Abstract
Paired-related transcription factors, PRRX1 and PRRX2, which are present in mesenchymal tissues and participate in mesenchymal cell differentiation, were recently found in the stem/progenitor cells of the pituitary gland of ectodermal origin. To clarify the role of PRRX1 and PRRX2 in the pituitary gland, the present study first aimed to identify transcription factors that regulate Prrx1 and Prrx2 expression. A promoter assay for the upstream regions of both genes was performed by co-transfection of the expression vector of several transcription factors, many of which are frequently found in the pituitary stem/progenitor cells. The results for the promoter activity of both genes showed expression in a cell type-dependent manner. Comprehensive comparison of transcriptional activity of several transcription factors was performed with CHO cells, which do not show Prrx1 and Prrx2 expression, and the results revealed the presence of common and distinct factors for both genes. Among them, KLF6 showed specific and remarkable stimulation of Prrx2 expression. In vitro experiments using an electrophoretic mobility shift assay and siRNA interference revealed a potential ability for regulation of Prrx2 expression by KLF6. Finally, immunohistochemistry confirmed the presence of KLF6 in the SOX2/PRRX2 double-positive stem/progenitor cells of the postnatal pituitary gland. Thus, the finding of KLF6 might provide a novel clue to clarify the maintenance of stem/progenitor cells of the postnatal pituitary gland.
Keywords: Differentiation, KLF6, Pituitary, PRRX2, Stem cell
The adenohypophysis (anterior and intermediate lobes), which develops from the oral ectoderm by an invagination at the early stage of the embryo, synthesizes and secretes various hormones to perform essential roles such as in growth, metabolism, reproduction, lactation, homeostasis and stress response. A number of transcription factors spatiotemporally appear and promote cell differentiation during pituitary organogenesis to allow production of each hormone in the respective specific cell that produces the hormones [1,2,3,4,5,6]. Stem/progenitor cells are crucially responsive for tissue maintenance as the cell resource of hormone-producing cells and nonhormone-producing cells, such as folliculo-stellate cells ([7] and references therein). We previously reported that the stem/progenitor marker SOX2 is expressed in all cells of the presumptive pituitary tissue and that the pituitary-specific transcription factor PROP1 starts to appear in SOX2-positive cells at rat embryonic day (E) 11.5, followed by occupation by all cells of the primordium Rathke’s pouch at E13.5 [8, 9]. Hence, it follows that all of the pituitary cells once received the effects of PROP1. On the other hand, we most recently demonstrated that paired-related homeobox transcription factor PRRX1 (same as Mhox1, PRX1, PHOX1, PMX1 and K2) is expressed in PROP1/SOX2 double-positive cells and replaced with PROP1 during pituitary development [10,11,12]. PRRX2 (same as S8, PRX2, PMX2), a cognate factor of PRRX1, was identified in postnatal SOX2-positive cells, emerging behind Prrx1 expression [13], indicating that the two cognate factors play their roles distinctively in the diverse stem/progenitor cells. Investigation of the function of the novel transcription factors PRRX1 and PRRX2 invariably served to unmask the pituitary stem/progenitor cells at the molecular level.
In previous studies, Prrx1 and Prrx2 were cloned as a binding protein for AT-rich elements in the muscle creatine kinase enhancer [14] and as a homeobox gene from the mouse adult spleen cDNA library [15], respectively. Double mutants of Prrx1/Prrx2 revealed important roles for both gene products in craniofacial and limb morphogenesis [16,17,18]. Prrx1 and Prrx2 transcripts were largely observed to have overlapping expression profiles in the derivatives from the lateral plate mesoderm and in the neural crest-derived mesenchyme, which is where epithelio-mesenchymal interactions occur [16]. Interestingly, the Prrx2 transcripts were first detected in the extraembryonic mesoderm at the primitive streak stage of the embryo, followed by strong expression in the initially formed intraembryonic mesoderm at the presomite stage [19]. Thus, PRRX1 and PRRX2 are likely involved in ontogenesis in a similar and/or distinct manner, but much less is known about their roles in the pituitary originating from the embryonic ectoderm.
In this study, we first examined whether both Prrx1 and Prrx2 are regulated similarly or distinctively by many pituitary transcription factors and found that Krüppel-like factor 6 (KLF6), which plays a role in the regulation of cell proliferation, differentiation and development, is characteristically responsible for only Prrx2 expression. Then, we demonstrated that KLF6 plays a role in the regulation of Prrx2 by binding to the proximal 5’ upstream region of Prrx2. Immunohistochemistry ultimately revealed that KLF6 is certainly present in PRRX2-positive pituitary stem/progenitor cells.
Materials and Methods
Animals and immunohistochemistry
Wistar-Imamichi and S100β-GFP transgenic Wistar-crlj strain (S100β-GFP) [20] rats were housed individually in a temperature-controlled room under a 12 h light/12 h dark cycle. The present study was approved by the Institutional Animal Care and Use Committee, Meiji University, based on the NIH Guidelines for the Care and Use of Laboratory Animals.
Frozen sections of rat pituitaries on embryonic day (E) 16.5 and postnatal day (P) 20–30 were prepared as described in a previous paper [10]. Immunohistochemistry was performed by reaction with antibodies for human SOX2, rat PRRX1 and rat PRRX2 as described previously [10] and with rat KLF6 (dilution 1:100 of sc-365633, Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by incubation with the corresponding fluorescence (Cy3, Cy5 and fluorescent isothiocyanate) conjugated secondary antibodies [10] and embedding in mounting medium containing 4’, 6- diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). Immunofluorescence was observed with a BZ-9000 fluorescence microscope (Keyence, Osaka, Japan).
Transfection reporter assays
Upstream regions of the mouse Prrx1 (NC_000067.6) and Prrx2 (NC_000068.7) genes were amplified using primers (Table 1) and were ligated to the secreted alkaline phosphatase (SEAP) plasmid vector pSEAP2-Basic (BD Biosciences Clontech, Palo Alto, CA, USA). This resulted in reporter vectors: Prrx1 (–2297/+103), Prrx1 (–1400/+103), Prrx1 (–1140/+103), Prrx1 (–450/+103), Prrx2 (–5060/+21), Prrx2 (–3567/+21), Prrx2 (–1752/+21), Prrx2 (–1091/+21) and Prrx2 (–372/+21). Mouse Klf6 full-length open reading frame (NM_011803.2) was ligated in frame into the mammalian expression vector pcDNA3.1Zeo+ (Invitrogen, Carlsbad, CA, USA) or pET32a (Novagen, Madison, WI, USA). In addition, the expression vectors of cDNAs coding several pituitary transcription factors, which were involved in differentiation, were also constructed in frame in the pcDNA3.1 vector (Table 2).
Table 1. List of primers used for construction of the 5’ upstream region of Prrx1 and Prrx2.
| Prrx1 | ||
| Forward primer | –2297 | 5’-aatacgcgtTCTAGAACAATGGGGGAG-3’ |
| –1400 | 5’-aatacgcgtAGTGGCTGGAGCGAGGCG-3’ | |
| –1140 | 5’-aatacgcgtTCTCATAGCTACAGGAGAAG-3’ | |
| –450 | 5’-aatacgcgtTCTCCGCCAAAACAAAGCTG-3’ | |
| Reverse primer | +103 | 5’-acacactcgcgaTCCACTTAATAGGAGCCTGTA-3’ |
| Prrx2 | ||
| Forward primer | –5060 | 5’-aatacgcgtAGGAGGATTTGTGTGGCTTG-3’ |
| –3567 | 5’-aatacgcgtGGGCACCAGGAACATTTGTA-3’ | |
| –1752 | 5’-aatacgcgtACACACCAGAAGAGGGCATC-3’ | |
| –1091 | 5’-aatacgcgtAGGCTCTGGACCTCACTTTG-3’ | |
| –372 | 5’-aatacgcgtCAAATTCGAGGCTAATCTGC-3’ | |
| Reverse primer | +21 | 5’-acacactcgcgaGTGCCGGATCTCAAGTCAGT-3’ |
The uppercase and lowercase letters indicate the sequence of the gene to be amplified and adaptor containing the recognition sequence for restriction enzymes (Mul I for the forward primer and Nru I for the reverse primer, respectively).
Table 2. List of primer sets to amplify full-length cDNA.
| Factor | Species | Accession Number | Forward primer | Reverse primer |
| Prop1 | Rat | NM_153627.1 | CGGAATTCGCCATGCGGTCACTACTCCAGCCA | CCGGCTCGAGTTACTCTTGCTTCCGTTGCTTAGCC |
| Prrx1 | Rat | NM_153821.1 | GAGAATTCGCGATGTTTTATTTCCACTG | GGAGCTCGAGTTCATCCTTGTGTCATATC |
| Prrx2 | Rat | NM_001105739.1 | CCGGGAATTCGGCATGGACAGCGCGGCCGCCGCCTT | CCGGCTCGAGTGTCAGTTCACTGTGGGCACC |
| SOX2 | Rat | NM_001109181.1 | GGAGAATTCGCCATGTATAACATGATGGAGACG | GGACTCGAGTCACATGTGCGACAGGGG |
| Lhx2 | Mouse | NM_010710.3 | AGAGAGAATTCGCGATGCTGTTCCACAGTC | ACACACTCGAGTTGGGGGGTGGCGAGTC |
| Lhx3 | Rat | XM_001059910 | GGAATTCGCCATGGAAGCTCGCGGGGAGCT | TCTCTCGAGTCAGAACTGAGCATGGTCTAC |
| Isl1 | Mouse | NM_021459.4 | AGAGACAATTGGATATGGGAGACATGGGCGA | TGTGTCTCGAGCCTCATGCCTCAATAGGACTGG |
| Otx2 | Mouse | NM_001286481.1 | AGAGAGAATTCAGCATGATGTCTTATCTAAAGCAACCGC | TGTGTCTCGAGACAGGTCTTCACAAAACCTGGAA |
| Pax6 | Mouse | NM_001244198.1 | GACAATTGAGCATGCAGAACAGTCACAGC | ACGCTCGAGCTCTCTCCTTCTCTCTTTA |
| Foxj1 | Mouse | NM_008240.3 | GACAATTGGACATGGCGGAGAGCTGGCTG | ACGCTCGAGACCTTTTACAAGAAGGCAC |
| Hey1 | Mouse | NM_010423.2 | GAGAATTCACCATGAAGAGAGCTCACCCAG | ACGCTCGAGCATCAGTTCTTTAGAAAGC |
| Hey2 | Mouse | NM_013904.1 | GAGAATTCGCGATGAAGCGCCCTTGTGAGG | ACGCTCGAGATTCAAGAATAAGTTAAAAG |
| Id3 | Mouse | NM_008321.2 | AGAGAGAATTCAACATGAAGGCGCTGAGCCC | TGTGTCTCGAGTTCGGGAGGTGCCAGGACGAC |
| Klf6 | Mouse | NM_011803.2 | GAGAATTCAGCATGAAACTTTCACCTGCG | ACGCTCGAGCCTACAGGATTCGTCCCTC |
| Nfib | Mouse | NM_008687.6 | GAGAATTCGTCATGATGTATTCTCCCATCTGTCTCACTCAGGATGAG | GCGTCGACCAAGCTAGCCCAGGTACC |
| Nkx3-1 | Mouse | NM_010921.3 | AGAGAGAATTCGGGATGCTTAGGGTAGCGG | TGTGTCTCGAGTTGCTACCAGAAAGATGGATGC |
| Runx1 | Mouse | NM_001111021.2 | GAGAATTCAAGAGTGCGAGTCTGCCTGTG | ACGCTCGAGTCCCTCGATGGCGATGGCGC |
| Sp1* | Human | BC062539.1 | GAGAATTCACCATGAGCGACCAAGATCACT | TCCTCGAGCTACCTTGCATCCCGGG |
| Tal1 | Mouse | NM_011527.3 | AGAGAGAATTCACGATGACGGAGCGGCCGCCGAGC | TGTGTCTCGAGTCACATAAGTCCATTGACCTGCTT |
| Tcf7l2 | Mouse | NM_001142918.1 | AGAGAGAATTCAAAATGCCGCAGCTGAACGGC | TGTGTCTCGAGCCTAGCAGATGCGGTG |
| Tead2 | Mouse | NM_001285498.1 | GAGAATTCAAGATGGGGGATCCCCGGACTG | ACGCTCGAGCTCCATCAGTCCCTGACCAGG |
| Zfp36 | Mouse | NM_011756.4 | GAGAATTCACCATGGATCTCTCTGCCATC | ACGCTCGAGGCACTTGTCACTCAGAGAC |
| Zfp36l1 | Mouse | NM_007564.5 | GAGAATTCAGCATGATCCGCGGCGCCCCGGCAC | ACGCTCGAAGTCAGTGTGAGGGCTACTGGCGGAG |
* The human SP1 cDNA clone was kindly provided by Dr. R. Tijan of the University of California at Berkeley.
The culture and transfection conditions for CHO cells generated from Chinese hamster ovary and NIH3T3 cells (mouse fibroblast cell line), which were obtained from RIKEN Cell Bank, Ibaraki, Japan, and for TtT/GF cells (TtT/GF, kindly provided by Dr K Inoue) have been described previously [21, 22]. The TtT/GF cell line, which was generated from a mouse pituitary tumor and is known to express S100, a Ca2+-binding protein and marker for non-endocrine pituitary cells such as folliculo-stellate cells. After incubation, each 5 µl of cultured medium was assayed for secreted alkaline phosphatase activity as described previously [21].
Electrophoretic mobility shift assay (EMSA)
A FAM-labelled probe of the Prrx2 5’ upstream (-372/+21) region was made by PCR using a set of primers (FAM-labelled Prrx2-372 forward primer and Prrx2 reverse primer in Table 1) with the same conditions as described previously [23]. Bacterial recombinant KLF6 proteins (rec-KLF6) were expressed and purified as described previously [24]. The binding reaction was carried out in a mixture containing 100 fmol FAM-labelled double-stranded (ds) nucleotides, 500 ng rec-KLF6 and 1 µg ds-poly(dIdC) in 10 μl of binding buffer (10 mM Hepes buffer, pH 7.9, containing 0.4 mM MgCl2, 0.4 mM DTT, 50 mM NaCl, 0.1 mM ZnCl2, 0.1 mM spermidine and 4% (v/v) glycerol) with or without an unlabelled fragment (–372/+21) by incubation at 37 C for 30 min, followed by electrophoresis on a 4% polyacrylamide gel at 100 V for 60 min.
Knockdown analysis of Klf6 mRNA and PCR analyses
Knockdown of Klf6-mRNAs was performed using siRNAs with the same conditions as described previously [11] using TtT/GF cells. siRNAs for Klf6 (MSS215606, MSS215607 and MSS215608) and control siRNA (12935-300) were purchased from Life Technologies (Carlsbad, CA, USA). Cell number was counted by staining of the nuclei with Diff-Quik (Sysmex, Kobe, Japan).
cDNAs were synthesized using total RNAs prepared from siRNA-treated TtT/GF cells, and quantitative real-time PCR was performed using a primer set for mouse Klf6 (NM_011803.2), 5’-ACCAGACACTTCCGAAAGCA-3’ and 5’-TCTTAGCCTGGAAGCCTCTTT-3’. The primer sets for mouse Prrx1, Prrx2 and TATA-box binding protein (Tbp) were the same as described previously [11]. Each datum measured by triplicate experiments was calculated by the comparative CT method (DDCT method) to estimate the gene copy number relative to Tbp as an internal standard. The DNA sequence of the PCR product of each sample was confirmed by nucleotide sequence determination (data not shown).
Statistical analysis
Differences between two groups were subjected to the Student’s t-test, and differences among the above three groups were subjected to one-way ANOVAs with Dunnett’s test. A P value less than 0.05 was considered statistically significant.
Results
Localization of PRRX1 and PRRX2 in the pituitary stem/progenitor cells
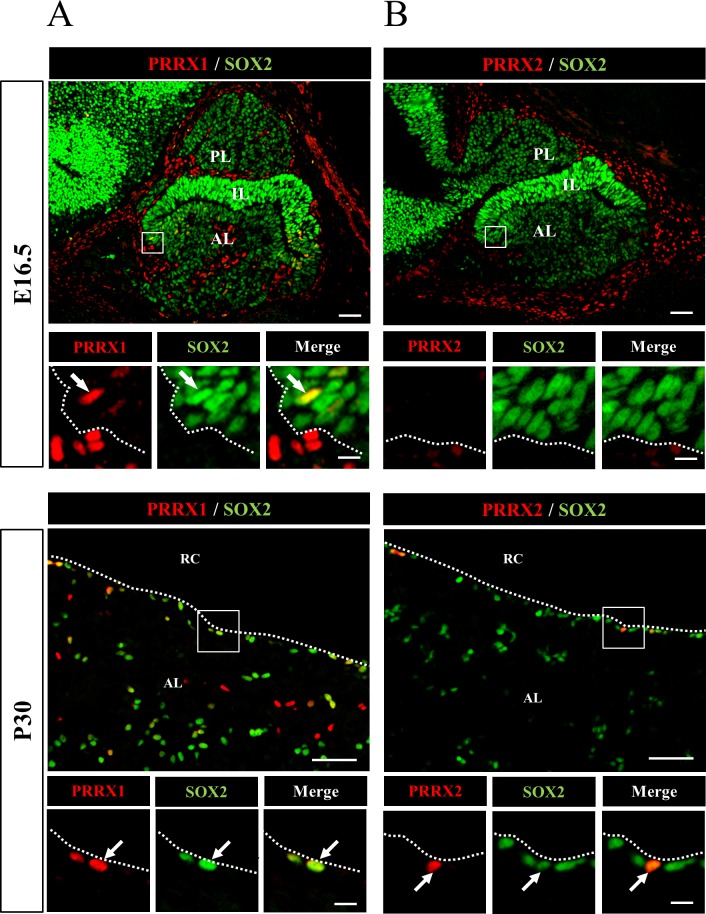
Immunohistochemistry of PRRX1 and PRRX2 was performed for embryonic (E16.5) and postnatal (P30) pituitaries. Double immunostaining with SOX2 and PRRX1 showed that PRRX1 was present in some of the SOX2-positive cells of both sections at E16.5 and P30, while cells positive for PRRX1 only were present in the periphery of the pituitary on E16.5 (Fig. 1A). On the other hand, PRRX2 was not present in the embryonic pituitary but rather was present in the SOX2-positive cells of the anterior lobe of the postnatal pituitary on P30 (Fig. 1B).
Fig. 1.
Localization of PRRX1 and PRRX2 in pituitary stem/progenitor cells. Immunohistochemistry for PRRXs (A, PRRX1; B, PRRX2) and SOX2 was performed using frozen sections of rat pituitary at embryonic day 16.5 (E16.5) and postnatal day 30 (P30). Areas of PRRXs and SOX2 in open boxes, which were visualized with Cy3 (red) and fluorescein isothiocyanate (green), were enlarged as shown below together with the merged image. The arrow and dotted line indicate cells double positive for PRRXs and SOX2 and the marginal cell layer, respectively. AL, anterior lobe; IL, intermediate lobe; PL, posterior lobe; RC, Rathke’s cleft. Scale bars: 50 µm and 10 µm (enlarged images).
Transcriptional activities of the 5’ upstream region of Prrx1 and Prrx2
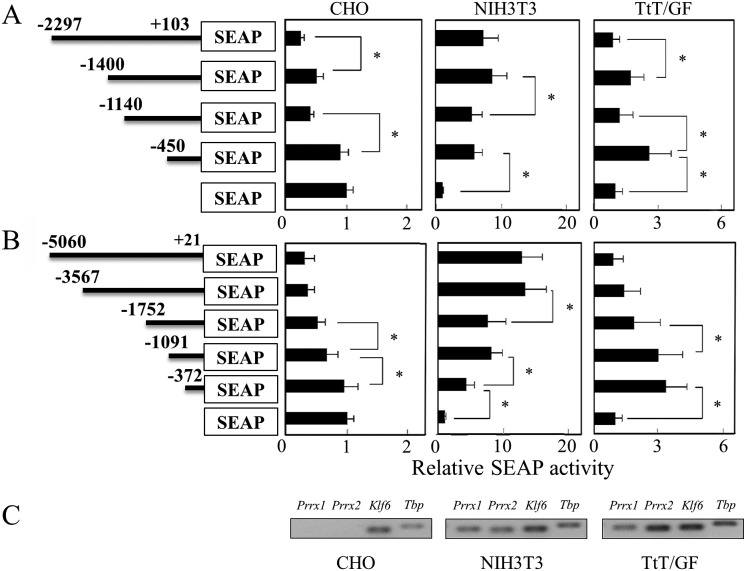
Reporter vectors harboring the 5’ upstream regions of Prrx1 (–2297 b) and Prrx2 (–5060 b) together with their truncated regions were assayed for their transcriptional activity by transfection into three cell lines, CHO, NIH3T3 and TtT/GF. In the CHO cells, the promoter activities of the upstream regions of Prrx1 and Prrx2 decreased along with the increased upstream length (Fig. 2A and B). On the other hand, in the NIH3T3 and TtT/GF cells, the promoter activities were higher than those of the pSEAP-basic vector used as a control. The regions up to –450 b of Prrx1 and –372 b of Prrx2, respectively, showed significantly high promoter activity, indicating the presence of basic promoter elements in the proximal region. Notably, the promoter activity of Prrx1 in the NIH3T3 cells did not show much alteration over –450 b, while that in the TtT/GF cells showed a different response depending on the upstream length (Fig. 2A). On the other hand, the promoter activity of Prrx2 showed an increase in the NIH3T3 cells and a decrease in the TtT/GF cells with different responses depending on the upstream length (Fig. 2B). These data indicate that Prrx2 expression is cell-type dependent with unique regulatory factors. We subsequently examined expression of Prrx1 and Prrx2 by RT-PCR and confirmed that NIH3T3 and TtT/GF cells express both genes but that CHO does not (Fig. 2C).
Fig. 2.
Transient transfection assay of mouse Prrx1 and Prrx2 promoters. Reporter vectors of Prrx1 (A) and Prrx2 (B) were transfected into CHO, NIH3T3 and TtT/GF cells. An aliquot of cultured medium was used for the SEAP assay. Reporter gene activities are indicated relative to that of the pcDNA3.1 vector. Representative data (mean ± SD) are shown from means of quadruplicate transfections from two independent experiments. Asterisks indicate statistical significance by one-way ANOVAs with Dunnett’s test (*P<0.05). (C) RT-PCR was performed for Prrx1, Prrx2, Klf6 and Tbp (TATA-box binding protein).
Next, we performed co-transfection with several transcription factors sorted by their characteristic expression and/or function for organogenesis from a microarray analyses of RNAs prepared from the rat pituitary on E15.5 and pituitary S100β-positive pituitary cells of an S100β-GFP rat (unpublished data). The SEAP activities for Prrx1 (–2297/+103), Prrx1 (–450/+103), Prrx2 (–5060/+21) and Prrx2 (–372/+21) were then assayed (Table 3). The results showed that transcription of Prrx1 is regulated repressively by FOXJ1, HEY1, NF1B and ZFP36 by less than 0.6-fold and stimulated by SOX2, LHX2, ISL1 and PAX6 by more than 1.9-fold. Transcription of Prrx2 was repressed by HEY1, HEY2, PROP1, OTX2, ID3, ZFP36 and ZFP36L1 by less than 0.6-fold and stimulated by PRRX2, SOX2, PAX6 and KLF6 by more than 1.9-fold. Among factors affecting commonly or distinctively both gene expressions, it was interesting that PRRX2 stimulated its own gene expression and that SOX2 had opposite effects for two genes. Thus, several transcription factors that participate in pituitary organogenesis have regulatory potency with respect to Prrx1 and Prrx2 expression. We were ultimately interested in KLF6, since it potently stimulated only Prrx2 and its role is little noticed in pituitary development. To verify evidence of correlation between PRRX2 and KLF6, we performed the following experiments.
Table 3. Transcriptional activity of the 5’ upstream region of Prrx1 and Prrx2 in the presence of transcription factors.
|
Prrx1 |
Prrx2 |
|||
| –2297 | –450 | –5060 | –372 | |
| PROP1 | 1.29 ± 0.20 | 0.87 ± 0.15 | 0.23 ± 0.29** | 0.42 ± 0.23** |
| PRRX1 | 0.80 ± 0.20 | 0.53 ± 0.17** | 0.90 ± 0.17 | 0.51 ± 0.20** |
| PRRX2 | 1.75 ± 0.21** | 0.94 ± 0.26 | 4.11 ± 0.22** | 0.79 ± 0.10 |
| SOX2 | 3.31 ± 0.13** | 0.89 ± 0.18 | 0.39 ± 0.10 | 0.46 ± 0.20** |
| LHX2 | 1.74 ± 0.17* | 1.31 ± 0.17 | 1.08 ± 0.22 | 0.93 ± 0.11 |
| LHX3 | 1.41 ± 0.21 | 1.54 ± 0.26* | 0.62 ± 0.18** | 1.44 ± 0.24 |
| ISL1 | 3.94 ± 0.16** | 1.67 ± 0.02** | 1.01 ± 0.26 | 1.12 ± 0.11 |
| OTX2 | 1.73 ± 0.11** | 1.41 ± 0.16* | 0.49 ± 0.04** | 0.60 ± 0.29* |
| PAX6 | 2.21 ± 0.17** | 1.39 ± 0.14 | 2.72 ± 0.12** | 1.40 ± 0.02** |
| FOXJ1 | 0.27 ± 0.11** | 0.61 ± 0.06** | 0.16 ± 0.10** | 0.53 ± 0.05** |
| HEY1 | 0.52 ± 0.16** | 0.59 ± 0.10** | 0.06 ± 0.16** | 0.39 ± 0.08** |
| HEY2 | 0.52 ± 0.08** | 0.94 ± 0.11 | 0.16 ± 0.27** | 0.43 ± 0.06** |
| ID3 | 1.09 ± 0.28 | 0.81 ± 0.25 | 0.43 ± 0.21** | 0.38 ± 0.11* |
| KLF6 | 0.75 ± 0.07** | 1.05 ± 0.13 | 18.23 ± 0.13** | 6.36 ± 0.21* |
| NFIB | 1.05 ± 0.16 | 0.55 ± 0.22** | 0.75 ± 0.23** | 1.07 ± 0.13 |
| NKX3-1 | 1.40 ± 0.22 | 0.83 ± 0.11 | 1.52 ± 0.20* | 0.80 ± 0.10 |
| RUNX1 | 0.87 ± 0.08 | 0.88 ± 0.21 | 0.80 ± 0.12 | 1.13 ± 0.20 |
| SP1 | 3.69 ± 0.20** | 2.21 ± 0.20** | 7.87 ± 0.16** | 4.83 ± 0.07** |
| TAL1 | 0.79 ± 0.17* | 0.80 ± 0.08* | 0.97 ± 0.20 | 0.89 ± 0.04 |
| TCF7L2 | 1.21 ± 0.11* | 0.83 ± 0.28 | 1.86 ± 0.06** | 0.88 ± 0.11 |
| TEAD2 | 0.58 ± 0.22** | 0.71 ± 0.14** | 0.31 ± 0.07** | 0.79 ± 0.24 |
| ZFP36 | 0.44 ± 0.18** | 0.39 ± 0.26** | 0.25 ± 0.14** | 0.69 ± 0.20** |
| ZFP36L1 | 0.63 ± 0.14** | 0.89 ± 0.24 | 0.40 ± 0.15** | 1.03 ± 0.04 |
| KLF6+SP1 | 1.04 ± 0.15 | 0.77 ± 0.07* | 26.27 ± 0.12** | 6.40 ± 0.14** |
Transcriptional activity was assayed in CHO cells by transfection of reporter vectors, Prrx1 (–2297/+103), Prrx1 (–405/+103), Prrx2 (–5060/+21) and Prrx2 (–372/+21), and an expression vector harboring cDNA encoding the transcription factors listed in the Table. Data are indicated as the relative activity against that without the expression vector, and representative data are shown as means ± SD of quadruplicate transfections in two independent experiments. Asterisks indicate statistical significance by Student’s t-test (*P<0.05; **P<0.01).
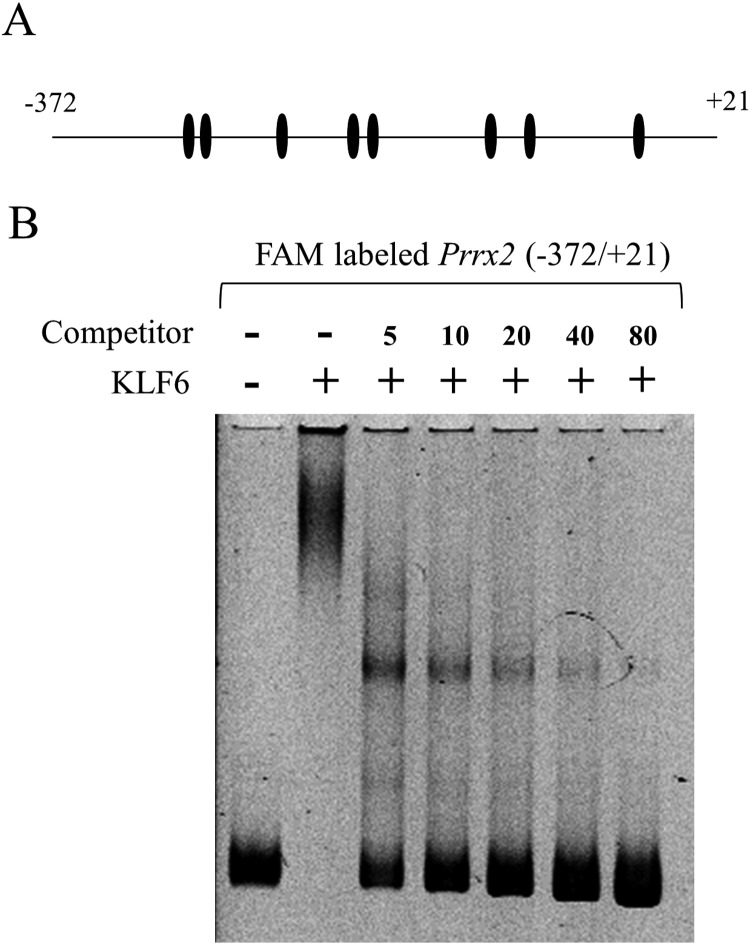
Binding of KLF6 on Prrx2 promoter region
Since reporter assays showed a specific regulation of KLF6 for the Prrx2 promoter, we surveyed the consensus recognition sequence of KLF6, CCNCNCCC including CACCC and GC-rich elements. As shown in Fig. 3A, the putative binding site for KLF6 was found in 8 positions in the 372 base length of the proximal upstream region. To confirm the KLF6 binding to the upstream region of Prrx2, EMSA was performed using a FAM labelled probe (–372/+21). The mixture of probe and protein gave broad shift bands with high molecular sizes in contrast to the high mobility band obtained with the probe alone (Fig. 3B), showing a potential ability of KLF6 with respect to Prrx2 regulation. When unlabelled competitors were added in increasing molar amounts, the molecular sizes and intensities of the shift bands decreased and those of the bands of the probe increased. Addition of an 80-molar excess amount caused most of the shift band to fade, indicating decomposition of the specific binding between KLF6 and the probe.
Fig. 3.
Electrophoretic mobility shift assay (EMSA) for KLF6. (A) A diagram of the 5’ upstream (–372/+21) region of Prrx2, which was used as a binding probe, is shown. A putative binding site (CCNCNCCN including GC element and CACCC) of KLF6 is shown with a closed ellipse. (B) EMSA was performed using a 100 fmol FAM-labelled fragment (–372/+21) without and with a 5–80 molar excess amount of non-labelled fragment (–372/+21) as a competitor to confirm the specific DNA/protein complex. Electrophoresis of the FAM-labelled fragment (probe) alone is shown at the left of the panel.
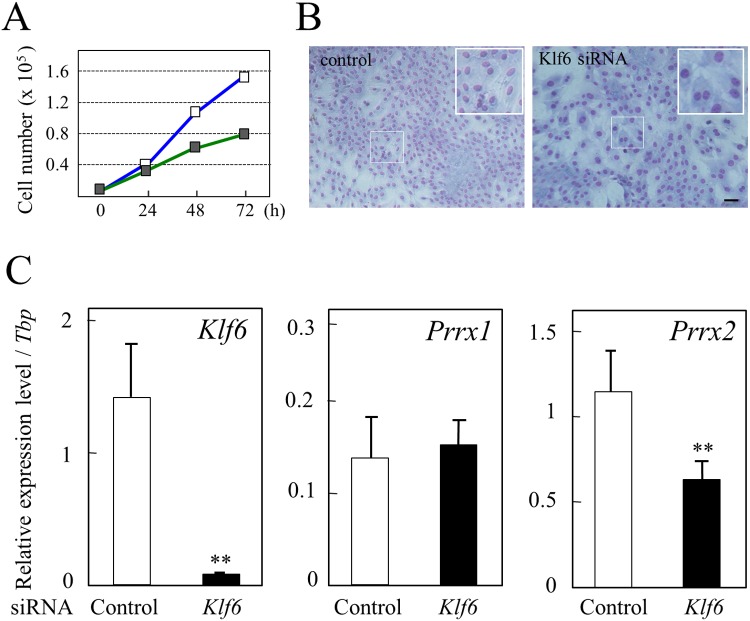
Knockdown with Klf6 siRNA
Knockdown of Klf6 mRNA using siRNAs in TtT/GF cells, which expresses Klf6 (Fig. 2C), was performed to confirm whether KLF6 modulates expression of Prrx2 and Prrx1. Cell proliferation was decreased by about 50% at 72 h after siRNA transfection (Fig. 4A). Microscopy after staining nuclei with Diff-Quik showed a decrease in cell density for the Klf6 siRNA-transfected cells in comparison with the control. In addition, we observed that siRNA treatment resulted in enlargement of nuclei and cytoplasm and a decreased number of cells (insets in Fig. 4B).
Fig. 4.
Knockdown analysis of the Klf6 mRNA level using siRNA. A: The Klf6 mRNA level was knocked down for TtT/GF cells by transfection of Klf6 siRNAs, and cell proliferation was measured. The respective cell numbers of the control (open boxes) and siRNA-transfected cells (closed boxes) were measured. Data are shown as means ± SD for two independent experiments. B: Microscopy observation of cells after 72 h of culture. Upper and lower panels show photographs of the control and siRNA-transfected cells, respectively. An enlarged image of the boxed area is shown at the upper right. Scale bars: 100 μm. C: Real-time PCR was performed for Klf6 (left panel), Prrx1 (middle panel) and Prrx2 (right panel) using total RNAs prepared from cells cultured for 24 h after transfection. All quantified data are shown as the relative expression level against that of TATA-box binding protein (Tbp). Open and closed bars indicate data of cells transfected with control siRNA and Klf6 siRNA, respectively. Asterisks indicate statistical significance by Student’s t-test (**P<0.01).
Quantitative real-time PCR of RNAs from cells confirmed knockdown of the Klf6 mRNA level by transfection of Klf6 siRNAs at about 90% efficiency (Fig. 4C). Then we measured the expression levels of Prrx1 and Prrx2 and observed that the expression level of Prrx1 did not change, whereas that of Prrx2 decreased significantly by about 55% (Fig. 4C), showing a Klf6 siRNA-dependent repression of Prrx2 expression.
Immunohistochemistry of KLF6
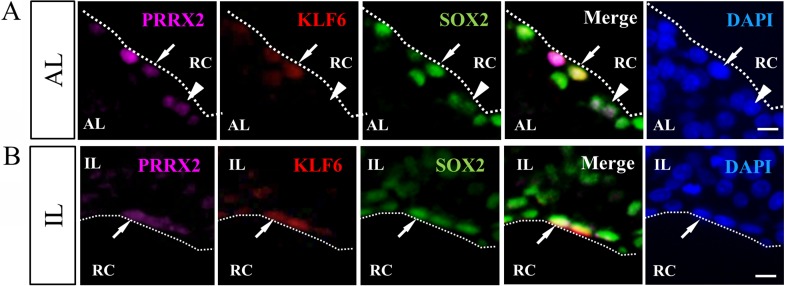
Finally, to verify whether KLF6 is present in the pituitary stem/progenitor cells, triple-immunostaining of KLF6, SOX2 and PRRX2 was performed for the adult rat pituitary (P30). KLF6 was found in the cells lining the marginal cell layer (MCL) of the anterior and intermediate lobes (Fig. 5A and B). Although KLF6-negative cells were found among PRRX2/SOX2 double-positive cells (Fig. 5A arrowhead), immunohistochemistry confirmed that all positive signals of KLF6 were observed in the PRRX2/SOX2 double-positive cells of pituitary stem/progenitor cells.
Fig. 5.
Immunohistochemistry of KLF6 together with PRRX2 and SOX2. Immunohistochemistry for KLF6 was performed using frozen sections of rat pituitary at P20. Note that immunohistochemical images of KLF6, PRRX2 and SOX2 were visualized with Cy3 (red), Cy5 (purple) and fluorescein isothiocyanate (green) and counterstained with DAPI (blue). Dotted lines indicate the marginal cell layer (MCL). AL, anterior lobe; IL, intermediate lobe; RC, Rathke’s cleft. Scale bars: 10 μm.
Discussion
The present study examined the transcriptional activity of several transcription factors for Prrx1 and Prrx2, which are present in pituitary stem/progenitor cells [10], and revealed that Prrx1 and Prrx2 expression are regulated stimulative or repressive with common factors, such as PAX6, FOXJ1, HEY1, TEAD2 and ZFP36; by contrast, Prrx1 is distinctly regulated by SOX2, LHX2 and ISL1, and Prrx2 is distinctly regulated by PRRX2, HEY2, ID3, KLF6 and TCF7L2, respectively, which are known to play crucial roles in pituitary stem/progenitor cells and organogenesis [1, 5, 6, 25]. We focused attention on KLF6, which participates in modulation of Prrx2 expression, using in vitro experiments and demonstrated by immunohistochemistry that KLF6 is specifically present in the PRRX2-positive stem/progenitor cells of the anterior lobe of the rat postnatal pituitary gland. The results provide important clues to uncovering the adult pituitary stem/progenitor cells at the molecular level.
In the present study, we confirmed the distinct temporospatial localizations of PRRX1 and PRRX2 in the embryonic and postnatal pituitaries. Recently, we observed the appearance of PRRX1 and PRRX2 in postnatal SOX2-positive cells but not postnatal PROP1-negative cells present in a stem cell niche of the pituitary, the marginal cell layer [13]. Interestingly, PRRX2 is not present in the embryonic pituitary. Our data showing that KLF6 co-localized with PRRX2 in cells of the marginal cell layer at the postnatal stage suggest that KLF6 might regulate Prrx2 expression in an adult pituitary stem cell.
KLF6 is a member of the Krüppel-like factors with three zinc fingers characteristically in its carboxyl region. The family consists of 17 proteins, 9 of which are included the SP1 superfamily [26]. KLF6 is known to play a role in differentiation and development of tissues, in addition to its role as a tumor suppressor gene [26]. Klf6 (Klf6-/-) knockout mice died by E12.5 and were associated with markedly reduced hematopoiesis and poorly organized yolk sac vascularization [27]. They generated Klf6-/- embryonic stem (ES) cells and demonstrated that Klf6-/--ES cells have significant hematopoietic defects associated with delayed expression of differentiation markers, Brachyury, Klf1 and Gata1, followed by differentiation into embryoid bodies (EBs), and that forced expression of KLF6 enhances the hematopoietic potential of wild-type EBs, implicating the role of Klf6 in ES-cell differentiation and hematopoiesis. Interestingly, the role of KLF6 was demonstrated by knockdown analysis, and it was found that this protein is likely involved in phosphorylation of retinoblastoma protein along with upregulation of cyclin D1 and cdk4, which are required for the cell cycle G1/S transition [28] however, cyclin D1 and cdk4 were not examined in this study. These data indicate that KLF6 plays roles in differentiation and development through cell cycle progression [26]. Taken together with the accumulated reports, our finding that KLF6 is present in pituitary stem/progenitor cells indicates its important role in maintaining stemness and/or progression of differentiation.
This study showed the remarkable regulatory activity of KLF6 on the promoter of Prrx2. Presumably, the presence of putative KLF-binding sites at 8 positions in the proximal region within the 372 b upstream might serve to massively bind and activate the expression of a reporter gene. Notably, it has been reported that KLF6 has the ability to interact with other transcription factors, such as Sp1, KLF4, p53 and aryl hydrocarbon receptor, in a cell type- and/or tissue-dependent manner [29,30,31]. We observed that expression of Sp1 is higher than that of Klf6 in the pituitary (data not shown), making it thereby sufficient for Prrx2 to be regulated with a KLF6-SP1 complex. However, SP1-dependent activation between KLF6 and SP1 was additive but not synergistic with respect to Prrx2 expression (Table 1). Assay of the promoter activity in the three cell lines showed that Prrx2 as well as Prrx1 is responsive with cell type- and the promoter length-dependent. Therefore, a further search for cooperative factors in pituitary stem/progenitor cells is important to clarify the function of KLF6.
EMSA showed an unexpected smear pattern with high molecular complexes between the labelled probe and KLF6, the specific binding of which was verified by addition of a non-labelled probe. However, the reason why the complex showed the smear pattern remains unclear. Previously, we observed that LIM-homeodomain factor resulted in a large molecular size for the complex [32]. In that case, the LIM domain formed protein-protein interactions, resulting in catenation of protein-DNA complexes. However, there is no report of a protein-protein interaction domain in KLF6. On the other hand, KLF6 has three C2H2-type Zn-fingers, which bound on CACCC and GC-elements. In addition, there are 8 binding sequences for KLF6 in the probe used. Thus, binding of 8 KLF6 molecules to the probe may result in a large molecular complex. An alternative explanation is that the binding between the three zinc fingers and 8 binding elements might form a large complex by multiple binding of KLF6 to the probe DNA and/or by catenation of the probe DNA through the three zinc fingers. At least, this massive KLF6 binding may induce marked activation of Prrx2 expression.
In summary, the present study demonstrated that the pituitary transcription factors PRRX1 and PRRX2 present in stem/progenitor cells are regulated by many transcription factors participating in pituitary organogenesis. Particularly, KLF6 was identified as stimulating distinctively Prrx2 expression but not Prrx1 expression. In addition, in vivo evidence of the localization of KLF6 and Prrx2 in the stem/progenitor cells of the pituitary gland was demonstrated. This KLF6 finding provides a novel clue to resolve the role of the pituitary stem/progenitor cells and maintenance of this tissue.
Acknowledgments
This work was partially supported by JSPS KAKENHI Grant Nos. 21380184 (to YK) and 24580435 (to TK), and by a research grant (A) to YK from the Institute of Science and Technology, Meiji University. This study was also supported by the Meiji University International Institute for BioResource Research (MUIIR) and by Research Funding for the Computational Software Support Program from Meiji University.
References
- 1.Zhu X, Gleiberman AS, Rosenfeld MG. Molecular physiology of pituitary development: signaling and transcriptional networks. Physiol Rev 2007; 87: 933–963 [DOI] [PubMed] [Google Scholar]
- 2.Brinkmeier ML, Davis SW, Carninci P, MacDonald JW, Kawai J, Ghosh D, Hayashizaki Y, Lyons RH, Camper SA. Discovery of transcriptional regulators and signaling pathways in the developing pituitary gland by bioinformatic and genomic approaches. Genomics 2009; 93: 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins-Chow DE, Camper SA. How many homeobox genes does it take to make a pituitary gland? Trends Genet 1998; 14: 284–290 [DOI] [PubMed] [Google Scholar]
- 4.Zhu X, Rosenfeld MG. Transcriptional control of precursor proliferation in the early phases of pituitary development. Curr Opin Genet Dev 2004; 14: 567–574 [DOI] [PubMed] [Google Scholar]
- 5.Davis SW, Castinetti F, Carvalho LR, Ellsworth BS, Potok MA, Lyons RH, Brinkmeier ML, Raetzman LT, Carninci P, Mortensen AH, Hayashizaki Y, Arnhold IJ, Mendonça BB, Brue T, Camper SA. Molecular mechanisms of pituitary organogenesis: In search of novel regulatory genes. Mol Cell Endocrinol 2010; 323: 4–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vankelecom H, Gremeaux L. Stem cells in the pituitary gland: A burgeoning field. Gen Comp Endocrinol 2010; 166: 478–488 [DOI] [PubMed] [Google Scholar]
- 7.Vankelecom H, Chen J. Pituitary stem cells: Where do we stand? Mol Cell Endocrinol 2014; 385; 2–17 [DOI] [PubMed] [Google Scholar]
- 8.Yoshida S, Kato T, Susa T, Cai L-Y, Nakayama M, Kato Y. PROP1 coexists with SOX2 and induces PIT1-commitment cells. Biochem Biophys Res Commun 2009; 385: 11–15 [DOI] [PubMed] [Google Scholar]
- 9.Yako H, Kato T, Yoshida S, Inoue K, Kato Y. Three-dimensional studies of Prop1-expressing cells in the rat pituitary primordium of Rathke’s pouch. Cell Tissue Res 2011; 346: 339–346 [DOI] [PubMed] [Google Scholar]
- 10.Higuchi M, Kato T, Chen M, Yako H, Yoshida S, Kanno N, Kato Y. Temporospatial gene expression of Prx1 and Prx2 is involved in morphogenesis of cranial placode-derived tissues through epithelio-mesenchymal interaction during rat embryogenesis. Cell Tissue Res 2013; 353: 27–40 [DOI] [PubMed] [Google Scholar]
- 11.Susa T, Kato T, Yoshida S, Yako H, Higuchi M, Kato Y. Paired-related homeodomain proteins Prx1 and Prx2 are expressed in embryonic pituitary stem/progenitor cells and may be involved in the early stage of pituitary differentiation. J Neuroendocrinol 2012; 24: 1201–1212 [DOI] [PubMed] [Google Scholar]
- 12.Yako H, Kato T, Yoshida S, Higuchi M, Chen M, Kanno N, Ueharu H, Kato Y. Three-dimensional studies of Prop1-expressing cells in the rat pituitary just before birth. Cell Tissue Res 2013; 354: 837–847 [DOI] [PubMed] [Google Scholar]
- 13.Higuchi M, Yoshida S, Ueharu H, Chen M, Kato T, Kato Y. PRRX1 and PRRX2 distinctively participate in pituitary organogenesis and a cell-supply system. Cell Tissue Res 2014; 357: 323–335 [DOI] [PubMed] [Google Scholar]
- 14.Cserjesi P, Lilly B, Bryson L, Wang Y, Sassoon DA, Olson EN. MHox: a mesodermally restricted homeodomain protein that binds an essential site in the muscle creatine kinase enhancer. Development 1992; 115: 1087–1101 [DOI] [PubMed] [Google Scholar]
- 15.Kongsuwan K, Webb E, Housiaux P, Adams JM. Expression of multiple homeobox genes within diverse mammalian haemopoietic lineages. EMBO J 1988; 7: 2131–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meijlink F, Beverdam A, Brouwer A, Oosterveen TC, Berge DT. Vertebrate aristaless-related genes. Int J Dev Biol 1999; 43: 651–663 [PubMed] [Google Scholar]
- 17.Mitchell JM, Hicklin DM, Doughty PM, Hicklin JH, Dickert JWJ, Jr, Tolbert SM, Peterkova R, Kern MJ. The Prx1 homeobox gene is critical for molar tooth morphogenesis. J Dent Res 2006; 85: 888–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balic A, Adams D, Mina M. Prx1 and Prx2 cooperatively regulate the morphogenesis of the medial region of the mandibular process. Dev Dyn 2009; 238: 2599–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Jong R, Meijlink F. The homeobox gene S8: mesoderm-specific expression in presomite embryos and in cells cultured in vitro and modulation in differentiating pluripotent cells. Dev Biol 1993; 157: 133–146 [DOI] [PubMed] [Google Scholar]
- 20.Itakura E, Odaira K, Yokoyama K, Osuna M, Hara T, Inoue K. Generation of transgenic rats expressing green fluorescent protein in S-100beta-producing pituitary folliculo-stellate cells and brain astrocytes. Endocrinology 2007; 148: 1518–1523 [DOI] [PubMed] [Google Scholar]
- 21.Susa T, Kato T, Kato Y. Reproducible transfection in the presence of carrier DNA using FuGENE6 and Lipofectamine2000. Mol Biol Rep 2008; 35: 313–319 [DOI] [PubMed] [Google Scholar]
- 22.Mitsuishi H, Kato T, Chen M, Cai LY, Yako H, Higuchi M, Yoshida S, Kanno N, Ueharu H, Kato Y. Characterization of a pituitary-tumor-derived cell line, TtT/GF, that expresses Hoechst efflux ABC transporter subfamily G2 and stem cell antigen 1. Cell Tissue Res 2013; 354: 563–572 [DOI] [PubMed] [Google Scholar]
- 23.Sato T, Kitahara K, Susa T, Kato T, Kato Y. Pituitary transcription factor Prop-1 stimulates porcine pituitary glycoprotein hormone alpha subunit gene expression. J Mol Endocrinol 2006; 37: 341–352 [DOI] [PubMed] [Google Scholar]
- 24.Susa T, Ishikawa A, Kato T, Nakayama M, Kato Y. Molecular cloning of paired related homeobox 2 (prx2) as a novel pituitary transcription factor. J Reprod Dev 2009; 55: 502–511 [DOI] [PubMed] [Google Scholar]
- 25.Vankelecom H. Pituitary stem/progenitor cells: embryonic players in the adult gland? Eur J Neurosci 2010; 32: 2063–2081 [DOI] [PubMed] [Google Scholar]
- 26.Andreoli V, Gehrau RC, Bocco JL. Biology of Krüppel-like factor 6 transcriptional regulator in cell life and death. IUBMB Life 2010; 62(Suppl 3): 896–905 [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, Keller G, Friedman SL. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood 2006; 107: 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sirach E, Bureau C, Péron JM, Pradayrol L, Vinel JP, Buscail L, Cordelier P. KLF6 transcription factor protects hepatocellular carcinoma-derived cells from apoptosis. Cell Death Differ 2007; 14: 1202–1210 [DOI] [PubMed] [Google Scholar]
- 29.Difeo A, Huang F, Sangodkar J, Terzo EA, Leake D, Narla G, Martignetti JA. KLF6-SV1 is a novel antiapoptotic protein that targets the BH3-only protein NOXA for degradation and whose inhibition extends survival in an ovarian cancer model. Cancer Res 2009; 69: 4733–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinstein M, Idelman G, Plymate SR, Narla G, Friedman SL, Werner H. Transcriptional activation of the insulin-like growth factor I receptor gene by the Kruppel-like factor 6 (KLF6) tumor suppressor protein: potential interactions between KLF6 and p53. Endocrinology 2004; 145: 3769–3777 [DOI] [PubMed] [Google Scholar]
- 31.Wilson SR, Joshi AD, Elferink CJ. The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharmacol Exp Ther 2013; 345: 419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato T, Ishikawa A, Yoshida S, Sano Y, Kitahara K, Nakayama M, Susa T, Kato Y. Molecular cloning of LIM homeodomain transcription factor Lhx2 as a transcription factor of porcine follicle-stimulating hormone beta subunit (FSHβ) gene. J Reprod Dev 2012; 58: 147–155 [DOI] [PubMed] [Google Scholar]