Abstract
STATs play crucial roles in a wide variety of biological functions, including development, proliferation, differentiation and migration as well as in cancer development. In the present study, we examined the impact of constitutive activation of Stat3 on behavior of keratinocytes, including keratinocyte stem cells (KSC) in vivo. BK5.Stat3C transgenic (Tg) mice, which express a constitutively active form of Stat3 (Stat3C) in the basal layer of the epidermis and in the bulge region KSCs exhibited a significantly reduced number of CD34+/α6 integrin+ cells compared to non-transgenic (NTg) littermates. There was a concomitant increase in the Lgr-6, Lrig-1, and Sca-1 populations in the Tg mice in contrast to the CD34 and Keratin-15 positive population. In addition, increased expression of c-myc, β-catenin, and epithelial–mesenchymal transition (EMT)-related genes as well as decreased expression of α6-integrin was observed in the hair follicles of Tg mice. Notably, Sca-1 was found to be a direct transcriptional target of Stat3 in keratinocytes. The current data suggest that elevated Stat3 activity leads to depletion of hair follicle KSCs along with a concomitant increase of stem/progenitor cells above the bulge region. Overall, the current data indicate that Stat3 plays an important role in keratinocyte stem/progenitor cell homeostasis.
Keywords: keratinocyte stem cells, Sca-1, Stat3C, bulge region, skin carcinogenesis
Introduction
The stem and progenitor cells in the hair follicle and the interfollicular epidermis (IFE) confer regenerative capacity to the epidermis [1]. It is widely recognized that a population of multi-potent stem cells resides in the niche called the bulge region of the hair follicle [2]. In addition to the bulge region keratinocyte stem cells (KSCs), there are various populations of follicular cells that form the stem/progenitor/transit amplifying (TA) cells, which may give rise to various progeny of each type. In addition to CD34 and Keratin-15 (K15), a number of stem/progenitor cell markers including MTS24/Plet1, Lgr5, Lgr6, Lrig-1, and Sca-1 are now known to be expressed in the mouse hair follicle. These various cell populations have been shown to possess colony forming ability in vitro and regenerative potential in a number of in vivo experimental settings [3–7].
Stat3 plays a critical role in maintaining the pluripotent stem cell phenotype as well as stem cell survival [8]. Studies using an inducible, active Stat3 expression construct showed that Stat3 activation is sufficient to maintain embryonic stem (ES) cells in an undifferentiated state [9]. Moreover, stable expression of a dominant negative Stat3 in ES cells induced differentiation even in the continuous presence of leukemia inhibitory factor (LIF) [10]. Several previous studies reported that Stat3 activation is required for LIF-driven ES cell self-renewal [11]. In addition, Stat3 appears to be one of the core regulators in mouse ES cells in addition to key stem cell regulators such as Oct4, Sox2, and Nanog [12]. Together, these results suggest that activation of Stat3 may play an important role, in collaboration with other proteins, in regulating ES cell behavior. Although Stat3 activation has been shown to maintain cells in an undifferentiated state, it should be noted that Stat3 activation has also been shown to be associated with differentiation in specific cell types [13–16].
Recently, there is emerging evidence on the importance of Stat3 in regulating cell migration, motility and invasion in both physiological and pathological situations. The first indication came from conditional knockout (KO) of Stat3 in the mouse skin. Deletion of Stat3 in keratinocytes compromised the wound-healing process in skin and inhibited migration of cultured keratinocytes [17]. A similar role of Stat3 in regulating cell migration has also been found in mouse embryonic fibroblasts and keloidderived fibroblasts [18,19]. In Drosophila, the JAK-STAT pathway has been shown to activate migratory and invasive behavior of ovarian epithelial cells in ovarian development [20]. Stat3 has also been found to control cell movement during zebrafish gastrulation [21]. In agreement with this finding Stat3-KO mice exhibited embryonic lethality during gastrulation [22]. Furthermore, promotion of tumor invasion and metastasis by Stat3 has been widely reported in various cancers, including ovarian carcinoma, melanoma, bladder, pancreatic, and prostate cancers [23–27]. These data suggest that regulation of cell movement could be a fundamental function of Stat3. However, exactly how Stat3 regulates cell migration remains largely unknown. Earlier reports have shown that Stat3 plays a role in cell movement associated with gastrulation, a key step in early embryogenesis involving morphogenetic changes and the specification of cell fate and may be playing similar roles during cell migration [21]. This function of Stat3 associated with migration is further confirmed by studies where keratinocytes deficient in Stat3 show impaired growth factor dependent migration in culture in a cell autonomous manner [17].
Previously, we reported that forced expression of a constitutive active form of Stat3 (Stat3C) leads to increased susceptibility to skin tumorigenesis by both two-stage as well as UV carcinogenesis protocols [28– 30]. In contrast, mice where Stat3 is deleted in the basal layer of the epidermis as well as in the outer root sheath (ORS) of hair follicles are highly resistant to both two-stage and UVB skin carcinogenesis [30,31]. To further understand the mechanisms underlying the role of Stat3 during skin tumorigenesis, we used the BK5.Stat3C mouse model to address the role of Stat3 in keratinocyte stem/progenitor cell behavior. The current data suggest that Stat3 plays an important role in behavior of hair follicle stem/progenitor cell populations.
Materials and Methods
Experimental Animals
BK5.Stat3C mice have been described previously [28]. Male and female mice, 7–14 wk of age, were used for the experiments. Non-transgenic (NTg) littermate controls were used for all experiments unless indicated otherwise.
Label Retaining Cell (LRC) Analysis
Ten-day-old pups were injected with 5-bromo-2′-deoxyuridine BrdU (50 mg/g body weight) i.p. every 12 h for 2 d. The pulse chase period was continued for 70 d. Mouse dorsal skin was harvested, fixed in formalin, embedded in paraffin and analyzed by immunohistochemistry for BrdU. The BrdU positive cell quantitation was done by two methods. Total number of follicles showing BrdU positive cells and number of BrdU positive cells/50 follicles in skin sections were quantified. At least three sections per animal and three to five animals per group were used for the analysis.
Total Hair Follicle Cell Isolation and Colony Forming Efficiency (CFE)
Mouse dorsal skin was harvested by established protocols. Fat underlying the subcutis was mechanically scraped and the skins were incubated in dispase (5 mg/ml) overnight at 4°C. Epidermis was harvested by mechanical scraping with a scalpel and the dermis was incubated in 1% collagenase for 2 h at 37°C. The resulting solution was centrifuged at 300g and subsequently at 52g for 5 min at 4°C. The supernatant was discarded and the pellet was homogenized in 0.25% Trypsin-ethylenediaminetetraacetic acid (EDTA) multiple times and incubated at 37°C for 12 min. The solution was pipetted multiple times and cells were strained through 70 and 40μm filters. Cells were analyzed for viability using trypan blue exclusion and the total number of viable cells were counted using hemocytometer. Five thousand cells from BK5. Stat3C and control mice were plated onto mitomycin C treated NIH3T3 cells and cultured for 4 wk. Holoclones (closely packed clones made up of atleast 5 cells) and mero/paraclones (loosely packed clones of atleast 5–8 cells) were counted.
Flow Cytometry Analysis
Total hair follicle cells were isolated using the above protocol. The isolated total hair follicle cells were labeled with biotin conjugated antibodies for CD34 and PE-α6-integrin. Cells were incubated on ice for 1 h. Hair follicle cells were then incubated with adenomatous polyposis coli (APC)-conjugated streptavidin secondary antibody for 30 min on ice. For conjugated antibodies like CD34-PE and Sca-1-FITC, cells were incubated with the antibody on ice for 1 h. Cells were fixed with a final concentration of 1% PFA and analyzed on a BD FACS Calibur or BD FACS Aria. Data analysis was done using Cell Quest and FlowJo analysis programs.
Immunostaining
For formalin-fixed, paraffin embedded sections, slides were deparaffinized and sodium citrate was used for antigen retrieval. Slides were blocked using goat/donkey serum for 1 h at room temperature, incubated with primary antibody for 1 h at room temperature or 4°C overnight and subsequently with secondary antibody for 30 min at room temperature. For OCT frozen sections, slides were air dried for 5–10 min, fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature, washed with Immunostain wash buffer (GeneTex, Irvine, CA), blocked with goat serum for 30–40 min at room temperature and immunostained with primary antibody for an hour and with the appropriate secondary antibody for 30 min. Slides were mounted using mounting media (Vectashield with DAPI).
Chromatin Immunoprecipitation Assay (ChIP Assay)
Mouse skin epidermis was cross-linked with formaldehyde followed by epidermal lysate preparation. A Pierce ChIP kit was used for these experiments (Thermo Scientific, Waltham, MA). Immunoprecipitations were done using Stat3 and β-catenin (Cell Signaling Technology, Danvers, MA) antibodies. DNA occupancy was then assessed by polymerase chain reaction (PCR) using primers spanning putative Stat3 binding sites of the indicated gene promoters.
Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
RNA was isolated from BK5.Stat3C transgenic (Tg) and NTg keratinocytes using a QIAGEN RNeasy Kit. First strand synthesis using random primers (Invitrogen, Grand Island, NY) was used for cDNA preparation. SYBR Green primers were used for quantitative real-time PCR, which was performed on the Applied Biosystems RT-PCR (Viia 7, Applied Biosystems, Carlsbad, CA).
Reagents and Antibodies
Trypsin-EDTA (Gibco, Grand Island, NY), Dispase (Gibco), Collagenase (Gibco), Biotin-CD34 (eBio-sciences, San Diego, CA), α6-integrin-PE (BD Biosciences, San Jose, CA), Streptavidin-APC (Invitrogen), Sca-1 (BD Biosciences), Myc (Santa Cruz, Santa Cruz, CA), Cyclin D1 (Cell Signaling Technology, Danvers, MA), β-catenin (Cell Signaling Technology), active β-catenin (Sigma, St. Louis, MO), Lgr6 (Santa Cruz), Lrig-1 (R&D), K15 (Neomarkers, Kalamazoo, MI), CD34 (BD Biosciences).
Statistical Analysis
Statistical analysis was conducted using Student's t-test. Data were considered significantly different when the P value was ≤0.05.
Results
BK5.Stat3C Mice Have Reduced Number of Bulge Region KSCs
To determine the role of Stat3 on bulge region KSC population, the LRC population in the bulge region of BK5.Stat3C Tg mice was compared to that of NTg mice. For these experiments, 10-d-old pups were pulse labeled with BrdU (given i.p. using the protocol described in Materials and Methods Section) and then chased for 70 d. As shown in Figure 1A, there was a significant decrease in the number of LRCs in the hair follicles of BK5.Stat3C Tg mice compared to NTg mice. The LRCs were quantitated by two independent methods. First, the percent hair follicles that contained BrdU positive cells were counted. Second, the average number of BrdU positive cells/50 hair follicles in both groups of mice was counted. Both methods revealed a statistically significant decrease in the number of LRCs in the Tg mice compared to the NTg mice (quantitation in Figure 1B).
Figure 1.
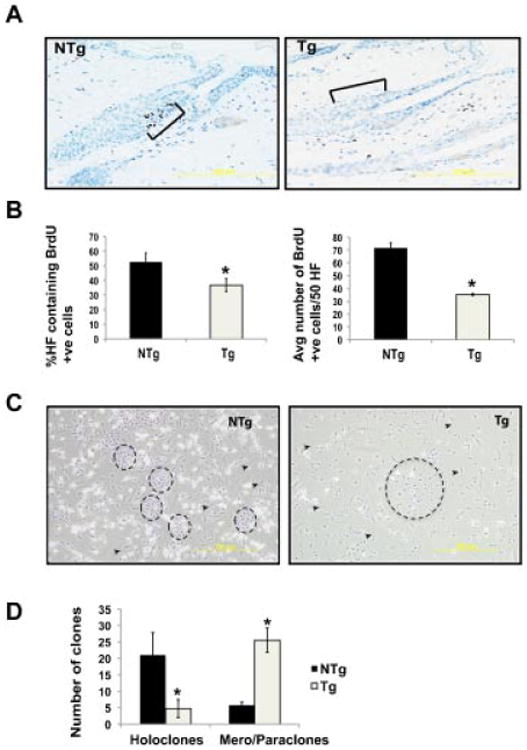
Constitutive overexpression of Stat3C leads to decreased LRCs and holoclones. (A) 10-day-old pups were given BrdU i.p. every 12 h for 2 d. Skins from BK5.Stat3C Tg and littermates (NTg) were harvested after 70 d, fixed in formalin and embedded in paraffin. Sections were immunostained for BrdU. Brackets indicate the bulge region of the hair follicle. (B) LRCs were quantified by (i) counting the percent hair follicles that show BrdU positive cells and (ii) counting the average number of BrdU positive cells/50 follicles. (C) Hair follicle cells were isolated from BK5.Stat3C Tg mice and littermates (NTg) and plated on mitomycin C treated NIH3T3 cells and cultured for 4 wk. (D) Clones were quantified by counting holoclones and mero/paraclones, *P< 0.05.
To determine if expression of a constitutively active form of Stat3 in the hair follicles led to an imbalance in the progenitor/stem cell populations, total hair follicle cells were isolated from both groups and used for clonal assays. Based on the classification by Barrandon and Green [32], normal keratinocytes have an intrinsic ability to form cell colonies in vitro which are classified into holoclones, meroclones, and paraclones. These clones consist of cells that represent stem (holoclones), early (meroclone), and late (paraclones) amplifying cells [32]. For the purposes of our studies, meroclones and paraclones were grouped together due to similar morphology. Closely packed holoclones and the more disperse paraclones/meroclones in both groups were counted (Figure 1C). Clones are marked by circles whereas arrowheads point to the fibroblasts that form the feeder layer. Hair follicle cells from BK5.Stat3C Tg mice had a decreased number of holoclones and an increased number of paraclones/meroclones compared to the hair follicle cells from NTg mice (Figure 1D). Taken together these data suggest that expression of a constitutively active form of Stat3 in the K5 compartment leads to a reduction of bulge region KSCs.
Expression of Hair Follicle Stem/Progenitor Cell Markers in the Hair Follicles
Bulge region KSCs are also marked by expression of CD34 and α6-integrin [33]. Therefore, the number of CD34 and α6-integrin double positive cells in the hair follicles of BK5.Stat3C Tg and NTg mice were analyzed by flow cytometry. A decreased number of double positive cells was observed in the hair follicles of BK5. Stat3C Tg mice compared to the NTg mice (Figure 2A). Moreover, the structure of the bulge region in the Tg mice was less well-defined (again see supplemental Figure 1), although the hair follicles in BK5.Stat3C Tg mice appeared to undergo hair cycle stages of anagen, catagen and telogen (supplemental Figure 2). The expression of K15 during both the anagen as well as the catagen/telogen stages of the hair cycle was also examined and found to be decreased during both of these hair cycle phases (Supplemental Figure 1). In addition, there was an increase in the number of Sca-1 positive cells in the hair follicles of Tg mice as compared to NTg mice assessed by flow cytometry analysis (Figure 2B).
Figure 2.
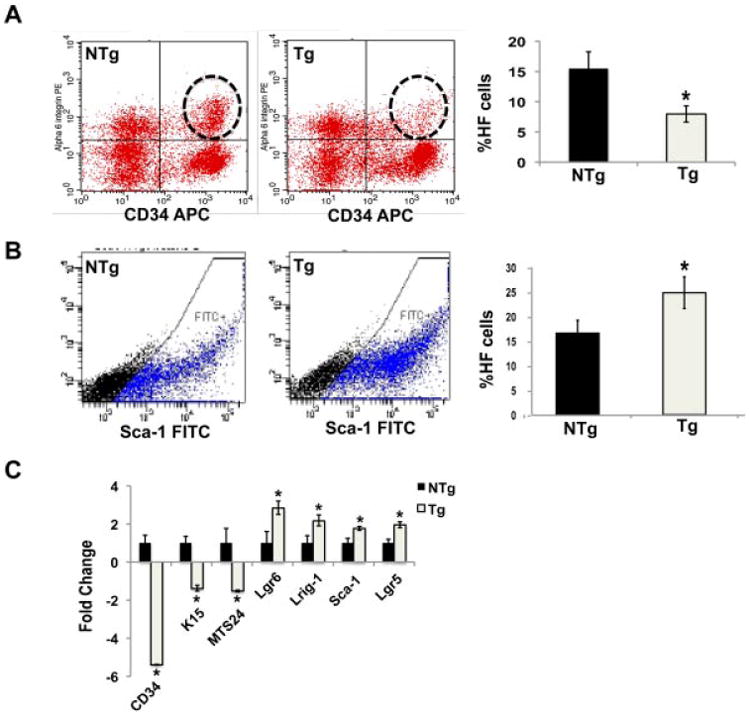
Expression of Stat3C causes an increase in the hair follicle population outside the bulge region at the expense of bulge stem cells. (A) Total hair follicle cells were subjected to flow cytometry analysis after staining for CD34 and a6-integrin. (B) Flow cytometry analysis of hair follicle cells for expression of Sca-1. (C) Epidermal keratinocytes were harvested from BK5.Stat3C Tg mice and nontransgenic littermates (NTg); RNA was isolated and lineage markers analyzed by qPCR, *P < 0.05.
Previous reports have shown that epidermal basal cells also express CD34 and other stem/progenitor cell markers [33]. Furthermore, it has been reported that under certain conditions bulge region KSCs can contribute to interfollicular epidermal keratinocytes [34,35]. To determine if Stat3C overexpression also influences stem/progenitor cells in the epidermis we analyzed stem/progenitor cell markers in interfollicular epidermal keratincoytes. As shown in the Figure 2C, quantitative polymerase chain reaction (qPCR) analyses showed statistically significant decreases in the mRNA levels of CD34, K15, and MTS24 and increased levels of mRNA for the cell populations marked by Lgr-6, Lrig-1, and Sca-1. In the hair follicle, immunohistochemical analysis showed that there was a decrease in staining for both CD34 and K15 in the bulge region of Tg mice as compared to NTg mice. Notably, there was an increase in expression of Sca-1 in the hair follicles of Tg mice consistent with the data shown in Figure 3A. Further analyses revealed an increase in the expression levels of Lrig-1 and Lgr6 in the hair follicle of Tg mice (Figure 3A and B, respectively). In addition, expression of another wellknown KSC marker, p63, was analyzed by immunofluorescence and no significant difference in either the number of cells expressing p63 or levels of p63 expression was seen between Tg and NTg mice (Figure 3C). Together, these data show that expression of a constitutively active form of Stat3 decreases the expression of stem cell markers in the bulge region of the hair follicles as well as interfollicular epidermal cells as well as an associated increase in the expression of stem/progenitor cell markers for cells that reside above the bulge region in the hair follicles of these mice.
Figure 3.
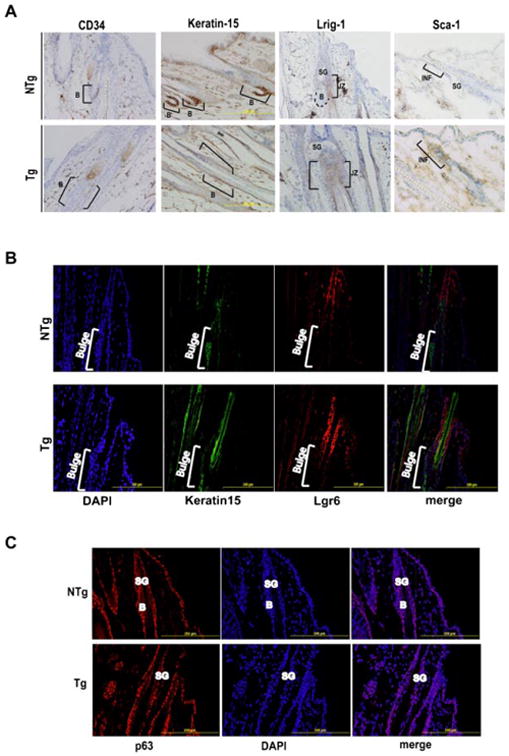
Stat3C expression leads to altered expression of hair follicle stem/progenitor populations in the hair follicle. Skins from BK5.Stat3C Tg mice and nontransgenic littermates (NTg) were fixed in formalin and embedded in paraffin or frozen in OCT. (A) Sections were stained for CD34, Keratin-15, Lrig-1 and Sca-1 and examined microscopically. Brackets and B refer to the bulge region of the hair follicle. SG, JZ, and INF refer to sebaceous gland, junctional zone, and infundibulum regions, respectively. Sections were immunostained for Keratin-15, Lgr-6 (B), and p63 (C) and examined microscopically. Brackets and B refer to the bulge region of the hair follicle. SG refers to sebaceous gland.
Stat3 Directly Regulates Sca-1 Gene Expression in Keratinocytes
Earlier reports suggested that the Sca-1 promoter has gamma-interferon activating sequences (GAS), which may be putative Stat3 binding sites [36]. A squamous cell carcinoma (SCC) cell line (JWF2) was used to examine the induction of Sca-1 mRNA by various Stat3 inducers and inhibitors. JWF2 cells were treated with Interleukin 6 (IL-6) (10 ng/ml) and epidermal growth factor (EGF) (50 ng/ml) and harvested 3 h later. As shown in Figure 4A, the level of Sca-1 mRNA was also increased following treatment with both EGF and IL-6 while Sca-1 mRNA levels induced by these ligands were reduced when the cells were pretreated with STA-21 (20 μM) for 1 h. Induction of Stat3 signaling was confirmed by analyzing the levels of Cyclin D1 mRNA, a known Stat3 target gene. Cyclin D1 levels were elevated 3 h after treatment with both EGF and IL-6 (Figure 4B). STA-21, which is an established Stat3 inhibitor [37], reduced both EGF and IL-6 induced cyclin D1 mRNA levels. A similar decrease was seen in the mRNA expression of Sca-1 when dorsal skins of BK5.Stat3C mice were harvested after treatment with STA-21 (Figure 4C). To determine if Stat3 could directly regulate Sca-1 expression, we analyzed the Sca-1 promoter and found that the Sca-1 promoter has general STAT binding sequences (TTN4-6AA). Therefore, ChIP analysis was performed to examine if Stat3 binds to the Sca-1 promoter. As shown in Figure 4D and E, the Stat3 signal relative to input was amplified in the BK5.Stat3C Tg mice as compared to the NTg controls. In contrast, no difference was observed between Tg and NTg mice in ChIP assays using β-catenin antibody as a negative control. These data suggest that Stat3 regulates the expression of Sca-1 by direct binding to its promoter region.
Figure 4.

Sca-1 is a direct target of Stat3. (A) JWF2 cells were treated with EGF or IL-6 for 3 h. For treatments that include STA-21, cells were pretreated with STA-21 for 1 h and then treated with EGF or IL-6 for 3 h. RNA isolation and cDNA preparation was done to analyze mRNA levels of Sca-1, *P < 0.05. (B) RNA isolation and cDNA preparation was done to analyze mRNA levels of cyclin D1. (C) BK5.Stat3C Tg and NTg mice were treated with 20 μM STA-21 topically twice every week for 2 wk. RNA isolation and cDNA preparation was done to analyze mRNA levels of Sca-1. (D) Epidermal scrapings from BK5.Stat3C Tg mice and littermates (NTg) were used to perform ChIP analysis. β-Catenin and Stat3 were used for immunoprecipitation and the DNA was amplified using primers spanning the Sca-1 DNA promoter sequence containing putative Stat3 binding sites. Input controls were used for each sample and IgG was used as a negative control. (E) Graphs representing quantification of Sca-1 positive bands normalized to input.
Impact of Stat3 Activation on Epidermal Differentiation Markers
Given that expression of Stat3C in the basal layer led to increased expression of hair follicle progenitor cell markers and decreased number of bulge region KSCs, markers of keratinocyte differentiation were analyzed to examine if Stat3C overexpression leads to an overall increase in differentiation. As shown in supplemental Figure 3A and B, there was no significant difference in the expression pattern or levels of classical differentiation markers including Keratin 1 (K1), Keratin 10 (K10), Loricrin and Involucrin in the IFE or hair follicles as analyzed by immunohistochemistry. These data suggest that Stat3C expression does not affect the bulge region KSCs through changes in the differentiation of hair follicle stem/progenitor cells.
Constitutive Activation of Stat3 Leads to Altered Expression of β-Catenin, c-Myc, and α6-Integrin in the Bulge Region
β-Catenin is known to play an important role in cell fate of KSCs including adult stem cell renewal and lineage selection (reviewed in [38]). More importantly, an increase in β-catenin expression has recently been implicated in reducing bulge region KSCs, similar to what we have observed in BK5.Stat3C Tg mice [39]. Therefore, both total as well as nuclear β-catenin expression levels were examined in the hair follicles of BK5.Stat3C Tg and compared to age-matched NTg mice. As shown in Figure 5A, hair follicles from NTg mice had a very well defined bulge region marked by K15 expression but little or no expression of β-catenin. On the other hand, β-catenin expression was readily observed throughout the ORS in Tg mice (again see Figure 5A). Nuclear β-catenin levels were also evaluated and increased nuclear localization of β-catenin was observed in the hair follicles of Tg mice (Figure 5B). In addition, Myc expression was also found to be elevated in BK5. Stat3C mice as observed by immunofluorescence analysis (Figure 5C). We have previously shown that Stat3 directly regulates Myc expression in the epidermis [31]. Earlier studies have shown that Myc is a direct transcriptional target of both Stat3 [40] and β-catenin [41], both of which are elevated in BK5.Stat3C mice (see again Figure 5). In addition, hair follicles of Tg mice had reduced expression of α6-integrin as analyzed by both flow cytometry and immunofluorescence staining of skin sections (Figure 5D). Thus, constitutive activation of Stat3 in the K5 compartment led to altered expression of both β-catenin and α6-integrin that may have contributed to the changes seen in hair follicle morphology and stem/progenitor cell populations. These results suggest that induction of Stat3 target gene products, Myc and β-catenin, may also contribute to the effects seen on the stem/ progenitor populations in BK5.Stat3C mice.
Figure 5.
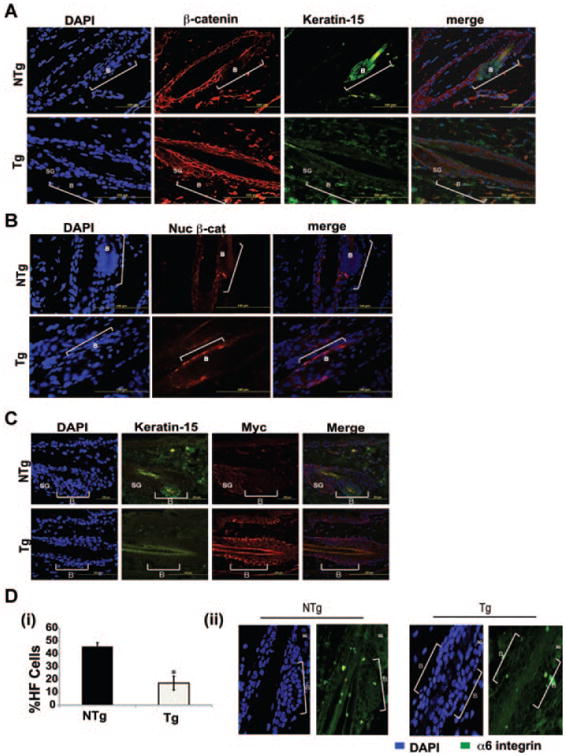
Expression of Stat3C leads to increased expression of β-catenin, and Myc and decreased expression of α6-integrin in the hair follicle. Skins from Tg mice and NTg littermates were fixed in formalin and embedded in paraffin. Sections were co-immunostained for Keratin-15 and total β-catenin (A) and co-immunostained for Keratin-15 and nuclear β-catenin (B) and examined microscopically (Green-Keratin-15, Red-β-catenin, Blue-DAPI). (C) Skins from BK5.Stat3C Tg and NTg mice were fixed in formalin and embedded in paraffin. Sections were coimmunostained with K15, and c-myc and analyzed microscopically. (D) (i) Flow cytometry analysis of a6-integrin positive cells in BK5.Stat3C Tg and NTg mice, *P < 0.05. (ii) Immunofluorescence analysis to analyze the expression of α6-integrin in the hair follicles of BK5.Stat3C Tg and NTg mice. Brackets and B refer to the bulge region of the hair follicle. SG refers to sebaceous gland.
Stat3C Expression in the K5 Compartment Induces EMT
To further understand the mechanism(s) underlying the decrease in CD34 positive KSCs, markers associated with epithelial–mesenchymal transition (EMT) were analyzed at the protein level by immunofluorescence and Western blotting and at the mRNA level by qPCR. As shown by immunofluorescence analysis in Figure 6A, there were increased levels of Vimentin in ORS hair follicle cells of Tg mice as compared to the NTg mice. As shown above, analysis of stem/progenitor cell markers of interfollicular keratinocytes correlated well with stem/progenitor cell markers in hair follicle cells. Likewise, Western blot analysis of epidermal protein lysates showed a significant increase in Vimentin but not E-cadherin (see Figure 6B). Furthermore, there was a marked increase in the expression levels of twist, snail, slug, fibronectin, vimentin, and α-smooth muscle actin (α-SMA) although again the levels of E-cadherin were not significantly changed as assessed by qPCR (Figure 6C).
Figure 6.
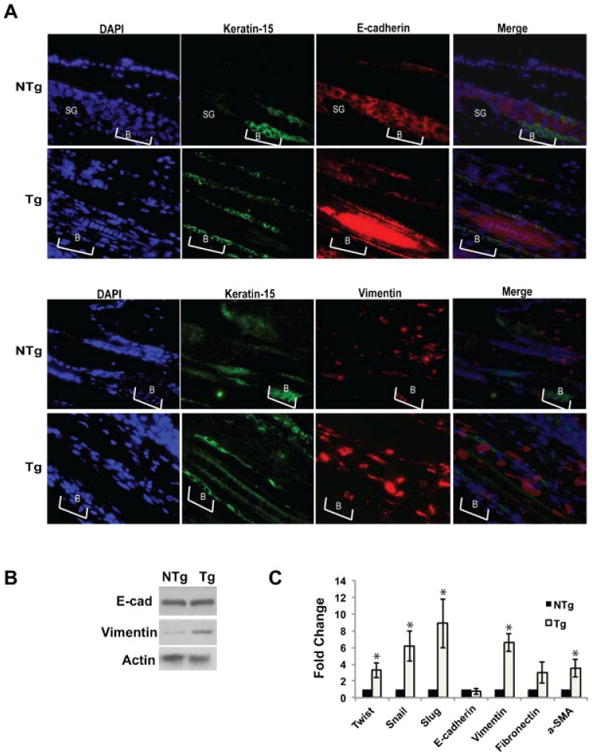
Expression of Stat3C in the K5 compartment alters expression of markers of epithelial-mesenchymal transition (EMT). (A) Skins from Tg mice and NTg littermates were fixed in formalin and embedded in paraffin. Sections were co-immunostained for Keratin-15, E-cadherin and Vimentin. Brackets and B refer to the bulge region of the hair follicle. SG refers to sebaceous gland. For the analysis of EMT markers by Western blotting (B) and qPCR (C) epidermal keratinocytes were harvested from BK5.Stat3C Tg and NTg mice and protein and RNA was isolated, respectively, *P < 0.05.
Discussion
In the present study, we have shown that constitutive activation of Stat3 significantly alters the behavior of keratinocyte stem/progenitor cells that reside in the hair follicle. In this regard, the expression of a constitutively active/dimerized form of Stat3 in the basal compartment of mouse epidermis (including the bulge region of the hair follicle) led to a dramatic effect on bulge region KSCs. BK5.Stat3C mice had reduced number of CD34/ α6-integrin positive cells and reduced number of LRCs. Although BK5.Stat3C Tg mice had a reduced number of bulge region KSCs, these mice had an increased number of other hair follicle stem/progenitor cell populations, including the Lgr-6, Lrig-1 and Sca-1 positive cell populations that reside above the bulge region. These FACS expression analyses showing a decrease in bulge region KSCs and an increase in other stem progenitor cells were consistent with hair follicle colony formation experiments. Total hair follicle cells from BK5. Stat3C mice grown on mitomycin C treated fibroblasts showed reduced number of holoclones compared to those from NTg mice, with an increase in mero/paraclones. In addition to a decreased number of CD34/ α6-integrin positive cells, there were also decreases in the expression of bulge region KSC marker K15 and MTS-24. Further analyses revealed changes in expression of β-catenin, c-myc, α6-integrin and several EMT-related genes. Overall, the current results demonstrate that Stat3 plays important roles in the behavior of bulge region KSCs and other stem/progenitor populations in the hair follicle as well as the IFE. The effects of Stat3 appear to be due to both direct effects on hair follicle stem/progenitor cell marker expression (e.g., Sca-1) as well as expression of multiple genes involved in stem/progenitor cell maintenance and migration (e.g., β-catenin c-myc and Vimentin).
Based on the current data, we hypothesize that increased expression of specific Stat3 regulated genes leads to maturation of bulge region KSCs and migration out of the bulge region niche. One of these Stat3 regulated genes appears to be Sca-1. The increase in Lrig-1 and Lrg6 positive cell populations that reside in the region of the hair follicle that lie between the CD34 positive and Sca-1 positive regions may be the result of upward migration induced as a result of elevated Sca-1 expression in BK5.Stat3C Tg mice. Recent evidence has indicated that Sca-1 is a target gene for Stat3 in the duct cells of the mouse submandibular gland [36,42]. In the current study, we confirmed by ChIP analysis that Sca-1 is a direct gene target of Stat3 in mouse keratinocytes. ChIP analyses were consistent with reduced gene expression levels of Sca-1 in epidermal cells treated with the Stat3-selective inhibitor, STA-21. Thus, increased expression of Sca-1 in bulge region KSCs and possibly other stem/progenitor populations may have facilitated movement upward toward the infundibulum leading to an increase in the number of cells in this compartment. Although suggestive of an effect of Stat3 on KSC maturation/migration, further experiments, such as lineage tracing of bulge region KSCs, will be required to confirm this hypothesis.
In previous studies, it was reported that deregulated expression of Myc under control of the K14 promoter led to alterations in stem cell maintenance and a 75% reduction in the number of stem cells in K14.Myc2 Tg mice [43]. Additional studies also showed that transient activation of c-myc in K14.MycER™ Tg mice drove keratinocytes from the stem cell to the TA compartment [44]. As shown in Figure 5, increased expression of Myc in the hair follicles of BK5.Stat3C mice in cooperation with elevated Sca-1 and other genes may have contributed to the migration of stem cells out of the bulge region of the hair follicles.
It is known that β-catenin stabilization is essential for promoting the transition between stem cell quiescence and conversion to proliferating TA progeny [45]. It is also known that ectopic expression of either stabilized β-catenin or a regulated form of β-catenin induces cell migration in different cell types. Also, a critical threshold of β-catenin signaling activated by cooperative mechanisms may be important during EMT and tumorigenesis [46]. Studies have shown that β-catenin expression levels may be critical in specifying cell fate decisions in stem cells. In addition, continued expression and stabilization of β-catenin may contribute to tumorigenesis [47,48]. In addition, α6-integrin is known to provide adhesive interactions between epithelial cells and the basement membrane [49]. In the current study, we analyzed the expression of both total β-catenin as well as active β-catenin in the nucleus of the hair follicle bulge cells in both the Tg as well as the NTg mice. We observed elevated levels of β-catenin protein in the bulge region of BK5.Stat3C Tg mice, which may have contributed to the increased migration of bulge region KSCs out of their niche. We also observed by both flow cytometry and immunofluorescence analysis that the Tg mice showed decreased expression of integrin (α6) in the bulge region, which may also have contributed to the migration of the KSCs out of the bulge region by decreasing the adhesive interactions between epithelial cells and the basement membrane. Thus, Stat3-induced changes in β-catenin and α6-integrin expression together with elevated levels of c-myc and Sca-1 may have cooperated in facilitating movement of KSCs out of the bulge-region. Consistent with our observations, Yang et al. [39] recently showed that elevated c-myc and β-catenin levels likely contributed to the depletion of bulge-region KSCs in Smad4 KO mice.
To better understand the mechanism(s) underlying the movement of stem cells into the progenitor populations, expression levels of EMT markers were also analyzed in the current study. As shown in Figure 6, qPCR analyses of epidermal keratinocyte RNA samples demonstrated increased mRNA levels for vimentin, fibronectin, and α-SMA as well as an increase in the expression of twist, snail, and slug. Although, interfollicular epidermal cells were used for the analyses in the aforementioned experiment, the data from the analysis of hair follicle cells using immunofluorescence for E-cadherin and vimentin were very similar. These data suggested that expression of Stat3C induced an EMT phenotype in the keratinocytes from the Tg mice. Previous studies from our lab demonstrated that skin tumors from BK5. Stat3C mice had increased levels of Twist [29]. Twist, a known target gene of Stat3 [50], regulates EMT by downregulating the expression of epithelial specific proteins, such as the E-cadherin and by upregulating the expression of mesenchymal markers such as vimentin and aSMA [51,52]. Thus, constitutive expression of Stat3C in these mice may have induced EMT by directly upregulating Twist thereby modulating expression of several EMT related genes and further facilitating the migration of the cells out of the bulge-region niche.
It is also interesting to note that in spite of decreased bulge region KSCs, the putative target cells for tumor formation in mouse skin [53], BK5.Stat3C mice were highly susceptible to both two-stage chemical and UV-induced skin carcinogenesis protocols as previously shown [29,30]. In particular, skin tumors that develop in BK5.Stat3C mice undergoing two-stage chemical carcinogenesis progress rapidly to invasive SCC [29]. It is also clear from the current results that there are still KSCs present in the bulge-region that could contribute to skin tumor development although overall they are significantly reduced in number. Furthermore, previous studies from our lab have shown that Stat3 plays an important role in the survival of bulge region KSCs after initiation with DMBA [54,55]. Thus, enhanced survival of remaining KSCs mediated by Stat3C expression could account for the observed enhanced susceptibility of these mice to two-stage carcinogenesis. In addition, the current findings also suggest that other hair follicle stem/ progenitor cell populations that reside above the bulge region, may also be involved in skin tumor development in BK5.Stat3C mice. Since the Sca-1 positive cells retain clonogenic capacity, they may be capable of multiple rounds of division and contribute to tumor development. Notably, it has been reported that several murine tumors including retinoblastoma, prostate and mammary gland tumors and chronic myeloid leukemia showed high Sca-1 expression levels that correlated with a highly malignant phenotype [56–60]. In the prostate, Akt overexpression in Sca-1 positive cells initiates tumorigenesis, with cancer progression correlating with increased Sca-1 cells [57]. In preliminary analyses, an increase in Sca-1 positive cells was observed in the skin tumors obtained from BK5.Stat3C mice subjected to the two-stage skin carcinogenesis protocol (D. Rao and J. DiGiovanni, unpublished studies). Studies are currently underway to test if Sca-1 positive cells are directly involved in skin tumor development or tumor progression.
In conclusion, the current data demonstrate that Stat3 plays an important role in regulating the behavior of keratinocyte stem/progenitor cells found in the hair follicle. Expression of a constitutively active form of Stat3 led to attrition of the bulge region stem cell population (i.e., CD34+/α6+/K15 population) and increases in stem/progenitor cell populations that reside above the bulge region. We hypothesize that Stat3 regulates multiple genes, including Sca-1, c-myc, β-catenin as well as several EMT related genes thus facilitates the maturation of bulge region KSCs and their migration out of the bulge region niche. Further study of the effects of Stat3 activation on KSCs and other keratinocyte cell populations will lead to a better understanding of the exact mechanisms for the current observations as well as the underlying mechanisms for the greater susceptibility of BK5.Stat3C mice to skin carcinogenesis.
Supplementary Material
Figure S1. Keratin-15 expression in the hair follicles of BK5.Stat3C and Ntg mice. Skins from K5.Stat3C Tg and NTg mice were fixed in formalin and embedded in paraffin. Sections were immunostained for Keratin-15.
Figure S2. Histological analysis to show hair stages in BK5.Stat3C transgenic and Ntg mice. Skins from BK5. Stat3C Tg and NTg mice were fixed in formalin and embedded in paraffin. Sections were stained with H&E and analyzed microscopically.
Figure S3. Analysis of classical differentiation marker expression by immunufluorescence. Skins from BK5. Stat3C Tg and NTg mice were fixed in formalin and embedded in paraffin. (A) Sections were immunostained with K1, K10, Loricrin and Involucrin and analyzed microscopically. (B) Sections were immunostained for K1, K10 and Involucrin and the hair follicles were analysed microscopically for expression of these differentiation markers.
Acknowledgments
The authors thank Erika Abel, Okkyung Rho and Mahipal Suraneni for excellent scientific suggestions during the course of this study. The authors also thank Nancy Otto and Jimi Lynn Brandon for excellent histology and tissue analysis services and Pamela Whitney and Richard Salinas for providing Flow Cytometry Core Services. This study was supported by National Cancer Institute (CA76520 to J. DiGio-vanni) and NIEHS Center (ES07784).
Grant sponsor: National Cancer Institute; Grant number: CA76520; Grant sponsor: NIEHS Center; Grant number: ES07784
Abbreviations
- IFE
interfollicular epidermis
- KSCs
keratinocyte stem cells
- K15
Keratin-15
- ES
embryonic stem
- KO
knockout
- ORS
outer root sheath
- NTg
non-transgenic
- LRC
label retaining cell
- PCR
polymerase chain reaction
- qRT-PCR
real time-polymerase chain reaction
- Tg
transgenic
- qPCR
quantitative polymerase chain reaction
- EMT
epithelial–mesenchymal transition
Footnotes
Supporting Information: Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- 1.Fuchs E. The tortoise and the hair: Slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumbar T, Guasch G, Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nijhof JG, Braun KM, Giangreco A, et al. The cell-surface marker MTS24 identifies a novel population of follicular keratinocytes with characteristics of progenitor cells. Development. 2006;133:3027–3037. doi: 10.1242/dev.02443. [DOI] [PubMed] [Google Scholar]
- 4.Jaks V, Barker N, Kasper M, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 5.Snippert HJ, Haegebarth A, Kasper M, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 6.Jensen KB, Collins CA, Nascimento E, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen UB, Yan X, Triel C, Woo SH, Christensen R, Owens DM. A distinct population of clonogenic and multipotent murine follicular keratinocytes residing in the upper isthmus. J Cell Sci. 2008;121:609–617. doi: 10.1242/jcs.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 9.Matsuda T, Nakamura T, Nakao K, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeuf H, Hauss C, Graeve FD, Baran N, Kedinger C. Leukemia inhibitory factor-dependent transcriptional activation in embryonic stem cells. J Cell Biol. 1997;138:1207–1217. doi: 10.1083/jcb.138.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T. Differentiation and growth arrest signals are generated through the cytoplasmic region of gp130 that is essential for Stat3 activation. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]
- 14.Bonni A, Sun Y, Nadal-Vicens M, et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 15.Yanagisawa M, Nakashima K, Taga T. STAT3-mediated astrocyte differentiation from mouse fetal neuroepithelial cells by mouse oncostatin M. Neurosci Lett. 1999;269:169–172. doi: 10.1016/s0304-3940(99)00447-4. [DOI] [PubMed] [Google Scholar]
- 16.Karras JG, Wang Z, Huo L, Howard RG, Frank DA, Rothstein TL. Signal transducer and activator of transcription-3 (STAT3) is constitutively activated in normal, self-renewing B-1 cells but only inducibly expressed in conventional B lymphocytes. J Exp Med. 1997;185:1035–1042. doi: 10.1084/jem.185.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sano S, Itami S, Takeda K, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim CP, Phan TT, Lim IJ, Cao X. Stat3 contributes to keloid pathogenesis via promoting collagen production, cell proliferation and migration. Oncogene. 2006;25:5416–5425. doi: 10.1038/sj.onc.1209531. [DOI] [PubMed] [Google Scholar]
- 19.Ng DC, Lin BH, Lim CP, et al. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol. 2006;172:245–257. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silver DL, Geisbrecht ER, Montell DJ. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development. 2005;132:3483–3492. doi: 10.1242/dev.01910. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita S, Miyagi C, Carmany-Rampey A, et al. Stat3 controls cell movements during zebrafish gastrulation. Dev Cell. 2002;2:363–375. doi: 10.1016/s1534-5807(02)00126-0. [DOI] [PubMed] [Google Scholar]
- 22.Takeda K, Noguchi K, Shi W, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei D, Le X, Zheng L, et al. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–329. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- 24.Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: Localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–3558. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- 25.Xie TX, Wei D, Liu M, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 26.Itoh M, Murata T, Suzuki T, et al. Requirement of STAT3 activation for maximal collagenase-1 (MMP-1) induction by epidermal growth factor and malignant characteristics in T24 bladder cancer cells. Oncogene. 2006;25:1195–1204. doi: 10.1038/sj.onc.1209149. [DOI] [PubMed] [Google Scholar]
- 27.Abdulghani J, Gu L, Dagvadorj A, et al. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol. 2008;172:1717–1728. doi: 10.2353/ajpath.2008.071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sano S, Chan KS, Carbajal S, et al. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat Med. 2005;11:43–49. doi: 10.1038/nm1162. [DOI] [PubMed] [Google Scholar]
- 29.Chan KS, Sano S, Kataoka K, et al. Forced expression of a constitutively active form of Stat3 in mouse epidermis enhances malignant progression of skin tumors induced by two-stage carcinogenesis. Oncogene. 2008;27:1087–1094. doi: 10.1038/sj.onc.1210726. [DOI] [PubMed] [Google Scholar]
- 30.Kim DJ, Angel JM, Sano S, DiGiovanni J. Constitutive activation and targeted disruption of signal transducer and activator of transcription 3 (Stat3) in mouse epidermis reveal its critical role in UVB-induced skin carcinogenesis. Oncogene. 2009;28:950–960. doi: 10.1038/onc.2008.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan KS, Sano S, Kiguchi K, et al. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Ghazizadeh S, Taichman LB. Multiple classes of stem cells in cutaneous epithelium: a lineage analysis of adult mouse skin. EMBO J. 2001;20:1215–1222. doi: 10.1093/emboj/20.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102:451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 36.Purwanti N, Karabasil MR, Matsuo S, et al. Induction of Sca-1 via activation of STAT3 system in the duct cells of the mouse submandibular gland by ligation of the main excretory duct. Am J Physiol Gastrointest Liver Physiol. 2011;301:G814–G824. doi: 10.1152/ajpgi.00408.2010. [DOI] [PubMed] [Google Scholar]
- 37.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci USA. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watt FM, Collins CA. Role of beta-catenin in epidermal stem cell expansion, lineage selection, and cancer. Cold Spring Harb Symp Quant Biol. 2008;73:503–512. doi: 10.1101/sqb.2008.73.011. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, Wang L, Yang X. Disruption of Smad4 in mouse epidermis leads to depletion of follicle stem cells. Mol Biol Cell. 2009;20:882–890. doi: 10.1091/mbc.E08-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 41.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 42.Purwanti N, Azlina A, Karabasil MR, et al. Involvement of the IL-6/STAT3/Sca-1 system in proliferation of duct cells following duct ligation in the submandibular gland of mice. J Med Invest. 2009;56:253–254. doi: 10.2152/jmi.56.253. [DOI] [PubMed] [Google Scholar]
- 43.Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28:165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- 44.Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11:558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 45.Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rastaldi MP, Ferrario F, Giardino L, et al. Epithelial–mesenchy-mal transition of tubular epithelial cells in human renal biopsies. Kidney Int. 2002;62:137–146. doi: 10.1046/j.1523-1755.2002.00430.x. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 48.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 49.de Pereda JM, Lillo MP, Sonnenberg A. Structural basis of the interaction between integrin alpha6beta4 and plectin at the hemidesmosomes. EMBO J. 2009;28:1180–1190. doi: 10.1038/emboj.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng GZ, Zhang WZ, Sun M, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem. 2008;283:14665–14673. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 52.Smit MA, Geiger TR, Song JY, Gitelman I, Peeper DS. A Twist-Snail axis critical for TrkB-induced epithelial–mesenchymal transition-like transformation, anoikis resistance, and metastasis. Mol Cell Biol. 2009;29:3722–3737. doi: 10.1128/MCB.01164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris RJ. A perspective on keratinocyte stem cells as targets for skin carcinogenesis. Differentiation. 2004;72:381–386. doi: 10.1111/j.1432-0436.2004.07208004.x. [DOI] [PubMed] [Google Scholar]
- 54.Kim DJ, Kataoka K, Rao D, Kiguchi K, Cotsarelis G, Digiovanni J. Targeted disruption of stat3 reveals a major role for follicular stem cells in skin tumor initiation. Cancer Res. 2009;69:7587–7594. doi: 10.1158/0008-5472.CAN-09-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kataoka K, Kim DJ, Carbajal S, Clifford JL, DiGiovanni J. Stage-specific disruption of Stat3 demonstrates a direct requirement during both the initiation and promotion stages of mouse skin tumorigenesis. Carcinogenesis. 2008;29:1108–1114. doi: 10.1093/carcin/bgn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 2007;13:823–832. [PMC free article] [PubMed] [Google Scholar]
- 57.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Batts TD, Machado HL, Zhang Y, Creighton CJ, Li Y, Rosen JM. Stem cell antigen-1 (sca-1) regulates mammary tumor development and cell migration. PLoS ONE. 2011;6:e27841. doi: 10.1371/journal.pone.0027841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Upadhyay G, Yin Y, Yuan H, Li X, Derynck R, Glazer RI. Stem cell antigen-1 enhances tumorigenicity by disruption of growth differentiation factor-10 (GDF10)-dependent TGF-beta signaling. Proc Natl Acad Sci USA. 2011;108:7820–7825. doi: 10.1073/pnas.1103441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Caro M, Cobaleda C, Gonzalez-Herrero I, et al. Cancer induction by restriction of oncogene expression to the stem cell compartment. EMBO J. 2009;28:8–20. doi: 10.1038/emboj.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Keratin-15 expression in the hair follicles of BK5.Stat3C and Ntg mice. Skins from K5.Stat3C Tg and NTg mice were fixed in formalin and embedded in paraffin. Sections were immunostained for Keratin-15.
Figure S2. Histological analysis to show hair stages in BK5.Stat3C transgenic and Ntg mice. Skins from BK5. Stat3C Tg and NTg mice were fixed in formalin and embedded in paraffin. Sections were stained with H&E and analyzed microscopically.
Figure S3. Analysis of classical differentiation marker expression by immunufluorescence. Skins from BK5. Stat3C Tg and NTg mice were fixed in formalin and embedded in paraffin. (A) Sections were immunostained with K1, K10, Loricrin and Involucrin and analyzed microscopically. (B) Sections were immunostained for K1, K10 and Involucrin and the hair follicles were analysed microscopically for expression of these differentiation markers.


