brain, and MMP9 RNA expression increases in blood leukocytes following ischemic stroke in humans. MMP9 levels are reported to correlate with brain amyloid burden, though no differences in activity in blood were reported in AD 13. MMP9 levels are elevated in CSF of AD patients and help differentiate AD from other diseases 14 and MMP9 protein and enzymatic activity are increased in AD brain. Variable blood findings in the literature may relate in part to whether levels of pre-mRNA, mRNA, pre-protein, active protein or MMP9 enzymatic activity were measured.
MME, membrane metallo-endopeptidase (Neprilysin, NEP), is a zinc metallopeptidase which is a major Aβ-degrading peptide. Genetic variations in the gene have been associated with increased AD risk in some but not all studies. Though systemic neprilysin can decrease peripheral beta amyloid levels, this has been reported to decrease brain Aβ in some studies 15 and not in others 16. The levels of the Aβ-degrading enzyme neprilysin are reduced in human dementia pugilistica cases. Though MME/neprilysin is high in blood in this and other studies, levels of NEP and NEP2 have been reported to be lower, higher or unchanged in AD brain 17. Notably, Neprilysin protects against cerebral amyloid angiopathy and Aβ-induced degeneration of cerebrovascular smooth muscle cells 18. In addition, peripheral delivery of a CNS targeted neprilysin reduces Aβ toxicity in one mouse model of Alzheimer’s disease 19 and decreases plaque formation but not memory deficits in another mouse model 20. Another key role of NEP in amyloid processing is emphasized by the fact that the intracellular domain of the amyloid precursor protein binds to the NEP promoter and induces its expression and NEP degrades both beta amyloid 42 and 40 21. NEP is expressed in amyloid plaques, dystrophic neurites and astrocytes but not microglia of sporadic AD but not familial AD cases 22.
TGF
Transforming growth factor signaling has been implicated in AD. TGF stimulates Aβ uptake into microglia via SMAD 3 and decreases amyloid plaque burden in AD mouse models 23. The CC genotype of TGFB1 may increase the risk of late-onset AD 24. TGFB1 levels are altered in human AD brain 25 and are increased in brain of transgenic AD mouse models 26. Levels of TGFB1 have been reported to be altered in AD blood but may relate to stage of the disease 27. Elevated CSF levels of TGFB1 have also been reported in AD 28. Amyloid-beta peptide suppresses TGFB1-induced MMP2 production via Smad 7, and TGFB1 potentiates amyloid-beta generation in astrocytes and transgenic mice. Moreover, TGFB1 over expressing mice develop an amyloid angiopathy similar to that seen in AD 29.
RNF213 has E3 ligase and AAA+ATPase activities and genetic variants (SNPs) in this gene have a strong association with Moya Moya disease. This is of interest since vascular abnormalities can precede development of amyloid plaques and behavioral deficits in AD patients and AD animal models 30. Knockdown of RNF213 in zebrafish causes irregular wall formation in large arteries and abnormal sprouting of vessels.
Occludin (OCLN) is highly down regulated in blood cells in this study. It is a key protein that forms part of the tight junctions between brain endothelial cells that control blood-brain barrier (BBB) function. Aging and high cholesterol decrease BBB function and OCLN levels in normal mouse brain. Aβ1–42 decreases OCLN expression in brain endothelial cells 31, though there is no direct evidence for changes of tight junction proteins in AD brain 32.
The Ataxia Telangiectasia mutated gene (ATM) is involved in controlling the cell cycle, and aberrant entry of neurons into the cell cycle has been one mechanism proposed for neuronal cell death in AD brain 33. Aβ1–42 and DNA-damaging drugs increase ATBF1 levels in cultured neurons which activates ATM signaling responsible for neuronal death through the binding of ATBF1 to phosphorylated ATM 33.
Transglutaminase 3 (TGM3) was up regulated over two fold in AD blood in this study. Transglutaminases (TGs) are Ca2+-dependent enzymes that catalyze modifications of glutaminyl (Q) residues including a number of amine-bearing compounds, including lysyl (K) residues and polyamines in brain. TGMs have been implicated in the pathogenesis of AD, Huntington’s and other neurodegenerative diseases. Both Aβ and tau are substrates of TGM cross-linking activity which modulates the protein aggregation process in AD brains 34.
IGFR2 and BMX
Insulin growth factor receptor 2 is altered in AD brain 35 and is significantly up regulated in blood in this study. Insulin signaling is reported to be altered in AD brain in general, and global decreases of brain blood flow and glucose metabolism precede symptoms in AD 36. There is significant evidence for aberrant IGFR1 signaling in AD brain 37. IGFR2 levels are specifically up regulated in neurons most vulnerable to forming amyloid plaques in mouse AD models 38. BMX is also markedly increased in AD blood in this study (1.9 fold), and it interacts with IGFR pathways and is elevated following brain trauma. NOV and NAP4K5 are both regulated in AD blood in this study and they are also involved in insulin signaling (Figure 3).
Of the canonical pathways significantly regulated in AD blood, it was notable that decreased regulation of transcription was among them. This would appear to be parsimonious with the decreases of blood flow and glucose metabolism that occur in AD brain.
CA4
Carbonic anhydrase 4 plays a critical role in pH regulation and long-term synaptic transformation, and is associated with mental retardation, AD and Down syndrome. CA4 is highly associated with brain blood vessels and not other vascular beds and in brain is the major astrocyte carbonic anyhydrase. Carbonic anyhydrase 4 is also one of the major RNAs that change expression in blood following ischemic stroke. Carbonic anyhydrase II (CA2) is decreased in brain of a mouse AD model, is increased in human AD brain 39, and carbonic anhydrase II is elevated in plasma of AD patients, particularly in males 40. Any relationship between CA2 and CA4 is unknown.
NOV/CCN3 (Nephroblastoma over expressed gene)
Matricellular proteins of the CCN family (CYR61/CTGF/NOV) regulate and are regulated by cytokines including TGF (see above), which is also regulated in AD blood in this study. NOV also regulates extracellular matrix enzymes via integrins and proteoglycans, and can up regulate MMP9 – which also increases in AD blood in this study.
ICAM-4 (intracellular adhesion molecule-4, Landsteiner-Wiener blood group) and NSMAF(Neutral sphingomyelinase (N-SMase) activation associated factor). ICAM-1 and sphingolipids have been proposed as blood-based biological marker candidates of microvascular pathology in AD, where plasma concentrations of ICAM-1 are elevated in AD and sphingolipids are significantly altered in mild AD or during the predementia stage of mild cognitive impairment 41. ICAM-4’s function is not well understood. It has structural relatedness to ICAM-1, -2 and -R, and in our data is down-regulated, contrary to the ICAMs associated with AD. The neutral sphyngomyelinase(N-SMase) activation associated factor (NSMAF) is up-regulated in our data. Sphingolipid metabolism has been associated with AD neuropathology, contributing to amyloid-beta production in AD 42.
ITGB3 (integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61). This gene showed up-regulation in AD in this study. It is a baseline platelet activation biomarker, where activated glycoprotein IIb-IIIa was shown to be significantly higher in patients with AD with fast vs slow cognitive decline during 1-year follow up 43.
5. Conclusions
This is the first study to consider possible confounding effects of WMH on the AD blood transcriptome. A list of 288 probe sets representing 188 genes expressed in whole blood differentiates AD from controls using several analytical methods irrespective of WMH status. A number of the genes regulated in this study have been reported to be regulated in other studies of both AD blood and AD brain, and strongly support a systemic effect of AD that includes altered RNA expression of peripheral immune cells. Further validation in an independent cohort to specifically test the sensitivity and specificity of the identified genes to predict AD is needed.
Supplementary Material
Supplementary Table 1A. Demographic information for AD and control subjects with and without WMH.
Supplementary Table 1B. Genes differentially regulated between AD+ subjects and AD-subjects.
Supplementary Table 1C. The enriched functional categories for the 288 probe sets (188 genes) that were differentially expressed in AD subjects versus non-AD subjects (p<0.005 and fold change >1.2). GO-Gene Ontology; BP-biological process; CC – cellular component.
Supplementary Table 1D. Pathways associated with AD-specific gene expression profile using a Fisher’s exact test.
Supplementary Table 1E. Differentially expressed genes overlapped with other studies of AD blood transcriptome.
Supplementary Table 1F. Differentially expressed genes overlapped with DEGs from AD brain transcriptome.
Supplementary Materials and Methods.
Subjects on the X axis, and probability of diagnosis on the Y axis as determined from Prediction Analysis of Microarrays (PAM). Diamonds represent AD and squares represent Non-AD.
Acknowledgments
We would like to thank the subjects who participated in this study. This work was supported by NIH P30 AG10129 (CD), R01 AG021028 (CD), RO1 NS056302 (FRS/CD) and the American Heart Association Bugher Foundation Center for Stroke Prevention Research (FRS). The authors report no conflicts of interest. Due to space limitations, additional references will be provided upon request.
Footnotes
The authors report no conflicts of interest.
References
- 1.Fehlbaum-Beurdeley P, Jarrige-Le Prado AC, Pallares D, Carriere J, Guihal C, Soucaille C, et al. Toward an alzheimer’s disease diagnosis via high-resolution blood gene expression. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2010;6:25–38. doi: 10.1016/j.jalz.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Rye PD, Booij BB, Grave G, Lindahl T, Kristiansen L, Andersen HM, et al. A novel blood test for the early detection of alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2011;23:121–129. doi: 10.3233/JAD-2010-101521. [DOI] [PubMed] [Google Scholar]
- 3.Lunnon K, Ibrahim Z, Proitsi P, Lourdusamy A, Newhouse S, Sattlecker M, et al. Mitochondrial dysfunction and immune activation are detectable in early alzheimer’s disease blood. Journal of Alzheimer’s disease : JAD. 2012;30:685–710. doi: 10.3233/JAD-2012-111592. [DOI] [PubMed] [Google Scholar]
- 4.Maes OC, Schipper HM, Chertkow HM, Wang E. Methodology for discovery of alzheimer’s disease blood-based biomarkers. J Gerontol A Biol Sci Med Sci. 2009;64:636–645. doi: 10.1093/gerona/glp045. [DOI] [PubMed] [Google Scholar]
- 5.Han G, Wang J, Zeng F, Feng X, Yu J, Cao HY, et al. Characteristic transformation of blood transcriptome in alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2013;35:373–386. doi: 10.3233/JAD-121963. [DOI] [PubMed] [Google Scholar]
- 6.Cuenco TK, Lunetta KL, Baldwin CT, McKee AC, Guo J, Cupples LA, et al. Association of distinct variants in sorl1 with cerebrovascular and neurodegenerative changes related to alzheimer disease. Arch Neurol. 2008;65:1640–1648. doi: 10.1001/archneur.65.12.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshita M, Fletcher E, DeCarli C. Current concepts of analysis of cerebral white matter hyperintensities on magnetic resonance imaging. Topics in magnetic resonance imaging : TMRI. 2005;16:399–407. doi: 10.1097/01.rmr.0000245456.98029.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Stamova B, Jickling G, Tian Y, Zhan X, Ander BP, et al. Distinctive rna expression profiles in blood associated with white matter hyperintensities in brain. Stroke; a journal of cerebral circulation. 2010;41:2744–2749. doi: 10.1161/STROKEAHA.110.591875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booij BB, Lindahl T, Wetterberg P, Skaane NV, Saebo S, Feten G, et al. A gene expression pattern in blood for the early detection of alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2011;23:109–119. doi: 10.3233/JAD-2010-101518. [DOI] [PubMed] [Google Scholar]
- 10.Scherzer CR, Eklund AC, Morse LJ, Liao Z, Locascio JJ, Fefer D, et al. Molecular markers of early parkinson’s disease based on gene expression in blood. Proc Natl Acad Sci U S A. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang D, Han G, Feng X, Sun J, Duan Y, Lei H. Concerted perturbation observed in a hub network in alzheimer’s disease. PLoS One. 2012;7:e40498. doi: 10.1371/journal.pone.0040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Feng X, Liang D, Duan Y, Lei H. Down-regulation of energy metabolism in alzheimer’s disease is a protective response of neurons to the microenvironment. Journal of Alzheimer’s disease : JAD. 2012;28:389–402. doi: 10.3233/JAD-2011-111313. [DOI] [PubMed] [Google Scholar]
- 13.Lim NK, Villemagne VL, Soon CP, Laughton KM, Rowe CC, McLean CA, et al. Investigation of matrix metalloproteinases, mmp-2 and mmp-9, in plasma reveals a decrease of mmp-2 in alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2011;26:779–786. doi: 10.3233/JAD-2011-101974. [DOI] [PubMed] [Google Scholar]
- 14.Bjerke M, Zetterberg H, Edman A, Blennow K, Wallin A, Andreasson U. Cerebrospinal fluid matrix metalloproteinases and tissue inhibitor of metalloproteinases in combination with subcortical and cortical biomarkers in vascular dementia and alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2011;27:665–676. doi: 10.3233/JAD-2011-110566. [DOI] [PubMed] [Google Scholar]
- 15.Guan H, Liu Y, Daily A, Police S, Kim MH, Oddo S, et al. Peripherally expressed neprilysin reduces brain amyloid burden: A novel approach for treating alzheimer’s disease. J Neurosci Res. 2009;87:1462–1473. doi: 10.1002/jnr.21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker JR, Pacoma R, Watson J, Ou W, Alves J, Mason DE, et al. Enhanced proteolytic clearance of plasma abeta by peripherally administered neprilysin does not result in reduced levels of brain abeta in mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:2457–2464. doi: 10.1523/JNEUROSCI.3407-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang JY, Hafez DM, James BD, Bennett DA, Marr RA. Altered nep2 expression and activity in mild cognitive impairment and alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2012;28:433–441. doi: 10.3233/JAD-2011-111307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miners JS, Kehoe P, Love S. Neprilysin protects against cerebral amyloid angiopathy and abeta-induced degeneration of cerebrovascular smooth muscle cells. Brain Pathol. 2011;21:594–605. doi: 10.1111/j.1750-3639.2011.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer B, Marr RA, Gindi R, Potkar R, Michael S, Adame A, et al. Peripheral delivery of a cns targeted, metalo-protease reduces abeta toxicity in a mouse model of alzheimer’s disease. PLoS One. 2011;6:e16575. doi: 10.1371/journal.pone.0016575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meilandt WJ, Cisse M, Ho K, Wu T, Esposito LA, Scearce-Levie K, et al. Neprilysin overexpression inhibits plaque formation but fails to reduce pathogenic abeta oligomers and associated cognitive deficits in human amyloid precursor protein transgenic mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:1977–1986. doi: 10.1523/JNEUROSCI.2984-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang JY, Bruno AM, Patel CA, Huynh AM, Philibert KD, Glucksman MJ, et al. Human membrane metallo-endopeptidase-like protein degrades both beta-amyloid 42 and beta-amyloid 40. Neuroscience. 2008;155:258–262. doi: 10.1016/j.neuroscience.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorfman VB, Pasquini L, Riudavets M, Lopez-Costa JJ, Villegas A, Troncoso JC, et al. Differential cerebral deposition of ide and nep in sporadic and familial alzheimer’s disease. Neurobiology of aging. 2010;31:1743–1757. doi: 10.1016/j.neurobiolaging.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyss-Coray T, Lin C, Yan F, Yu GQ, Rohde M, McConlogue L, et al. Tgf-beta1 promotes microglial amyloid-beta clearance and reduces plaque burden in transgenic mice. Nat Med. 2001;7:612–618. doi: 10.1038/87945. [DOI] [PubMed] [Google Scholar]
- 24.Caraci F, Bosco P, Signorelli M, Spada RS, Cosentino FI, Toscano G, et al. The cc genotype of transforming growth factor-beta1 increases the risk of late-onset alzheimer’s disease and is associated with ad-related depression. Eur Neuropsychopharmacol. 2012;22:281–289. doi: 10.1016/j.euroneuro.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto K, Horio J, Satoh H, Sue L, Beach T, Arita S, et al. Expression profiles of cytokines in the brains of alzheimer’s disease (ad) patients compared to the brains of non-demented patients with and without increasing ad pathology. Journal of Alzheimer’s disease : JAD. 2011;25:59–76. doi: 10.3233/JAD-2011-101815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salins P, He Y, Olson K, Glazner G, Kashour T, Amara F. Tgf-beta1 is increased in a transgenic mouse model of familial alzheimer’s disease and causes neuronal apoptosis. Neurosci Lett. 2008;430:81–86. doi: 10.1016/j.neulet.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Motta M, Imbesi R, Di Rosa M, Stivala F, Malaguarnera L. Altered plasma cytokine levels in alzheimer’s disease: Correlation with the disease progression. Immunol Lett. 2007;114:46–51. doi: 10.1016/j.imlet.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Rota E, Bellone G, Rocca P, Bergamasco B, Emanuelli G, Ferrero P. Increased intrathecal tgf-beta1, but not il-12, ifn-gamma and il-10 levels in alzheimer’s disease patients. Neurol Sci. 2006;27:33–39. doi: 10.1007/s10072-006-0562-6. [DOI] [PubMed] [Google Scholar]
- 29.Buckwalter M, Pepper JP, Gaertner RF, Von Euw D, Lacombe P, Wyss-Coray T. Molecular and functional dissection of tgf-beta1-induced cerebrovascular abnormalities in transgenic mice. Ann N Y Acad Sci. 2002;977:87–95. doi: 10.1111/j.1749-6632.2002.tb04801.x. [DOI] [PubMed] [Google Scholar]
- 30.Farkas E, Luiten PG. Cerebral microvascular pathology in aging and alzheimer’s disease. Prog Neurobiol. 2001;64:575–611. doi: 10.1016/s0301-0082(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 31.Kook SY, Hong HS, Moon M, Ha CM, Chang S, Mook-Jung I. Abeta(1)(−)(4)(2)-rage interaction disrupts tight junctions of the blood-brain barrier via ca(2)(+)-calcineurin signaling. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:8845–8854. doi: 10.1523/JNEUROSCI.6102-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viggars AP, Wharton SB, Simpson JE, Matthews FE, Brayne C, Savva GM, et al. Alterations in the blood brain barrier in ageing cerebral cortex in relationship to alzheimer-type pathology: A study in the mrc-cfas population neuropathology cohort. Neurosci Lett. 2011;505:25–30. doi: 10.1016/j.neulet.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 33.Jung CG, Uhm KO, Miura Y, Hosono T, Horike H, Khanna KK, et al. Beta-amyloid increases the expression level of atbf1 responsible for death in cultured cortical neurons. Mol Neurodegener. 2011;6:47. doi: 10.1186/1750-1326-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilhelmus MM, Grunberg SC, Bol JG, van Dam AM, Hoozemans JJ, Rozemuller AJ, et al. Transglutaminases and transglutaminase-catalyzed cross-links colocalize with the pathological lesions in alzheimer’s disease brain. Brain Pathol. 2009;19:612–622. doi: 10.1111/j.1750-3639.2008.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guttula SV, Allam A, Gumpeny RS. Analyzing microarray data of alzheimer’s using cluster analysis to identify the biomarker genes. Int J Alzheimers Dis. 2012;2012:649456. doi: 10.1155/2012/649456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Candeias E, Duarte AI, Carvalho C, Correia SC, Cardoso S, Santos RX, et al. The impairment of insulin signaling in alzheimer’s disease. IUBMB Life. 2012;64:951–957. doi: 10.1002/iub.1098. [DOI] [PubMed] [Google Scholar]
- 37.Moloney AM, Griffin RJ, Timmons S, O’Connor R, Ravid R, O’Neill C. Defects in igf-1 receptor, insulin receptor and irs-1/2 in alzheimer’s disease indicate possible resistance to igf-1 and insulin signalling. Neurobiology of aging. 2010;31:224–243. doi: 10.1016/j.neurobiolaging.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Amritraj A, Hawkes C, Phinney AL, Mount HT, Scott CD, Westaway D, et al. Altered levels and distribution of igf-ii/m6p receptor and lysosomal enzymes in mutant app and app + ps1 transgenic mouse brains. Neurobiology of aging. 2009;30:54–70. doi: 10.1016/j.neurobiolaging.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Sultana R, Boyd-Kimball D, Cai J, Pierce WM, Klein JB, Merchant M, et al. Proteomics analysis of the alzheimer’s disease hippocampal proteome. Journal of Alzheimer’s disease : JAD. 2007;11:153–164. doi: 10.3233/jad-2007-11203. [DOI] [PubMed] [Google Scholar]
- 40.Jang BG, Yun SM, Ahn K, Song JH, Jo SA, Kim YY, et al. Plasma carbonic anhydrase ii protein is elevated in alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2010;21:939–945. doi: 10.3233/JAD-2010-100384. [DOI] [PubMed] [Google Scholar]
- 41.Mielke MM, Lyketsos CG. Alterations of the sphingolipid pathway in alzheimer’s disease: New biomarkers and treatment targets? Neuromolecular medicine. 2010;12:331–340. doi: 10.1007/s12017-010-8121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haughey NJ, Bandaru VV, Bae M, Mattson MP. Roles for dysfunctional sphingolipid metabolism in alzheimer’s disease neuropathogenesis. Biochimica et biophysica acta. 2010;1801:878–886. doi: 10.1016/j.bbalip.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stellos K, Panagiota V, Kogel A, Leyhe T, Gawaz M, Laske C. Predictive value of platelet activation for the rate of cognitive decline in alzheimer’s disease patients. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2010;30:1817–1820. doi: 10.1038/jcbfm.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1A. Demographic information for AD and control subjects with and without WMH.
Supplementary Table 1B. Genes differentially regulated between AD+ subjects and AD-subjects.
Supplementary Table 1C. The enriched functional categories for the 288 probe sets (188 genes) that were differentially expressed in AD subjects versus non-AD subjects (p<0.005 and fold change >1.2). GO-Gene Ontology; BP-biological process; CC – cellular component.
Supplementary Table 1D. Pathways associated with AD-specific gene expression profile using a Fisher’s exact test.
Supplementary Table 1E. Differentially expressed genes overlapped with other studies of AD blood transcriptome.
Supplementary Table 1F. Differentially expressed genes overlapped with DEGs from AD brain transcriptome.
Supplementary Materials and Methods.
Subjects on the X axis, and probability of diagnosis on the Y axis as determined from Prediction Analysis of Microarrays (PAM). Diamonds represent AD and squares represent Non-AD.