Abstract
Nitrate is an important nutrient and electron acceptor for microorganisms, having a key role in nitrogen (N) cycling and electron transfer in anoxic sediments. High-nitrate inputs into sediments could have a significant effect on N cycling and its associated microbial processes. However, few studies have been focused on the effect of nitrate addition on the functional diversity, composition, structure and dynamics of sediment microbial communities in contaminated aquatic ecosystems with persistent organic pollutants (POPs). Here we analyzed sediment microbial communities from a field-scale in situ bioremediation site, a creek in Pearl River Delta containing a variety of contaminants including polybrominated diphenyl ethers (PBDEs) and polycyclic aromatic hydrocarbons (PAHs), before and after nitrate injection using a comprehensive functional gene array (GeoChip 4.0). Our results showed that the sediment microbial community functional composition and structure were markedly altered, and that functional genes involved in N-, carbon (C)-, sulfur (S)-and phosphorus (P)- cycling processes were highly enriched after nitrate injection, especially those microorganisms with diverse metabolic capabilities, leading to potential in situ bioremediation of the contaminated sediment, such as PBDE and PAH reduction/degradation. This study provides new insights into our understanding of sediment microbial community responses to nitrate addition, suggesting that indigenous microorganisms could be successfully stimulated for in situ bioremediation of POPs in contaminated sediments with nitrate addition.
Keywords: elevated nitrate, functional gene, sediment microbial community, in situ bioremediation
Introduction
With the rapid development of industry and urbanization, aquatic systems have been polluted with a variety of contaminants, including persistent organic pollutants. When present, contaminants in an aquatic ecosystem may be adsorbed onto suspended particles and eventually settle to the sediment. As a result, aquatic sediments are repositories of physical and biological debris acting as sinks for a wide variety of pollutants (Zoumis et al., 2001). Even when external pollution sources are controlled, internal loading of pollutants from sediment into the water column may continue for decades, delaying the recovery of degraded aquatic ecosystems (Jeppesen et al., 2005; SØNdergaard et al., 2007).
Nitrate is one of the essential nitrogen components in the biosphere, serving as an important nutrient and electron acceptor for microorganisms. Nitrate input can alleviate nitrogen (N) limitation, influence N-cycling processes and subsequently promote a cascade of biogeochemical processes, which are almost exclusively mediated by microorganisms (Galloway et al., 2008). As one of the more thermodynamically favorable electron acceptors for anaerobic respiration, nitrate can be reduced by a large number of microorganisms associated with the oxidation of organic matter and other reduced substrates. Over the years, a number of studies have demonstrated that the addition of nitrate could enhance in situ bioremediation of contaminated sediments by promoting organic carbon (C) degradation (Hutchins et al., 1998; Cunningham et al., 2001), suppressing hydrogen sulfide production (Jenneman et al., 1986) and preventing phosphorus (P) release (Foy, 1986; Yamada et al., 2012).
N cycling is almost exclusively mediated by microorganisms, and understanding how sediment microbial communities respond to nitrate addition will help us elucidate potential nitrate reduction mechanisms, and remediate contaminated sediments in situ. Few studies have examined the response of microbial communities to nitrate inputs. However, distinctly different results about microbial community responses to elevated nitrate were observed, with some reporting significant changes in the microbial community (Carrino-Kyker et al., 2013) and others showing no effect (Bowen et al., 2011; Carrino-Kyker et al., 2012). The complexity of N cycling and methodologies used for these studies could be one of major reasons for this disparity. In addition, within the complex network of N cycling, some key enzymes are shared and they may be simultaneously influenced by the input of N compounds, and/or linked with other biogeochemical processes, such as C and sulfur (S) cycling (Kraft et al., 2011). Moreover, many nitrate-reducing bacteria posses versatile metabolisms, able to use a variety of electron acceptors for respiration when coupled with the degradation of organic compounds, allowing for survival under the fluctuating redox environments when nitrate is unavailable (Hauck et al., 2001; Castelle et al., 2013; Zhao et al., 2013). A limited view of how nitrate addition affects microbial processes may be provided if only a subset of N-cycling enzymes are studied. In addition, experimental scale and sampling time points may lead to biases in the evaluation of microbial community responses to elevated nitrate. Therefore, a comprehensive survey of microbial community responses to elevated nitrate input into the sediment system is necessary.
Previous studies have shown that functional gene arrays are able to examine up to thousands of functional genes simultaneously, and have become routine molecular tools to analyze the functional composition, structure and dynamics of microbial communities from a variety of ecosystems (Denef et al., 2003; Taroncher-Oldenburg et al., 2003; Rhee et al., 2004; Chandler et al., 2006; Iwai et al., 2008; He et al., 2010a; 2012a, 2012b). GeoChip 4.0 is the most comprehensive functional gene array so far, containing more than 120 000 distinct probes targeting key genes involved in C, N, S and P cycling, and other environmental processes, providing a comprehensive survey of microbial structure, functional potential and environmental processes (Tu et al., 2014), thus overcoming these limitations mentioned above. For example, GeoChips have been successfully used for tracking and studying the responses of microbial communities to different environmental changes, such as elevated temperature (Zhou et al., 2012), CO2 and/or O3 (He et al., 2010b, 2013; Li et al., 2013; Xu et al., 2013) and contaminants (Xu et al., 2010; Yang et al., 2013).
Considering the key role of nitrate and the complexity of N cycling in biosphere, as well as the metabolic versatility of the nitrate-reducing bacteria, GeoChip 4.0 could be the promising tool to study the effect of elevated nitrate on sediment microbial communities. In this study, we hypothesize (i) that sediment microbial community composition and structure would be altered by the injection of nitrate into a contaminated sediment through the consequent changes in sediment microenvironmental conditions, (ii) that various microbial functional groups would change differentially over time in response to nitrate addition, especially those related to N-cycling networks and available electron acceptors/donors in the sediment and (iii) that sediment microorganisms with metabolic versatility would be efficiently stimulated, possibly resulting in the remediation of contaminated sediments, such as the degradation of polybrominated diphenyl ethers (PBDEs) and polycyclic aromatic hydrocarbons (PAHs) after nitrate addition. To test these hypotheses, we analyzed sediment microbial communities from a field-scale site for in situ bioremediation of complexly contaminated creek in Pearl River Delta, one of the world's most dynamic industrial zones, by nitrate injection using GeoChip 4.0 in concert with traditional microbiological analyses. This study provides new insights into our understanding of sediment microbial community responses and possible bioremediation of persistent organic pollutants in contaminated sediments by nitrate injection.
Materials and methods
The following is the summary of methods used in this study. More detailed information is provided in the Supplementary Information.
Site description and sampling
The pilot-scale sediment system for in situ bioremediation of a contaminated creek site by nitrate injection was located in a typical industry zone at Ronggui town (22°45′ N, 113°15′ E) in the Pearl River Delta. Water locks were located upstream and downstream of the creek, respectively, for controlling the water depth. In the experimental system, the sediment was composed of compact mud ∼1 m in depth. Calcium nitrate solution was injected at a dose of 45.3 g N m−2 into the sediment at a depth of 30 cm by pump every month. Three sampling sites with three subcores at each site were set perpendicular to the flow direction of the creek, and the distance between two adjacent sites was 4 m. During the experiment, the water depth was ∼100 cm, and the concentrations of dissolved oxygen dropped to near zero at a water depth of 80 cm. Sediment temperature was 25 °C and sediment dissolved oxygen was maintained at zero. Sediment samples were taken from the top 50-cm layer of these three sites using piston-column sediment sampler (4.0 cm internal diameter, XDB0204, Beijing New Landmark Soil Equipment Co., Ltd., Beijing, China) before nitrate injection (0 h), and 24 h, 48 h, 96 h, 144 h and 1 month after nitrate injection, with each sample composited from the three sediment cores at each treatment site. All samples were immediately transported to an anaerobic chamber where they were homogenized, sieved to remove the detritus (>5 mm) and split into subsamples for a variety of analyses including molecular microbial analyses, sediment property detections and microcosm experiments. Sample transport, homogenization and subpackage were performed at 25 °C within 2 h.
Geochemical analysis
To better define the geochemical properties of the sediment samples, several sets of variables were measured: (i) the concentrations of nitrate, nitrite, ammonium, sulfate, phosphate, total nitrogen (TN) and total phosphorus in sediments and porewaters; (ii) mixed liquor volatile suspended solid values, the contents of total organic carbon (TOC), PBDEs, PAHs and the partitioning of heavy metal species in sediments; and (iii) the concentrations of the gaseous ammonia and hydrogen sulfide at the sampling sites.
DNA preparation and GeoChip 4.0 analysis
DNA was extracted and purified according to the procedures described previously (Zhou et al., 1996). DNA labeling, as well as the purification of labeled DNA, was carried out as described previously (Xu et al., 2010). GeoChip 4.0 (Tu et al., 2014) synthesized by NimbleGen (Madison, WI, USA) was used to analyze the functional structure of the microbial communities and functional gene abundances. The hybridization and scanning procedure and instrumental analysis conditions were the same as in our previous study (Xu et al., 2013). Signal intensities were measured on the basis of scanned images, and spots with signal-to-noise ratios lower than 2.0 were removed before statistical analysis, as described previously (He et al., 2010b).
Statistical analysis
Preprocessed GeoChip data were further analyzed with different statistical methods: (i) hierarchical clustering for microbial community structure and composition; (ii) detrended correspondence analysis combined with analysis of similarities, nonparametric multivariate analysis of variance (Adonis) and multi-response permutation procedure for determining the overall functional changes in the microbial communities; (iii) microbial diversity index, significant Pearson's linear correlation (r) analysis, as well as analysis of variance; (iv) percentage changes of functional genes by nitrate addition were calculated using the following formula: (Ti−T0) × 100/T0, where T0 and Ti were the average signal intensities of genes detected by GeoChip 4.0 in the samples collected before or after i-h nitrate treatment; and (v) Mantel and redundancy analysis, as well as their partial analyses, were used to link the functional structure of microbial communities with environmental variables.
Results
Effects of nitrate addition on sediment chemical properties
Nitrate concentrations in the sediments and porewaters were measured as nitrate was injected, and it was found that the changes in nitrate concentration between these two compartments were not concordant (Table 1). After injection of calcium nitrate solution for 24 h, the sedimentary nitrate concentrations increased significantly (P<0.05) to 34.74 mg l−1 and then decreased back to the ambient nitrate concentration (∼1.2 mg l−1) after 1 month (data not shown), whereas no significant change was observed in the porewaters of sediment. Porewater sulfate concentrations increased at 48 h (P<0.05), with the highest level detected at 96 h after nitrate addition. Although no significant change was observed for the concentrations of ammoniacal-N and TN during the 144-h treatment process, obvious increases were detected for both at 48 h after nitrate injection. After 1 month of treatment, ∼88% of the ammoniacal-N was removed and the concentrations of TN were returned to initial levels (data not shown). In addition, significant (P<0.05) increases in sulfate were detected after 96 h of operation, which was consistent with a significantly (P<0.05) decreased concentration of S element in the treated sediments after more than a 4-month operation, and the color of the sediments changed from dark brown to yellowish-orange following nitrate injection (data not shown). In addition, significant (P<0.05) increases were also detected in the mixed liquor volatile suspended solid values of sediment after 48 h, and the addition of nitrate also promoted the removal of TOC in sediments. The percentage of TOC in the sediment significantly decreased from 5.33±0.06% to 4.90±0.09% (P<0.05) after 24 h (Table 1) and to 2.55±0.17% (P<0.01) after 120 days (data not shown), respectively. These results were well supported by the reduction of PAHs and PBDEs after 120 days of operation (Supplementary Figure S1). Simultaneously, the concentrations of hydrogen sulfide and gaseous ammonia in the sampling sites were decreased from 0.024 mg m−3 and 0.465 mg m−3 to 0.005 mg m−3 and 0.274 mg m−3, respectively. Such changes of chemical properties in the sediment may affect the sediment microbial community composition and structure.
Table 1. Major geochemical properties of the sediments and porewaters.
| Sample ID | Unit | 0 h | 24 h | 48 h | 96 h | 144 h |
|---|---|---|---|---|---|---|
| Porewater parameters | ||||||
| Nitrate-N | mg l−1 | 0.15±0.03a | 1.18±0.14a | 1.14±0.02a | 2.68±0.14a | 1.13±0.01a |
| Nitrite-N | mg l−1 | 0.05±0.00a | 0.02±0.03a | 0.03±0.03a | 0.42±0.05a | 0.02±0.02a |
| Ammoniacal-N | mg l−1 | 51.89±13.14a | 30.96±12.72a | 41.89±2.55a | 29.11±6.76a | 48.74±20.43a |
| TN | mg l−1 | 35.53±14.93a | 39.35±15.47a | 65.53±8.27a | 30.44±14.95b | 50.37±18.99a |
| TP | mg l−1 | 0.47±0.13b | 0.19±0.06b | 2.24±0.91a | 0.55±0.36b | 0.76±0.49b |
| Phosphate | mg l−1 | 0.13±0.14a | 0.05±0.00a | 0.03±0.02a | 0.02±0.01a | 0.02±0.02a |
| Sulfate | mg l−1 | 3.59±1.23b | 3.02±0.01b | 4.73±1.49a,b | 17.55±4.17a | 9.18±2.95a,b |
| Sediment parameters | ||||||
| Nitrate-N | mg kg−1 | 7.25±1.16b,c | 32.74±4.02a,c | 28.83±3.40b | 23.83±3.03a,c | 14.50±4.91d |
| Ammoniacal-N | mg kg−1 | 348.07±188.21a | 359.81±187.98a | 452.44±109.05a | 312.10±115.75a | 234.49±22.57a |
| TP | g kg−1 | 1.05±0.65a | 0.55±0.03a | 0.94±0.34a | 0.55±0.14a | 0.95±0.29a |
| TN | g kg−1 | 0.52±0.03a | 0.96±0.22a | 0.85±0.17a | 0.88±0.04a | 0.70±0.24a |
| TOC | % | 5.33±0.06a | 4.89±0.09b | 4.02±0.01c | 4.20±0.24c | 3.95±0.37c |
| MLVSS | % | 4.16±0.93b | 4.70±1.64a,b | 6.20±1.68a | 4.97±1.52a | 3.52±1.47b |
Abbreviations: MLVSS, mixed liquor volatile suspended solid; TN, total nitrogen; TOC, total organic carbon; TP, total phosphorus.
All data are presented as mean±s.e., n=3. Significances among the different treatment time points were analyzed by multi-way analysis of variance. The significances of a, b, c and d are at P<0.05 level.
Overall review of sediment microbial community responses to nitrate addition
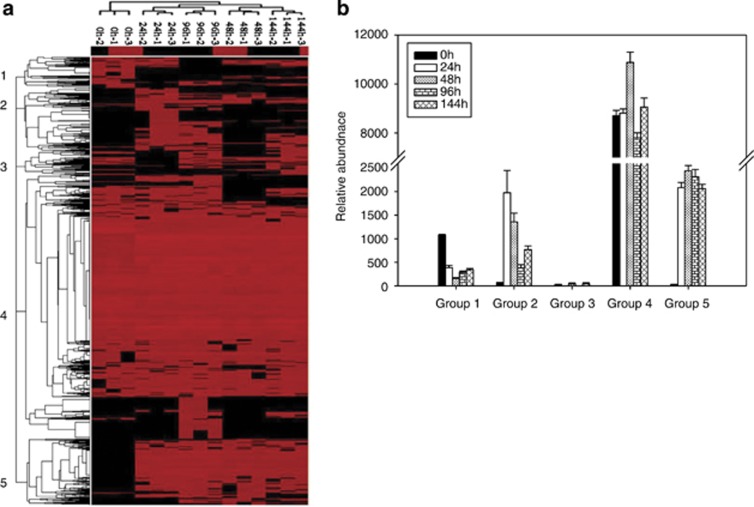
To assess the response of microbial communities to nitrate addition, GeoChip 4.0 was used to examine the important biogeochemical processes, especially key functional genes involved in C, N, P and S cycling. As revealed by multi-response permutation procedure analysis, a significant (P<0.001) difference was detected for the overall microbial community functional structure (Supplementary Table S1). When we compared the overall gene number and abundance detected between the initial and treatment stages, significant (P<0.01) increases were observed in all of the treatment time points, peaking by 96 h (Supplementary Table S2), although there were some fluctuations in different time points. After 24 h of nitrate addition, 29% of overlapping genes showed significant (P<0.05) changes with 14% increase and 15% decrease, and a total of 33.6% unique genes were detected. The percentages of unique genes from the treatment samples following nitrate amendment, ranging from 22.5% to 37.1%, were significantly (P<0.05) higher than those from the initial stage (0 h), ranging from 7.9% to 16.7% (Supplementary Table S2). The overall microbial functional diversity was also significantly (P<0.01) higher in the samples treated with nitrate based on the Shannon–Weiner (H') and Simpson's (1/D) indices. In addition, significantly (P<0.01) positive correlations were observed among the abundances of different functional genes, especially those involved in N, C, S and P cycling (Supplementary Table S3). Hierarchical clustering analysis showed that all samples treated with nitrate were clustered together and were well separated from samples taken before nitrate addition (Figure 1a). Five major gene groups could be visualized in the color tree, and considerable variability in functional gene distribution was observed among different samples; in addition, a number of functional genes were found to be unique to nitrate treatment samples. For example, group 4 with 74.1% of all genes detected, largely involved in organic remediation, C degradation and denitrification, was generally detected in all samples, whereas groups 2 and 5 with 7.5% and 14.6%, respectively, of all genes detected were largely found in the nitrate treatment samples.
Figure 1.
Cluster analysis of functional genes detected using GeoChip 4.0. The figure was generated using CLUSTER and visualized in TREEVIEW (http://rana.lbl.gov/EisenSoftware.htm). Black indicates signal intensities below the threshold value, and red indicates a positive hybridization signal. The color intensity indicates differences in signal intensity. The samples from different sampling points were clearly separated. Five different gene patterns were observed and indicated by numbers in the tree (a), and they were represented as group 1 to 5 on axis x and the relative abundances of the gene signal intensity contained in different groups were presented on axis y in the graphs (b).
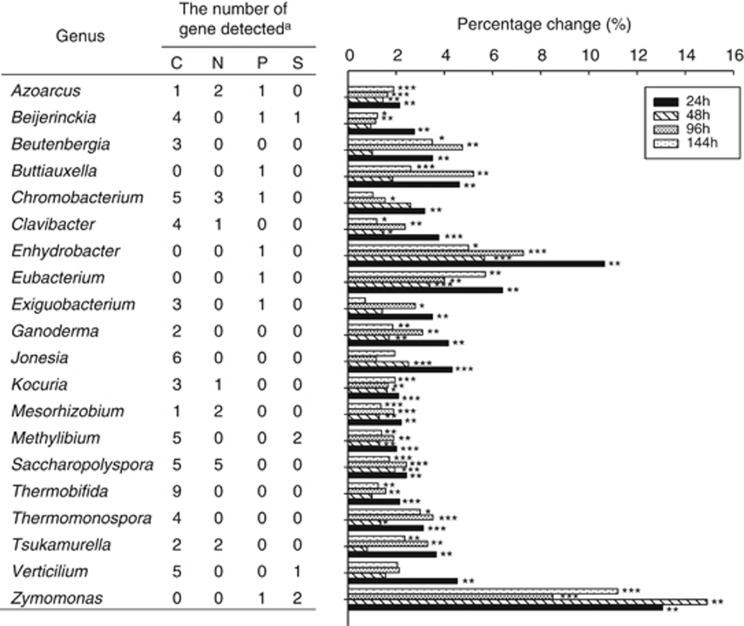
In total, the genes detected by GeoChip were based on the probes from 633 known microbial genera, 20 of which showed significant increases at 24 h after nitrate injection and almost maintained at all time points. Within those 20 genera, 18 were bacterial origin, with 8 each from the Actinobacteria and Proteobacteria phyla, respectively, and 2 from Firmicutes phylum. Interestingly, most of the genera detected are known to be metabolically versatile, involved in multiple biogeochemical cycling processes (Figure 2). For example, the gene families detected from genera Azoarcus and Chromobacterium, typical β-Proteobacterial denitrifiers, are involved in C-, N- and P-cycling processes, whereas those from the genus Beijerinckia, belonging to α-Proteobacteria, are involved in C-, P- and S-cycling processes. The gene families from Actinobacteria genera, Clavibacter, Kocuria, Saccharopolyspora and Tsukamurella are involved in C and N cycling. The average functional gene abundances from these microorganisms increased between 2 and 13 times within the initial 24 h following nitrate addition (Figure 2). Therefore, all these results indicate that the functional characteristics of sediment microbial communities were significantly altered and the functional genes from indigenous microorganisms with metabolic versatility were more efficiently stimulated after nitrate addition in this site.
Figure 2.
The commonly significantly increased functional genes from the microorganisms with metabolic versatility. aThe number of gene family detected for C-, N-, P- and S-cycling processes. The calculation of percentage change was based on the average signal intensity of each microorganism. Significances between the samples from the initial stage and different time points following nitrate amendment were performed by the Student t-test. ***P<0.01; **P<0.05; *P<0.1.
Changes of microbial functional gene abundances in response to nitrate addition
N-cycling genes
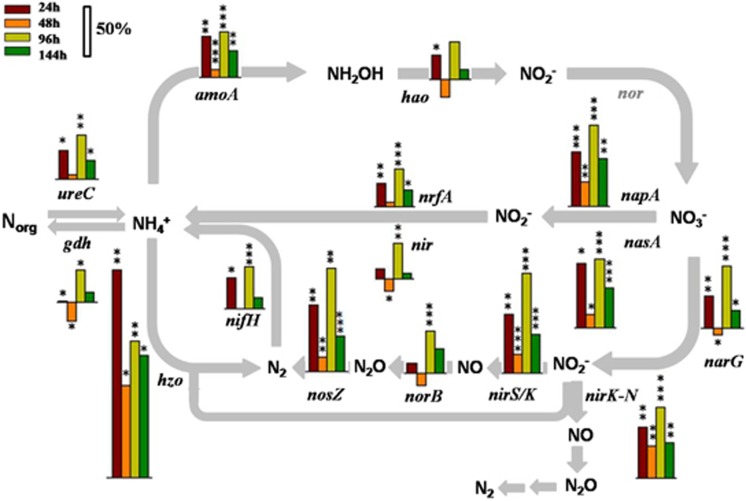
Totally, ∼50% of the detected N-cycling genes showed a significant (P<0.05) increase in abundance at 24 h after nitrate addition. Among the 18 gene families involved in assimilatory N reduction, dissimilatory N reduction to ammonium, N2 fixation, denitrification, ammonification and anaerobic ammonium oxidation, 10 of them increased significantly (P<0.05), with the highest for hzo (Figure 3). A small drop was observed in the samples collected at 48 h, in which only five gene families showed significant changes: an increase for nirS/K, napA, nrfA and a decrease for norB and hao. After 96 h, changes of the gene families involved in N cycling had rebounded, with 15 of them showing significant increases (Figure 3). Most of denitrification genes, such as nirS, nirK and nosZ, as well as nitrifier nirK, were significantly (P<0.05) stimulated and maintained at all time points after nitrate injection. The genes encoding ammonia monooxygenase (amoA) for nitrification exhibited significantly (P<0.05) higher signal intensities in the treatment samples, whereas no significant change was observed in hao encoding hydroxylamine oxidase. Interestingly, although most ammonia oxidizers are chemoautotrophic in natural environments, 9 of the top 10 significantly increased amoA genes were from bacteria with metabolic versatility (that is, Desulfovibrio magneticus, Mycobacterium gilvum, Rhodopseudomonas palustris and Agrobacterium tumefaciens), and only one (156572306) from Archaea (Supplementary Figure S2). These results indicated that nitrate addition might alter the N-cycling processes by changing the N compound composition and the abundance of N-cycling genes, especially those from microorganisms with diverse metabolic capabilities.
Figure 3.
Effects of nitrate addition on abundances of functional genes involved in N cycling. The upward bars present the positive percentage changes, whereas the downward bars present the negative percentage changes. Significances between the samples from the initial stage and different time points following nitrate amendment were performed by the Student t-test. ***P<0.01; **P<0.05; *P<0.1.
C-cycling genes
Among the 3565 C-cycling genes detected, ∼77.8% of them were involved in C compound degradation, such as starch, pectin, hemicelluloses, cellulose, aromatic, chitin and lignin. Most of these genes showed significantly (P<0.05) higher abundance (Table 2), although a small drop was observed at 48 h after nitrate addition. All of the four enzymes for lignin degradation, glyoxal oxidase (glx), lignin peroxidase (lip), manganese peroxidase (mnp) and phenol oxidase (lcc), showed significant increases after nitrate addition. The highest increases (130.97–273.89%) were observed for the important aromatic degradation gene family limEH encoding limonene-1,2-epoxide hydrolase. Another important aromatic degradation gene family vanA encoding vanillate demethylase also showed a significant increase after nitrate addition, and 5 of the top 10 increased vanA genes were from the important halogenated organic compound degradation organism Sphingomonas wittichii RW1 (Supplementary Figure S3). Furthermore, most of the gene abundance for C degradation was positively correlated with those for nitrate reduction significantly (P<0.01) (Supplementary Table S4). Consistently, the contents of TOC, PAHs and PBDEs in sediments were obviously reduced after nitrate addition. Significant increases were observed in the number and abundance of the functional genes involved in methane oxidation and production in this study. The numbers of mmoX gene, encoding soluble methane monooxygenase hydroxylase component α-subunit, were significantly (P<0.05) increased, ∼2.5 times at 24 h after nitrate injection, peaking ∼3.5 times at 96 h after nitrate injection and maintained at 2 times after 144 h. Specifically, mmoX genes from Methylomonas sp. KSWIII and Frankia sp. CcI3 were detected after nitrate addition and commonly existed in all of the sampling time points. Similar results were detected for mcrA encoding the α-subunit of methyl coenzyme M reductase, which increased from 19–21 genes in the 0-h samples to 24–46 genes after nitrate addition. The mcrA gene from Methanocorpusculum labreanum Z was detected in all samples after nitrate addition. Different trends were observed for pmoA encoding the α-subunit of particular methane monooxygenase. Significantly (P<0.05) higher gene number and abundance were detected for pmoA at 24 and 96 h after nitrate injection, whereas significant (P<0.05) decreases at 48 h and no change at 144 h after nitrate injection were also observed. Totally, more than 81.67% of the genes detected for C cycling showed significant increases at 24 h following nitrate addition, indicating that the functional genes for C cycling were very sensitive to elevated nitrate and most of the genes could be enriched by nitrate addition.
Table 2. Effects of nitrate amendment on abundances of key functional genes involved in C cycling.
| Gene or enzyme | Functional process |
Percentage change (%)a |
P-valueb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 96 h | 144 h | 24 h | 48 h | 96 h | 144 h | ||
| aclB | 42.89 | 0.31 | 51.95 | −13.34 | 0.0473 | 0.9623 | 0.0409 | 0.2076 | |
| CODH | C fixation | 48.65 | 8.83 | 66.43 | 32.83 | 0.0085 | 0.0998 | 0.0037 | 0.0027 |
| pcc | 29.29 | 7.91 | 46.07 | 26.98 | 0.0196 | 0.0428 | 0.0015 | 0.0092 | |
| Rubisco | 24.92 | 7.38 | 39.82 | 19.34 | 0.0781 | 0.0235 | 0.0264 | 0.0165 | |
| FTHFS | Acetogenesis | 97.25 | 35.53 | 80.38 | 65.53 | 0.0090 | 0.0578 | 0.0083 | 0.0001 |
| aceA | Glyoxylate cycle | 40.44 | 13.01 | 68.90 | 28.10 | 0.0664 | 0.0442 | 0.0058 | 0.0266 |
| aceB | 27.54 | 2.31 | 46.12 | 24.24 | 0.0007 | 0.2094 | 0.0085 | 0.0067 | |
| amyA | Starch degradation | 28.83 | 8.09 | 40.79 | 19.38 | 0.0193 | 0.0104 | 0.0051 | 0.0162 |
| amyX | 102.83 | 63.52 | 203.70 | 198.37 | 0.0003 | 0.1954 | 0.0001 | 0.0000 | |
| apu | −1.96 | −11.77 | 19.82 | −0.70 | 0.9257 | 0.3792 | 0.2239 | 0.4434 | |
| cda | 18.37 | 1.06 | 37.14 | 10.51 | 0.0421 | 0.5999 | 0.0014 | 0.0694 | |
| Glucoamylase | 60.06 | 23.36 | 75.32 | 72.07 | 0.0380 | 0.0933 | 0.0012 | 0.0072 | |
| Isopullulanase | 24.15 | 75.51 | 59.70 | −0.77 | 0.3201 | 0.0529 | 0.2274 | 0.9830 | |
| nplT | −5.17 | −16.66 | 0.86 | −10.71 | 0.0752 | 0.0448 | 0.8270 | 0.1264 | |
| pulA | 30.42 | 4.38 | 33.63 | 10.05 | 0.0425 | 0.4943 | 0.0077 | 0.0316 | |
| Pectinase | Pectin degradation | 47.13 | 10.22 | 74.07 | 38.62 | 0.0278 | 0.3659 | 0.0064 | 0.0191 |
| ara-bacteria | Hemicellulose degradation | 28.85 | −0.83 | 35.11 | 9.85 | 0.0175 | 0.3228 | 0.0081 | 0.0687 |
| ara-fungi | 79.24 | 25.60 | 65.72 | 17.79 | 0.0336 | 0.0547 | 0.0105 | 0.0021 | |
| Mannanase | 8.94 | −7.71 | 10.71 | −6.20 | 0.0910 | 0.3421 | 0.2831 | 0.1448 | |
| xylA | 42.91 | 19.04 | 57.43 | 28.69 | 0.0213 | 0.0258 | 0.0180 | 0.0089 | |
| Xylanase | 42.48 | 18.60 | 43.89 | 24.50 | 0.0259 | 0.1569 | 0.0056 | 0.1333 | |
| CDH | Cellulose degradation | 25.64 | −3.09 | 32.72 | 13.43 | 0.0492 | 0.4167 | 0.1075 | 0.0322 |
| Cellobiase | 55.83 | 9.16 | 63.08 | 35.26 | 0.0102 | 0.1360 | 0.0003 | 0.0141 | |
| Endoglucanase | 39.66 | 14.25 | 54.13 | 28.45 | 0.0168 | 0.0035 | 0.0011 | 0.0176 | |
| Exoglucanase | 58.24 | 28.49 | 81.21 | 49.73 | 0.0017 | 0.0129 | 0.0006 | 0.0050 | |
| limEH | Aromatic degradation | 193.84 | 130.97 | 273.89 | 195.00 | 0.0056 | 0.0140 | 0.0053 | 0.0010 |
| vanA | 35.91 | 17.33 | 49.72 | 27.34 | 0.0384 | 0.0317 | 0.0037 | 0.0338 | |
| vdh | 19.94 | −2.60 | 48.07 | 22.80 | 0.1412 | 0.6355 | 0.0023 | 0.1045 | |
| Acetylglucosaminidase | Chitin degradation | 43.69 | 6.90 | 53.97 | 20.15 | 0.0210 | 0.1768 | 0.0088 | 0.1139 |
| Endochitinase | 43.79 | 19.69 | 62.48 | 25.89 | 0.0144 | 0.0100 | 0.0136 | 0.0027 | |
| Exochitinase | 61.05 | 42.99 | 74.46 | 53.91 | 0.0084 | 0.0097 | 0.0040 | 0.0218 | |
| glx | Lignin degradation | 15.06 | 4.69 | 23.69 | 11.45 | 0.0491 | 0.1740 | 0.0003 | 0.1185 |
| lip | 88.05 | 52.75 | 98.86 | 59.77 | 0.0011 | 0.0041 | 0.0054 | 0.0032 | |
| mnp | 44.13 | 13.14 | 61.96 | 42.04 | 0.0087 | 0.1876 | 0.0016 | 0.0164 | |
| Phenol oxidase | 47.64 | 20.48 | 55.61 | 27.11 | 0.0075 | 0.0177 | 0.0095 | 0.0107 | |
| mmoX | Methane oxidation | 121.45 | 46.18 | 166.61 | 79.85 | 0.0001 | 0.0235 | 0.0188 | 0.0199 |
| pmoA | 15.97 | −14.51 | 71.06 | 13.32 | 0.0418 | 0.0492 | 0.0188 | 0.1148 | |
| mcrA | Methanogenesis | 109.16 | 41.99 | 119.74 | 49.22 | 0.0190 | 0.0838 | 0.0004 | 0.0275 |
Percentage change was calculated using the following formula: (Ti−T0) × 100/T0, where T0 was the average signal intensities of genes detected from the initial samples without nitrate addition by GeoChip 4.0, Ti was those from different time points following nitrate addition.
P-values were the Student t-test results between the samples from the initial stage and different time points following nitrate addition.
The bold P-values indicate significant (P<0.05) changes.
S- and P-cycling genes
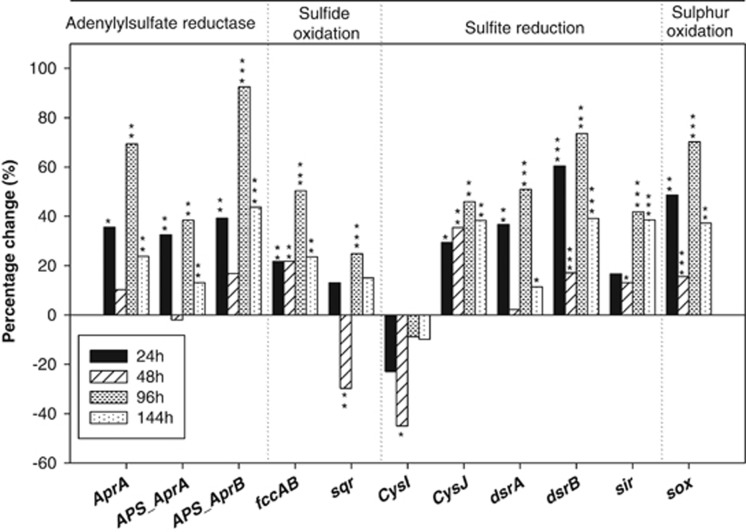
The changes in abundance of most S-cycling genes (Figure 4) were similar to those for C- and N-cycling genes with a general increase after nitrate addition. Within the 11 gene families involved in S cycling, most were significantly increased after nitrate addition, with the exception of the gene encoding β-subunit of sulfite reductase (cysI), which showed no significant change (Figure 4). However, significant increases were observed in the important sulfite reduction genes, cysJ, dsrA/B and sir, with dsrA/B being more sensitive to nitrate addition. A total of 108 genes involved in sulfite reduction process were detected after nitrate addition and commonly existed in all of the time points. Within these genes, 46.3% of them were dsrA/B genes for dissimilatory sulfite reductase and all from uncultured microorganisms. Significantly (P<0.05 or 0.01) positive increases were observed in two important sulfite oxidation genes, sox (sulfur oxidation gene cluster) and fccAB (flavo cytochrome c sulfide dehydrogenase), in all of the time points after nitrate addition. The nine commonly significantly increased sox genes were related to metabolic versatile microorganisms (Supplementary Figure S4), out of which seven belonged to α-Proteobacteria and three of them were derived from a methane-consuming bacterium, Methylobacterium populi BJ001. In addition, the sox gene (120593543) from the efficient PAH degradation organism Polaromonas naphthalenivorans CJ2 was also simulated after nitrate addition. Furthermore, a fair number of functional genes from nitrate-reducing sulfide oxidation bacteria were detected and enriched after nitrate addition, such as sqr and sox genes from Thiobacillus denitrificans. The evidence that nitrate reduction coupled with sulfide oxidation was also provided by the microcosm experiments inoculated with the sediments, in which the nitrate consumptions were coupled with the generations in sulfate and the increases in the abundance of nitrate-reducing sulfide oxidation bacteria after 24 h of incubation.
Figure 4.
Effects of nitrate addition on abundances of functional genes involved in S cycling. Significances between the samples from the initial stage and different time points following nitrate amendment were performed by the Student t-test. ***P<0.01; **P<0.05; *P<0.1.
The abundance of genes encoding polyphosphate kinase (ppk) for polyphosphate synthesis from ATP significantly increased with P<0.05 at 24 h and P <0.01 at 48 h after nitrate addition. Genes encoding exopolyphosphates (ppx) for inorganic polyphosphate degradation and phytase for phytate degradation were detected with significantly (P<0.05) increased abundances at 24 h, no significant change at 48 h, and significantly (P<0.01) enriched again at 96 h after nitrate addition (Supplementary Figure S5A). Similar to those genes involved in N and S cycling, most of the top 10 commonly significantly increased genes for P utilization were from the metabolically versatile microorganisms, including iron cycle contributors (that is, Magnetospirillum magneticum AMB-1 and Leptothrix cholodnii SP-6) and Archaea Haloferax volcanii with broad electron acceptor capabilities (Supplementary Figure S5B). In addition, the number and abundance of cytochrome c gene were significantly (P<0.05 or 0.01) enriched after nitrate injection (Supplementary Figure S6) and showed significant (P<0.001) correlations with the functional genes involved in P utilization. Similar to N- and C-cycling genes, these results suggested that nitrate addition might modify S and P cycling by changing the abundance of key functional genes, especially those from the microorganisms with metabolic versatility.
Relationships between functional structure and environmental variables
To explore possible linkages between the functional structure of sediment microbial communities and environmental factors, GeoChip data and environmental factors including ammonium, nitrate, nitrite, TN, TOC, total phosphorus, phosphate, sulfate in the porewaters and sediments were used for Mantel or partial Mantel tests and redundancy analysis. First, Mantel tests of all environmental factors and the signal intensity of all detected genes showed significant correlations only between five individual functional genes (pmoA, gdh, nirK, fccAB and CDH genes) and those porewater variables, but none of those genes was significantly correlated with the sediment variables (Supplementary Table S5). Second, redundancy analysis was used to further explore what environmental factors largely shaped the functional structure of sediment microbial communities. Consistent with Mantel tests, the functional genes detected did not have significant relationships with the 12 selected environmental factors. However, significant (P<0.01) correlations were detected between all functional genes, C-, N-, P- or S-cycling genes, and five selected environmental factors, including four porewater factors (the concentrations of ammonium, TN, total phosphorus and sulfate) and the only sediment factor (nitrate concentration) (Table 3). Based on the model, 55.3% of the total variance could be explained by all canonical axes (F=2.231, P=0.001) with the first axis explained 45.1% (F=2.249, P=0.001) for all of the detected functional genes. The functional genes involved in C, N, P and S cycling could be totally explained 58.9%, 58.8%, 60.8% and 59.8% by these five factors, respectively (Table 3). The samples from the same time points were clustered together and were well separated from the 0-h samples by the first axis. In addition, the sediment nitrate concentration was identified as the main environmental factor for shaping the functional microbial communities at 24, 48 and 144 h after nitrate addition, whereas the sediment TN was the first environmental factor for those at 96 h (Table 4). These results indicate that sediment nitrate concentration could be a dominant factor shaping microbial community functional structure and potentially regulating microbial functional processes in the sediment.
Table 3. The relationships between the functional genes detected and the environmental factors selecteda by RDA.
| Functional process |
The first canonical axis |
The sum of all canonical axes |
||||
|---|---|---|---|---|---|---|
| Eigenvalue | F-value | P-value | Eigenvalue | F-value | P-value | |
| All | 0.451 | 2.249 | 0.001 | 0.553 | 2.231 | 0.001 |
| C cycling | 0.230 | 2.688 | 0.029 | 0.589 | 2.579 | 0.001 |
| N cycling | 0.212 | 2.415 | 0.059 | 0.588 | 2.565 | 0.001 |
| P utilization | 0.244 | 2.898 | 0.013 | 0.608 | 2.791 | 0.001 |
| S cycling | 0.232 | 2.713 | 0.025 | 0.598 | 2.673 | 0.001 |
Abbreviation: RDA, redundancy analysis.
The five environmental factors selected include four interstitial water factors (the concentrations of ammonium, total nitrogen, total phosphorus and sulfate) and one sediment factor (nitrate concentration).
Table 4. The first environmental factor shaping the functional microbial community structure analyzed by RDA.
| Samplea | Functional process | First environmental factor | F-value | P-value |
|---|---|---|---|---|
| 24 h | C cycling | Sediment nitrate concentration | 6.20 | 0.062 |
| N cycling | Sediment nitrate concentration | 5.89 | 0.065 | |
| P utilization | Sediment nitrate concentration | 7.04 | 0.029 | |
| S cycling | Sediment nitrate concentration | 6.44 | 0.065 | |
| 48 h | C cycling | Sediment nitrate concentration | 5.22 | 0.037 |
| N cycling | Sediment nitrate concentration | 4.79 | 0.027 | |
| P utilization | Sediment nitrate concentration | 5.15 | 0.074 | |
| S cycling | Sediment nitrate concentration | 5.60 | 0.016 | |
| 96 h | C cycling | Sediment TN | 8.82 | 0.048 |
| N cycling | Sediment TN | 7.71 | 0.056 | |
| P utilization | Sediment TN | 7.55 | 0.001 | |
| S cycling | Sediment TN | 8.36 | 0.001 | |
| 144 h | C cycling | Sediment nitrate concentration | 6.59 | 0.033 |
| N cycling | Sediment nitrate concentration | 5.74 | 0.011 | |
| P utilization | Sediment nitrate concentration | 5.75 | 0.001 | |
| S cycling | Sediment nitrate concentration | 6.42 | 0.001 |
Abbreviations: RDA, redundancy analysis; TN, total nitrogen.
The sample contains the GeoChip data from the samples of the initial stage and different time point following nitrate addition.
Discussion
Understanding the functional diversity, composition, structure and dynamics of sediment microbial communities is critical for designing bioremediation strategies in contaminated sites. Coexisting within contaminated sediments are a variety of chemical compounds, such as ammonia, S compounds, persistent organic pollutants and metal ions, some of which can be used as electron acceptors or donors. Under such conditions, biogeochemical processes largely depend on the microbial community and their metabolic potential. In this study, we showed that the functional composition and structure of the sediment microbial community were markedly altered, with key functional genes involved in C, N, S and P cycling being highly enriched after nitrate injection. In addition, some of those key genes were largely derived from microorganisms with diverse metabolic capabilities, possibly leading to potential in situ bioremediation of contaminants (for example, PBDEs, PAHs) in the sediment. The results generally support our hypotheses and provide new insights into our understanding of sediment microbial community responses to nitrate addition.
One hypothesis is that the sediment microbial community composition and structure would be altered with nitrate injection, consequently resulting in changes of sediment microenvironmental conditions. Previous studies have found that distinct microbial community compositions and redox zones were obtained from markedly different sedimentary settings, and significant correlation between changes in geochemical composition and changes in community structure was detected (Tankéré et al., 2002; Jorgensen et al., 2012). It was suggested that hydrological regime was a very important factor for the structure of nitrate-reducing bacterial communities (Bougon et al., 2009), and the water content of sediment significantly influenced the nitrate diffusion coefficient (Fang et al., 2008). Our previous study also showed that the mass transfer rate of the available electron donor shaped the structures of microbial community and the profiles of substrates and products in sediments (Xu et al., 2010). In this study, the diversity and abundance of sediment microorganisms were significantly increased and the coexistence, instead of a competition, between nitrate reduction and other electron acceptor reductions within the sediment was observed, with some important geochemical factors in the sediment, such as the concentrations of TOC, TN, total phosphorus, ammoniacal-N and sulfate, being subsequently changed, suggesting that different microenvironmental conditions had been formed after nitrate injection due to the spatial separation in the sediment matrix. These results well support the above hypothesis, whereas such changes may be reflected in the change of abundances of key functional genes, thus subsequently regulating ecosystem functioning.
Our second hypothesis is that various microbial functional groups would change considerably over time in response to nitrate addition, especially those related to N-cycling networks and available electron acceptors/donors in the sediment. One of the important factors to regulate biochemical processes, and thus the community composition and structure, was the ratio of electron donor to electron acceptor. Previous studies showed that the microbial community structure exhibited a considerable shift over the sediment remediation phases examined by adding electron acceptors or donors (Van Nostrand et al., 2011; Louati et al., 2013), and introducing nitrate into contaminated subsurface site as an electron acceptor could enhance in situ biodegradation and sometimes was capable of partially or completely removing organic contaminants (Thomas et al., 1997; Hutchins et al., 1998; Cunningham et al., 2001; Tang et al., 2005). More experiments indicated that dissimilatory N reduction to ammonium is the dominant pathway of dissimilatory nitrate reduction in sediments that are rich in electron donors, such as labile organic carbon and sulfide (Brunet and Garcia-Gil, 1996; Porubsky et al., 2009), and the formation of ammonia can be linked to the use of reduced S compounds as electron donors during the nitrate reduction process (Brunet and Garcia-Gil, 1996). In this study, key genes involved in N cycling were increased after nitrate addition, and the gene abundance of nrfA, the marker gene for dissimilatory N reduction to ammonium, showed significant (P<0.01 or 0.05) positive correlations with a considerable proportion of the genes involved in labile organic C degradation and S oxidization after nitrate injection. However, no significant increase in ammonium concentration was observed following nitrate injection, and most of them were even removed after 1 month of treatment, which could be due to the simultaneous stimulation of anaerobic ammonium oxidation. Indeed, the gene encoding hydrazine oxidoreductase (hzo), which has been proposed as a useful marker for the analysis of anaerobic ammonium oxidation bacteria (Francis et al., 2007), was identified as the most significantly increased gene among the N-cycling genes detected after nitrate addition in this study. There are a few possibilities: (1) the addition of nitrate alleviated the limitation of nitrite and increased the efficiency of anaerobic ammonium oxidation; (2) dissimilatory N reduction to ammonium generated the ammonium pool through nitrate reduction that fostered the ammonium oxidation reactions. These explanations are well supported by the significantly positive correlation between the abundance of hzo and of the nitrate (narG and napA) and nitrite (nrfA) reductase genes in this site. The stimulation of most S-oxidation genes after nitrate addition was well supported by the increases of sulfate concentrations in porewater and the reduced hydrogen sulfide emission. The changes of sediment color from dark brown to yellowish-orange also could be due to the nitrate-mediated sulfide oxidation of the dark-color metal-bound sulfides (FeS and FeS2) (Gu et al., 2012). These results confirm our second hypothesis and show a direct correlation between the genes involved in the N cycle and other important biogeochemical cycles.
As a result, we also predicted that sediment microorganisms with metabolic versatility would be efficiently stimulated owing to the coexistence of different electron acceptors and donors in the contaminated sediment, resulting in in situ bioremediation of contaminated sediments. Although it is predicted theoretically that the presence of nitrate would inhibit sulfate reduction because nitrate has higher reduction potential than sulfate under anaerobic condition, increasing evidences showed that the reduction of electron acceptors with lower reduction potential was not inhibited and even was enhanced by the presence of nitrate (Canfield et al., 2010; Gu et al., 2012; Zhao et al., 2013). Several genera with broader electron-accepting capabilities have been enriched in systems containing multiple electron acceptors, such as Bacillus sp. and Ochrobactrum sp. from the radioactively contaminated sediment with high nitrate concentration (Thorpe et al., 2012) and Sulfuricurvum sp. and Desulfovibrio sp. from the system containing O2, ClO4−, sulfate and nitrate (Zhao et al., 2013). Similar results were obtained in this study. For example, the amoA gene (239798446) from Desulfovibrio sp. and the van gene (198265864) from Ochrobactrum sp. were ranked in the top 10 stimulated genes after nitrate addition. Most of the significantly stimulated C degradation genes were from the important halogenated organic compound degradation organism Sphingomonas wittichii RW1 (Hong et al., 2002), which could contribute a lot to the removal of PAHs and PBDEs from sediments. The coexistence of S with nitrate, methane and PAHs could be the reason for the frequent detection of the commonly significantly increased sox genes from the nitrate-reducing sulfide oxidation bacteria Thiobacillus denitrificans ATCC 25259, the methane-consuming bacterium Methylobacterium populi BJ001 and the PAHs degradation bacterium Polaromonas naphthalenivorans CJ2. As the reactive soluble P often prefers to be immobilized by the oxidized iron particle after nitrate injection (Foy, 1986; McAuliffe et al., 1998; Yamada et al., 2012), most of the significantly increased P utilization genes were from microorganisms with iron cycling capabilities. Even the significantly increased amoA genes were frequently detected from metabolically diverse microorganisms. These results support the hypothesis very well and suggest that the efficient stimulation of microorganisms exhibiting diverse physiological and metabolic properties could make great contributions to in situ bioremediation of contaminated sediments after nitrate addition.
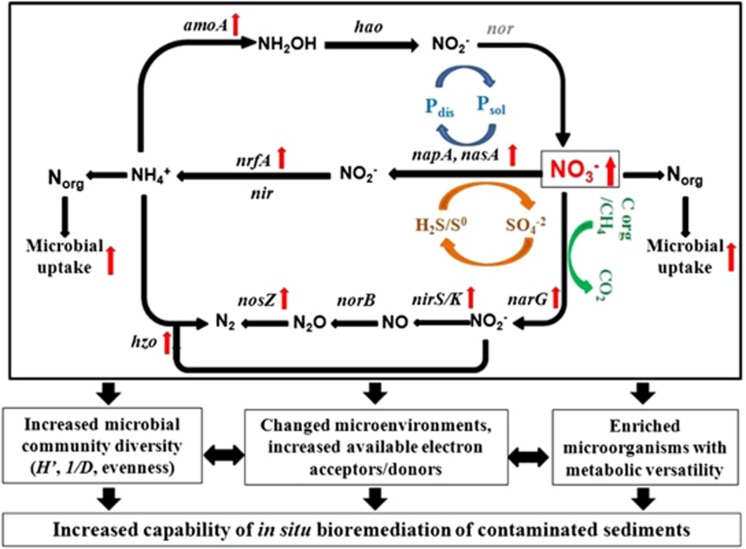
In summary, the results allow us to form a conceptual model for further understanding the effect of nitrate injection on sediment microbial communities (Figure 5). First, sediment microbial communities were very sensitive to the changes of nitrate concentrations, and nitrate could serve as a nutrient and stimulate microbial growth demonstrated by mixed liquor volatile suspended solid values in the sediments. Second, GeoChip data showed that microbial functional genes involved in key N-cycling processes were enriched, such as narG, nirS/K and nosZ for denitrification, napA/nasA and nrfA for nitrate and nitrite reduction and hzo for ammmox. In addition, those N-cycling processes would be linked with other nutrient cycling pathways, such as C degradation (for example, vanA for aromatic degradation), S cycling (for example, sox for sulfite oxidation) and P cycling, and all those key genes were found to be largely derived from microorganisms with diverse physiological and metabolic properties. In addition, the changes in key functional processes were also reflected in increased microbial community diversity, enriched microorganisms with metabolic versatility and increased availability of electron acceptors/donors, as well as changed microenvironmental conditions (for example, redox potential). Finally, all those changes may create favorable conditions for indigenous microbial communities to remediate contaminated sediments in situ, such as the degradation of PBDEs and PAHs (Figure 5). However, to further understand and evaluate the potential impact of elevated nitrate on bioremediation of contaminated sediment ecosystems, it is essential to launch an integrated and comprehensive monitoring program to track the dynamics and adaptive responses of microbial communities together with physical, chemical and biological analyses of ecosystem properties and functions, especially the degradation of persistent organic pollutants.
Figure 5.
A concept model for effects of nitrate on microbial functional communities in complexly contaminated sediments.
Acknowledgments
We thank Dr Daiyong Deng for field work and Dr James W Voordeckers for improving the English language. This research was supported by the National Basic Research Program of China (973 Program) (2012CB22307), the National Natural Science Foundation of China (31170470), Guangdong Province—Chinese Academy of Sciences Strategic Cooperative Project (2012B091100257), the International Cooperation Projects of Guangdong Province (2011B050400005), Guangdong Provincial Programs for Science and Technology Development (2012A061100009), Guangdong Provincial Innovative Development of Marine Economy Regional Demonstration Projects (GD2012-D01-002), the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (OBER), ‘Genomics: GTL Foundational Science through the ENIGMA Project (DE-AC02-05CH11231) (as part of ENIGMA, a Scientific Focus Area) and the OBER Biological Systems Research on the Role of Microbial Communities in Carbon Cycling Program through DE-SC0004601, and by the U.S. National Science Foundation MacroSystems Biology Program through NSF EF-1065844.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bougon N, Aquilina L, Briand MP, Coedel S, Vandenkoornhuyse P. Influence of hydrological fluxes on the structure of nitrate-reducing bacteria communities in a peatland. Soil Biol Biochem. 2009;41:1289–1300. [Google Scholar]
- Bowen JL, Ward BB, Morrison HG, Hobbie JE, Valiela I, Deegan LA, et al. Microbial community composition in salt marsh sediments resists perturbation by nutrient enrichment. ISME J. 2011;5:1540–1548. doi: 10.1038/ismej.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet RC, Garcia-Gil LJ. Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol Ecol. 1996;21:131–138. [Google Scholar]
- Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, Delong EF, et al. A cryptic sulfur cycle in oxygen-minimum–zone waters off the Chilean coast. Science. 2010;330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- Carrino-Kyker S, Smemo K, Burke D. The effects of pH change and NO3- pulse on microbial community structure and function: a vernal pool microcosm study. FEMS Microbiol Ecol. 2012;81:660–672. doi: 10.1111/j.1574-6941.2012.01397.x. [DOI] [PubMed] [Google Scholar]
- Carrino-Kyker S, Smemo K, Burke D. Shotgun metagenomic analysis of metabolic diversity and microbial community structure in experimental vernal pools subjected to nitrate pulse. BMC Microbiol. 2013;13:78. doi: 10.1186/1471-2180-13-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle CJ, Hug LA, Wrighton KC, Thomas BC, Williams KH, Wu D, et al. Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat Commun. 2013;4:2120. doi: 10.1038/ncomms3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler DP, Jarrell AE, Roden ER, Golova J, Chernov B, Schipma MJ, et al. Suspension array analysis of 16S rRNA from Fe- and SO42- reducing bacteria in uranium-contaminated sediments undergoing bioremediation. Appl Environ Microbiol. 2006;72:4672–4687. doi: 10.1128/AEM.02858-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JA, Rahme H, Hopkins GD, Lebron C, Reinhard M. Enhanced in situ bioremediation of BTEX-contaminated groundwater by combined injection of nitrate and sulfate. Environ Sci Technol. 2001;35:1663–1670. doi: 10.1021/es001722t. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Park J, Rodrigues JLM, Tsoi TV, Hashsham SA, Tiedje JM. Validation of a more sensitive method for using spotted oligonucleotide DNA microarrays for functional genomics studies on bacterial communities. Environ Microbiol. 2003;5:933–943. doi: 10.1046/j.1462-2920.2003.00490.x. [DOI] [PubMed] [Google Scholar]
- Fang HHP, Zhang M, Zhang T, Chen J. Predictions of nitrate diffusion in sediment using horizontal attenuated total reflection (HATR) by Fourier transform infrared (FTIR) spectrometry. Water Res. 2008;42:903–908. doi: 10.1016/j.watres.2007.08.038. [DOI] [PubMed] [Google Scholar]
- Foy RH. Suppression of phosphorus release from lake sediments by the addition of nitrate. Water Res. 1986;20:1345–1351. [Google Scholar]
- Francis CA, Beman JM, Kuypers MMM. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 2007;1:19–27. doi: 10.1038/ismej.2007.8. [DOI] [PubMed] [Google Scholar]
- Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science. 2008;320:889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- Gu C, Laverman AM, Pallud C. Environmental controls on nitrogen and sulfur cycles in surficial aquatic sediments. Front Microbiol. 2012;3:45. doi: 10.3389/fmicb.2012.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck S, Benz M, Brune A, Schink B. Ferrous iron oxidation by denitrifying bacteria in profundal sediments of a deep lake (Lake Constance) FEMS Microbiol Ecol. 2001;37:127–134. [Google Scholar]
- He Z, Deng Y, Zhou J. Development of functional gene microarrays for microbial community analysis. Curr Opin Biotechnol. 2012;23:49–55. doi: 10.1016/j.copbio.2011.11.001. [DOI] [PubMed] [Google Scholar]
- He Z, Van Nostrand JD, Zhou J. Applications of functional gene microarrays for profiling microbial communities. Curr Opin Biotechnol. 2012;23:460–466. doi: 10.1016/j.copbio.2011.12.021. [DOI] [PubMed] [Google Scholar]
- He Z, Xiong J, Kent DA, Deng Y, Xue K, Wang G, et al. Distinct responses of soil microbial communities to elevated CO2 and O3 in a soybean agro-ecosystem. ISME J. 2013;8:714–726. doi: 10.1038/ismej.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Deng Y, Van Nostrand JD, Tu QC, Xu M, Hemme CL, et al. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010;4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- He Z, Xu M, Deng Y, Kang SH, Kellogg L, Wu L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010;13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- Hong H-B, Chang Y-S, Nam I-H, Fortnagel P, Schmidt S. Biotransformation of 2,7-dichloro- and 1,2,3,4-tetrachlorodibenzo-p-dioxin by Sphingomonas wittichii RW1. Appl Environ Microbiol. 2002;68:2584–2588. doi: 10.1128/AEM.68.5.2584-2588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins SR, Miller DE, Thomas A. Combined laboratory/field study on the use of nitrate for in situ bioremediation of a fuel-contaminated aquifer. Environ Sci Technol. 1998;32:1832–1840. [Google Scholar]
- Iwai S, Kurisu F, Urakawa H, Yagi O, Kasuga I, Furumai H. Development of an oligonucleotide microarray to detect di- and monooxygenase genes for benzene degradation in soil. FEMS Microbiol Ecol. 2008;285:111–121. doi: 10.1111/j.1574-6968.2008.01223.x. [DOI] [PubMed] [Google Scholar]
- Jenneman GE, McInerney MJ, Knapp RM. Effect of nitrate on biogenic sulfide production. Appl Environ Microbiol. 1986;51:1205–1211. doi: 10.1128/aem.51.6.1205-1211.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen E, SØNdergaard M, Jensen JP, Havens KE, Anneville O, Carvalho L, et al. Lake responses to reduced nutrient loading – an analysis of contemporary long-term data from 35 case studies. Freshw Biol. 2005;50:1747–1771. [Google Scholar]
- Jorgensen SL, Hannisdal B, Lanzén A, Baumberger T, Flesland K, Fonseca R, et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci USA. 2012;109:E2846–E2855. doi: 10.1073/pnas.1207574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft B, Strous M, Tegetmeyer H. Microbial nitrate respiration - genes, enzymes and environmental distribution. J Biotechnol. 2011;155:104–117. doi: 10.1016/j.jbiotec.2010.12.025. [DOI] [PubMed] [Google Scholar]
- Li XY, Deng Y, Li Q, Lu CY, Wang JJ, Zhang HW, et al. Shifts of functional gene representation in wheat rhizosphere microbial communities under elevated ozone. ISME J. 2013;7:660–671. doi: 10.1038/ismej.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louati H, Said O, Got P, Soltani A, Mahmoudi E, Cravo-Laureau C, et al. Microbial community responses to bioremediation treatments for the mitigation of low-dose anthracene in marine coastal sediments of Bizerte lagoon (Tunisia) Environ Sci Pollut Res Int. 2013;20:300–310. doi: 10.1007/s11356-012-0860-x. [DOI] [PubMed] [Google Scholar]
- McAuliffe TF, Lukatelich RJ, McComb AJ, Qiu S. Nitrate applications to control phosphorus release from sediments of a shallow eutrophic estuary: an experimental evaluation. Mar Freshw Res. 1998;49:463–473. [Google Scholar]
- Porubsky WP, Weston NB, Joye SB. Benthic metabolism and the fate of dissolved inorganic nitrogen in intertidal sediments. Estuar Coast Shelf Sci. 2009;83:392–402. [Google Scholar]
- Rhee S-K, Liu X, Wu L, Chong SC, Wan X, Zhou J. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl Environ Microbiol. 2004;70:4303–4317. doi: 10.1128/AEM.70.7.4303-4317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SØNdergaard M, Jeppesen E, Lauridsen TL, Skov C, Van Nes EH, Roijackers R, et al. Lake restoration: successes, failures and long-term effects. J Appl Ecol. 2007;44:1095–1105. [Google Scholar]
- Tang YJ, Carpenter S, Deming J, Krieger-Brockett B. Controlled release of nitrate and sulfate to enhance anaerobic bioremediation of phenanthrene in marine sediments. Environ Sci Technol. 2005;39:3368–3373. doi: 10.1021/es040427w. [DOI] [PubMed] [Google Scholar]
- Tankéré SPC, Bourne DG, Muller FLL, Torsvik V. Microenvironments and microbial community structure in sediments. Environ Microbiol. 2002;4:97–105. doi: 10.1046/j.1462-2920.2002.00274.x. [DOI] [PubMed] [Google Scholar]
- Taroncher-Oldenburg G, Griner EM, Francis CA, Ward BB. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl Environ Microbiol. 2003;69:1159–1171. doi: 10.1128/AEM.69.2.1159-1171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JM, Bruce CL, Gordy VR, Duston KL, Hutchins SR, Sinclair JL, et al. Assessment of the microbial potential for nitrate-enhanced bioremediation of a JP-4 fuel-contaminated aquifer. J Ind Microbiol Biotech. 1997;18:213–221. [Google Scholar]
- Thorpe CL, Law GTW, Boothman C, Lloyd JR, Burke IT, Morris K. The synergistic effects of high nitrate concentrations on sediment bioreduction. Geomicrobiol J. 2012;29:484–493. [Google Scholar]
- Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD, et al. 2014GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis Mol Ecol Resoure-pub ahead of print 12 February 2014doi: 10.1111/1755-0998.12239 [DOI] [PubMed]
- Van Nostrand JD, Wu L, Wu W-M, Huang Z, Gentry TJ, Deng Y, et al. Dynamics of microbial community composition and function during in situ bioremediation of a uranium-contaminated aquifer. Appl Environ Microbiol. 2011;77:3860–3869. doi: 10.1128/AEM.01981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wu W-M, Wu L, He Z, Van Nostrand JD, Deng Y, et al. Responses of microbial community functional structures to pilot-scale uranium in situ bioremediation. ISME J. 2010;4:1060–1070. doi: 10.1038/ismej.2010.31. [DOI] [PubMed] [Google Scholar]
- Xu M, He Z, Deng Y, Wu L, van Nostrand J, Hobbie S, et al. Elevated CO2 influences microbial carbon and nitrogen cycling. BMC Microbiol. 2013;13:124. doi: 10.1186/1471-2180-13-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada TM, Sueitt AP, Beraldo DA, Botta CM, Fadini PS, Nascimento MR, et al. Calcium nitrate addition to control the internal load of phosphorus from sediments of a tropical eutrophic reservoir: microcosm experiments. Water Res. 2012;46:6463–6475. doi: 10.1016/j.watres.2012.09.018. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu M, He Z, Guo J, Sun G, Zhou J. Microbial electricity generation enhances decabromodiphenyl ether (BDE-209) degradation. PLoS One. 2013;8:e70686. doi: 10.1371/journal.pone.0070686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H-P, Ilhan ZE, Ontiveros-Valencia A, Tang Y, Rittmann BE, Krajmalnik-Brown R. Effects of multiple electron acceptors on microbial interactions in a hydrogen-based biofilm. Environ Sci Technol. 2013;47:7396–7403. doi: 10.1021/es401310j. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xue K, Xie JP, Deng Y, Wu L, Cheng XH, et al. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat Clim Change. 2012;2:106–110. [Google Scholar]
- Zoumis T, Schmidt A, Grigorova L, Calmano W. Contaminants in sediments: remobilisation and demobilisation. Sci Total Environ. 2001;266:195–202. doi: 10.1016/s0048-9697(00)00740-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.