Abstract
Cervicovaginal microbiota not dominated by lactobacilli may facilitate transmission of HIV and other sexually transmitted infections (STIs), as well as miscarriages, preterm births and sepsis in pregnant women. However, little is known about the exact nature of the microbiological changes that cause these adverse outcomes. In this study, cervical samples of 174 Rwandan female sex workers were analyzed cross-sectionally using a phylogenetic microarray. Furthermore, HIV-1 RNA concentrations were measured in cervicovaginal lavages of 58 HIV-positive women among them. We identified six microbiome clusters, representing a gradient from low semi-quantitative abundance and diversity dominated by Lactobacillus crispatus (cluster R-I, with R denoting ‘Rwanda') and L. iners (R-II) to intermediate (R-V) and high abundance and diversity (R-III, R-IV and R-VI) dominated by a mixture of anaerobes, including Gardnerella, Atopobium and Prevotella species. Women in cluster R-I were less likely to have HIV (P=0.03), herpes simplex virus type 2 (HSV-2; P<0.01), and high-risk human papillomavirus (HPV; P<0.01) and had no bacterial STIs (P=0.15). Statistically significant trends in prevalence of viral STIs were found from low prevalence in cluster R-I, to higher prevalence in clusters R-II and R-V, and highest prevalence in clusters R-III/R-IV/R-VI. Furthermore, only 10% of HIV-positive women in clusters R-I/R-II, compared with 40% in cluster R-V, and 42% in clusters R-III/R-IV/R-VI had detectable cervicovaginal HIV-1 RNA (Ptrend=0.03). We conclude that L. crispatus-dominated, and to a lesser extent L. iners-dominated, cervicovaginal microbiota are associated with a lower prevalence of HIV/STIs and a lower likelihood of genital HIV-1 RNA shedding.
Keywords: bacterial vaginosis, cervicovaginal HIV-1 RNA, cervicovaginal microbiome, HIV, sexually transmitted infections, Rwanda
Introduction
A healthy cervicovaginal microbiome consists predominantly of lactobacilli, which are thought to restrict growth of pathogenic bacteria by maintaining a low vaginal pH (Hickey et al., 2012, Petrova et al., 2013). Clinical conditions associated with an altered cervicovaginal microbiome include bacterial vaginosis (BV) and vaginal candidiasis. BV is typically diagnosed using Amsel criteria (Amsel et al., 1983) or Gram stain Nugent scoring (Nugent et al., 1991) and vaginal candidiasis by microscopy and/or culture of Candida species.
The human cervicovaginal microbiome has an important role in protecting women and neonates from diseases, such as pelvic inflammatory disease (Taylor et al., 2013), preterm birth (Li et al., 2012) and sepsis (Verani et al., 2010). Current epidemiological evidence using the above-mentioned diagnostic methods also suggests that any deviation from a lactobacilli-dominated cervicovaginal microbiome increases women's susceptibility to HIV and, in HIV-positive women, genital HIV shedding (Sha et al., 2005; van de Wijgert et al., 2008; Hayes et al., 2010). Furthermore, relationships between BV by Nugent scoring and herpes simplex virus type 2 (HSV-2; Cherpes et al., 2003), Neisseria gonorrhoeae, Chlamydia trachomatis (Wiesenfeld et al., 2003) and Trichomonas vaginalis (Rathod et al., 2011) have been described.
In the last decade, phylogenetic analyses of cervicovaginal samples (mostly bacterial 16S ribosomal RNA gene sequencing) have shown that bacterial communities in the vagina are more complex than previously thought (Hummelen et al., 2010; Ling et al., 2010; Ravel et al., 2011; Schellenberg et al., 2011; Srinivasan et al., 2012). American studies identified five microbiome clusters (dominated by Lactobacillus iners, L. crispatus, L. gasseri, L. jensenii and a mixture of anaerobic bacteria, respectively) in asymptomatic women (Ravel et al., 2011; Gajer et al., 2012) and multiple clusters consisting of mixtures of anaerobic bacteria, not dominated by a single taxon, in women with BV (Srinivasan et al., 2012). These studies have led to the view that BV is a condition characterized by polymicrobial dysbiosis (Hickey et al., 2012).
Molecular methods to study the cervicovaginal microbiome have not yet been used in large epidemiological studies with clinical outcomes due to limited availability and high costs. A few exploratory studies have described differences between HIV-positive and HIV-negative women, and most found trends toward decreased lactobacilli and increased bacterial diversity (Spear et al., 2008; Mitchell et al., 2009; Dols et al., 2011; Pépin et al., 2011; Schellenberg et al., 2011; Dols et al., 2012; Mitchell et al., 2013).
The long-term consequences of cervicovaginal dysbiosis are the most prevalent in resource-poor countries, African countries in particular, but comprehensive molecular microbiome data from these countries are scarce. We, therefore, sought to describe cervicovaginal microbiome compositions of women at high risk of HIV in Kigali, Rwanda, using a phylogenetic microarray, and to correlate these compositions with the presence of HIV, other sexually transmitted infections (STIs), clinical signs and symptoms, and demographic and behavioral characteristics.
Materials and methods
Study design
The Kigali HIV Incidence Study (KHIS) was a prospective cohort study conducted in 2007–2008 at Rinda Ubuzima, a non-governmental research clinic in Kigali, Rwanda. The study was approved by the National Ethics Committee, Rwanda, and the Columbia University Medical Center Review Board, USA. All participants provided written informed consent. The study design and procedures were described elsewhere (Braunstein et al., 2011; Supplementary Information). Briefly, 800 female sex workers were screened for HIV and 397 HIV-negative women were retested every three months for 1 year to determine the HIV incidence. All cohort participants, plus 141 women who tested HIV-positive at screening, were seen again once during the second year. Socio-demographic, behavioral and clinical data were collected by face-to-face interviewing at all visits. Pelvic examinations, including cervical sampling for microarray analyses, were done at the month-6 (M6) and year-2 (Yr2) visits. Diagnostic tests for STIs, BV (Amsel and Nugent), candidiasis (microscopy), pregnancy and cervical cytology were conducted at regular intervals throughout the study (Supplementary Information). All participants received counseling and non-spermicidal condoms free of charge. Women who tested positive for curable STIs were treated by study clinicians. Women who tested HIV-positive received a CD4 count. Women who tested positive for HIV or pregnancy, or had abnormal cervical cytology, were referred to public clinics providing appropriate care.
Cervical samples were taken from all women who attended the M6 and Yr2 visits, and 202 of these 719 samples were selected for microarray testing. To increase our statistical power for comparing microbiota of women with and without genital pathogens, we oversampled women with HIV and other STIs, but we also included a random sample of women without genital pathogens. All women contributed one sample, except for 28 women who contributed two samples each. In total, 6 out of 202 samples did not show appropriate positive control signals, resulting in the availability of 196 samples of 174 women for analysis. All 196 samples were used for the clustering and ecological analyses, but only one sample per participant (for a total of 174 samples) was used for all other analyses.
Microarray design
The microarray contained 461 DNA hybridization probes targeting microorganisms and 164 positive (16S conserved regions) and negative controls (Supplementary Information) (Dols et al., 2011, 2012). Of the 251 probes that generated a consistent signal with a signal/background (S/B) ratio >5, 66 16S probes were species-specific, 56 16S probes targeted multiple bacterial species within one genus, 36 16S probes were specific at family or order level, 69 targeted higher taxonomic levels, 5 were groEL probes, 16 were 18S probes and 3 were viral probes. We focused our clustering analyses on these 251 probes, and all additional analyses on the 122 16S probes generating species or genus-specific signals. A probe targeting a bacterium classified by the Ribosomal Database Project as an uncultured bacterium in the Lachnospiraceae family matched perfectly with a bacterium recently named BV-associated bacterium 1 (BVAB1) in Genbank (Genbank entry AY724739.1) (Fredricks et al., 2005). We refer to it as BVAB1 and included it in the 122 species/genus-specific probes. We made a few other probe assumptions on the basis of existing knowledge about bacterial presence in the vagina (Supplementary Information).
We used normalized S/B ratios to estimate bacterial loads, referred to from here onward as ‘semi-quantitative abundance' or ‘abundance'. For probes targeting different species within one genus, S/B ratios were summed to calculate genera-based microbial ecology parameters (see further). For probes targeting the same genus, we chose one probe that targeted the most species within that genus. For total abundance per sample, we summed genus-specific abundance of all genera present in that sample.
Microarray testing and data processing
DNA was extracted using the AGOWA mag Mini DNA isolation kit (AGOWA GmbH, Berlin, Germany) and bead beating in a BeadBeater (BioSpec Products Inc., Bartlesville, OK, USA; Supplementary Information). Microarray sample preparation, labeling, amplification and hybridization were described previously (Dols et al., 2011; Supplementary Information).
Imagene 5.6 software (BioDiscovery, Marina del Rey, CA, USA) was used to quantify the signal (S) by calculating the mean of all pixel values in the spot, as well as the background (B) surrounding the spot. If S was not confidently above B (S>B+2*s.d. of B) (Quackenbush, 2002), the S/B ratio was set at 1. Samples for which the positive controls showed a low S/B ratio (mean S/B ratio >2*s.d. lower than the mean level of positive controls of all samples) were excluded from the analysis. Lowess smoothing was performed for slide normalization (Quackenbush, 2002).
HIV-1 RNA in cervicovaginal lavages
The pelvic examination at the Y2 visit included a cervicovaginal lavage (CVL): the left and right fornix and cervical os were irrigated twice with 5 ml normal saline, which was aspirated after 30 s; a median volume of 5.5 ml (range 3.8–7.5 ml) was recovered. The CVL fluid was immediately placed on ice or at 2–8 °C, and centrifuged at 1000 rpm for 10 min within 4 h of collection. Cell pellets and aliquots of supernatant were stored at −80 °C until testing. Of the 64 HIV-positive women with a microarray result, 61 had a CVL supernatant sample for HIV-1 RNA viral load testing available, and 58 had valid test results. In addition, 50 HIV-positive women without a microarray result had a valid HIV-1 RNA test result. In total, 500 μl CVL fluid was mixed with 500 μl PBS and subjected to nucleic acid isolation, amplification and detection using COBAS AmpliPrep/COBAS Taqman v2.0 according to the manufacturer's instructions (Roche Molecular Systems, Branchburg, NJ, USA).
Statistical analysis
Statistical analyses were performed using Python 2.7 (Python Software Foundation, Beaverton, OR, USA), MATLAB (R2012a, The MathWorks, Natick, MA, USA), STATA release 12 (StataCorp, College Station, TX, USA) and MS Excel (Microsoft, Redmond, WA, USA).
We used neighborhood co-regularized multi-view spectral clustering of normalized log2-transformed S/B ratios to identify cervicovaginal microbiome clusters (Ng et al., 2001; Kumar et al., 2011; Tsivtsivadze et al., 2013). This modified spectral method generates and combines multiple clustering possibilities and leads to a result that is more robust compared with that of standard clustering techniques. Furthermore, the method captures complex neighborhood-based interactions in the dataset. We used Gaussian similarity measures to calculate a co-occurrence matrix and to identify the number of clusters in the dataset (Strehl and Ghosh, 2003). For additional analyses using the microbiome clusters as the outcome or main predictor, we only included women who had more than 70% probability of belonging to a cluster as determined by probabilistic decomposition of the co-occurrence matrix (147 of 174 women).
We used previously described methods to determine microbial ecology parameters per sample: richness, defined as the median number of genera per sample (Marzorati et al., 2008); the Shannon diversity index (Shannon, 1948); and evenness, expressed as a Co-value with 0 representing complete evenness and 100 as complete unevenness (Marzorati et al., 2013). We focused the evenness calculations on the five most abundant bacteria in each cluster to reduce the influence of the long tail of minority species (Marzorati et al., 2013). To compare cumulative Co-values per cluster, an average sample per cluster was generated by calculating median S/B ratios per genus across the samples in that cluster.
To assess positive and negative correlations between bacteria, we first determined Spearman correlation coefficients (with Bonferroni correction) between genera, followed by those between species within the genera that were statistically significantly correlated plus species that are not yet classified at genus level, such as BVAB1. To correlate microarray findings with Nugent scores, the S/B ratios of 12 Lactobacillus probes (covering 70 species) were summed, the S/B ratios of one G. vaginalis probe and one Bacteroides fragilis probe were summed and one M. mulieris probe was used.
Characteristics of the total study population, women included in the microarray experiments and women in each cervicovaginal microbiome cluster were described using the median and interquartile range (IQR) for continuous data and counts and percentages for categorical data. Women in clusters R-III, R-IV and R-VI (each characterized by a high diversity of anaerobic bacteria) were pooled in some analyses to increase statistical power. Differences in correlates between clusters, and between samples assigned and those not assigned to clusters, were assessed by two-sided Kruskal–Wallis test for continuous data, Fishers' exact test for categorical data, and the ‘nptrend' function in STATA, an extension of the Wilcoxon rank-sum test, for trends (Cuzick, 1985).
Determinants of detectable HIV-1 RNA in CVL samples, including microarray clusters, were assessed by logistic regression. In these analyses, women in clusters R-I and R-II were pooled because only one woman in cluster R-I was HIV-positive. All models were adjusted for plasma CD4 count because advanced HIV infection (characterized by low-CD4 count) is associated with high plasma and genital viral load.
Results
Study sample
The prevalence of HIV in the KHIS study at screening was 24%, and 19 participants seroconverted during follow-up (Braunstein et al., 2011). Other viral and bacterial STIs, BV and candidiasis were common (Table 1). By design, the 174 women selected for microarray analyses had a higher prevalence of HIV, STIs and pregnancy than the original sample of 800 women. The prevalence of BV by Amsel criteria or Nugent scoring was not different between the microarray sample and the original sample.
Table 1. Participant characteristics: total study population versus microarray sample.
| Total study population at screening (n=800) | Microarray sample (n=174) | |
|---|---|---|
| Demographics | ||
| Median age (range); years | 25 (18–45) | 27 (18–47) |
| Marital status: married or steady partner | 278 (34.9) | 68 (39.1) |
| Divorced/separated/widowed | 210 (26.3) | 40 (23) |
| Never married | 584 (73.0) | 131 (75.3) |
| Median income per week (IQR); × 1000 RWFa | 15 (8–23) | 12 (8–20) |
| Education levela: no education | 166 (20.9) | 43 (24.7) |
| Some primary school but not completed | 326 (41.1) | 71 (40.8) |
| Completed primary and/or started secondary | 301 (38.0) | 60 (34.5) |
| Currently pregnant | 61 (7.6) | 22 (12.6) |
| Median number of lifetime pregnancies (IQR)a | 2 (1–3) | 2 (1–3) |
| Recent change in reproductive intentionsb | 7 (1.8) | 7 (4.0) |
| Sexual risk taking | ||
| Median years in sex worka,c (IQR) | 3 (2–5) | 3 (2–5) |
| Currently working as sex worker | 787 (99.4) | 148 (85.1) |
| Washed inside vagina in last 3 monthsb | 355 (92.9) | 160 (92) |
| Median frequency of washing inside vagina in week prior to enrollment (IQR)a | 7 (7–12) | 7 (7–7) |
| Median number of sex acts in past month (IQR) | 40 (20–64) | 20 (12–40) |
| Condom use at last sex acta | 593 (74.1) | 127 (73.0) |
| Consistent condom useb,d | 173 (46.5) | 60 (37.3) |
| Clinical characteristics | ||
| Genitourinary symptoms in month prior to enrollmenta | 155 (19.8) | 39 (23.1) |
| Genitourinary symptoms in last 3 monthsb | 50 (13.0) | 33 (19.0) |
| Pelvic examb: abnormal vaginal discharge | 44 (11.7) | 23 (13.2) |
| Abundant cervical mucus | 15 (4.0) | 7 (4.0) |
| Vaginal pH >4.5 | 303 (81.7) | 137 (79.2) |
| Antibiotics use during past 14 days | 1 (0.1) | 1 (0.6) |
| Laboratory diagnoses | ||
| Any viral STI | 510 (65.6) | 154 (88.5) |
| HIV | 192 (24.0) | 64 (36.8) |
| HSV-2 | 485 (60.6) | 137 (78.7) |
| HPV | 183 (48.8) | 83 (47.7) |
| Any bacterial STI (HIV-negative women only)b,e | 105 (28.9) | 38 (51.4) |
| Treponema pallidum | 25 (6.6) | 6 (8.7) |
| Neisseria gonorrhoeae | 29 (7.9) | 12 (16.7) |
| Chlamydia trachomatis | 17 (4.6) | 9 (12.3) |
| Trichomonas vaginalis | 61 (16.2) | 22 (26.5) |
| Abnormal cervical cytologyb,f | 10 (2.8) | 10 (6.2) |
| Wet mountb | ||
| Yeasts | 17 (4.5) | 11 (6.3) |
| Positive whiff test | 25 (6.6) | 8 (4.6) |
| Clue cells >20% | 65 (17.2) | 26 (14.9) |
| Bacterial vaginosis by Nugent scoringb,g | ||
| Negative (0–3) | 159 (44.8) | 73 (42.4) |
| Intermediate (4–6) | 53 (14.9) | 19 (11) |
| Positive (7–10) | 143 (40.3) | 80 (46.5) |
Abbreviations: HPV, human papillomavirus; HSV, herpes simplex virus; IQR, interquartile range; RWF, Rwandan francs; STI, sexually transmitted infection.
Cells represent number and percentage unless stated otherwise.
Baseline values are reported.
Only assessed during follow-up; values reported for the HIV-negative cohort at M6 (n=387).
47/800 values of total population and 11/174 of microarray-tested women missing.
Sexually active women only.
Only tested in HIV-negative cohort at M6 (n=363, of which 83 microarray-tested).
12/174 values of microarray-tested women missing.
32/387 values missing.
Most of the selected women (median age 27, range 18–47) had never been married (75%) and did not complete primary school (66%) (Table 1). Although almost all women (99%) self-identified as sex worker and had worked as a sex worker for a median of 3 years (IQR=2–5), 14% left sex work during the follow-up (Ingabire et al., 2012). Almost all women (92%) reported vaginal finger-cleansing using water or water and soap, and 37% reported consistent condom use. Twenty-two women (13%) were pregnant at the time of sampling.
Microarray probe hybridization
The 122 species/genus-specific 16S probes represented 314 species from 51 genera, 32 families and 17 orders. On average, samples contained species from 11 genera (range 0–25). Most samples contained lactobacilli (98% of samples), including L. iners (74%), L. crispatus (16%), L. jensenii/L. salivarius/other (6%), L. gasseri/L. johnsonii/other (6%) and L. vaginalis/other (21%) (see Supplementary Information for other Lactobacillus spp. targeted by these probes). Leptotrichia (94%), Prevotella (91%), Corynebacterium (90%) and Gardnerella species (82%) were also present in most samples. However, lactobacilli other than L. iners and L. crispatus, Leptotrichia and Corynebacterium species were present in low abundance, and did therefore not play a significant role in the clustering (see further). The overall presence of common BV-associated anaerobes was Atopobium (65% of samples), Dialister (61%), BVAB1 (50%), Mobiluncus (48%), Sneathia (47%) and Megasphaera (44%), but their presence was low in the lactobacilli-dominated clusters and approached 100% in BV-associated clusters (see further). No or low signals were obtained for the Bifidobacteriaceae family other than Gardnerella, Bacteroides, Escherichia coli, Enterococcus, Streptococcus and Staphylococcus species.
Cervicovaginal microbiome clusters
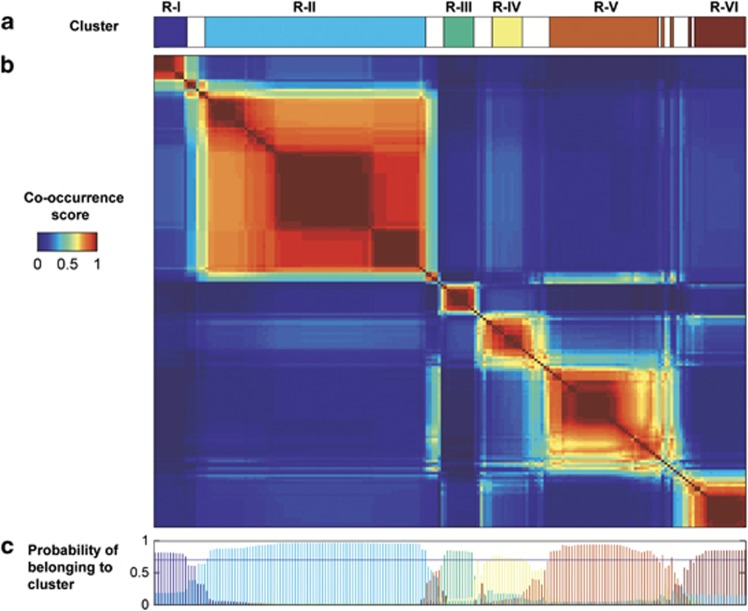
We identified six cervicovaginal microbiome clusters by neighborhood co-regularized multi-view spectral clustering, using data from 196 samples and 251 probes as described above (Figure 1a). The co-occurrence matrix (Figure 1b), which shows how often samples co-occur in multiple spectral clustering analysis runs, visualizes these six clusters. We refer to these clusters as R-I to R-VI (with the R denoting ‘Rwanda'). The probabilistic decomposition of this matrix (Figure 1c), which returns the probability of each sample belonging to a specific cluster, confirms the presence of these six clusters. A total of 36 samples from 27 women had a probability of <70% of belonging to one of these six clusters. Samples that were not assigned (shown in white on the top bar of Figure 1a) fell between any two clusters and had little in common. The 27 women in the unassigned group had fewer lifetime pregnancies, and reported consistent condom use more often, but were otherwise similar to the 147 women in the other groups (Supplementary Table S1).
Figure 1.
Co-occurrence matrix and microbiome clustering. (a) shows the six clusters obtained by Neighborhood Co-regularized Multi-view Spectral Clustering of microarray data. The white spaces between the clusters represent samples with less than 70% probability of belonging to a cluster (see also c). The co-occurrence score reflects how many times samples co-occurred in different clustering configurations. The matrix (b) shows all 196 samples on both the x- and y-axis in the same order. Only samples with at least 70% probability of belonging to a cluster are assigned to a cluster (c).
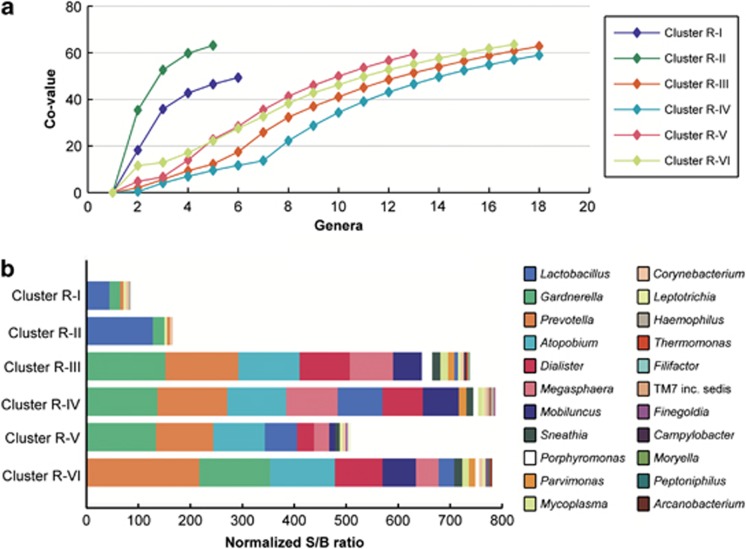
We used microbial ecological parameters (Figure 2) and a heatmap (Figure 3) to characterize each cluster. Cluster R-1 was dominated by L. crispatus and cluster R-II by L. iners. Clusters R-III to R-VI were not dominated by one taxa, but contained several (facultative) anaerobes, with or without L. iners, in different compositions (see further). This was reflected in the evenness of the five most abundant genera, which was lower in clusters R-I and R-II compared with the other clusters (cumulative Co-values of 47 and 63, compared with 12, 9, 23 and 22, respectively; Figure 2a). When comparing clusters R-I and R-II to clusters R-III, R-IV, R-V and R-VI, the total semi-quantitative abundances and richness were lower (a median of 5 and 6 genera per sample compared with 18, 18, 14 and 17 genera, respectively; Figure 2b).
Figure 2.
Distribution of the most abundant genera per cluster. (a) shows the cumulative Co-values for the most abundant genera in each cluster in descending order. Note that BVAB1 is not included because it is not yet described at genus level. Although the final cumulative Co-value of each cluster is around 60, clusters R-I and R-II have the most uneven distribution of their most abundant genera. (b) shows these most abundant genera per cluster, and highlights the dominance of lactobacilli in clusters R-I and R-II compared with a more even distribution of anaerobic genera in the other clusters. Also, large differences in richness and total S/B ratio (abundance) are apparent, with clusters R-I and R-II containing fewer genera and lower abundance than the other clusters.
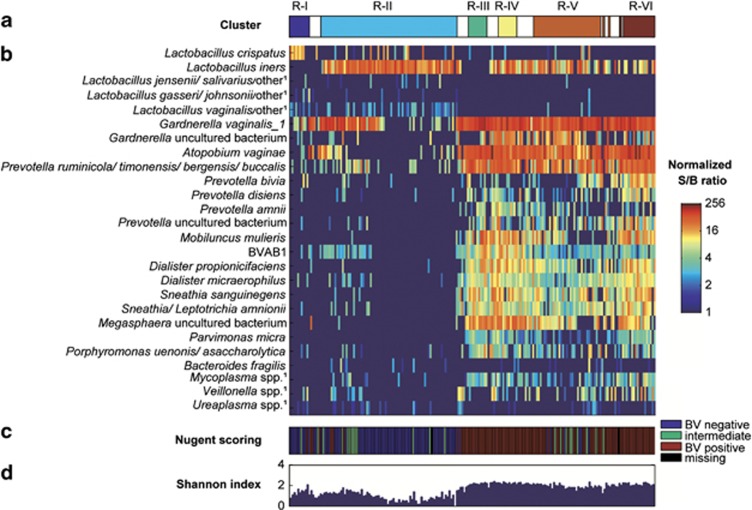
Figure 3.
Bacterial composition of the microbiome clusters. (a) shows the six clusters obtained by Neighborhood Co-regularized Multi-view Spectral Clustering of microarray data. The white spaces between the clusters represent samples with less than 70% probability of belonging to a cluster. (b) Heatmap, showing normalized S/B ratios on a log2 logarithmic scale of the most abundant species/genera per cluster, as well as species/genera that have traditionally been associated with BV and have been frequently reported in the literature. (c) BV-status of the women by Nugent score at the time of sampling. The color key is on the right. (d) The Shannon diversity index for each sample. 1Abbreviated probe name; additional targeted species in the same genus are listed in Supplementary Information.
Clusters R-III to R-VI contained high abundances of Gardnerella, Prevotella and Atopobium species and lower abundances of Dialister, Megasphaera and Mobiluncus species and BVAB1 (Figures 2 and 3). BVAB1 was mostly present in clusters R-III and R-IV. Cluster R-III's unique feature was the presence of a lower abundance of L. iners than the other mixed anaerobic clusters. Clusters R-IV and R-VI included the above-mentioned anaerobes and L. iners, but R-IV was the only cluster containing high abundance of an uncultured bacterium in the Gardnerella genus (see Supplementary Information) and R-VI contained the highest levels of Prevotella species, including P. bivia. Cluster R-V had a lower total bacterial abundance than the other mixed anaerobic clusters.
Correlations between species and with diagnostic test results
Most anaerobic bacteria were positively correlated with each other but negatively correlated with Lactobacillus species (Supplementary Figure S1; Supplementary Information).
The microarray clusters correlated poorly with the Amsel criteria (Table 2). Only 14–33% of women in the mixed anaerobic microbiome clusters had >20% clue cells present on wet mount, and 11–14% had a positive whiff test. Furthermore, while 97–100% of women in the mixed anaerobic clusters had a vaginal pH >4.5, 55–60% of women in lactobacilli-dominated clusters also did. In contrast, the clusters correlated well with Nugent scores (Table 2). Clusters R-I and R-II were statistically significantly associated with a normal Nugent score of 0–3 and clusters R-III to R-VI with a BV Nugent score of 7–10. Women with intermediate microbiota (Nugent score 4–6) did not cluster together but were mostly included in clusters R-II and R-V. The probes representing the morphotypes included in Nugent scoring (G. vaginalis and Bacteroides fragilis combined, M. mulieris and 12 lactobacilli probes covering 70 species) also correlated well with the Nugent scores (all P<0.001; Supplementary Figure S2).
Table 2. Amsel criteria and Nugent scores by cervicovaginal microbiome cluster.
| R-I (n=11) | R-II (n=69) | R-III (n=9) | R-IV (n=9) | R-V (n=35) | R-VI (n=14) | P-value | |
|---|---|---|---|---|---|---|---|
| Amsel criteria | |||||||
| Clue cells >20% | 0 (0) | 5 (7.2) | 2 (22.2) | 3 (33.3) | 5 (14.3) | 2 (14.3) | 0.105 |
| Positive whiff test | 0 (0) | 0 (0) | 1 (11.1) | 1 (11.1) | 1 (2.9) | 2 (14.3) | 0.018 |
| Vaginal pH >4.5 | 6 (54.6) | 41 (60.3) | 9 (100) | 9 (100) | 34 (97.1) | 14 (100) | <0.001 |
| Nugent scores | |||||||
| Normal (0–3) | 9 (81.8) | 55 (80.9) | 0 (0) | 0 (0) | 6 (17.1) | 0 (0) | <0.001 |
| Intermediate (4–6) | 1 (9.1) | 10 (14.7) | 0 (0) | 0 (0) | 8 (22.9) | 0 (0) | |
| BV (7–10) | 1 (9.1) | 3 (4.4) | 9 (100) | 9 (100) | 21 (60) | 14 (100) | |
Abbreviations: BV, bacterial vaginosis; R, Rwanda.
Cells represent number and percentage. Differences between clusters were tested for statistical significance using Fishers' exact test.
Socio-demographic, behavioral and HIV/STI correlates of cervicovaginal microbiome clusters
Women belonging to each of the six clusters had similar socio-demographic and behavioral characteristics, except that women in the lactobacilli clusters were more likely to ever having been married than women in the mixed anaerobic clusters (Table 3 and Supplementary Table S1). Women in the pooled cluster R-III/R-IV/R-VI were significantly more likely to report genitourinary symptoms on the day of sampling than women in the other clusters. No significant differences in pelvic exam findings were found.
Table 3. Cervicovaginal microbiome cluster correlates.
| Demographics | R-I (n=11) | R-II (n=69) | R-V (n=35) | R-III/IV/VI (n=32) | Pa | Ptrendb |
|---|---|---|---|---|---|---|
| Median age (range); years | 29 (23–37) | 27 (19–47) | 26 (18–42) | 27 (20–44) | 0.44 | 0.28 |
| Marital status: married or steady partner | 5 (45.5) | 30 (43.5) | 18 (51.4) | 7 (21.9) | 0.07 | 0.1 |
| Divorced/separated/widowed | 2 (18.2) | 17 (24.6) | 6 (17.1) | 7 (21.9) | 0.87 | 0.8 |
| Never married | 7 (63.6) | 48 (69.6) | 27 (77.1) | 29 (90.6) | 0.08 | 0.02 |
| Median income/week (IQR); × 1000 RWFc | 10.5 (4–17) | 12 (7.5–22) | 15 (8–20) | 10 (6.5–15) | 0.46 | 0.93 |
| Education:cNo education | 2 (18.9) | 14 (20.3) | 10 (28.6) | 9 (28.1) | 0.96 | 0.38 |
| Some primary but not completed | 5 (45.5) | 30 (43.5) | 13 (37.1) | 13 (40.6) | ||
| Completed primary and/or started secondary | 4 (36.4) | 25 (36.2) | 12 (34.3) | 10 (31.3) | ||
| Currently pregnant | 0 (0) | 12 (17.4) | 4 (11.4) | 2 (6.5) | 0.33 | 0.47 |
| Median # lifetime pregnancies (IQR)c | 3 (2–4) | 2 (1–3) | 2 (1–3) | 2 (1–3) | 0.14 | 0.09 |
| Sexual risk taking | ||||||
| Median years in sex work (IQR)d | 3 (2–3) | 3 (2–5) | 3 (2–5) | 3 (2–4) | 0.59 | 0.85 |
| Currently working as sex worker | 10 (90.9) | 57 (82.6) | 31 (88.6) | 27 (84.4) | 0.87 | 0.97 |
| Washed inside vagina in last 3 months | 9 (81.8) | 62 (89.9) | 33 (94.3) | 31 (96.9) | 0.31 | 0.09 |
| Consistent condom usee | 5 (45.5) | 18 (28.6) | 10 (29.4) | 13 (46.4) | 0.29 | 0.39 |
| Clinical characteristics | ||||||
| Genitourinary symptoms in last 3 months: | ||||||
| Genital itching | 3 (27.3) | 7 (10.1) | 3 (8.6) | 6 (18.8) | 0.23 | 0.87 |
| Other specific genital complaintsf | 1 (9.1) | 5 (7.3) | 3 (8.6) | 3 (9.4) | 0.97 | 0.79 |
| Dysuria | 0 (0) | 1 (1.4) | 2 (5.7) | 0 (0) | 0.45 | 0.9 |
| Genitourinary symptoms at day of samplingg | 0 (0) | 0 (0) | 0 (0) | 3 (12.5) | 0.06 | 0.02 |
| Pelvic exam: abnormal vaginal discharge | 1 (9.1) | 9 (13) | 5 (14.3) | 4 (12.5) | 1 | 0.87 |
| Abundant cervical mucus | 0 (0) | 2 (2.9) | 3 (8.6) | 2 (6.3) | 0.56 | 0.23 |
| Antibiotics use during past 14 days | 0 (0) | 1 (1.5) | 0 (0) | 0 (0) | 0.87 | 0.47 |
| Laboratory diagnoses | ||||||
| Any viral STI | 4 (36.4) | 62 (89.9) | 33 (94.3) | 30 (93.8) | <0.01 | <0.01 |
| HIV | 1 (9.1) | 23 (33.3) | 12 (34.3) | 18 (56.3) | 0.03 | <0.01 |
| HSV-2 | 4 (36.4) | 54 (78.3) | 29 (82.9) | 28 (87.5) | <0.01 | <0.01 |
| Any HPV type | 1 (9.1) | 29 (42) | 21 (60) | 17 (53.1) | 0.02 | 0.02 |
| High-risk HPV | 0 (0) | 26 (37.7) | 19 (54.3) | 15 (46.9) | <0.01 | 0.02 |
| Any bacterial STIh | 0 (0) | 20 (60.6) | 6 (37.5) | 5 (55.6) | 0.15 | 0.94 |
| Treponema pallidum | 0 (0) | 4 (12.9) | 1 (5.9) | 1 (14.3) | 0.79 | 0.95 |
| Neisseria gonorrhoeae | 0 (0) | 5 (15.6) | 2 (12.5) | 2 (25.0) | 0.86 | 0.47 |
| Chlamydia trachomatis | 0 (0) | 6 (18.8) | 2 (11.8) | 1 (12.5) | 0.93 | 0.82 |
| Trichomonas vaginalis | 0 (0) | 9 (25) | 2 (10.5) | 4 (40) | 0.28 | 0.45 |
| Candidiasis by wet mount | 1 (9.1) | 6 (8.7) | 3 (8.6) | 0 (0) | 0.29 | 0.15 |
| Abnormal cervical cytologyi | 0 (0) | 2 (3) | 4 (13.3) | 3 (10.3) | 0.17 | 0.07 |
Abbreviations: HPV, human papillomavirus; HSV, herpes simplex virus; IQR, interquartile range; RWF, Rwandan francs; STI, sexually transmitted infection.
Cells represent number and percentage unless stated otherwise.
Kruskall–Wallis test for continuous data and Fishers' exact test for categorical data.
Trends in the order R-I, R-II, R-V, R-III/R-IV/R-VI (see methods).
Baseline values reported.
10/147 values missing.
Sexually active women only.
Genital burning/pain/ulcers/sores/blisters/unusual pain during sex.
Only reported at Yr2 (n=86).
Only tested at M6 in the HIV-negative cohort (n=58, 68, 59 and 60, respectively).
12/147 values missing.
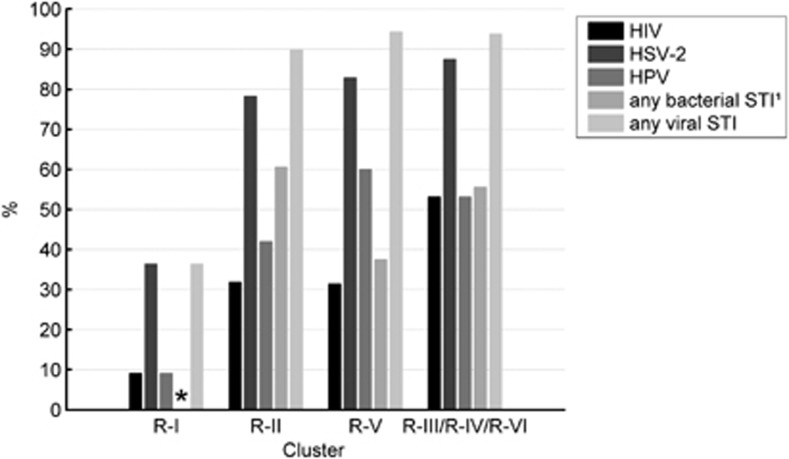
Women in the L. crispatus-dominated cluster R-I were statistically significantly less likely to have HIV, HSV-2, any HPV and high-risk HPV than women in the other clusters, and had no bacterial STIs (Table 3; Figure 4). Only one woman in cluster R-I was HIV-positive (9%) compared with 33–56% of women in the other clusters (P=0.03). The percentages were 36% versus 78–88% for HSV-2 (P<0.01), 9% versus 42–60% for any HPV (P=0.02), 0% versus 38–54% for high-risk HPV (P<0.01) and 0% versus 38–61% for bacterial STIs (T. pallidum, N. gonorrhoeae, C. trachomatis and T. vaginalis combined; P=0.15). Statistically significant trends in prevalence of HIV and other STIs were found from low prevalence in cluster R-I to higher prevalence in clusters R-II and R-V, and highest prevalence in the pooled cluster R-III/R-IV/R-VI (Table 3; Figure 4).
Figure 4.
Associations between microbiome clusters and HIV/STI prevalence. Women in the Lactobacillus crispatus-dominated cluster R-I had a lower prevalence of viral STIs than women in the other clusters (P<0.01) and had no bacterial STIs (P=0.15). A trend of increasing prevalence of viral STIs in clusters with increasing bacterial diversity was found, with the lowest prevalence in cluster R-I and increasing prevalence in clusters R-II, R-V and R-III/R-IV/R-VI, respectively (Ptrend<0.01). *No bacterial STIs were found in women assigned to cluster R-I. 1Only tested at M6 in the HIV-negative cohort; data available for 61 women.
Microbiome clusters and HIV-1 RNA genital tract shedding
The prevalence of HIV-1 RNA in CVLs of HIV-positive women also increased with increasing microbiome diversity: 10% (two women) in the pooled cluster R-I/R-II, 40% (four women) in cluster R-V and 42% (eight women) in the pooled cluster R-III/R-IV/R-VI (Ptrend=0.03). The adjusted odds of having detectable HIV-1 RNA (adjusted for CD4 count) was 8.78 (95% confidence interval (CI)=1.12–69.09) times higher for women in cluster R-V and 5.29 (95% CI=0.91–30.67) times higher for women in the pooled cluster R-III/R-IV/R-VI compared with women in the pooled cluster R-I/R-II. After adjusting for CD4 count, positive correlations were found between HIV-1 RNA concentrations and abundance of several BV-associated probes and a negative association with a Lactobacillus genus probe (Supplementary Information).
In bi-variable logistic regression models using data from 108 HIV-positive women with an available HIV-1 RNA measurement, other factors associated with detectable genital HIV-1 RNA levels were genital itching (odds ratio (OR)=17.55, 95% CI=2.10–146.46), other specific genital symptoms (100 versus 31%, P<0.001), abundant cervical mucus on pelvic exam (OR=8.00, 95% CI=0.86–74.37), a BV Nugent score of 7–10 (OR=4.67, 95% CI=1.44–15.16), not using antiretroviral therapy (ART, data available for 101 women, OR=8.71, 95% CI=2.76–27.5) and HPV infection (OR=3.62, 95% CI=1.25–10.48), but not HSV-2 antibodies. Similar proportions of women in each cluster used ART (42%, 43% and 31% of women in R-I/R-II, R-V and R-III/R-IV/R-VI, respectively; Ptrend 0.5) and clustering was not associated with ART.
Discussion
We identified six cervicovaginal microbiome clusters in Rwandan sex workers. To date, 10 studies have described clusters on the basis of next generation sequencing data (Forney et al., 2010; Hummelen et al., 2010; Ravel et al., 2011; Schellenberg et al., 2011; Frank et al., 2012; Martin et al., 2012; Smith et al., 2012; Srinivasan et al., 2012; Drell et al., 2013; Lee et al., 2013). Even though these studies included different study populations and employed a variety of molecular and clustering procedures, consistent clustering patterns can be discerned. The majority of studies, including ours, found one cluster dominated by L. crispatus and one by L. iners. Clusters dominated by L. jensenii or L. gasseri were reported less frequently, and we did not find them in our study. All studies identified at least one cluster that was not dominated by a single taxon, but contained mixtures of anaerobes with or without Lactobacillus species. Clusters dominated by facultative anaerobic organisms, including streptococci, staphylococci and E. coli/Shigella species, were rarely reported, but the above-mentioned next generation sequencing studies suggest that these taxa are present in up to 30% of women in low abundance. This is also in agreement with our findings. Based on the above, we consider BV (represented by our clusters III to VI) as a state of polymicrobial dysbiosis. We speculate that whether the dysbiosis is symptomatic or not depends on the degree and nature of the dysbiosis, total bacterial loads, and the intensity and nature of the host's immune responses. Although most BV-associated bacteria are not pathogenic in immune-competent hosts, we also speculate that some (e.g., streptococci and E. coli) might lead to invasive disease when present at sufficiently high levels. However, none of this has been definitively shown in clinical studies to date.
Our study showed that women in the L. crispatus cluster had the lowest prevalence of HIV/STIs, with a slight increase in the L. iners cluster and a significant increase in the dysbiotic clusters. A similar trend was found for HIV-1 RNA shedding in the genital tract of HIV-positive women. These findings are in agreement with the majority of studies that have also investigated these associations (Spear et al., 2008; Mitchell et al., 2009; Dols et al., 2011; Pépin et al., 2011; Schellenberg et al., 2011; Dols et al., 2012; Mitchell et al., 2013), but we are the first to report these associations for multiple STI pathogens and to demonstrate a dose-response relationship. It is important to note that the L. crispatus cluster only contained 11 women, and that the temporality of our findings is unclear because our study was cross-sectional. Our results, if confirmed in prospective studies, might imply that not only symptomatic dysbiosis should be treated, but also asymptomatic dysbiosis (defined here as a microbiome not dominated by lactobacilli) in certain risk groups. These risk groups might include women at high risk for HIV/STIs or adverse pregnancy outcomes and women with recurrent dysbiosis.
Laboratory studies also suggest that L. crispatus might protect against pathogens. L. crispatus is an efficient lactic acid producer (Hickey et al., 2012), produces antimicrobial compounds (Graver and Wade, 2011; Hickey et al., 2012; Aldunate et al., 2013; Petrova et al., 2013) and inhibits inflammation (Rose et al., 2012; Petrova et al., 2013). The latter is particularly important in the context of HIV transmission, as HIV infects CD4+-immune cells that are recruited to the genital mucosa when inflammation is present. In contrast, BV-associated bacteria could increase HIV-infection risk and HIV replication in the genital mucosa of HIV-infected women, by provoking local immune activation and/or disruption of the vaginal epithelium (Sha et al., 2005; Marconi et al., 2013; Mitchell et al., 2013; Petrova et al., 2013). In vitro studies have indeed shown that some BV-associated bacteria can enhance HIV expression, translation and/or replication (Klebanoff and Coombs, 1991; Hashemi et al., 1999; Ahmed et al., 2010).
Our data support the hypothesis that L. iners is less efficient than L. crispatus in preventing BV and other adverse reproductive health outcomes (Verstraelen et al., 2009; Srinivasan et al., 2010; Gajer et al., 2012; Jespers et al., 2012; Santiago et al., 2012). Recent genomic and transcriptomic studies suggest that L. iners is highly adapted to the vaginal compartment (Macklaim et al., 2011). However, it differentially expresses over 10% of its genome in dysbiotic compared with healthy states, with increased expression of a cytolysin, mucin, glycerol transport and related metabolic enzymes (Macklaim et al., 2013). These changes likely result in the production of succinate and other short-chain fatty acids as the end product of metabolism as opposed to lactic acid, leading to an increased vaginal pH. L. iners might also be the first Lactobacillus species to recover after dysbiosis (Gajer et al., 2012), which suggests a bidirectional relationship between L. iners and vaginal pathogens or dysbiosis. Like lactobacilli, G. vaginalis and Prevotella species are almost always present in the vaginal microbiome, but in much higher abundance in BV; some studies have noted a synergistic effect between them, perhaps due to metabolic dependencies (Ling et al., 2010; Zozaya-Hinchliffe et al., 2010; Ravel et al., 2011; Jespers et al., 2012). Several subspecies of G. vaginalis have been described, with different levels of epithelial adhesion capacity (Paramel Jayaprakash et al., 2012; Castro et al., 2013). The latter is the first step toward biofilm formation, which is thought to be an important mechanism of BV persistence (Machado et al., 2013). A. vaginae, which is also present in high abundance in BV, most likely has an important role in biofilm formation as well (Machado et al., 2013).
Some limitations of our study (such as its cross-sectional nature) and the field in general (heterogeneity in populations and methodology) have already been mentioned. Other limitations of our study include small sample sizes of some of the comparison groups (most notably the limited number of bacterial STI cases despite oversampling of these cases), imprecise timing of certain behaviors around the time of sampling, lack of a control group of women at low risk for HIV and STIs and the fact that we only used cervical samples (including endo- and ectocervix). Although small differences between the cervical and vaginal microbiome have been described, the types and relative quantities of the most abundant bacteria are similar between the two sampling sites and bacterial community compositions are also similar (Nikolaitchouk et al., 2008; Kim et al., 2009; Ravel et al., 2011; Smith et al., 2012).
We used a microarray instead of next generation sequencing or quantitative PCRs for several reasons. First, microarrays can assess the presence of multiple bacteria in a semi-quantitative manner. The number of probes included on the microarray can be reduced as knowledge about optimal and suboptimal vaginal microbiota increases, which may eventually result in a diagnostic tool. The main limitations of microarrays are their inability to detect ‘new' species that are not included a priori on the microarray and the fact that they are not fully quantitative. However, to obtain fully quantitative data, one would have to perform multiple qPCR assays, which is time-consuming and expensive.
Now that vaginal microbiota of women with and without BV in different parts of the world have been well described, and molecular techniques have become more accessible and affordable, we believe that the time has come to incorporate these techniques into larger epidemiological studies with clinical outcomes. These studies should investigate the temporal relationships between cervicovaginal microbiota and adverse reproductive health outcomes, including adverse pregnancy outcomes and invasive infections in women and their neonates. They should also address other unanswered clinical questions, such as the role of bacterial loads of different types of bacteria in these adverse outcomes. At the same time, laboratory studies should further investigate the functional characteristics of different microbiome communities to improve our understanding of the etiology of dysbiosis and the pathogenesis of its clinical consequences. Eventually, interventions that restore and maintain lactobacilli-dominated microbiota, and particularly L. crispatus-dominated microbiota, should continue to be optimized and tested. If successful and affordable interventions are identified, they could potentially have a significant public health impact. For example, while studies have shown that the increased risk for HIV acquisition by cervicovaginal dysbiosis is only about 1.5–2.0 (compared to about 2.0–3.0 for STI pathogens), the overall population impact would be large because of the very high prevalence of dysbiosis (30–60%) in areas with generalized HIV epidemics (Hayes et al., 2010). Our study and other recent molecular vaginal microbiome studies have provided important new insights into the cervicovaginal microenvironment, and as a result, potential public health interventions can now be properly evaluated.
Acknowledgments
We thank the study participants, the Rinda Ubuzima study team for KHIS study implementation and Nynke van Berkum and TNO analysts for microarray testing. This work was funded by the Aids Fonds Netherlands (project number 201102), European and Developing Countries Clinical Trials Partnership (project number CT.2005.33070.001) and the European Commission (CHAARM consortium).
Disclosure
The findings and conclusions in this paper are those of the authors and do not necessarily represent the views of the funders.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ahmed N, Hayashi T, Hasegawa A, Furukawa H, Okamura N, Chida T, et al. Suppression of human immunodeficiency virus type 1 replication in macrophages by commensal bacteria preferentially stimulating Toll-like receptor 4. J Gen Virol. 2010;91:2804–2813. doi: 10.1099/vir.0.022442-0. [DOI] [PubMed] [Google Scholar]
- Aldunate M, Tyssen D, Johnson A, Zakir T, Sonza S, Moench T, et al. Vaginal concentrations of lactic acid potently inactivate HIV. J Antimicrob Chemother. 2013;68:2015–2025. doi: 10.1093/jac/dkt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsel R, Totten PA, Spiegel CA, Chen KCS, Eschenbach D, Holmes KK. Nonspecific vaginitis—diagnostic-criteria and microbial and epidemiologic associations. Am J Med. 1983;74:14–22. doi: 10.1016/0002-9343(83)91112-9. [DOI] [PubMed] [Google Scholar]
- Braunstein SL, Ingabire CM, Kestelyn E, Uwizera AU, Mwamarangwe L, Ntirushwa J, et al. High human immunodeficiency virus incidence in a cohort of Rwandan female sex workers. Sex Transm Dis. 2011;38:385–394. doi: 10.1097/olq.0b013e31820b8eba. [DOI] [PubMed] [Google Scholar]
- Castro J, Henriques A, Machado A, Henriques M, Jefferson KK, Cerca N. Reciprocal interference between Lactobacillus spp. and Gardnerella vaginalis on initial adherence to epithelial cells. Int J Med Sci. 2013;10:1193–1198. doi: 10.7150/ijms.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherpes TL, Meyn LA, Krohn MA, Lurie JG, Hillier SL. Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis. Clin Infect Dis. 2003;37:319–325. doi: 10.1086/375819. [DOI] [PubMed] [Google Scholar]
- Cuzick J. A Wilcoxon-type test for trend. Stat Med. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
- Dols JA, Reid G, Kort R, Schuren FHJ, Tempelman H, Bontekoe TR, et al. PCR-based identification of eight Lactobacillus species and 18 hr-HPV genotypes in fixed cervical samples of South African women at risk of HIV and BV. Diagn Cytopathol. 2012;40:472–477. doi: 10.1002/dc.21786. [DOI] [PubMed] [Google Scholar]
- Dols JA, Smit PW, Kort R, Reid G, Schuren FHJ, Tempelman H, et al. 2011Microarray-based identification of clinically relevant vaginal bacteria in relation to bacterial vaginosis Am J Obstet Gynecol 204305e1–e7. [DOI] [PubMed] [Google Scholar]
- Drell T, Lillsaar T, Tummeleht L, Simm J, Aaspollu A, Vain E, et al. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One. 2013;8:e54379. doi: 10.1371/journal.pone.0054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forney LJ, Gajer P, Williams CJ, Schneider GM, Koenig SS, McCulle SL, et al. Comparison of self-collected and physician-collected vaginal swabs for microbiome analysis. J Clin Microbiol. 2010;48:1741–1748. doi: 10.1128/JCM.01710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Manigart O, Leroy V, Meda N, Valéa D, Zhang W, et al. Altered vaginal microbiota are associated with perinatal mother-to-child transmission of HIV in African women from Burkina Faso. J Acquir Immune Defic Syndr. 2012;60:299–306. doi: 10.1097/QAI.0b013e31824e4bdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- Gajer P, Brotman RM, Bai GY, Sakamoto J, Schuette UME, Zhong X, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graver MA, Wade JJ. The role of acidification in the inhibition of Neisseria gonorrhoeae by vaginal lactobacilli during anaerobic growth. Ann Clin Microbiol Antimicrob. 2011;10:8. doi: 10.1186/1476-0711-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi FB, Ghassemi M, Roebuck KA, Spear GT. Activation of human immunodeficiency virus type 1 expression by Gardnerella vaginalis. J Infect Dis. 1999;179:924–930. doi: 10.1086/314674. [DOI] [PubMed] [Google Scholar]
- Hayes R, Watson-Jones D, Celum C, van de Wijgert J, Wasserheit J. Treatment of sexually transmitted infections for HIV prevention: end of the road or new beginning. AIDS. 2010;24:S15–S26. doi: 10.1097/01.aids.0000390704.35642.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. Understanding vaginal microbiome complexity from an ecological perspective. Transl Res. 2012;160:267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummelen R, Fernandes AD, Macklaim JM, Dickson RJ, Changalucha J, Gloor GB, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One. 2010;5:e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingabire C, Mitchell K, Veldhuijzen N, Umulisa M, Nyinawabega J, Kestelyn E, et al. Joining and leaving sex work: experiences of women in Kigali, Rwanda. Cult Health Sex. 2012;14:1037–1047. doi: 10.1080/13691058.2012.713120. [DOI] [PubMed] [Google Scholar]
- Jespers V, Menten J, Smet H, Poradosu S, Abdellati S, Verhelst R, et al. Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol. 2012;12:83. doi: 10.1186/1471-2180-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Thomas SM, Ho MF, Sharma S, Reich CI, Frank JA, et al. Heterogeneity of vaginal microbial communities within individuals. J Clin Microbiol. 2009;47:1181–1189. doi: 10.1128/JCM.00854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanoff SJ, Coombs RW. Virucidal effect of Lactobacillus acidophilus on human immunodeficiency virus type 1: possible role in heterosexual transmission. J Exp Med. 1991;174:289–292. doi: 10.1084/jem.174.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Rai P, Daume H., III . Proceedings of Advances in Neural Information Processing Systems 24, Granada, Spain. Curran Associates, Inc.: NY, USA; 2011. Co-Regularized Multi-View Spectral Clustering; pp. 1413–1421. [Google Scholar]
- Lee JE, Lee S, Lee H, Song Y-M, Lee K, Han MJ, et al. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One. 2013;8:e63514. doi: 10.1371/journal.pone.0063514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, McCormick J, Bocking A, Reid G. Importance of vaginal microbes in reproductive health. Reprod Sci. 2012;19:235–242. doi: 10.1177/1933719111418379. [DOI] [PubMed] [Google Scholar]
- Ling ZX, Kong JM, Liu F, Zhu HB, Chen XY, Wang YZ, et al. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics. 2010;11:488. doi: 10.1186/1471-2164-11-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado A, Jefferson KK, Cerca N. Interactions between Lactobacillus crispatus and bacterial vaginosis (BV)-associated bacterial species in initial attachment and biofilm formation. Int J Mol Sci. 2013;14:12004–12012. doi: 10.3390/ijms140612004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklaim JM, Fernandes AD, Di Bella JM, Hammond J, Reid G, Gloor G. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome. 2013;1:12. doi: 10.1186/2049-2618-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklaim JM, Gloor GB, Anukam KC, Cribby S, Reid G. At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc Natl Acad Sci USA. 2011;108 (Suppl 1:4688–4695. doi: 10.1073/pnas.1000086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi C, Donders GG, Bellen G, Brown DR, Parada CM, Silva MG. Sialidase activity in aerobic vaginitis is equal to levels during bacterial vaginosis. Eur J Obstet Gynecol Reprod Biol. 2013;167:205–209. doi: 10.1016/j.ejogrb.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Martin DH, Zozaya M, Lillis R, Miller J, Ferris MJ. The microbiota of the human genitourinary tract: trying to see the forest through the trees. Trans Am Clin Climatol Assoc. 2012;123:242–256. [PMC free article] [PubMed] [Google Scholar]
- Marzorati M, Maignien L, Verhelst A, Luta G, Sinnott R, Kerckhof FM, et al. Barcoded pyrosequencing analysis of the microbial community in a simulator of the human gastrointestinal tract showed a colon region-specific microbiota modulation for two plant-derived polysaccharide blends. Antonie Van Leeuwenhoek. 2013;103:409–420. doi: 10.1007/s10482-012-9821-0. [DOI] [PubMed] [Google Scholar]
- Marzorati M, Wittebolle L, Boon N, Daffonchio D, Verstraete W. How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ Microbiol. 2008;10:1571–1581. doi: 10.1111/j.1462-2920.2008.01572.x. [DOI] [PubMed] [Google Scholar]
- Mitchell C, Balkus JE, Fredricks D, Liu CZ, McKernan-Mullin J, Frenkel LM, et al. Interaction between lactobacilli, bacterial vaginosis-associated bacteria, and HIV type 1 RNA and DNA genital shedding in U.S. and Kenyan women. AIDS Res Hum Retroviruses. 2013;29:13–19. doi: 10.1089/aid.2012.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C, Moreira C, Fredricks D, Paul K, Caliendo AM, Kurpewski J, et al. Detection of fastidious vaginal bacteria in women with HIV infection and bacterial vaginosis. Infect Dis Obstet Gynecol. 2009;2009:236919. doi: 10.1155/2009/236919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AY, Jordan MI, Weiss Y. Proceedings of Advances In Neural Information Processing Systems 14, Vancouver, Canada. MIT Press: London, UK; 2001. On Spectral Clustering: Analysis And An Algorithm; pp. 849–856. [Google Scholar]
- Nikolaitchouk N, Andersch B, Falsen E, Stroembeck L, Mattsby-Baltzer I. The lower genital tract microbiota in relation to cytokine-, SLPI- and endotoxin levels: application of checkerboard DNA-DNA hybridization (CDH) APMIS. 2008;116:263–277. doi: 10.1111/j.1600-0463.2008.00808.x. [DOI] [PubMed] [Google Scholar]
- Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J Clin Microbiol. 1991;29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramel Jayaprakash T, Schellenberg JJ, Hill JE. Resolution and characterization of distinct cpn60-based subgroups of Gardnerella vaginalis in the vaginal microbiota. PLoS One. 2012;7:e43009. doi: 10.1371/journal.pone.0043009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pépin J, Deslandes S, Giroux G, Sobéla F, Khonde N, Diakité S, et al. The complex vaginal flora of West African women with bacterial vaginosis. PLoS One. 2011;6:e25082. doi: 10.1371/journal.pone.0025082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S. Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol Rev. 2013;37:762–792. doi: 10.1111/1574-6976.12029. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Rathod SD, Krupp K, Klausner JD, Arun A, Reingold AL, Madhivanan P. Bacterial vaginosis and risk for Trichomonas vaginalis infection: a longitudinal analysis. Sex Transm Dis. 2011;38:882–886. doi: 10.1097/OLQ.0b013e31821f91a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose WA, McGowin CL, Spagnuolo RA, Eaves-Pyles TD, Popov VL, Pyles RB. Commensal bacteria modulate innate immune responses of vaginal epithelial cell multilayer cultures. PLoS One. 2012;7:e32728. doi: 10.1371/journal.pone.0032728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago GLD, Tency I, Verstraelen H, Verhelst R, Trog M, Temmerman M, et al. Longitudinal qPCR study of the dynamics of L. crispatus, L. iners, A. vaginae, (sialidase positive) G. vaginalis, and P. bivia in the vagina. PLoS One. 2012;7:e45281. doi: 10.1371/journal.pone.0045281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg JJ, Links MG, Hill JE, Dumonceaux TJ, Kimani J, Jaoko W, et al. Molecular definition of vaginal microbiota in East African commercial sex workers. Appl Environ Microbiol. 2011;77:4066–4074. doi: 10.1128/AEM.02943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha BE, Zariffard MR, Wang QJ, Chen HY, Bremer J, Cohen MH, et al. Female genital-tract HIV load correlates inversely with Lactobacillus species but positively with bacterial vaginosis and Mycoplasma hominis. J Infect Dis. 2005;191:25–32. doi: 10.1086/426394. [DOI] [PubMed] [Google Scholar]
- Shannon CE.1948A mathematical theory of communication Bell Syst Tech J 27379–423.623–656. [Google Scholar]
- Smith BC, McAndrew T, Chen ZG, Harari A, Barris DM, Viswanathan S, et al. The cervical microbiome over 7 years and a comparison of methodologies for its characterization. PLOS One. 2012;7:e40425. doi: 10.1371/journal.pone.0040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear GT, Sikaroodi M, Zariffard MR, Landay AL, French AL, Gillevet PM. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis. 2008;198:1131–1140. doi: 10.1086/591942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, et al. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One. 2012;7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Liu CZ, Mitchell CM, Fiedler TL, Thomas KK, Agnew KJ, et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS One. 2010;5:e10197. doi: 10.1371/journal.pone.0010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehl A, Ghosh J. Cluster ensembles—a knowledge reuse framework for combining multiple partitions. J Mach Learn Res. 2003;3:583–617. [Google Scholar]
- Taylor BD, Darville T, Haggerty CL. Does bacterial vaginosis cause pelvic inflammatory disease. Sex Transm Dis. 2013;40:117–122. doi: 10.1097/OLQ.0b013e31827c5a5b. [DOI] [PubMed] [Google Scholar]
- Tsivtsivadze E, Borgdorff H, van de Wijgert JHHM, Schuren FHJ, Verhelst R, Heskes T.2013Neighborhood Co-Regularized Multi-View Spectral Clustering Of Microbiome DataIn: Zhou Z, Schwenker F (eds) Partically Supervised LearningSecond IAPR International Workshop, Nanjing, China, May 2013. Revised Selected PapersSpringer: Berlin; 80–90. [Google Scholar]
- van de Wijgert J, Morrison C, Cornelisse P, Munjoma M, Moncada J, Kaewvichit R, et al. Bacterial vaginosis and vaginal yeast, but not vaginal cleansing, increase HIV-1 acquisition in African women. J Acquir Immune Defic Syndr. 2008;8:203–210. doi: 10.1097/QAI.0b013e3181743936. [DOI] [PubMed] [Google Scholar]
- Verani JR, McGee L, Schrag SJ. Prevention of perinatal group B streptococcal disease–revised guidelines from CDC, 2010. MMWR Recomm Rep. 2010;59:1–36. [PubMed] [Google Scholar]
- Verstraelen H, Verhelst R, Claeys G, De Backer E, Temmerman M, Vaneechoutte M. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 2009;9:116. doi: 10.1186/1471-2180-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenfeld HC, Hillier SL, Krohn MA, Landers DV, Sweet RL. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis. 2003;36:663–668. doi: 10.1086/367658. [DOI] [PubMed] [Google Scholar]
- Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 2010;48:1812–1819. doi: 10.1128/JCM.00851-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.