Abstract
Determining the composition and function of subgingival dental plaque is crucial to understanding human periodontal health and disease, but it is challenging because of the complexity of the interactions between human microbiomes and human body. Here, we examined the phylogenetic and functional gene differences between periodontal and healthy individuals using MiSeq sequencing of 16S rRNA gene amplicons and a specific functional gene array (a combination of GeoChip 4.0 for biogeochemical processes and HuMiChip 1.0 for human microbiomes). Our analyses indicated that the phylogenetic and functional gene structure of the oral microbiomes were distinctly different between periodontal and healthy groups. Also, 16S rRNA gene sequencing analysis indicated that 39 genera were significantly different between healthy and periodontitis groups, and Fusobacterium, Porphyromonas, Treponema, Filifactor, Eubacterium, Tannerella, Hallella, Parvimonas, Peptostreptococcus and Catonella showed higher relative abundances in the periodontitis group. In addition, functional gene array data showed that a lower gene number but higher signal intensity of major genes existed in periodontitis, and a variety of genes involved in virulence factors, amino acid metabolism and glycosaminoglycan and pyrimidine degradation were enriched in periodontitis, suggesting their potential importance in periodontal pathogenesis. However, the genes involved in amino acid synthesis and pyrimidine synthesis exhibited a significantly lower relative abundance compared with healthy group. Overall, this study provides new insights into our understanding of phylogenetic and functional gene structure of subgingival microbial communities of periodontal patients and their importance in pathogenesis of periodontitis.
Keywords: functional gene array, Illumina sequencing, periodontitis, subgingival dental plaque
Introduction
Periodontitis is a chronic inflammatory disease of the tooth supporting tissues. The progressive destruction of the periodontal tissues (gingival, periodontal membrane and alveolar bone) results in pocket formation, bone resorption and ultimately in tooth loss if it is untreated. Periodontitis is the leading cause of tooth loss worldwide (Petersen et al., 2005; Pihlstrom et al., 2005). Furthermore, it has been proven to be a significant risk factor of systematic diseases, including coronary heart disease (Dietrich et al., 2008), stroke (Sfyroeras et al., 2012), obesity (Suvan et al., 2011), atherosclerosis (Tonetti, 2009), preterm birth and low birth weight in infants (Rakoto-Alson et al., 2009).
It is well accepted that periodontitis is initiated by bacteria embedded in biofilms known as subgingival dental plaque and involves complex interactions of bacteria with the host (Tatakis and Kumar, 2005; He and Shi, 2009; Darveau, 2010). Macromolecules synthesized by periodontal prokaryotes, such as fimbriae, hydrolytic and proteolytic enzymes, as well as metabolic end products (sulfur-containing compounds, ammonia, short-chain fatty acids and so on) play important roles in the destruction of tooth supporting tissues (Socransky and Haffajee, 1992). These molecules, on one hand, are extremely toxic even at moderate concentration and can cause host tissue damage directly. On the other hand, by stimulating the host inflammatory cascade that involves chemokines, cytokines, products of arachidonic acid metabolism and matrix metalloproteinases, they could promote connective tissue destruction indirectly (Graves et al., 2000).
Traditionally, studies on pathogenesis of periodontal microbiota involve a ‘reductionist' approach to studying complex subgingival microbial communities by analyzing individual bacterial species. As a result, a small group of predominantly Gram-negative anaerobic bacteria and spirochetes, notably Porphyromonas gingivalis, Tannerella forsythensis and Treponema denticola, have long been regarded as the main putative pathogens of periodontitis (Holt and Ebersole, 2005; Ertugrul et al., 2013; Fischer et al., 2013). However, more recent studies have shown that, instead of a single pathogen, periodontitis may be a poly-microbial infectious disease caused by potentially pathogenic microbial communities, in which microorganisms interact in a synergistic or cooperative manner, leading to pathogenesis (Darveau, 2010; Hajishengallis and Lamont, 2012; Griffen et al., 2012). This pathogenic pattern revealed that each individual within the community might be involved in the disease occurrence and progression, and even members in low abundance could still act as critical species affecting complex community behaviors. Consequently, an increased interest in using a ‘holistic' approach to define the complex regulatory networks that contribute to this disease has emerged with advances in metagenomic technologies, for example, high-throughput microarrays (Colombo et al., 2009, 2012) and next-generation sequencing technology (Colombo et al., 2009, 2012; Liu et al., 2012; Griffen et al., 2012).
Functional gene array technology is one of several recently developed high-throughput metagenomic approaches for analyzing microbial communities from different sources (He et al., 2012a, 2012b). Various types of functional gene arrays have been developed for analyzing microbial community structure (He et al., 2011). For example, GeoChip 4.0 contains ∼83 992 probes and targets ∼142 000 genes from 410 functional gene families involved in nitrogen, carbon, sulfur and phosphorus cycling, antibiotic resistance, virulence factors and bacterial phage-mediated lysis (Tu et al., 2014a). Another type of functional gene array, termed HuMiChip 1.0, was constructed for analyzing human microbiomes that contains 36 802 probes targeting ∼50 000 coding sequences from 139 key functional genes in various metabolic pathways such as the metabolism of amino acids, carbohydrates, energy, lipids, glycan, cofactors, vitamins and nucleotides (Tu et al., 2014b). As human microbiomes have some general functions that GeoChip targets, combining these two functional gene arrays would make it a promising tool to analyze the functional composition and structure of human microbiomes.
In this study, the subgingival dental plaque samples from periodontitis patients and healthy controls were analyzed with Illumina MiSeq (San Diego, CA, USA) sequencing and GeoChip 4.0+HuMiChip 1.0. We aimed to (1) elucidate the microbial taxonomical composition changes in periodontitis patients and identify key microbial populations; and (2) identify key metabolic genes and pathways contributing to periodontal microbial community pathogenicity. Our results indicated that the microbial community composition and structure in periodontitis patients was dramatically altered and key metabolic genes and pathways could be directly or indirectly involved in periodontitis pathogenicity.
Materials and methods
Subjects and sampling
Subjects with moderate periodontitis (n=12), severe periodontitis (n=13) and sex- and age-matched healthy controls (n=12) were recruited from the West China Hospital of Stomatology, Sichuan University. Inclusion criteria of periodontitis patients were (Tanner et al., 2007): (1) age between 20 and 70 years; (2) medically healthy; (3) no previous periodontal treatment and no antibiotics use within the past 6 months; and (4) willing to consent to the clinical examination and microbial sampling. Periodontal healthy subjects had (1) no pockets and clinical attachment loss; (2) no alveolar bone absorption on X-ray examination; and (3) <15% of sites with bleeding on probing or redness. The periodontal patients were subdivided into moderate periodontitis and severe periodontitis groups based on the definition as following: moderate periodontitis: 4 mm<probe depth ⩽6 mm, attachment loss 3–5 mm, 1/3 root length< alveolar bone destruction <1/2 root length; severe periodontitis: probe depth ⩾6 mm, attachment loss >5 mm, alveolar bone destruction >1/2 root length (Meng, 2008). To obtain sufficient genomic DNA, only generalized periodontitis patients (>30% of teeth are involved; Armitage, 1999) were enrolled. The subgingival plaque samples were collected after patients had signed an informed consent and the sampling procedure was approved by the local ethics committee of West China Hospital of Stomatology, Sichuan University (Chengdu, China). After the supragingival plaque removal, the subgingival plaque was collected from selected periodontal pockets by inserting one sterile endodontic paper point (Caulk-DENTSPLY, Milford, DE, USA) into each pocket for 10 s. Samples from the same volunteer were pooled, placed in 700 μl of Tris-EDTA buffer, carried to laboratory within 4 h and stored at −80 °C before further processing. For periodontal healthy individuals, the same method was used to collect plaque from the bottom of the gingival sulcus.
DNA extraction
Bacteria were separated from the paper points by vortexing. The paper points were discarded and community DNA was extracted using the QIAamp DNA micro Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions and adding a lysozyme (3 mg ml−1, 1.5 h) treatment step. The DNA quality was evaluated by the absorbance ratios at A260/A280 and A260/A230 using spectrophotometry (NanoDrop 1000, Thermo Scientific Inc., Wilmington, DE, USA) and final DNA concentrations were quantified with the Pico-Green kit (Invitrogen, Carlsbad, CA, USA). Only DNA samples with A260/A280 >1.7 and A260/A230 >1.8 were used for further analysis. The extracts were stored at −20 °C until use.
Illumina sequencing analysis of 16S rRNA gene amplicons
The barcoded PCR amplification was performed and the 16S rRNA gene PCR amplicons were sequenced with the Illumina MiSeq technology at the Institute for Environmental Genomics, University of Oklahoma (Norman, OK, USA). Details on microbial 16S rRNA gene amplification, highly paralleled DNA sequencing and preprocessing of reads are included in the Supplementary Materials online. The sequence data were further analyzed with the following statistical methods. (1) Unweighed principal coordinate analysis was used to compare the phylogenetic structures of healthy and disease control. In addition, three different nonparametric analyses for multivariate data were also used to examine the community difference between healthy and periodontal diseases group: analysis of similarities (ANOSIM) (Clarke, 1993), nonparametric multivariate analysis of variance (adonis) using distance matrices (Anderson, 2001) and multiresponse permutation procedure (MRPP). (2) The diversity indices including Shannon–Weaver H index, Simpson and Simpson Evenness were calculated using R 2.9.1 (http://www.r-project.org/) with the vegan package. (3) Circular maximum likelihood phylogenetic tree at genus level was constructed with MEGA 5 using neighbor-joining method with a bootstrap value of 1000 and the tree was displayed using iTOL (Letunic and Bork, 2011). (4) The relative abundances of bacterial taxon at the levels of phylum and genus were calculated and typically presented as mean±s.d. Permutation unpaired t-test was used to identify statistically significant differences. (5) The number of operational taxonomical units (OTUs) within each phylum were also calculated and presented as mean±s.d. Permutation unpaired t-tests were used to identify statistically significant differences.
Functional gene array analysis
The GeoChip 4.0 and HuMiChip 1.0 existed in the same array with ∼135K probes totally, among which HuMiChip 1.0 contains 36 802 probes (Tu et al., 2014b), GeoChip 4.0 has 83 992 probes (Tu et al., 2014a) and the rest of them are control probes for quality control, normalization and evaluation. The protocols for GeoChip 4.0 and HuMiChip 1.0 array analysis are the same. For details of target DNA labeling, hybridization and imaging, please refer to the Supplementary Materials. The signal intensities were digitized based on microarray images using NimbleScan (NimbleGen, Madison, WI, USA) as raw data that were uploaded to the laboratory microarray data manager pipeline (http://ieg.ou.edu/microarray/) and then preprocessed with the following major steps. (1) Spots flagged as poor quality by ImaGene 6.1 and with a signal-to-noise ratio ((signal intensity−background)/s.d. of background) of <2.0 and signal intensity of <1000 were removed. (2) Signal intensities were normalized to the Cy5-labeled universal standard across different arrays. (3) The normalized intensity of each spot was calculated by dividing the signal intensity of each spot by the mean intensity of the effective spots of the array. (4) All samples were separated into three groups, and a probe was removed if it was only detected in two or fewer samples in each group. All of the above are standard protocols for GeoChip-based analysis that have been described elsewhere. Preprocessed data were used for further statistical analysis. (1) The diversity indices including Shannon–Weaver H index, Simpson and Simpson Evenness were calculated using R 2.9.1 with the vegan package. The overlapped/unique numbers of genes were calculated with the following two steps: (a) all detected genes from all replicates for each group (healthy controls, moderate periodontitis or severe periodontitis) were counted to generate a pooled gene list; and (b) the pooled gene lists were used to calculate the overlapped/unique gene numbers between any two groups. (2) ANOSIM, adonis and MRPP were also used for community functional structure comparison. (3) Hierarchical clustering analysis was performed with CLUSTER 3.0 (http://rana.lbl.gov/EisenSoftware.htm) and visualized by TREEVIEW (de Hoon et al., 2004). (4) The normalized signal intensity for each functional gene was the average of the total signal intensity from all the replicates and all data are presented as mean±s.d. Permutation unpaired t-test were used to compare the significance of differences in relative abundance of each gene category or family between healthy and diseased groups.
Results
Changes in phylogenetic composition and structure in periodontal microbial communities
To understand the change in phylogenetic composition and structure of periodontal microbial communities, we sequenced 16S rRNA gene amplicons from periodontitis (n=25) and healthy (n=12) samples with Illumina MiSeq technology. Sequencing produced 548 989 raw sequences with average length of 252 bp. After preprocessing, 514 224 high-quality reads remained and were assigned to individual OTUs at 3% dissimilarity using the Uclust program (Edgar, 2010). In total, 1001 OTUs were detected in these 37 volunteers, with 800 phylogenetically derived from 12 known bacterial phyla and 201 from unclassified phenotypes.
The phylogenetic structure of all 37 samples were analyzed by unweighed principal coordinate analysis. As shown in Supplementary Figure S1, periodontitis and healthy subjects were well separated. The dissimilarity tests, including ANOSIM, adonis and MRPP, showed significant differences between periodontitis and healthy subjects (P<0.05). However, no significant differences between moderate periodontitis and severe periodontitis were detected, and hence we combined these two groups into the periodontitis group for further analysis.
As shown in Supplementary Table S1, significantly higher α-diversity (Shannon and Simpson) and evenness (Simpson Evenness) was observed in the periodontal group. A slightly higher richness (number of OTUs detected) was found in the periodontal patients, but no statistical significance was detected (P>0.05).
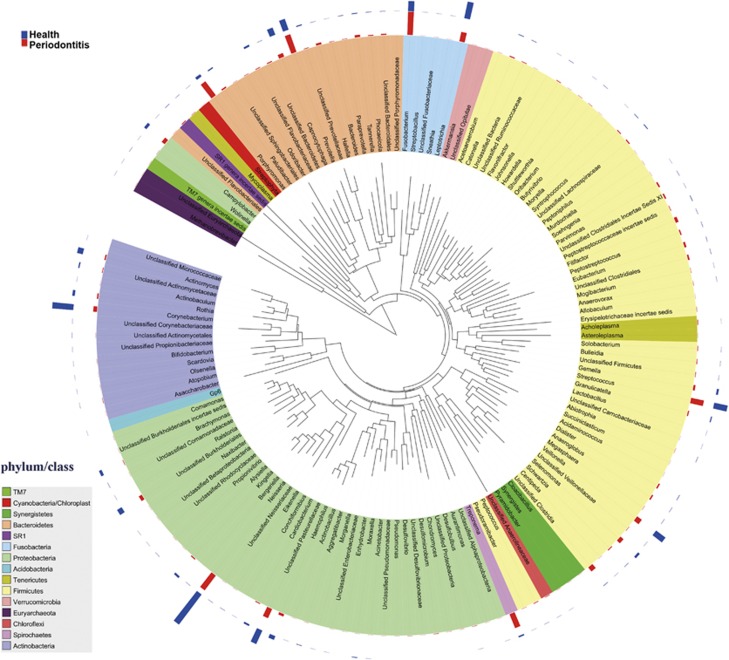
A phylogenetic tree constructed with MEGA 5 at the genus level is shown in Figure 1. Overall abundance and the magnitude of the difference between healthy and periodontitis groups are indicated by bars. A total of 108 genera were detected and most were rare taxa with abundance <0.3%. Fusobacterium, Neisseria, Prevotella, Streptococcus, Leptotrichia, Veillonella and Capnocytophaga were the predominant genera in both periodontal and healthy groups and they were also ranked top 10 positions in each group in terms of relative abundance.
Figure 1.
Circular maximum likelihood phylogenetic tree at the genus level. The tree was constructed with MEGA 5 using neighbor-joining method with a bootstrap value of 1000 and displayed using iTOL (Letunic and Bork, 2011). The bars in the outer band represent the relative abundance of bacterial genus in the healthy (blue) and the periodontal disease (red) groups.
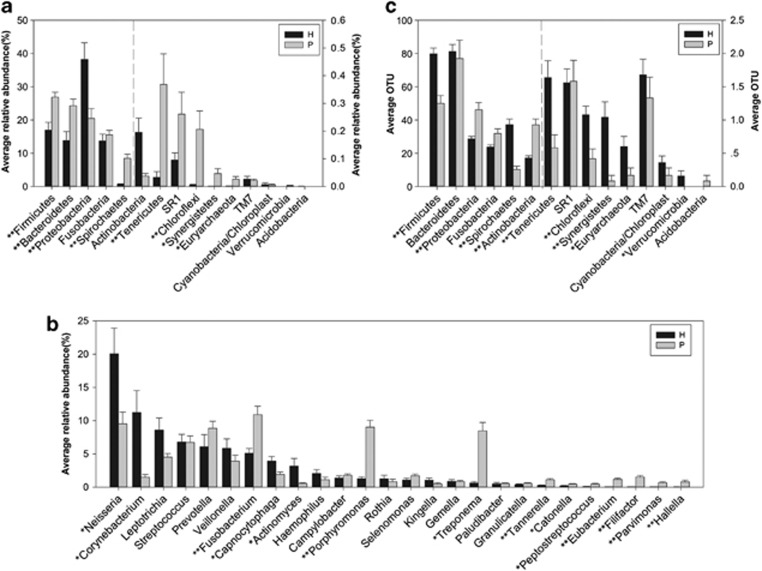
Figure 2 shows statistical difference at the levels of the most abundant phyla, genera and OTUs. At the phylum level, Firmicutes, Bacteroidetes, Spirochaetes, Tenericutes, Chloroflexi, Synergistetes and Euryarchaeota were highly associated with disease, whereas Proteobacteria were found at higher levels in healthy controls (Figure 2a). At the genus level, we found 39 genera distributed differently between healthy and periodontitis groups, and only the genera with average relative abundance >0.4% in periodontitis group are demonstrated in Figure 2b. Fusobacterium, Porphyromonas, Treponema, Filifactor, Eubacterium, Tannerella, Hallella, Parvimonas, Peptostreptococcus and Catonella showed higher abundances in the periodontitis group, whereas the genera Neisseria, Corynebacterium, Capnocytophaga and Actinomyces had lower abundances (Figure 2b). Prevotella had no difference between the two groups. Then, we compared the distribution of OTUs in periodontitis patients with that of healthy subjects. Fewer OTUs in phyla Firmicutes, Tenericutes, Chloroflexi, Synergistetes and Euryarchaeota and more OTUs in Proteobacteria and Actinobacteria were detected in periodontal patients with an unpaired t-test (Figure 2c).
Figure 2.
Differences between healthy (H) and periodontitis (P) subjects at the phylum, genus and OTU levels. (a) Phylum level. (b) Genus level. The genera with average relative abundance >0.4% in the periodontitis group are present. (c) Difference of average OTUs in each phylum; *P<0.05 and **P<0.001.
Shifts of functional gene composition and structure
To understand whether functional gene composition and structure of oral microbial communities change with periodontitis, we analyzed samples using functional microarrays. Unique and overlapped genes as well as diversity indexes among three groups of subjects were calculated based on data obtained from GeoChip 4.0+HuMiChip 1.0 (Table 1). Periodontal healthy subjects exhibited the highest number and percentage of unique genes (9964 and 23.1%, respectively), whereas only 1.0% of unique genes were found in both moderate and severe periodontitis patients. In addition, moderate and severe periodontitis shared the maximum overlapped genes (29 903 and 87.0%, respectively). Based on the Shannon, Simpson and Simpson Evenness indices, a lower functional gene diversity was observed in moderate and severe periodontitis with the order: severe periodontitis<moderate periodontitis<healthy control. Meanwhile, significantly fewer genes were detected in the periodontitis groups (P<0.05) as compared with the control group.
Table 1. Percentages of unique and overlapping genes, diversity indices and total genes detected for each group.
|
Unique and overlapping genes |
Diversity indices |
Total genes detected | |||||
|---|---|---|---|---|---|---|---|
| H | MP | SP | Shannon | Simpson | Simpson E | ||
| H | 9964 (23.1) | 32 035 (73.0) | 30 680 (70.0) | 10.3±0.2 | 30 016.7±6499.9 | 0.09±0.02 | 30 269±6583 |
| MP | 312 (1.0) | 29 903 (87.0) | 10.1±0.2 | 23 575.3±4276.8 | 0.07±0.01 | 23 758±4323 | |
| SP | 311 (1.0) | 9.9±0.2 | 21 171.8±4731.4 | 0.06±0.01 | 21 320±4781 | ||
The bold entries stand for the unique genes and italic for overlap genes between any two of those three groups (H, MP and SP).
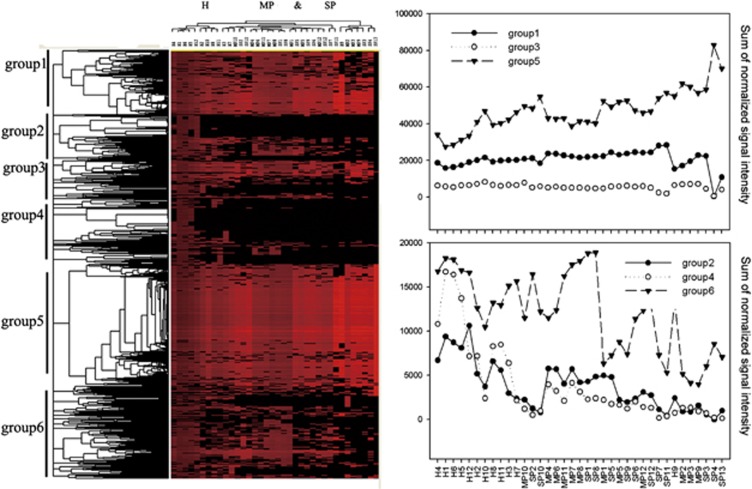
Significant differences (P<0.05) in the microbial functional gene structure were observed between healthy control and moderate periodontitis, or between healthy control and severe periodontitis, using three nonparametric multivariate analysis methods, although no significant differences between moderate and severe periodontitis were detected (P>0.05, Table 2). According to hierarchical clustering analysis, samples from periodontitis subjects clustered together and were well separated from healthy controls, whereas moderate and severe periodontitis could not be well separated (Figure 3). Although some functional genes were common to all samples, the variability in functional gene distribution among different samples was noticeable (Figure 3). For example, genes in groups 1, 3 and 5, with 15.1%, 6.2% and 28.4% of all genes detected, respectively, that are largely involved in amino acid synthesis and metabolism, central carbon metabolic pathways, complex carbohydrates and purine metabolism, feeder pathways to glycolysis and glycan biosynthesis, were generally detected in all samples. However, groups 2, 4 and 6, with 9.6%, 19.0% and 21.6% of all genes detected, respectively, were mainly found in the healthy samples.
Table 2. The overall microbial community structure comparison using three nonparametric statistical methods.
| Groups | MRPP | P-value | ANOSIM | P-value | Adonis | P-value |
|---|---|---|---|---|---|---|
| Among three groupsa | 0.351 | 0.001 | 0.224 | 0.001 | 4.145 | 0.001 |
| H vs MPa | 0.336 | 0.002 | 0.301 | 0.001 | 4.785 | 0.001 |
| H vs SPa | 0.363 | 0.001 | 0.362 | 0.001 | 6.160 | 0.001 |
| MP vs SP | 0.352 | 0.111 | 0.025 | 0.216 | 1.446 | 0.155 |
Abbreviations: Adonis, nonparametric multivariate analysis of variance (MANOVA) with the adonis function; ANOSIM, analysis of similarity; H, healthy control; MP, moderate periodontitis; MRPP, multiresponse permutation procedure; SP, severe periodontitis.
Indicates significant difference (P<0.05).
Figure 3.
Hierarchical clustering analysis of all detected genes in all 37 samples. Results were generated by CLUSTER and visualized by TREEVIEW. Red indicates signal intensities above background, whereas black indicates signal intensities below background. Brighter red coloring indicates higher signal intensities. The groups 1–6 contained functional genes from different functional gene families, but showed different patterns between healthy and diseased groups. The y axes in the right panel indicate the sum of normalized signal intensity of each group of genes detected. H, healthy control; MP, moderate periodontitis; SP, severe periodontitis.
As there was no significant difference detected between moderate and severe periodontitis groups by dissimilarity and clustering analysis in terms of overall functional structure, we combined these two groups into the periodontitis group for further analysis. Because most of the functional genes targeted by GeoChip 4.0 play important roles in biogeochemical processes in the natural environment, but not in the human body, we focused more on the HuMiChip 1.0. Hence, the microarray data listed below were obtained from HuMiChip 1.0 except virulence factors category.
Shifts of functional gene abundances in key metabolic processes
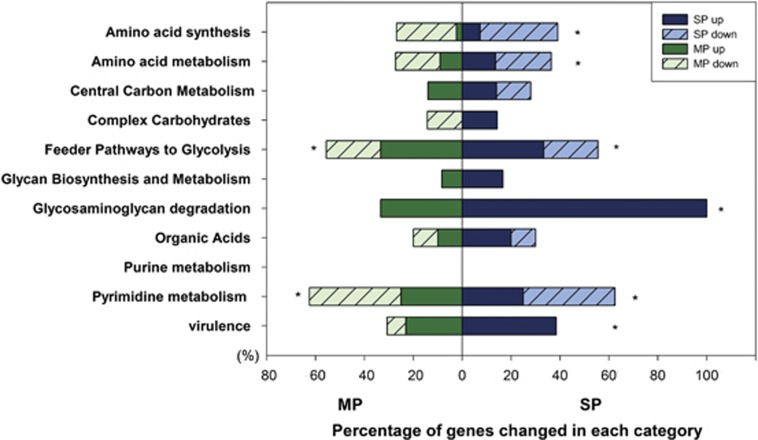
Genes showing significant higher abundance in periodontitis were mainly involved in virulence, amino acid metabolism, glycosaminoglycan (GAG) degradation, pyrimidine metabolism, glycolysis and organic acid biosynthesis. The relative abundance of genes involved in amino acid synthesis, however, was lower. A positive correlation was observed between the changes in gene patterns and the severity of inflammation in the above categories; that is, a higher percentage of functional genes changed in severe periodontitis than in moderate periodontitis as compared with the healthy group (Figure 4).
Figure 4.
Shifts of functional genes in periodontal communities for key metabolic pathways. For each pathway, first, the relative abundance of genes between healthy controls (H) and moderate periodontitis (MP) or severe periodontitis (SP) was compared with analysis of variance (ANOVA; α=0.05). Then, the numbers of genes with statistical difference were counted. The bars represented the percentage (%) of these genes in total genes in this category and * indicated more than 40% genes altered in this category.
Virulence factors
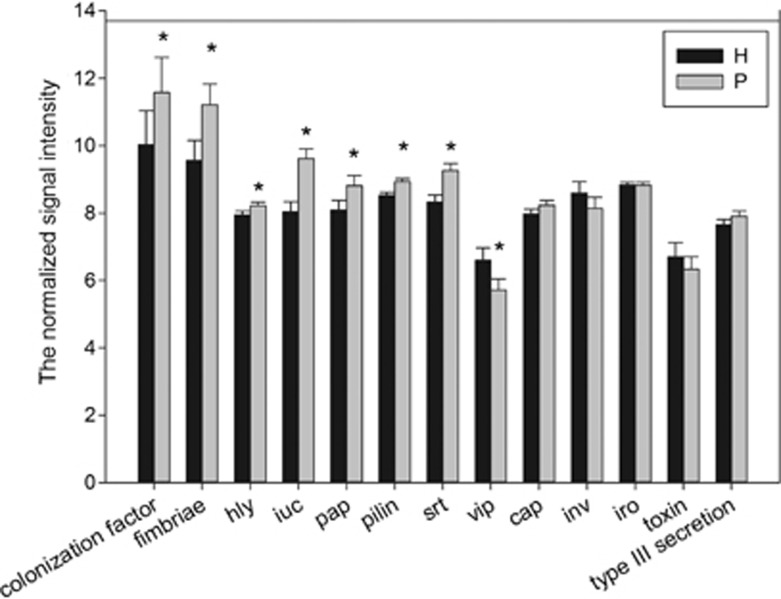
Virulence factors are molecules expressed and secreted by pathogens that enable to achieve the colonization of a niche in the host, to invade epithelial and endothelial cells and/or to evade host defenses (Graves et al., 2000). Colonization factor (encoded by colonization factor), fimbriae (encoded by fimbriae r), pili (encoded by pilin), P pili (encoded by pap) and sortase (encoded by srt) are all able to promote bacterial attachment to host tissues; hemolysins (encoded by hly) are functionally defined by the ability to lyse erythrocytes and they also have the capability of lysing a variety of other cell types, including mast cells, neutrophils and polymorphonuclear cells (Braun and Focareta, 1991); aerobactin (encoded by iuc) is a siderophore-dependent iron acquisition system and iron is an essential nutrient for bacterial growth (Marx, 2002). One of the most striking results in this study was that more than half of the targeted virulence genes in the periodontitis group, including colonization factor, fimbriae, hly, iuc, pap, pilin and srt, had a higher relative abundance compared with the healthy group (Figure 5).
Figure 5.
The normalized signal intensity of virulence genes. The signal intensity for each functional gene was the average of the total signal intensity from all the replicates. All data are presented as mean±s.e. (*P<0.05). Half of virulence genes in periodontitis (P) had a higher abundance than those in healthy (H) subjects.
Amino acid degradation and synthesis
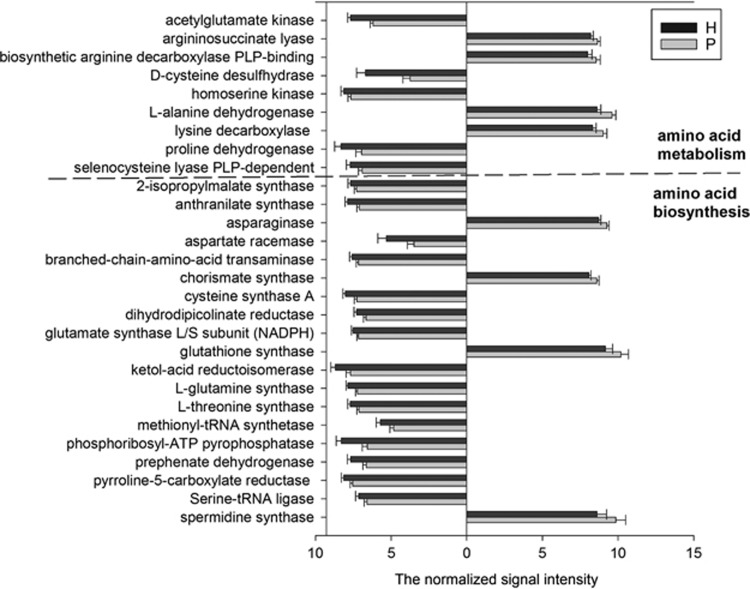
Out of 22 genes involved in amino acid metabolism, 9 showed distinct abundances between periodontitis and healthy groups (Figure 6). Of these, four genes exhibited higher relative abundances in periodontitis, including those encoding lysine decarboxylase, L-alanine dehydrogenase, biosynthetic arginine decarboxylase PLP-binding and argininosuccinate lyase, that play an indispensable role in lysine, alanine and arginine metabolism. Similarly, most probes of these four genes showed higher signal intensity in the periodontitis group than in the healthy group. For example, 177 probes were designed to detect the genes encoding lysine decarboxylase. Among 17 probes with positive hybridization signals, 11 probes showed higher abundances in the periodontitis group (Supplementary Figure S2). Another five genes encoding acetylglutamate kinase, D-cysteine desulfhydrase, homoserine kinase, proline dehydrogenase and selenocysteine lyase PLP-dependent showed lower abundances in the periodontitis group (Figure 6). Within amino acid synthesis genes, 19 of 41 genes had significantly different signal intensities, and 15 of these showed lower signal intensities in periodontitis.
Figure 6.
The normalized signal intensity of genes involved in amino acid metabolism and synthesis. All data are presented as mean±s.e. and genes with no significant difference are not shown (P>0.05). Bars on the left of y axis represent genes with lower relative abundance in periodontitis, whereas bars on the right indicate genes with higher abundance in periodontitis. Out of 22 genes involved in amino acid metabolism, 9 showed distinct abundances between periodontitis (P) and healthy (H) subjects. As for amino acid synthesis, 19 out of 41 genes changed and 15 of these genes showed lower relative abundance in the periodontitis group.
GAG degradation
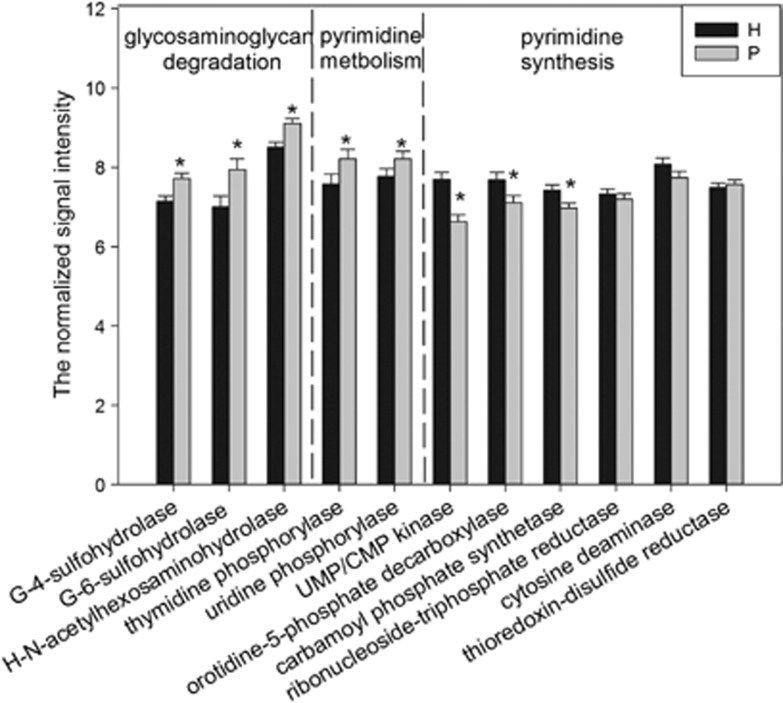
GAGs are a family of amino sugars including hyaluronic acid, chondroitin-4-sulfate (C-4-S), chondroitin-6-sulfate, heparan sulfate and keratan sulfate. In this study, the relative abundance of all three genes involved in GAG degradation was significantly higher in the periodontitis group (Figure 7). These genes encode N-acetyl-D-galactosamine-4-sulfate 4-sulfohydrolase, β-N-acetyl-D-hexosaminide N-acetylhexosaminohydrolase and N-acetyl-D-glucosamine-6-sulfate 6-sulfohydrolase, respectively.
Figure 7.
The normalized signal intensity of genes involved in glycosaminoglycan degradation, pyrimidine metabolism and pyrimidine biosynthesis. The signal intensity for each functional gene was the average of the total signal intensity from all replicates. All data were presented as mean±s.e. *P<0.05. G-4-sulfohydrolase, N-acetyl-D-galactosamine-4-sulfate 4-sulfohydrolase (involved in chondroitin sulfate biosynthesis); G-6-sulfohydrolase, N-acetyl-D-glucosamine-6-sulfate 6-sulfohydrolase (involved in keratan sulfate biosynthesis); H-N-acetylhexosaminohydrolase, β-N-acetyl-D-hexosaminide N-acetylhexosaminohydrolase (involved in keratan sulfate biosynthesis).
Pyrimidine metabolism and synthesis
The abundance of pyrimidine metabolism-related genes encoding thymidine phosphorylase and uridine phosphorylase were higher in periodontitis (Figure 7). The former catalyzes thymidine into thymine, and the latter catalyzes uridine into uracil. On the contrary, pyrimidine biosynthesis genes showed significant lower relative abundance in periodontitis, including those encoding carbamoyl phosphate synthetase, orotidine-5-phosphate decarboxylase (for uridine-5′-phosphate biosynthesis) and UMP/CMP kinase (for pyrimidine biosynthesis). Genes related to purine synthesis and metabolism did not show obvious abundance changes.
Organic acids
Genes encoding β-ketoacyl-ACP synthase III, butyrate kinase and butyrate-acetoacetate CoA-transferase were found to be more abundant in periodontitis (Supplementary Figure S3). The overexpression of β-ketoacyl-ACP synthase III contributes to a high level of fatty acids and the other two to butyrate biosynthesis. No significant difference was observed for other genes in this category.
Carbohydrate degradation
Carbohydrate degradation consists of four categories: glycolysis, central carbon metabolism, complex carbohydrate degrading and glycan biosynthesis and metabolism. More than half of the genes (5/9) in the glycolysis category had significantly different signal intensities, but they were not involved in metabolism of several key sugars (such as maltose, glucose and fructose) (Supplementary Figure S4). In the gene family of central carbon metabolism, only genes encoding carbon monoxide dehydrogenase and KDPG aldolase exhibited significantly higher abundance (data not shown). No significant differences were observed for complex carbohydrate degradation, glycan biosynthesis and metabolism genes (Figure 4).
Discussion
Using Illumina MiSeq sequencing and microarray (GeoChip 4.0 and HuMiChip 1.0) technologies, we showed significant phylogenetic and functional gene differences of subgingival microbial communities between periodontitis patients and healthy individuals. In general, the 16S rRNA analysis indicated phylogenetic structure of the oral microbiome was distinctly different between periodontal and healthy individuals. According to the functional gene analysis, the functional gene diversity decreased as the periodontal inflammation severity increased. Also, the relative abundance of some key genes involved in virulence factors, amino acid metabolism, GAG degradation as well as pyrimidine metabolism was higher in patients, and this can be explained by the following several possible reasons.
Significantly higher α-diversity and evenness were observed in the periodontal group, consistent with previous observations (Griffen et al., 2012; Liu et al., 2012; Abusleme et al., 2013). The significantly increased α-diversity without significantly increased richness indicated that the increased diversity mainly resulted from a more even subgingival community. Being different from the phylogenetic richness, significantly less functional richness (functional gene number) was observed in the periodontitis groups. No statistical correlation between OTUs and gene numbers (Pearson's test, Pearson's correlation coefficient=0.52, P=0.12, data not shown) was observed, indicating they were not closely related. It could be partially explained by the fact that the phylogenetic richness index did not take the phylogenetic assignment of OTUs into consideration and each species harbors specific genome. However, this result should be taken carefully because functional microarray are close-format metagenomic methodology and some genes might be omitted because of the probe design, and studies employing shot-gun sequencing of subgingival metagenome are needed to cross-validate it.
Significant differences of oral microbiomes were detected between periodontitis and healthy individuals at the phylum and genus levels, suggesting phylogenetic structure shifts of the subgingival microbial community. The present study confirmed the association between periodontitis and some microbial taxa, including the phylum Firmicutes, Bacteroidetes, Spirochaetes, Tenericutes, Chloroflexi, Synergistetes and Euryarchaeota and the genus Fusobacterium, Porphyromonas, Treponema, Filifactor, Eubacterium, Tannerella, Hallella, Parvimonas, Peptostreptococcus and Catonella. The taxonomic enrichment in these taxa, to a certain extent, might contribute to the functional shift of subgingival microbial community in periodontitis population as detected by GeoChip 4.0+HuMiChip 1.0.
Based upon the pathogenicity of the periodontal bacteria, Socransky et al. (1998) classified several complexes of bacteria, dividing them in groups and labeled by colors. The ‘red complex', including P. gingivalis (belongs to the phylum Bacteroidetes and the genus Porphyromonas), T. denticola (belongs to the phylum Spirochaetes and the genus Treponema) and Tannerella forsythia (belongs to the phylum Bacteroidetes and the genus Tannerella), are recognized as the most virulent pathogens in adult periodontal disease. As molecular-based approaches to microbial detection become increasingly facile and as studies employing culture-independent methods for periodontal microbiota analysis become abundant, some newly recognized organisms are found to show as good correlation with disease as the classical red complex (Kumar et al., 2005, 2006), calling into question the concept of color-coded complexes. However, whether the newly identified taxa will play an equal or more important role in periodontal pathogenesis than that of the species in color-coded complexes requires further investigation.
The genera with significant associations with periodontitis deserved further research. For example, Filifactor appeared to be prevalent and strongly disease associated. Research is warranted on this type of microorganisms as they may play an important role in development of periodontitis. As a matter of fact, Filifactor alocis, a fastidious, Gram-positive, obligate anaerobic rod, has been proven to harbor characteristics in common with established periodontal pathogens and has the potential to contribute to periodontal tissue destruction (Aruni et al., 2011; Moffatt et al., 2011).
Moreover, it is very interesting that much less species could be detected from the periodontitis groups in the phyla Firmicutes, Spirochaetes, Tenericutes, Chloroflexi, Synergistetes and Euryarchaeota. However, these phyla showed higher relative abundance in periodontal patients. As bacteria in the dental biofilm interact with each other actively, these results indicated that when the disease advances, strains with more virulent properties predominate the subgingival niches and they might inhibit the colonization, reproduction and growth of other microbiota, even those belonging to the same phylum. In addition, although more species-level phenotypes belonging to Proteobacteria could be detected in the periodontitis, the relative abundance of this phylum was lower than that in the healthy group.
The first key step for pathogenesis of periodontal bacteria is subgingival dental plaque (that is, biofilm) formation. Successful establishment of a biofilm heavily relies on the ability of bacteria to adhere to host tissues. Microorganisms use different strategies ranging from single monomeric proteins to intricate multimeric macromolecules for cell adhesion. The colonization factor, fimbriae, pili and P pili are crucial for bacteria's colonization on host tissues. Among them, pili, P pili and fimbriae are among the most important cellular components needed for adhesion of periodontal bacteria (Amano, 2010). The present study showed a significantly higher abundance of genes encoding pili, P pili and fimbriae (fim, pap and pilin, respectively) in periodontitis samples. Similarly, srt, encoding sortase, also showed a higher relative abundance in the periodontitis group. Sortase is a protease that catalyzes cell wall anchoring of surface proteins containing a LPXTG motif (Cossart and Jonquières, 2000) and assembly of pili in Gram-positive bacteria is sortase dependent (Cossart and Jonquières, 2000; Telford et al., 2006). The high level of genes encoding pili, P pili and fimbriae should be given specific attention because their roles in bacterial adhesion as well as adhesion-binding events may activate complex signal transduction cascades in host cells that could attenuate innate immune clearance of bacteria, thus facilitating bacterial invasion. For example, the long fimbriae of P. gingivalis could bind to Toll-like receptor 2 and to CD14 that induced the activation of complement receptor 3 in leukocytes. After being activated, complement receptor 3 interacted with long fimbriae and led to the downregulation of interleukin-12 p70, a crucial cytokine involved in intracellular bacterial clearance (Amano, 2010).
Periodontal tissues are rich in proteins. Periodontal microbes and host-derived proteases could degrade host periodontal proteins and proteins involved in the inflammatory and immune reactions into peptides and amino acids that the microorganisms can absorb and used as nutritional resources (Sandholm, 1986; Eley and Cox, 2003). Studies have shown elevated levels of peptides and amino acids in periodontal population (Rai et al., 2008; Barnes et al., 2011). The increased peptides and amino acids would supply as a richer energy pool for the expansion of some specific organisms relying on oligopeptides as carbon and energy sources (Takahashi and Sato, 2002) that ultimately influences the microbial and functional structure. In the present study, genes encoding lysine decarboxylase, L-alanine dehydrogenase, biosynthetic arginine decarboxylase PLP-binding and argininosuccinate lyase exhibited a higher abundance in periodontitis that might lead to the increasing of many amino acid metabolites in periodontal patients (Barnes et al., 2009, 2011). Among the genes involved in amino acid synthesis with distinct abundance, 15 out of 19 genes showed lower relative abundance in periodontitis, indicating that subgingival community of periodontal group might utilize some ammonia absorbed directly for physiological activities instead of biosynthesizing themselves.
The connective tissue and bone destruction is one of the most prominent features in periodontal diseases, more specifically degradation of extracellular matrix. A majority of the studies are concerned with collagen (the main protein in periodontal tissues and bone) breakdown and less attention has been paid on the noncollagenous components of extracellular matrix during periodontal disease. GAGs, the long linear polysaccharides consisting of a repeating disaccharide unit, are major noncollagenous components of extracellular matrix. This group of macromolecules has received the attention of dental researchers as a potential indicator for periodontal diseases (Last et al., 1985; Smith et al., 1995; Giannobile et al., 2003). The abundance of all three genes involved in GAG degradation was dramatically elevated in periodontitis as demonstrated previously. Of particular interest to us is the gene encoding N-acetyl-D-galactosamine-4-sulfate 4-sulfohydrolase associated with C-4-S degradation because the gingival crevicular fluid content of C-4-S exhibited significant correlations with probing depth and attachment loss (Last et al., 1985; Smith et al., 1995) and C-4-S is regarded as a biomarker for bone degradation as it accounts for 94% of GAGs in alveolar bone (Waddington et al., 1989; Armitage, 2004). The higher concentrations of C-4-S and other GAGs in periodontitis (Last et al., 1985; Smith et al., 1995; Giannobile et al., 2003) seem to contradict the observation that degradation genes were enriched. Reasons might be because, first, the functional gene composition is a reflection of a shift in the community composition and does not necessarily represent the overexpression of GAG degradation enzymes. Second, GAGs are released from the inflammatory tissues at a higher speed than that of degradation.
Periodontal cells are damaged by the periodontal microbiota directly or by the host immune system indirectly, followed by nucleic acid releasing. Nucleosides and nucleobases represent an important nutrient source for bacteria and could be used for nucleic acid biosynthesis or be catabolized as carbon and energy sources. We observed lower level of pyrimidine biosynthesis genes (encoding carbamoyl phosphate synthetase, orotidine-5-phosphate decarboxylase and UMP/CMP kinase) in the periodontitis group. However, no significant difference was detected in the relative abundance of the purine synthesis-associated genes. By using an in vitro multispecies model, Kuboniwa et al. (2009) also found that P. gingivalis reduced its pyrimidine biosynthesis but not purine biosynthesis-associated protein abundance when it was cultured with F. nucleatum and S. gordonii. These two investigations together indicated that bacterial purine biosynthesis pathway is more resistant to the community microbial structure alternation. However, this hypothesis requires further validation and more investigations are needed to reveal the mechanisms. The genes involved in purine metabolism show a similar relative abundance, which could partially explain why no increment of purine degradation metabolites was detected from the saliva of generalized chronic periodontitis patients (Aimetti et al., 2012).
Short-chain fatty acids can disrupt host defense systems by different mechanisms, such as induction of apoptosis in immune cells (Stehle et al., 2001; Abe, 2012) and gingival epithelial cells (Tsuda et al., 2010) and alteration of cell function and gene expression in human gingival fibroblasts (Chang et al., 2013). Previous studies have demonstrated higher concentrations of short-chain fatty acids in the gingival crevicular fluid of periodontitis patients (Lu et al., 2008; Qiqiang et al., 2012). In this study, we observed an increased abundance of genes responsible for production of short-chain fatty acids. Specially, butyrate kinase and butyrate-acetoacetate CoA-transferase genes exhibited significantly higher abundance in periodontitis patients, suggesting butyrate may be a key end product toxic to human host tissues. In this study, no difference was observed in the abundance of genes encoding acetate kinase, L-lactate dehydrogenase and propionate kinase, although some differences were found for those genes by Alonso De La Peña et al. (2007) and Aimetti et al. (2012). However, at the probe level, some of these genes had significantly higher signal intensities in periodontitis patients than in healthy subjects.
It is hypothesized that oral microorganisms do not rely heavily on carbohydrate metabolism to promote periodontitis development. In this study, functional genes involved in carbohydrate metabolism were relatively similar across all individuals. These results were consistent with the findings of Mazumdar et al. (2009), in which computational construction and analysis of the genome-scale metabolic network of P. gingivalis were employed. Even though the relative abundance of three genes involved in glycolysis was increased and that of the other two genes was decreased in periodontitis samples, the alteration of maltose, glucose, fructose and xylose metabolism genes was not evident, and this disagrees with other studies carried on saliva, gingival crevicular fluid and single pathogenic bacterium P. gingivalis Strain W83 (Nelson et al., 2003; Barnes et al., 2009, 2011).
In conclusion, our results revealed distinctly different microbial community composition and structures of subgingival dental plaque samples between periodontitis and healthy subjects, as well as the shift of key functional genes that may boost the progression of periodontitis. Furthermore, these enriched or reduced genes and taxa in patients might be used as targets for future antiperiodontitis therapy. To further understand and evaluate oral microbial communities and other associated diseases, further development of functional gene arrays will target more sequenced genomes, as well as metagenomes, and develop strain/species-specific probes for strain/species identification.
Acknowledgments
This work was supported by the International Science and Technology Cooperation Program of China (Grant Number: 2011DFA30940), the National Basic Research Program of China (‘973 Pilot Research Program', Grant Number: 2011CB512108), National Key Technologies R&D Program of the Twelve Five-Year Plan, the Ministry of Science and Technology of China (Grant 2012BAI07B03), the National Natural Science Foundation of China (81170959, 30901689 and 81172579), Sichuan Provincial Department of science and technology project (2013SZ0039) and by Oklahoma Applied Research Support (OARS), Oklahoma Center for the Advancement of Science and Technology (OCAST), the State of Oklahoma through the Project AR11-035. We are also grateful to Tong Yuan, Xiaofei Lu, Feifei Liu, Qichao Tu, Zhou Shi, Kai Xue, Lei Cheng (OU researchers) and Arthur Escalas (PhD student of Université Montpellier) for their great help on this project.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abe K. Butyric acid induces apoptosis in both human monocytes and lymphocytes equivalently. J Oral Sci. 2012;54:7–14. doi: 10.2334/josnusd.54.7. [DOI] [PubMed] [Google Scholar]
- Abusleme L, Dupuy A, Dutzan N, Silva N, Burleson J, Strausbaugh L, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimetti M, Cacciatore S, Graziano A, Tenori L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics. 2012;8:465–474. [Google Scholar]
- Alonso De La Peña V, Diz Dios P, Tojo Sierra R. Relationship between lactate dehydrogenase activity in saliva and oral health status. Arch Oral Biol. 2007;52:911–915. doi: 10.1016/j.archoralbio.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Amano A. Bacterial adhesins to host components in periodontitis. Periodontol 2000. 2010;52:12–37. doi: 10.1111/j.1600-0757.2009.00307.x. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- Armitage GC. Analysis of gingival crevice fluid and risk of progression of periodontitis. Periodontol 2000. 2004;34:109–119. doi: 10.1046/j.0906-6713.2002.003427.x. [DOI] [PubMed] [Google Scholar]
- Aruni AW, Roy F, Fletcher HM. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun. 2011;79:3872–3886. doi: 10.1128/IAI.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes VM, Teles R, Trivedi HM, Devizio W, Xu T, Mitchell MW, et al. Acceleration of purine degradation by periodontal diseases. J Dent Res. 2009;88:851–855. doi: 10.1177/0022034509341967. [DOI] [PubMed] [Google Scholar]
- Barnes VM, Ciancio SG, Shibly O, Xu T, Devizio W, Trivedi HM, et al. Metabolomics reveals elevated macromolecular degradation in periodontal disease. J Dent Res. 2011;90:1293–1297. doi: 10.1177/0022034511416240. [DOI] [PubMed] [Google Scholar]
- Braun V, Focareta T. Pore-forming bacterial protein hemolysins (cytolysins) Crit Rev Microbiol. 1991;18:115–158. doi: 10.3109/10408419109113511. [DOI] [PubMed] [Google Scholar]
- Chang MC, Tsai YL, Chen YW, Chan CP, Huang CF, Lan WC, et al. Butyrate induces reactive oxygen species production and affects cell cycle progression in human gingival fibroblasts. J Periodontal Res. 2013;48:66–73. doi: 10.1111/j.1600-0765.2012.01504.x. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Colombo APV, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the Human Oral Microbe Identification Microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo APV, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–1287. doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Jonquières R. Sortase, a universal target for therapeutic agents against Gram-positive bacteria. Proc Natl Acad Sci USA. 2000;97:5013–5015. doi: 10.1073/pnas.97.10.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Micro. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Dietrich T, Jimenez M, Krall Kaye EA, Vokonas PS, Garcia RI. Age-dependent associations between chronic periodontitis/edentulism and risk of coronary heart disease. Circulation. 2008;117:1668–1674. doi: 10.1161/CIRCULATIONAHA.107.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eley BM, Cox SW. Proteolytic and hydrolytic enzymes from putative periodontal pathogens: characterization, molecular genetics, effects on host defenses and tissues and detection in gingival crevice fluid. Periodontol 2000. 2003;31:105–124. doi: 10.1034/j.1600-0757.2003.03107.x. [DOI] [PubMed] [Google Scholar]
- Ertugrul AS, Arslan U, Dursun R, Hakki SS. Periodontopathogen profile of healthy and oral lichen planus patients with gingivitis or periodontitis. Int J Oral Sci. 2013;5:92–97. doi: 10.1038/ijos.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CL, Walters KS, Drake DR, Dawson DV, Blanchette DR, Brogden KA, et al. Oral mucosal lipids are antibacterial against Porphyromonas gingivalis, induce ultrastructural damage, and alter bacterial lipid and protein compositions. Int J Oral Sci. 2013;5:130–140. doi: 10.1038/ijos.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannobile WV, Al-Shammari KF, Sarment DP. Matrix molecules and growth factors as indicators of periodontal disease activity. Periodontol 2000. 2003;31:125–134. doi: 10.1034/j.1600-0757.2003.03108.x. [DOI] [PubMed] [Google Scholar]
- Graves DT, Jiang Y, Genco C. Periodontal disease: bacterial virulence factors, host response and impact on systemic health. Curr Opin Infect Dis. 2000;13:227–232. doi: 10.1097/00001432-200006000-00005. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012a;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XS, Shi WY. Oral microbiology: past, present and future. Int J Oral Sci. 2009;1:47–58. doi: 10.4248/ijos.09029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Nostrand J, Deng Y, Zhou J. Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities. Front Environ Sci Engin China. 2011;5:1–20. [Google Scholar]
- He Z, Deng Y, Zhou J. Development of functional gene microarrays for microbial community analysis. Curr Opin Biotechnol. 2012a;23:49–55. doi: 10.1016/j.copbio.2011.11.001. [DOI] [PubMed] [Google Scholar]
- He Z, Van Nostrand JD, Zhou J. Applications of functional gene microarrays for profiling microbial communities. Curr Opin Biotechnol. 2012b;23:460–466. doi: 10.1016/j.copbio.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex', a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Kuboniwa M, Hendrickson E, Xia Q, Wang T, Xie H, Hackett M, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last KS, Stanbury JB, Embery G. Glycosaminoglycans in human gingival crevicular fluid as indicators of active periodontal disease. Arch Oral Biol. 1985;30:275–281. doi: 10.1016/0003-9969(85)90045-7. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Faller L, Klitgord N, Mazumdar V, Ghodsi M, Sommer D, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu RF, Meng HX, Gao XJ, Feng L, Xu L. Analysis of short chain fatty acids in gingival crevicular fluid of patients with aggressive periodontitis. Chinese J Stomatol. 2008;43:664–667. [PubMed] [Google Scholar]
- Marx JJM. Iron and infection: competition between host and microbes for a precious element. Best Pract Res Cl Ha. 2002;15:411–426. [PubMed] [Google Scholar]
- Mazumdar V, Snitkin ES, Amar S, Segrè D. Metabolic network model of a human oral pathogen. J Bacteriol. 2009;191:74–90. doi: 10.1128/JB.01123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H. Periodontology. People's Medical Publishing House: Peking; 2008. [Google Scholar]
- Moffatt CE, Whitmore SE, Griffen AL, Leys EJ, Lamont RJ. Filifactor alocis interactions with gingival epithelial cells. Mol Oral Microbiol. 2011;26:365–373. doi: 10.1111/j.2041-1014.2011.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis Strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S, Ndiaye C. The global burden of oral diseases and risks to oral health. Bull World Health Organ. 2005;83:661–669. [PMC free article] [PubMed] [Google Scholar]
- Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- Qiqiang L, Huanxin M, Xuejun G. Longitudinal study of volatile fatty acids in the gingival crevicular fluid of patients with periodontitis before and after nonsurgical therapy. J Periodontal Res. 2012;47:740–749. doi: 10.1111/j.1600-0765.2012.01489.x. [DOI] [PubMed] [Google Scholar]
- Rai B, Kharb S, Jain R, Anand SC. Biomarkers of periodontitis in oral fluids. J Oral Sci. 2008;50:53–56. doi: 10.2334/josnusd.50.53. [DOI] [PubMed] [Google Scholar]
- Rakoto-Alson S, Tenenbaum H, Davideau J-L. Periodontal diseases, preterm births, and low birth weight: findings from a homogeneous cohort of women in Madagascar. J Periodontol. 2009;81:205–213. doi: 10.1902/jop.2009.090351. [DOI] [PubMed] [Google Scholar]
- Sandholm L. Proteases and their inhibitors in chronic inflammatory periodontal disease. J Clin Periodontol. 1986;13:19–26. doi: 10.1111/j.1600-051x.1986.tb01409.x. [DOI] [PubMed] [Google Scholar]
- Sfyroeras GS, Roussas N, Saleptsis VG, Argyriou C, Giannoukas AD. Association between periodontal disease and stroke. J Vasc Surg. 2012;55:1178–1184. doi: 10.1016/j.jvs.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Smith AJ, Addy M, Embery G. Gingival crevicular fluid glycosaminoglycan levels in patients with chronic adult periodontitis. J Clin Periodontol. 1995;22:355–361. doi: 10.1111/j.1600-051x.1995.tb00161.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Stehle HW, Leblebicioglu B, Walters JD. Short-chain carboxylic acids produced by gram-negative anaerobic bacteria can accelerate or delay Polymorphonuclear leukocyte apoptosis in vitro. J Periodontol. 2001;72:1059–1063. doi: 10.1902/jop.2001.72.8.1059. [DOI] [PubMed] [Google Scholar]
- Suvan J, D'Aiuto F, Moles DR, Petrie A, Donos N. Association between overweight/obesity and periodontitis in adults. A systematic review. Obes Rev. 2011;12:381–404. doi: 10.1111/j.1467-789X.2010.00808.x. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Sato T. Dipeptide utilization by the periodontal pathogens Porphyromonas gingivalis, Prevotella intermedia, Prevotella nigrescens and Fusobacterium nucleatum. Oral Microbiol Immunol. 2002;17:50–54. doi: 10.1046/j.0902-0055.2001.00089.x. [DOI] [PubMed] [Google Scholar]
- Tanner ACR, Kent R, Kanasi E, Lu SC, Paster BJ, Sonis ST, et al. Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. J Clin Periodontol. 2007;34:917–930. doi: 10.1111/j.1600-051X.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- Tatakis DN, Kumar PS. Etiology and pathogenesis of periodontal diseases. Dent Clin North Am. 2005;49:491–516. doi: 10.1016/j.cden.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in Gram-positive pathogens. Nat Rev Micro. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- Tonetti MS. Periodontitis and risk for atherosclerosis: an update on intervention trials. J Clin Periodontol. 2009;36:15–19. doi: 10.1111/j.1600-051X.2009.01417.x. [DOI] [PubMed] [Google Scholar]
- Tsuda H, Ochiai K, Suzuki N, Otsuka K. Butyrate, a bacterial metabolite, induces apoptosis and autophagic cell death in gingival epithelial cells. J Periodontal Res. 2010;45:626–634. doi: 10.1111/j.1600-0765.2010.01277.x. [DOI] [PubMed] [Google Scholar]
- Tu Q, He Z, Li Y, Chen Y, Deng Y, Lin L, et al. Development of HuMiChip for Functional Profiling of Human Microbiomes. PLoS One. 2014b;9:e90546. doi: 10.1371/journal.pone.0090546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD, et al. 2014aGeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis Mol Ecol Resoure-pub ahead of print 12 February 2014doi: 10.1111/1755-0998.12239 [DOI] [PubMed]
- Waddington RJ, Embery G, Last KS. Glycosaminoglycans of human alveolar bone. Arch Oral Biol. 1989;34:587–589. doi: 10.1016/0003-9969(89)90100-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.