Abstract
The coincidental theory of virulence predicts that bacterial pathogenicity could be a by-product of selection by natural enemies in environmental reservoirs. However, current results are ambiguous and the simultaneous impact of multiple ubiquitous enemies, protists and phages on virulence evolution has not been investigated previously. Here we tested experimentally how Tetrahymena thermophila protist predation and PNM phage parasitism (bacteria-specific virus) alone and together affect the evolution of Pseudomonas aeruginosa PAO1 virulence, measured in wax moth larvae. Protist predation selected for small colony types, both in the absence and presence of phage, which showed decreased edibility to protists, reduced growth in the absence of enemies and attenuated virulence. Although phage selection alone did not affect the bacterial phenotype, it weakened protist-driven antipredatory defence (biofilm formation), its associated pleiotropic growth cost and the correlated reduction in virulence. These results suggest that protist selection can be a strong coincidental driver of attenuated bacterial virulence, and that phages can constrain this effect owing to effects on population dynamics and conflicting selection pressures. Attempting to define causal links such as these might help us to predict the cold and hot spots of coincidental virulence evolution on the basis of microbial community composition of environmental reservoirs.
Keywords: coevolution, coincidental virulence, experimental evolution, disease ecology, predation, trade-off
Introduction
Virulence traits are typically believed to be selected for because they aid parasitic exploitation by increasing within-host growth, ultimately helping parasites to transmit more efficiently between hosts (Frank, 1996). Although this theory fits well with specialised host–parasite interactions, it does not explain the evolution of virulence with opportunistic pathogens whose generalist lifestyle is less tightly connected to specific hosts or environments (Brown et al., 2012). Virulence traits might then be maintained because they provide a selective advantage in ecological contexts unrelated to host exploitation (Levin and Eden, 1990; Levin, 1996; Matz and Kjelleberg, 2005; Casadevall and Pirofski, 2007; Brown et al., 2012).
One indirect benefit of bacterial virulence is the ability of bacteria to resist protist predation (Matz and Kjelleberg, 2005; Brown et al., 2012; Friman et al., 2013a, 2013b). Protists are an important class of bacterial predators found across diverse environments, and correlational evidence suggests that resistance to protist predation could preadapt some environmental bacteria to survive and proliferate within human macrophages (Cirillo et al., 1999; Harb et al., 2000; Matz and Kjelleberg, 2005; Rasmussen et al., 2005; Brussow, 2007). Moreover, it has been shown that a Shiga toxin of Escherichia coli O157:H7 increases its survival in the presence of protozoan predators (Steinberg and Levin, 2007; Lainhart et al., 2009), whereas protist predation can increase the frequency of Pseudomonas strains that produce virulence-enhancing ‘public goods' (Harrison et al., 2006; Jousset et al., 2009; Kohler et al., 2009; Rumbaugh et al., 2009; Friman et al., 2013a). However, direct experimental evolution tests of this hypothesis are more ambiguous: for example, serial passaging bacteria through macrophages can actually lead to either enhanced or reduced growth within amoeba (Ensminger et al., 2012), whereas P. aeruginosa long-term adaptation in the lungs of cystic fibrosis (CF) patients leads to both lowered acute virulence and defence against the ciliate protist Tetrahymena pyriformis (Friman et al., 2013b). Also, predation by T. thermophila has been shown to lead to a reduction in virulence of Serratia marcescens through pleiotropic effects of reduced bacterial growth and colonization ability (Friman et al., 2009; Mikonranta et al., 2012). It is thus unclear if any generalizations can be made.
The impact of another ubiquitous environmental selection pressure, lytic phages (Pernthaler, 2005; Brussow, 2007; Selezska et al., 2012), on virulence evolution is similarly ambiguous. As with protist predation, phages can maintain bacterial virulence via production of public goods (Morgan et al., 2012), or by selecting for upregulation of bacterial virulence factors (Hosseinidoust et al., 2013). However, bacterial resistance to phages is also frequently associated with pleiotropic costs in terms of reduced growth rate (Lenski, 1988; Bohannan and Lenski, 1999; Bohannan et al., 2002; Buckling et al., 2006) or reduced colonization ability (Laanto et al., 2012) that can result in reductions in virulence in insect (Friman et al., 2011) or fish hosts (Laanto et al., 2012).
Selection pressures imposed by either protist or phage enemies may be affected by the presence of the other enemy they interact with. For example, resistance to phages can constrain resistance to protists (Friman and Buckling, 2012). This kind of pleiotropy between different defence strategies could also constrain the effects of resistance against enemies on the correlated evolution of bacterial virulence. Although the above studies have shown that trait correlations can be important drivers of coincidental bacterial virulence in simple communities, it is still unclear how pleiotropic effects between defensive and virulence traits shape bacterial virulence in multienemy communities.
Here we tested experimentally how concurrent protist- (T. thermophila; bacteriovorous predator) and phage- (PNM; bacteria-specific virus) imposed selection shape the coincidental evolution of P. aeruginosa virulence (as measured in wax moth larvae), through correlations with other associated life-history traits. Even in the context of the widely studied opportunistic pathogen P. aeruginosa, the impact of single enemy-imposed selection on virulence is unclear. On the one hand, the evolution of resistance to both T. thermophila protist and PNM phage is associated with reduce growth rate, and hence, potentially lowered virulence (Friman and Buckling, 2012; Hall et al., 2012). On the other hand, short-term selection by T. pyriformis protist can favour intact quorum-sensing systems, which is associated with increased virulence (Friman et al., 2013a). In some cases, long-term selection within the lungs of CF patients has been shown to correlate with both reduced virulence and lowered resistance to phage and protist enemies (Friman et al., 2013b). Our results show that protist selection can lead to attenuation of P. aeruginosa virulence through pleiotropic effects with reduced growth ability. Interestingly, phages constrained this effect, and the data suggest that it is because phages reduced the strength of protist selection through competition for the same prey (leading to decrease in protist numbers) and by increasing bacterial edibility for protists. In the light of previous studies (Friman et al., 2009; Mikonranta et al., 2012; Friman et al., 2013a), we suggest that ciliate predation could be an important driver of bacterial virulence evolution in natural microbial communities, whereas this effect could be dampened by parasitic bacteriophages.
Materials and methods
Study species, culture conditions and selection experiment
We used a full factorial experimental design where PAO1 P. aeruginosa bacterium was cultured alone and in the presence of either a phage, protist or both for approximately 1 month. We used serial transfer (transfer of small volumes of experimental populations to fresh media) to renew resources in our batch culture microcosms every fourth day. At every transfer, bacterial, protist and phage densities were measured from a subsample extracted from microcosms. After the last transfer, bacteria were isolated from protists and phages by plating on agar plates and various phenotypic measures (defence against both protist and phage enemies, defence-associated pleiotropic growth cost and bacterial virulence in wax moth larvae) of individual bacterial clones were determined.
In more detail, twenty 25 ml glass vials (microcosms), each containing 6 ml of 1% King's Medium B (100-fold diluted KB; M9 salt solution supplemented with 10 g l−1 glycerol and 20 g l−1 proteose peptone), were inoculated with approximately 105 cells of P. aeruginosa isolate PAO1 (ATCC, Manassas, VA, USA; no. 15692). Five of the microcosms were inoculated with 3.6 × 103 clonal particles of PNM phage (Isolated from Mtkvari River, Georgia, 1999; Podoviridae C1 (Merabishvili et al., 2007); phage treatment), five with 246 cells of T. thermophila protist (CCAP strain 1630/1U; protist treatment), five with the same number of both enemies (protist–phage treatment), whereas five microcosms contained only bacterium (bacterium-alone, i.e. control treatment). Both phages and ciliates are commonly found in natural environments (Elliott, 1959; Merabishvili et al., 2009), where they cause high bacterial mortality and hence strong selection for resistance (Faruque et al., 2005; Pernthaler, 2005; Selezska et al., 2012). The microcosms were propagated at 28 °C in non-shaken conditions. One microliter of each culture was transferred to 24 ml of fresh medium every fourth day for a total of five transfers (24 days). Microcosms were vortexed for 1 min before sampling and transfered to fresh media to homogenise cultures. Each selection line was maintained for 24 days and populations were frozen at −80 °C in 20% glycerol at each sampled transfer.
Bacterial densities were determined with a spectrophotometer at every transfer (Biotek, optical density (OD) of 600 nm; Bio-Tek Instruments Inc., Winooski, VT, USA) and later by plating bacterial dilutions after the last transfer onto KB agar plates and counting the number of colony-forming units after 24 h incubation at 28 °C. To isolate phages from bacteria, 100 μl of chloroform was first added to 900 μl of subsamples, vortexed to lyse the bacterial cells and then centrifuged at 11 000 g for 3 min to pellet the bacteria debris, leaving a suspension of phage in the supernatant. Phage population densities were measured by plating phage dilutions onto soft agar plates containing the ancestral PAO1 bacterial cells and counting the number of plaque-forming units after 24 h at 28 °C. Protist cells were counted directly under a microscope (Leica DM IL Led, Leica Mikrosysteme Vertrieb GmbH, Wetzlar, Germany; × 10 magnification) as described previously (Friman and Buckling, 2012).
Measuring bacterial defence against protists
To measure changes in bacterial defence against protists, we randomly isolated 12 independent bacterial colonies from the last serial transfer and inoculated selected colonies into 1% KB medium for overnight incubation at 28 °C, and freezing in 20% glycerol. Note that in some cases, independently isolated colonies may have been genetically identical. The 12 independent bacterial colonies per replicate population (a total of 240 individual colonies) were first grown to similar densities in 96-well plates (24 h, 28 °C and in 200 μl of 1% KB medium; OD of 0.214±0.004; Biotek, OD of 600 nm) before we added 30 μl of ancestral protist inocula to each well (approximately 2800 cells ml−1). By adjusting initial bacterial densities to be approximately the same across wells, subsequent protist growth was not affected by bacterial growth differences but only by differences in the strength of bacterial antipredatory defence (Friman and Buckling, 2012). After 24 h of cocultivation at 28 °C, protist cell numbers were counted under a microscope (Leica DM IL Led; × 10 magnification). Bacterial defence was determined indirectly from the number of protist cells: the fewer the number of protist cells, the higher the bacterial defence (Friman and Laakso, 2011; Friman and Buckling, 2012). Mechanistically, bacterial defence against T. thermophila protist has previously been connected to enhanced biofilm formation, that is, enhanced formation of cell aggregates and attachment on culture vessels' surfaces (Mikonranta et al., 2012). Thus, we measured bacterial biofilm formation by growing individual colonies first for 24 h in 1% KB, and then 50 μl of 1% crystal violet solution was added to the microplate wells and later rinsed off with distilled water after 10 min. Crystal violet-stained bacteria were dissolved in 96% ethanol and the amount of biofilm measured as OD at 600 nm (O'Toole and Kolter, 1998).
Measuring bacterial resistance to phages
To measure changes in bacterial resistance and phage infectivity over time, we isolated 12 independent bacterial colonies from every population at every second transfer as described above (transfers 2, 4 and 6). Similarly, phage populations were isolated from bacteria with chloroform treatment, as described above. Bacterial resistance and phage infectivity were measured at the population level as the proportion of resistant bacteria (Buckling and Rainey, 2002). More specifically, resistance of a bacterial population was determined by streaking 12 independently isolated bacterial colonies across a line of phage (40 μl) that had previously streaked and dried on a KB agar plate. A colony was scored as resistant if there was no inhibition of growth by the phage. We used ‘time-shift' assays to measure bacteria–phage coevolution (Buckling and Rainey, 2002; Brockhurst et al., 2003). Specifically, we estimated the resistance of bacterial populations at transfers 2, 4 and 6 to their own phages (originating from the same population as the measured bacteria), which had been isolated from the current transfer (e.g. resistance of t2 bacterium to t2 phage), two transfers from the past (resistance of t2 bacterium to t0 phage) and two transfers from the future (resistance of t2 bacterium to t4 phage), allowing resistance and infectivity evolution of bacteria and phage to be determined (Friman and Buckling, 2012). Bacterial resistance against ancestral phage was also measured.
Measuring bacterial pleiotropic growth cost
To determine potential pleiotropic costs of defence, bacterial growth was measured in the absence of enemies after the sixth transfer (OD of 600 nm, 24 h, 28 °C and in 200 μl of 1% KB medium). The same bacterial colonies that were used for phage resistance and protist defence assays were also used for these growth measurements.
Measuring bacterial virulence
Change in bacterial virulence was measured using wax moth larvae (Galleria mellonella, Lepidoptera; Pyralidae; Livefood UK Ltd, Rooks Bridge, UK). It has been shown previously that bacterial virulence measured in G. mellonella correlates with virulence measured in mammals and mammalian cell cultures, making it a useful model host for determining virulence (Jander et al., 2000). Bacterial virulence was measured at the level of individual colonies as described earlier (Friman et al., 2009; Mikonranta et al., 2012). Before infections, we first grew bacteria for 24 h and subsequently diluted all colonies to approximately similar densities (OD of 0.2 at 600 nm equalling approximately 2.4 × 109 cells ml−1; treatment: F3,16=1.6, P=0.4). Two ancestral-like colonies were chosen if populations contained only this one colony phenotype (all populations in bacteria-alone and bacteria–phage treatments), and two ancestral-like and two small colonies were used when populations contained both ancestral-like and small colony phenotypes (all populations in bacteria–protist and bacteria–phage–protist treatments). As a result, a total of 60 individual colonies were used for infection experiments. Virulence of every individual colony was tested in eight independent wax moth larvae. We also infected 60 larvae with an ancestral colony, and additionally, 60 larvae with M9 salt solution to control for the damage caused by the injection itself. The larvae were injected with 30 μl of bacterial solution or M9 buffer between the abdominal segments six and seven with an 1 ml Terumo syringe. The larvae were infected during four consecutive days in constant laboratory conditions and there was no difference in larval weight between treatments (F3,156=1.13, P=0.34). After infection, larvae were placed on individual wells of 24-well cell culture plates and the survival was monitored at 2-h intervals for 3 days at 28 °C. Larvae were scored as dead when they did not respond to touch with forceps. Larvae that were still alive after 3 days from the infection were given a time of death of 120 h.
Statistical analyses
Bacterial, phage and protist population densities and colony frequency data were analysed as general linear mixed models, where replicate populations were fitted as a random factor, and time and community type as fixed factors. Colony frequencies were arcsine-transformed before the analysis. Bacterial growth in the absence of enemies (indicative of pleiotropic cost of defence) and defence against protist was analysed, first, at the level of treatments and, second, at the level of different colony types as general linear mixed models. In both analyses, replicate population were fitted in the model as random factors. In the case of defence assays, bacterial initial density was included into the model as a covariate to control statistically for the slight (but nonsignificant; treatment: F3,19, P=0.044) differences in bacterial densities before the addition of protist inocula. Changes in bacterial virulence were analysed as a general linear mixed model: mean time of death after injection (colony means within treatment) was explained with treatment and colony type, and replicate population was fitted as a random factor in the model. We also used multivariate, explanatory factor analysis to find underlying factors that best describe evolved prey types in different treatments. Analysis was conducted on whole colony data (total of 240 individual colonies) and was based on four measured phenotypic traits: bacterial growth in the absence of enemies, bacterial colony type, bacterial defence against protists and bacterial biofilm formation. Varimax rotation and Kaiser criterion was used to only include factors with eigenvalues over 1.
Results
Population dynamics and protist-driven bacterial diversification
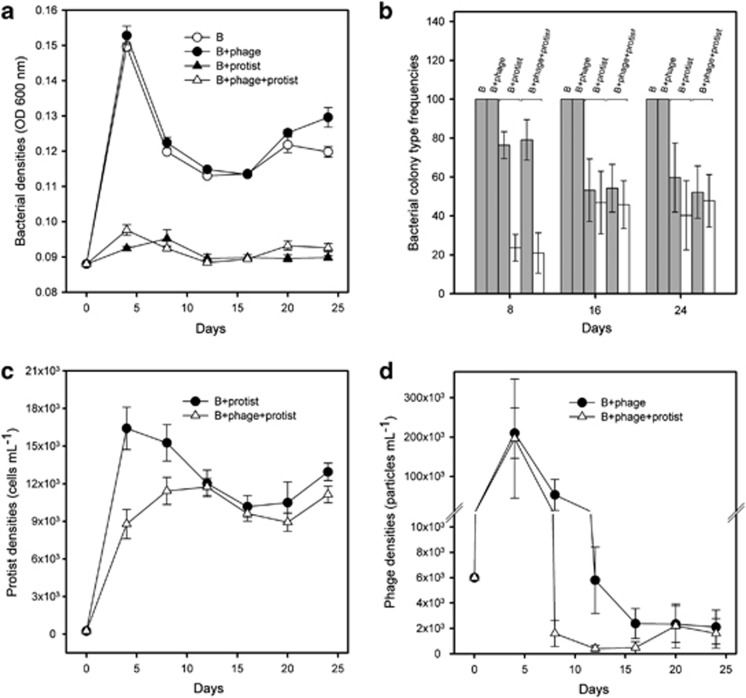
Compared with bacterium-alone treatment, protist predation decreased bacterial densities to a similar extent in the absence and presence of phages (protist: F1,16=2362, P<0.001; protist × phage: F1,16=2.7, P=0.12; Figure 1a), whereas phages slightly increased bacterial densities in the absence of protist (phage: F1,16=11.6, P=0.004; Figure 1a). Note that these bacterial OD measures corresponded well with bacterial colony-forming units; CFU ml−1 obtained at the last transfer: protists clearly decreased CFU ml−1, regardless of the presence of phage (protist: F1,16=118, P<0.001; protist × phage: F1,16=0.7, P=0.42), whereas phage alone did not affect bacterial densities (pairwise comparison P=0.3; Supplementary Figure 1).
Figure 1.
Bacterial population densities measured as OD (600 nm) (a), bacterial colony-type frequencies (b), protist cell densities (c) and phage densities (d) in bacterium-alone (B), bacterium–phage (B+phage), bacterium–protist (B+protist) and bacterium–phage–protist (B+phage+protist) treatments. Grey bars in panel (b) depict frequency of ancestral-like, and white bars frequency of small colony types. Please note, small colony types did not emerge in the bacterium-alone and bacterium–phage treatments, and thus no white bars are shown in panel (b). All panels show means of five independent replicates per treatment (N=5) and error bars in all panels denote ±1 s.e.m.
The presence of phages decreased protist densities compared with protist-only treatment (phage: F1,8=27.1, P=0.001; Figure 1c), whereas phage densities decreased to very low levels through time both in the absence and presence of protists (protist: F1,8=0.16, P=0.7; time: F5,8=15.5, P=0.001; Figure 1d). However, phages went to undetectable densities in three out of the five selection lines by the third transfer in the presence of protists, whereas phages persisted to the end of the experiment in all the populations in the absence of protists.
Smooth and large ancestral-like P. aeruginosa colony types were found in all treatments, whereas small and white colony types were detected only in the presence of protists, regardless of the presence of phages (protist: F1,16=22.28, P<0.001; Figure 1b). Phages had no effect on bacterial diversification (phage: F1,16=0, P=0.98; Figure 1b). Taken together, these results suggest that both enemies reduced each other's densities, protists had a greater effect on phages than vice versa and only protists affected P. aeruginosa phenotypic diversification.
Evolution of bacterial defence against protist and phage enemies
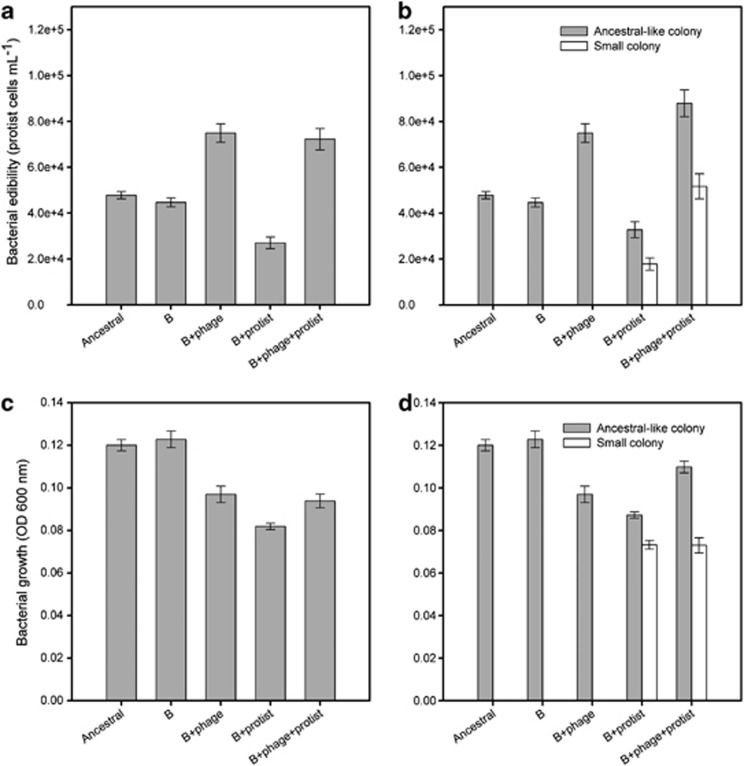
Bacteria that were previously preyed upon by protists supported less protist growth compared with bacteria that had evolved in the absence of protists (treatment: F3,16=15.3, P<0.001; pairwise comparison P=0.049; Figure 2a). By contrast, bacteria that were previously preyed upon by phages supported higher protist growth compared with bacteria that had evolved in the absence of protists, regardless of whether the bacteria originated from bacterium–phage or bacterium–phage–protist treatments (pairwise comparisons P<0.01; Figure 2a). We next analysed antipredatory defence of different colony types, and found that small colony types supported less protist growth compared with ancestral-like colony types within both bacterium–protist and bacterium–protist–phage treatments (colony type: F1,7.9=17.8, P=0.003; Figure 2b). Moreover, small colony types from bacterium–protist treatment supported less protist growth compared with small colony types from bacterium–protist–phage treatment (pairwise comparison: F1,2.4=25.5, P=0.024; Figure 2b). Interestingly, exposure to phages during the selection experiment resulted in only the ancestral-like colony types supporting higher protist densities (treatment: F3,14.3=11.5, P<0.001; pairwise comparisons P<0.01; Figure 2b).
Figure 2.
Bacterial edibility (i.e., defence) for protist (a and b) and bacterial growth in the absence of enemies (c and d) between ancestral (A), bacterium-alone (B), bacterium–phage (B+phage), bacterium–protist (B+protist) and bacterium–phage–protist (B+phage+protist) treatments. Panels (a and c) show differences at the level of treatments (means of 12 colonies per replicate; means of five independent replicates per treatment, N=5), and panels (b and d) at the level of colony types within treatments (means of small and ancestral-like colony types per replicate; five independent replicates per treatment). Twelve measurement replicates were used for the ancestor (A). Error bars in all panels denote ±1 s.e.m.
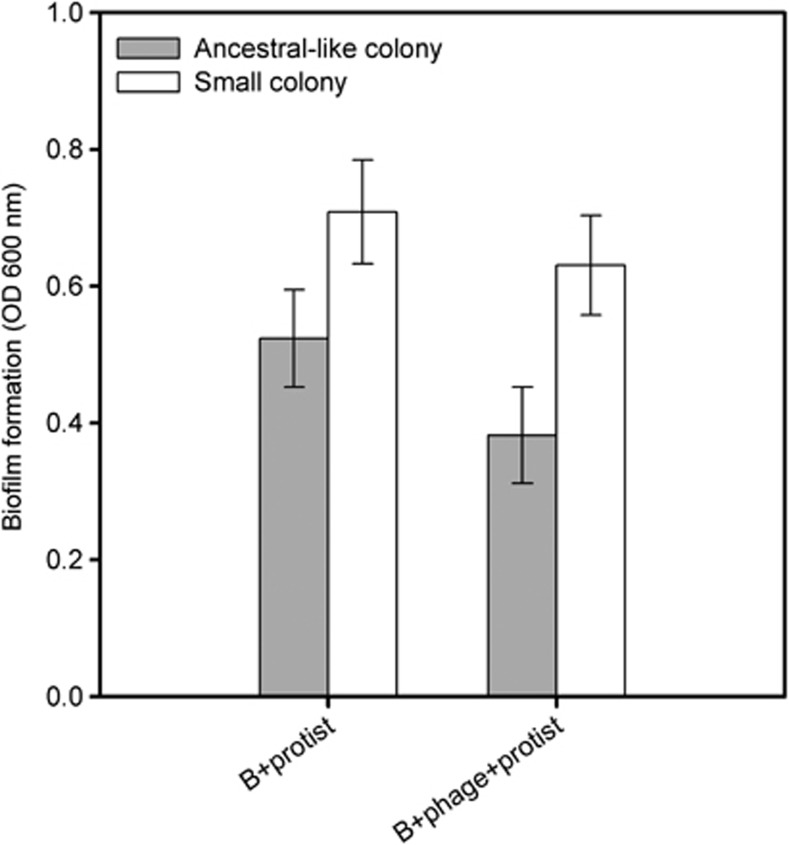
Although the mean bacterial biofilm formation did not differ between protist–bacterium and protist–phage–bacterium treatments (treatment: F1,16=2.8, P=0.07), small colony types formed more biofilm compared with ancestral-like colony types within both of these treatments (colony type: F1,16.2=8.9, P=0.009; colony type × treatment: F1,16.2=0.2, P=0.67; Figure 3). These results suggest that increased antipredatory defence was likely connected to increased biofilm formation.
Figure 3.
Bacterial biofilm formation for different colony types within bacterium–protist (B+protist) and bacterium–protist–phage (B+phage+protist) treatments (five independent replicates per treatment; means of ancestral-like and small colonies per replicate). Error bars denote ±1 s.e.m.
Surprisingly, none of the bacteria (irrespective of whether they had evolved in the absence or presence of protist during the selection experiment) evolved resistance against ancestral phages, or phage isolated from contemporary, past or future transfers (data not shown).
Taken together these results suggest that P. aeruginosa phenotypic diversification was connected to increased defence against protist predation and increased biofilm formation, whereas the level of evolved defence against protists was constrained by phage-imposed selection. Moreover, bacteria were unable to coevolve with phages in these experimental conditions.
Pleiotropic growth cost measured in the absence of enemies
When grown in the absence of any enemies, bacteria that had been experimentally evolved in the presence of protists reached lower maximum densities compared with bacteria from the bacterium-only evolution treatment (maximum densities reached within 24 h; protist: F1,16=9.5, P=0.007). Evolution in the presence of phages did not change bacterial maximum densities when compared with bacteria originating from the bacterium-alone treatment (maximum densities reached within 24 h; phage: F1,16=0.94, P=0.34; Figure 2c). Moreover, evolution in the presence of protists reduced bacterial growth to a greater extent when phages were absent during evolution (protist × phage: F1,16=7, P=0.017; Figure 2c). The reduction in bacterial growth was especially pronounced for small colony types (colony type: F1,26=7, P=0.017; Figure 2d), regardless of whether they originated from bacterium–protist or bacterium–protist–phage evolution treatments (phage: F1,8.2=0.03, P=0.85; Figure 2d). We infer this growth cost to be a consequence of pleiotropic changes that affected bacterial defence. Note that bacterial growth differences did not affect the defence measurements because bacterial initial densities were adjusted to approximately same before adding protist inocula (for more detail see Materials and methods).
Multivariate analysis of bacterial life-history trade-offs
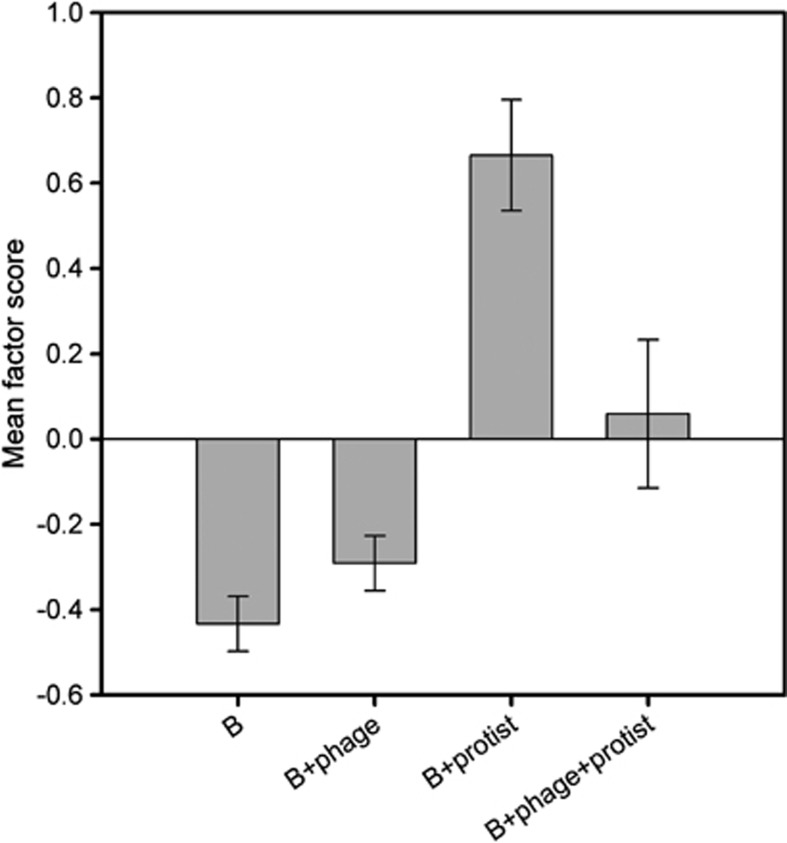
Multivariate factor analysis was able to identify only one factor on the basis of the four included traits. This factor loaded positively on biofilm formation (0.428) and colony type (0.832), and negatively on bacterial growth (−0.722) and edibility for protists (−0.513). As a result, positive factor scores on this factor are indicative of small colony types that form lots of biofilm, are relatively inedible to protists and grow slowly; hence, this factor could be described as a ‘defence' factor. The factor scores of P. aeruginosa colonies clearly differed between different treatments (treatment: F3,16=3.84, P=0.03; Figure 4). Evolution with protists resulted in high factor scores in bacterium–protist treatment (t(59)=5.114, P<0.001, Figure 4). However, this effect disappeared when also evolved in the presence of phage; factor scores were close to zero in bacterium–phage–protist treatment (t(59)=0.342, P=0.734, Figure 4). Colonies from both bacterium-alone and bacterium–phage treatments had negative mean factor scores (t(59)=−6.697, P<0.001 and t(59)=−4.521, P<0.001; Figure 4, respectively), suggesting that these colonies were generally poor at forming biofilm, had an ancestral-like phenotype, grew fast and were highly edible to protists. These results support the previous univariate analyses, suggesting that antipredatory defence incurred pleiotropic growth cost and was mechanistically linked to enhanced biofilm formation. However, evolution of these trait correlations was dampened in the presence of phage.
Figure 4.
Bacterial mean scores for defence factor between bacterium-alone (B), bacterium–phage (B+phage), bacterium–protist (B+protist) and bacterium–phage–protist (B+phage+protist) treatments (factoring is based on trait values of 12 independent colonies per replicate per treatment resulting in 240 individual colonies). The positive factor loadings denote for small colony types that form lots of biofilm, are relatively inedible to protists and show reduced growth. The negative factor loadings denote ancestral-like colony types that form little biofilm, are relatively edible to protists and grow well. Error bars denote ±1 s.e.m.
Evolutionary changes in bacterial virulence
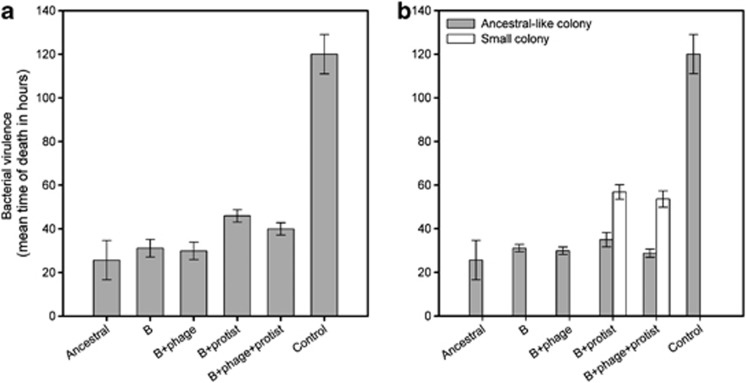
Bacteria that had evolved in the bacterium–protist treatment showed attenuated mean virulence compared with both bacteria evolved in the absence of enemies and ancestral bacteria (treatment: F5,58=19.4, P<0.001; pairwise comparisons: P=0.004 and P=0.035, respectively; Figure 5a). While in bacteria the bacterium–phage–protist treatment did not significantly differ in virulence from the bacterium–protist treatment (P=0.143), pairwise comparisons between the bacterium–phage–protist treatment with both the bacterium-alone and ancestral treatments were nonsignificant (P=0.077 and 0.133, respectively; Figure 5a). This suggests that evolution with protists did not lead to such a large attenuation of bacterial virulence when also evolved in the presence of phages.
Figure 5.
Bacterial virulence (mean time of death of wax moth larvae after bacterial infection (in h)) between ancestral (A), bacterium-alone (B), bacterium–phage (B+phage), bacterium–protist (B+protist), bacterium–phage–protist (B+phage+protist) and control (M9 buffer) treatments (a and b). (a) Differences between treatment means (five replicates per treatment; mean of eight infected larvae per colony; colony means per replicate, N=5). (b) Differences between different colony type means within treatments (five replicates per treatment; mean of eight infected larvae per colony; means of ancestral-like and small colonies per replicate). Moreover, 60 measurement replicates (larvae) were used for ancestral and control treatments. In both panels, error bars in all panels denote ±1 s.e.m.
Despite there being treatment differences in virulence, ancestral-like colony types isolated from different treatments did not differ in virulence (treatment: F1,18.3=0.98, P=0.44; Figure 5b). Small colony types had attenuated virulence irrespective of whether they originated from bacterium–protist or bacterium–phage–protist treatments (colony type: F1,28.8=63, P<0.001; colony type × phage: F1,28.8=0.28, P=0.6; Figure 5b).
Discussion
Here we studied how protist and phage enemies, alone and in combination, affected the evolution of bacterial virulence. We found that protist selection diversified bacterial populations into two different colony types: virulent ancestral-like types and less virulent but more defensive, small colony types. The net result of evolving with protists was reduced bacterial virulence. Phages had no direct effect on the evolution of bacterial diversification, phage resistance or virulence. However, evolving with phages attenuated bacterial defence against protists and its associated growth cost, which is also likely to explain why bacterial virulence was reduced to a lesser extent when concurrently exposed to both protists and phages. Taken together, our results suggest that the evolution of trait correlations is dependent on the ecological context and complexity of the surrounding community. In the case of coincidental evolution of virulence, enemy-mediated effects on virulence are likely to be less predictable in multienemy communities.
Protist predation has previously been shown to result in the evolution of both increases and decreases in bacterial virulence (Cirillo et al., 1999; Matz and Kjelleberg, 2005; Rasmussen et al., 2005; Steinberg and Levin, 2007; Friman et al., 2009; Lainhart et al., 2009; Mikonranta et al., 2012). Here we found that protist-imposed selection resulted in reduced virulence, which is in line with a previous study where T. thermophila was observed to select for highly defensive but less virulent small colony types of another opportunistic bacterial pathogen, S. marcescens (Friman et al., 2008, 2009; Mikonranta et al., 2012). In this previous study, bacterial defence was connected to S. marcescens' ability to form protozoan-resistant biofilm that could not be consumed by T. thermophila (Mikonranta et al., 2012). We found a similar pattern in our experiment: small colony types isolated from protist-present treatments formed biofilms more efficiently compared with ancestral-like P. aeruginosa colony types. Moreover, enhanced biofilm formation correlated negatively with P. aeruginosa growth, potentially providing a causal link between biofilm formation-mediated antipredatory defence, pleiotropic growth costs and reduced virulence (Mikonranta et al., 2012). Although it has been shown previously that protist predation can affect multiple bacterial virulence factors (Matz and Kjelleberg, 2005), reduced growth rate, as observed in vitro, is the most parsimonious explanation for reduced P. aeruginosa virulence in this experiment (Harrison et al., 2006; Friman et al., 2013b).
Interestingly, bacteria and phage did not coevolve during the experiment, and as a result, no correlation between phage-only selection and evolved virulence was observed. This lack of evolved resistance to phages is in contrast with a previous experiment (Hall et al., 2012), where P. aeruginosa evolved very high levels of resistance to PNM phage within few weeks. A possible explanation for this discrepancy is the use of different resource concentrations. In this experiment, we chose to use very diluted KB concentration (1% KB instead of 100% KB used in Hall et al., 2012), which is more reminiscent of productivity levels observed in external aquatic environments, and crucially, allows coexistence of protists and bacteria (Friman et al., 2013a). It has been shown that phage–bacteria coevolution can be reduced in low nutrient environments because the strength of selection is constrained in small populations owing to low mutation supply rate and infrequent encounter rates (Lopez-Pascua and Buckling, 2008).
Despite no apparent effect of phages on the evolution of phage resistance, the presence of phages constrained the evolution of bacterial antiprotist defence, leading to smaller pleiotropic growth cost and less pronounced reductions in bacterial virulence. We are aware of two non-mutually exclusive explanations for this attenuation. First, phages reduced protist densities, in turn reducing protist-imposed selection for defence. Second, there was a pleiotropic trade-off between phage and protist resistance, as observed for the closely related bacterium P. fluorescens (Friman and Buckling, 2012), such that defence against protists increases susceptibility to phages. As bacteria did not evolve intrinsic resistance to phages in any treatment, this hypothesised trade-off between phage and protist resistance may have been mediated by phages transmitting more effectively in the denser biofilms that were selected by protists. Consistent with this view, phage-imposed selection tended (P=0.07) to reduce the extent of biofilm formation by bacteria evolved in the presence of protists.
Small P. aeruginosa colony types have also been frequently detected in infections of CF patients (Malone et al., 2010). Similar to our study, these small colony types were able to form highly adhesive biofilm structures, were efficient at resisting nematode predation and scavenging by macrophages and could persist for weeks in a mouse infection model despite their considerable fitness disadvantage in vitro (Malone et al., 2010). This raises an important question: do small P. aeruginosa colony types originate from environmental reservoirs owing to protist predation, or evolve independently within the CF patient lungs owing to similar selective pressure exerted by macrophages? In the case of CF, small colony variants (SCVs) are associated with chronic, long-term, infections (Starkey et al., 2009; Malone et al., 2010; Folkesson et al., 2012), which suggest that they arise independently in the later stages of infection within the lungs of CF patients. These SCVs also often show elevated capacity to form biofilms (Starkey et al., 2009). Thus, even though environmental reservoirs may not be the source for SCVs in P. aeruginosa CF infections, it is possible that the traits that enforce P. aeruginosa persistence in CF lungs have been originally selected in environmental contexts under protist predation. In the future, these hypotheses could be tested by measuring changes in P. aeruginosa virulence in in vitro and in vivo mammalian models, to study directly if selection by both protists and macrophages result in parallel evolutionary changes in bacteria. It would be interesting to study if phenotypic similarities manifest at the molecular level. For example, protist-selected and clinical SCVs often show similar changes in terms of reduced growth, reduced virulence and enhanced biofilm formation (Malone et al., 2010; Friman and Laakso, 2011; Mikonranta et al., 2012; Friman et al., 2013b), which could be driven by mutations in electron transport and ATP synthesis, leading to lesser investment in cell-wall biosynthesis, and hence, reduced growth and pigment expression (Proctor et al., 2006). Currently, lots of genomic information is available for clinical P. aeruginosa SCVs owing to recent advances in whole genome sequencing. This knowledge could serve as a starting point to look for signs of parallel molecular evolution with protist-selected SCVs (e.g. similar mutation in c-di-GMP levels regulating yfiBNR operon, las and PQS quorum-sensing systems and lipopolysaccharide modification operons such as arnBCADTEF (Proctor et al., 2006)).
In the light of this and previous experiments (Friman et al., 2009; Mikonranta et al., 2012), we propose that predation by T. thermophila ciliates might consistently select for highly defended but poorly growing and less virulent SCVs across different opportunistic pathogens (S. marcescens and P. aeruginosa). Ciliate predation could thus potentially drive parallel evolutionary changes among bacteria in diverse microbial communities. However, the presence of other species, like bacteriophages, might weaken ciliate-imposed selection, resulting in less clear correlative changes between bacterial life-history traits (Friman and Buckling, 2012). Establishing these kind of causal links could allow us to use microbial community composition as a predictor for environments that might act as hot spots for coincidental selection of bacterial virulence. Moreover, determining how virulence traits and adaptations related to abiotic factors are correlated could further help in understanding the environmental circumstances where opportunistic bacteria turn into notorious pathogens.
Acknowledgments
This study was funded by Marie-Curie Intra-European Fellowship within the 7th European Community Framework Programme (VF), Wellcome Trust Institutional Strategic Support Fund fellowship (VF), Imperial College Junior Research Fellowship (VF), NERC (AB), Royal Society (AB) and BBSRC (AB).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bohannan BJ, Kerr B, Jessup CM, Hughes JB, Sandvik G. Trade-offs and coexistence in microbial microcosms. Antonie van Leeuwenhoek. 2002;81:107–115. doi: 10.1023/a:1020585711378. [DOI] [PubMed] [Google Scholar]
- Bohannan BJM, Lenski RE. Effect of prey heterogeneity on the response of a model food chain to resource enrichment. Am Naturalist. 1999;153:73–82. doi: 10.1086/303151. [DOI] [PubMed] [Google Scholar]
- Brockhurst MA, Morgan AD, Rainey PB, Buckling A. Population mixing accelerates coevolution. Ecol Lett. 2003;6:975–979. [Google Scholar]
- Brown SP, Cornforth DM, Mideo N. Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 2012;20:336–342. doi: 10.1016/j.tim.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H. Bacteria between protists and phages: from antipredation strategies to the evolution of pathogenicity. Mol Microbiol. 2007;65:583–589. doi: 10.1111/j.1365-2958.2007.05826.x. [DOI] [PubMed] [Google Scholar]
- Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Wei Y, Massey RC, Brockhurst MA, Hochberg ME. Antagonistic coevolution with parasites increases the cost of host deleterious mutations. Proc Biol Sci. 2006;273:45–49. doi: 10.1098/rspb.2005.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6:2169–2174. doi: 10.1128/EC.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo JD, Cirillo SL, Yan L, Bermudez LE, Falkow S, Tompkins LS. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect Immun. 1999;67:4427–4434. doi: 10.1128/iai.67.9.4427-4434.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott AM. Biology of Tetrahymena. Annu Rev Microbiol. 1959;13:79–96. [Google Scholar]
- Ensminger AW, Yassin Y, Miron A, Isberg RR. Experimental evolution of Legionella pneumophila in mouse macrophages leads to strains with altered determinants of environmental survival. PLoS Pathogens. 2012;8:e1002731. doi: 10.1371/journal.ppat.1002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque SM, Naser IB, Islam MJ, Faruque AS, Ghosh AN, Nair GB, et al. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci USA. 2005;102:1702–1707. doi: 10.1073/pnas.0408992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- Frank SA. Models of parasite virulence. Q Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- Friman V-P, Diggle SP, Buckling A. Protist predation can favour cooperation within bacterial species. Biol Lett. 2013a;9:20130548. doi: 10.1098/rsbl.2013.0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friman V-P, Ghoul M, Molin S, Johansen HK, Buckling A. Pseudomonas aeruginosa adaptation to lungs of cystic fibrosis patients leads to lowered resistance to phage and protist enemies. PLoS One. 2013b;8:e75380. doi: 10.1371/journal.pone.0075380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friman V-P, Buckling A. Effects of predation on real-time host–parasite coevolutionary dynamics. Ecol Lett. 2012;16:39–46. doi: 10.1111/ele.12010. [DOI] [PubMed] [Google Scholar]
- Friman VP, Hiltunen T, Jalasvuori M, Lindstedt C, Laanto E, Ormala AM, et al. High temperature and bacteriophages can indirectly select for bacterial pathogenicity in environmental reservoirs. PLoS One. 2011;6:e17651. doi: 10.1371/journal.pone.0017651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friman VP, Hiltunen T, Laakso J, Kaitala V. Availability of prey resources drives evolution of predator–prey interaction. Proc Biol Sci R Soc. 2008;275:1625–1633. doi: 10.1098/rspb.2008.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friman VP, Laakso J. Pulsed-resource dynamics constrain the evolution of predator–prey interactions. Am Naturalist. 2011;177:334–345. doi: 10.1086/658364. [DOI] [PubMed] [Google Scholar]
- Friman VP, Lindstedt C, Hiltunen T, Laakso J, Mappes J. Predation on multiple trophic levels shapes the evolution of pathogen virulence. PLoS One. 2009;4:e6761. doi: 10.1371/journal.pone.0006761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AR, De Vos D, Friman VP, Pirnay JP, Buckling A. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl Environ Microbiol. 2012;78:5646–5652. doi: 10.1128/AEM.00757-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb OS, Gao LY, Abu Kwaik Y. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ Microbiol. 2000;2:251–265. doi: 10.1046/j.1462-2920.2000.00112.x. [DOI] [PubMed] [Google Scholar]
- Harrison F, Browning LE, Vos M, Buckling A. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 2006;4:21. doi: 10.1186/1741-7007-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinidoust Z, van de Ven TG, Tufenkji N. Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl Environ Microbiol. 2013;79:6110–6116. doi: 10.1128/AEM.01421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Rahme LG, Ausubel FM. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset A, Rochat L, Pechy-Tarr M, Keel C, Scheu S, Bonkowski M. Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J. 2009;3:666–674. doi: 10.1038/ismej.2009.26. [DOI] [PubMed] [Google Scholar]
- Kohler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci USA. 2009;106:6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laanto E, Bamford JK, Laakso J, Sundberg LR. Phage-driven loss of virulence in a fish pathogenic bacterium. PLoS One. 2012;7:e53157. doi: 10.1371/journal.pone.0053157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainhart W, Stolfa G, Koudelka GB. Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J Bacteriol. 2009;191:5116–5122. doi: 10.1128/JB.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE. Experimental studies of pleiotropy and epistasis in Escherichia coli.1. Variation in competitive fitness among mutants resistant to virus-T4. Evolution. 1988;42:425–432. doi: 10.1111/j.1558-5646.1988.tb04149.x. [DOI] [PubMed] [Google Scholar]
- Levin BR. The evolution and maintenance of virulence in microparasites. Emerg Infect Dis. 1996;2:93–102. doi: 10.3201/eid0202.960203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Eden CS. Selection and evolution of virulence in bacteria—an ecumenical excursion and modest suggestion. Parasitology. 1990;100:S103–S115. doi: 10.1017/s0031182000073054. [DOI] [PubMed] [Google Scholar]
- Lopez-Pascua LC, Buckling A. Increasing productivity accelerates host–parasite coevolution. J Evol Biol. 2008;21:853–860. doi: 10.1111/j.1420-9101.2008.01501.x. [DOI] [PubMed] [Google Scholar]
- Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C, et al. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathogen. 2010;6:e1000804. doi: 10.1371/journal.ppat.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Kjelleberg S. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 2005;13:302–307. doi: 10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One. 2009;4:e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabishvili M, Verhelst R, Glonti T, Chanishvili N, Krylov V, Cuvelier C, et al. Digitized fluorescent RFLP analysis (fRFLP) as a universal method for comparing genomes of culturable dsDNA viruses: application to bacteriophages. Res Microbiol. 2007;158:572–581. doi: 10.1016/j.resmic.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Mikonranta L, Friman VP, Laakso J. Life history trade-offs and relaxed selection can decrease bacterial virulence in environmental reservoirs. PLoS One. 2012;7:e43801. doi: 10.1371/journal.pone.0043801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AD, Quigley BJ, Brown SP, Buckling A. Selection on non-social traits limits the invasion of social cheats. Ecol Lett. 2012;15:841–846. doi: 10.1111/j.1461-0248.2012.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- Rasmussen MA, Carlson SA, Franklin SK, McCuddin ZP, Wu MT, Sharma VK. Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect Immun. 2005;73:4668–4675. doi: 10.1128/IAI.73.8.4668-4675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, West SA. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19:341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- Selezska K, Kazmierczak M, Musken M, Garbe J, Schobert M, Haussler S, et al. Pseudomonas aeruginosa population structure revisited under environmental focus: impact of water quality and phage pressure. Environ Microbiol. 2012;14:1952–1967. doi: 10.1111/j.1462-2920.2012.02719.x. [DOI] [PubMed] [Google Scholar]
- Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg KM, Levin BR. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc Biol Sci R Soc. 2007;274:1921–1929. doi: 10.1098/rspb.2007.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.