Abstract
This study aimed to investigate the association between POU class5 homeobox 1 pseudogene 1 gene (POU5F1P1) rs10505477 polymorphism and the prognosis of Chinese gastric cancer patients, who received cisplatin-based chemotherapy after surgical resection. POU5F1P1 rs10505477 was genotyped using the SNaPshot method in 944 gastric cancer patients who received gastrectomy. The association of rs10505477 G > A polymorphism with the progression and prognosis in gastric cancer patients was statistically analyzed using the SPSS version 18.0 for Windows. The results reveal that rs10505477 polymorphism has a negatively effect on the overall survival of gastric cancer patients in cisplatin-based chemotherapy subgroup (HR = 1.764, 95% CI = 1.069–2.911, p = 0.023). Our preliminary study indicates for the first time that POU5F1P1 rs10505477 is correlated with survival of gastric cancer patients who receving cisplatin-based chemotherapy after gastrectomy. Further studies are warranted to investigate the mechanism and to verify our results in different populations.
Keywords: gastric cancer, POU5F1P1 rs10505477 polymorphism, single nucleotide polymorphism (SNP), cisplatin-based chemotherapy
1. Introduction
As the fourth most common cancer and the second leading cause of cancer-related death worldwide, gastric cancer (GC) contributes to a significant burden of disease, particularly in economically less-developed countries [1,2]. Although both its morbidity and mortality have been declining in the latest decade [2], GC patients still have a poor 5-year survival rate [3]. In recent years, several studies have demonstrated that GC is a stem cell disease [4,5,6,7]. This viewpoint offers us a brilliant insight to understand the molecular mechanism of gastric cancer and to identify new diagnostic and therapeutic targets for gastric cancer. As far as is known, tumor stem cells are under control of numerous regulatory factors, among them transcription factors should be considered as one of the most important regulatory factors [8]. Hence, it is warranted to identify potential markers of gastric cancer stem cell and related regulatory factors. Furthermore, exploration of the genetic variants in regulatory factor genes involved in the progress and prognosis of gastric cancer is also very important.
POU5F1 (also called OCT4 or OCT3) is a central gene in the regulation of stem cell pluripotency [9,10,11]. Some investigators [12,13] found that the over-expression of POU5F1 is significantly associated with the invasion and metastasis of GC. POU5F1P1 (also called POU5F1B) gene is classified as a highly homologous pseudo-gene of POU5F1 [14]. Panagopoulos and his colleagues have reported that POU5F1P1 produces a protein with similar function to POU5F1 [15]. So we hypothesize that the variants of POU5F1P1 may play a part in the tumorigenesis and progression of gastric cancer through influencing the function of POU5F1. POU5F1P1 is located in 8q24.21 region 3. Preliminary GWAS (Genome Wide Association Studies) and follow-up studies were carried out to reveal functional signal nucleotide polymorphisms (SNPs) of POU5F1P1 involved in cancer. Pal et al. [16] have found strong evidence of the association of POU5F1P1 rs871135 G > T polymorphism with prostate cancer and Wei et al. [17] revealed POU5F1P1 rs7014346 G > A polymorphism was significantly associated with breast cancer. Most studies of POU5F1P1 gene polymorphism concern the loci rs10505477; studies have shown that the rs10505477 C > T polymorphism plays an important role in the oncogenesis and progression of colorectal cancer (CRC) [18,19,20,21,22], but not in ovarian cancer [23]. However the association of rs10505477 with GC is poorly understood. In 2011, Paul et al.’s research first detected that there was no significant association of rs10505477 with upper gastrointestinal cancer in Caucasians [24]. As genetic variation is geographically structured, an allele tends to become more frequent in one population but not in another. Therefore we performed this genotyping study to see if the relationship between gastric cancer and POU5F1P1 rs10505477 G > A polymorphism in a Chinese Han population would be consistent with Paul’s study.
2. Results
2.1. Patients’ Characteristics
Nine-hundred and nine samples were included in this study after excluding those patients with failed genotyping. The patients’ characteristics and clinical information are summarized in Table 1. All patients received surgical resections, among which 291 had undergone chemotherapy. There were 700 males (77.0%) and 209 females (23.0%), with the median age of 61 years ranging from 28 to 83 years. In the follow-up period of 119 months (last follow-up in March 2009), we observed that a sum of 418 (46.0%) patients died. The maximum survival time was 119.0 months and the median survival time was 70.0 months. Our study confirmed that clinicopathologic characteristics, including tumor size, histological types, depth of invasion, lymph node metastasis, and TNM (Tumor, Node, Metastasis) stage were closely related to survival time (log-rank p < 0.05). Specifically, patients with tumor size > 5 cm (median survival time (MST), 51 months) had a 40.9% significantly higher risk of death (HR = 1.409, 95% CI = 1.161–1.710) compared with those with tumor size ≤ 5 cm (MST, 74 months), and the diffuse-type gastric cancer patients (MST, 52 months) had a 45.3% significantly higher risk of death (HR = 1.453, 95% CI = 1.189–1.776) than those intestinal-type patients (MST, 77 months). In addition, as the depth of invasion and TNM stage increased, the risk of death for gastric cancer showed a significant increase in a dose-dependent manner (log-rank p < 0.001).
Table 1.
Association between clinicopathological features and survival of gastric cancer.
| Variables | Patients, n = 909 | Deaths, n = 418 | MST (Months) | log-Rank p | HR (95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | |||||||||||
| ≤60 | 429 | 195 | 97 | 0.354 | 1.000 | ||||||
| >60 | 480 | 223 | 62 | 1.095 (0.903–1.327) | |||||||
| Sex | |||||||||||
| Male | 700 | 320 | 74 | 0.57 | 1.000 | ||||||
| Female | 209 | 98 | 67 | 1.067 (0.851–1.338) | |||||||
| Tumor size | |||||||||||
| ≤5 cm | 564 | 236 | 74 | <0.001 | 1.000 | ||||||
| >5 cm | 345 | 182 | 51 | 1.409 (1.616–1.710) | |||||||
| Location | |||||||||||
| Non-Cardia | 601 | 280 | 70 | 0.371 | 1.000 | ||||||
| Cardia | 308 | 138 | 77 | 0.912 (0.744–1.118) | |||||||
| Histological types | |||||||||||
| Intestinal | 387 | 149 | 77 | <0.001 | 1.000 | ||||||
| Diffuse | 518 | 266 | 52 | 1.453 (1.189–1.776) | |||||||
| Others | 4 | 3 | 11 | 2.732 (0.871–8.571) | |||||||
| Differentiation a | |||||||||||
| Well-to-moderate | 297 | 125 | 80 | 0.49 | 1.000 | ||||||
| Poorly | 472 | 228 | 62 | 1.158 (0.931–1.441) | |||||||
| Mucinous/signet-ring cell | 65 | 32 | 62 | 1.202 (0.815–1.772) | |||||||
| Others | 75 | 33 | 67 | 0.986 (0.671–1.448) | |||||||
| Depth of invasion b | |||||||||||
| T1 | 177 | 57 | N/A1 | <0.001 | 1.000 | ||||||
| T2 | 130 | 56 | 78 | 1.452(1.004–2.101) | |||||||
| T3 | 6 | 3 | 70 | 1.427(0.447–4.559) | |||||||
| T4 | 578 | 291 | 52 | 1.839(1.383–2.446) | |||||||
| Lymph node metastasis c | |||||||||||
| N0 | 359 | 128 | N/A1 | <0.001 | 1.000 | ||||||
| N1/N2/N3 | 529 | 277 | 48 | 1.731 (1.403–2.136) | |||||||
| Distant metastasis | |||||||||||
| M0 | 891 | 407 | 74 | 0.296 | 1.000 | ||||||
| M1 | 16 | 9 | 40 | 1.417 (0.732–2.743) | |||||||
| TNM stage | |||||||||||
| I | 239 | 80 | N/A1 | <0.001 | 1.000 | ||||||
| II | 195 | 77 | N/A1 | 1.241 (0.907–1.698) | |||||||
| III | 447 | 244 | 41 | 1.993 (1.547–2.568) | |||||||
| IV | 22 | 11 | 47 | 1.823 (0.970–3.424) | |||||||
| Chemotherapy | |||||||||||
| No | 618 | 293 | 62 | 0.344 | 1.000 | ||||||
| Yes | 291 | 125 | 98 | 0.904 (0.734–1.115) | |||||||
| Chemotherapy regimes | |||||||||||
| l-OHP | 109 | 38 | 60 | 0.082 | 1.000 | ||||||
| DDP | 179 | 89 | 51 | 1.398 (0.954–2.048) | |||||||
| Smoking | |||||||||||
| Non-smoker | 833 | 386 | 67 | 0.432 | 1.000 | ||||||
| Smoker | 76 | 32 | 97 | 0.866 (0.604–1.243) | |||||||
| Drinking | |||||||||||
| Non-drinker | 850 | 389 | 70 | 0.691 | 1.000 | ||||||
| Drinker | 59 | 29 | 63 | 1.079 (0.740–1.574) |
Abbreviations: MST, median survival time; HR, hazard ratio; CI, confidence interval; TNM, Tumor, Node and Metastasis; l-OHP, oxaliplatin; DDP, cisplatin. a Partial data were not available, and statistics were based on available data; b The information about the depth of invasion was not available for two patients; invaded depth of tumor was classified according to the criteria of American Joint Commission on Cancer (AJCC) 7th; c Lymph nodes were staged according to tumor-node-metastasis classification of the 7th edition of AJCC in which the number of lymph nodes with a metastasis of 1, 2, 3, 6 and 7 were classified as N1, N2 and N3, respectively. N/A1, Mean the median survival time could not be measured.
2.2. Associations of POU5F1P1 rs10505477 with Prognosis of Gastric Cancer (GC) Patients
Among 944 GC patients with complete clinical follow-up information, rs10505477 was successfully genotyped in 909 specimens. The frequency of each genotype was 34.7% (315 specimens) for the GG variant, 47.7% (434 specimens) for the GA variant, 17.6% (160 specimens) for the AA variant. Cox regression analysis was used to detect the association of rs10505477 polymorphism with gastric cancer survival in various genetic models. Regrettably, there was no association between POU5F1P1 rs10505477 G > A polymorphisms and the survival of GC patients in either genotype models (log-rank p = 0.185 for co-dominant model; log-rank p = 0.177 for dominant model; log-rank p = 0.478 for recessive model; as present in Table 2).
Table 2.
Association between rs10505477 polymorphism and overall survival of gastric cancer.
| Genetic Model | Genotypes | Patients | Deaths | MST (Months) | log-Rank p | HR (95% CI) * |
|---|---|---|---|---|---|---|
| Codominant model | GG | 315 | 136 | 77 | 0.185 | 1.000 |
| GA | 434 | 215 | 60 | 1.200 (0.968–1.488) | ||
| AA | 160 | 67 | 69 | 1.014 (0.757–1.359) | ||
| Dominant model | GG | 315 | 136 | 77 | 0.177 | 1.000 |
| GA/AA | 594 | 282 | 63 | 1.150 (0.937–1.411) | ||
| Recessive model | GG/GA | 749 | 351 | 67 | 0.478 | 1.000 |
| AA | 160 | 67 | N/A1 | 0.910 (0.701–1.182) |
Abbreviations: MST, median survival time; HR, hazard ratio; CI, confidence interval; * Hazard Ratio (HR) adjusted for age, sex, Tumor, Node and Metastasis (TNM) stage; N/A1, Mean the median survival time could not be measured.
We further assessed the association of POU5F1P1 rs10505477 polymorphisms with gastric cancer survival by stratified analysis of tumor size, tumor site, histological type, depth of invasion, lymph node metastasis, distant metastasis, TNM stage and chemotherapy. The results are shown in Table 3. In the different subgroups of patients, there was no significant association between genotypes and survival of GC patients in any genetic models.
Table 3.
Stratified analysis of association between rs10505477 polymorphism and overall survival of gastric cancer.
| Variables | Genotypes (Dominant Model) | HR (95% CI) a | p Heterogeneity | |
|---|---|---|---|---|
| GG | GA/AA | |||
| Total (n = 909) | 315 | 594 | 1.150 (0.937–1.411) | 0.177 |
| Tumor size | ||||
| ≤5 cm | 202 | 362 | 1.248 (0.949–1.640) | 0.112 |
| >5 cm | 113 | 232 | 0.989 (0.726–1.348) | 0.945 |
| Tumor site | ||||
| Non-Cardia | 207 | 394 | 1.226 (0.951–1.581) | 0.116 |
| Cardia | 108 | 200 | 1.038 (0.733–1.469) | 0.835 |
| Lauren classification | ||||
| Intestinal type | 145 | 242 | 0.996 (0.715–1.387) | 0.98 |
| Diffuse type | 170 | 352 | 1.224 (0.941–1.593) | 0.132 |
| Differentiation | ||||
| Well to moderate | 114 | 183 | 0.988 (0.689–1.418) | 0.949 |
| Poorly | 160 | 312 | 1.102 (0.835–1.454) | 0.494 |
| Mucinous/signet-ring cell | 19 | 46 | 2.075 (0.850–5.065) | 0.109 |
| Others | 22 | 53 | 1.674 (0.754–3.718) | 0.206 |
| Depth of invasion | ||||
| T1 | 66 | 111 | 1.072 (0.634–1.813) | 0.796 |
| T2 | 47 | 83 | 1.502 (0.842–2.680) | 0.168 |
| T3 | 2 | 4 | 0.627 (0.138–2.837) | 0.544 |
| T4 | 194 | 384 | 1.115 (0.872–1.425) | 0.385 |
| Lymph node metastasis | ||||
| N0 | 137 | 222 | 1.181 (0.842–1.655) | 0.335 |
| N1/N2/N3 | 170 | 369 | 1.135 (0.877–1.469) | 0.336 |
| Distant metastasis | ||||
| M0 | 309 | 582 | 1.130 (0.914–1.398) | 0.259 |
| M1 | 6 | 10 | 1.544 (0.721–3.305) | 0.264 |
| TNM stage | ||||
| I | 93 | 149 | 1.238 (0.789–1.942) | 0.352 |
| II | 64 | 195 | 1.032 (0.640–1.664) | 0.896 |
| III | 150 | 299 | 1.080 (0.826–1.412) | 0.574 |
| IV | 8 | 14 | 2.301 (0.600–8.818) | 0.224 |
| Chemotherapy | ||||
| No | 208 | 410 | 1.144 (0.893–1.466) | 0.286 |
| Yes | 107 | 184 | 1.414 (0.792–1.645) | 0.478 |
Abbreviations: HR, hazard ratio; CI, confidence interval; a Hazard Ratio (HR) adjusted for age, sex.
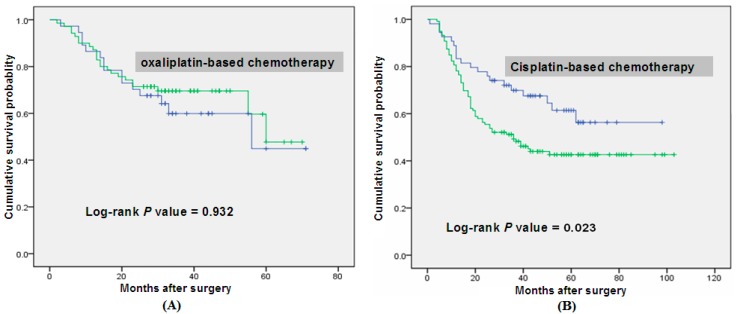
Then we stratified patients by chemotherapy regimens (based on cisplatin and oxaliplatin) and performed the Cox regression, Kaplan–Meier survival curves and the log-rank test to evaluate the association of rs10505477 genotypes with survival in stratified patients. Exhilaratingly, in dominant models, GA/AA genotypes had negative effect on overall survival of patients receiving chemotherapy based on cisplatin (HR = 1.764, 95% CI = 1.069–2.911, p = 0.023, Table 4). But no similar results were found in subgroup with chemotherapy based on oxaliplatin (l-OHP). And the survival curve was shown in Figure 1. It demonstrated that compared with the G allele, the A allele was a risk factor for the prognosis of these patients having chemotherapy based on cisplatin (CDDP).
Table 4.
Association between the dominant model of rs10505477 and overall survival of gastric cancer among chemotherapy regimen subgroup.
| Chemotherapy Based on l-OHP | |||||
|---|---|---|---|---|---|
| Genotype | Patients, n =108 | Deaths, n = 38 | MST (Months) | log-Rank p | HR (95% CI) a |
| GG | 39 | 14 | 55 | 0.932 | 1.000 |
| GA/AA | 69 | 24 | 60 | 2.038 (0.954–3.041) | |
| Chemotherapy Based on CDDP | |||||
| Genotype | Patients, n =173 | Deaths, n = 86 | MST (Months) | log-Rank p | HR (95% CI) a |
| GG | 54 | 20 | N/A1 | 0.023 | 1.000 |
| GA/AA | 119 | 66 | 36 | 1.764 (1.069–2.911) | |
Abbreviations: HR, hazard ratio; CI, confidence interval; MST, median survival time. a HR adjusted for age, sex, TNM stage; l-OHP, oxaliplatin; CDDP, cisplatin. N/A1, mean the median survival time could not be measured.
Figure 1.
Overall survival curve in relation to Pit-Oct-Unic Class 5 Homeobox 1 Pseudogene 1 Gene (POU5F1P1) rs10505477 polymorphism in gastric cancer patients in dominant model. (A) demonstrates that when compared with the GG genotype, GA/AA genotypes had nodifference on overall survival in oxaliplatin-based chemotherapy subgroup (p = 0.932); (B) demonstrates that the GA/AA genotypes had negative effects on overall survival in the subgroups of patients receiving cisplatin-based chemotherapy (p = 0.023).
Finally, stepwise Cox regression analysis was performed to obtain the association between included demographic characteristics, clinical features, the rs10505477 SNP and gastric cancer patients’ survival. As shown in Table 5, one variable (regimens: oxaliplatin vs. cisplatin) was included in the Cox regression model with a significance level for p < 0.05 entering and p > 0.10 for removing a variable (p = 0.048).
Table 5.
Stepwise Cox regression analysis on the survival of gastric cancer.
| Variables | B | SE | HR | 95% CI | p Value |
|---|---|---|---|---|---|
| Age | 0.316 | 0.189 | 1.371 | 0.947–1.985 | 0.094 |
| Sex | 0.267 | 0.228 | 1.306 | 0.835–2.042 | 0.242 |
| Histological types | −0.145 | 0.193 | 0.865 | 0.592–1.264 | 0.454 |
| Regimes (l-OHP vs. DDP) | 0.391 | 0.198 | 1.479 | 1.003–2.180 | 0.048 |
| Dominant model (GG vs. GA/AA) | 0.17 | 0.121 | 1.393 | 0.929–2.089 | 0.109 |
Abbreviations: B, relative risk rate; SE, Standard error; HR, hazard ratio; CI, confidence interval; l-OHP, oxaliplatin; DDP, cisplatin.
3. Discussion
In the present study, TNM stage and invasion depth were identified as independent prognostic factors, which is consistent with conclusions from previous studies [24,25,26,27]. Further, we found for the first time that POU5F1P1 rs10505477 GA/AA genotypes indicated poorer overall survival of gastric cancer in patients undergoing chemotherapy based on CDDP, compared with the GG genotype. This finding had never been demonstrated by other researchers before. However, no association between rs10505477 and survival of gastric cancer in either genotype was observed for oxaliplatin therapy.
Gastric cancer is a stem cell disease [4,5,6,7]; tumor stem cells have been identified with characteristics of pluripotency and self-renewal. Normally, stem cells exist in their own micro-ecological environment, maintaining the stability of the body through proliferation and differentiation. With genetic changes or alteration in the microenvironment, the regulatory mechanisms of stem cell proliferation and differentiation is disrupted, and, as a result, tumors may form [9,28].
POU5F1, a member of the POU (Pit-Oct-Unic) transcription factor family, is one of the most important transcription factors for maintaining the stem cells’ pluripotent and self-renewing state [9,10,11]. POU5F1 is expressed not only in embryonic stem cells and germ cells but also in various types of solid tumor cells, including gastric cancer [12,13]. It has been confirmed in some reports that POU5F1 plays an important role in gastrointestinal malignancy through WNT/β-catenin, TGF-β, JAK3/AKT and STAT3/Survivin pathway [29,30,31,32]. POU5F1P1 gene was classified as a highly homologous pseudo-gene of POU5F1. It has been reported that POU5F1P1 produces a protein with similar function to POU5F1 and that it is associated with prostate cancer [16,33], breast cancer [17] and colorectal cancer [19,20,21,22], whereas the association of POU5F1P1 with gastric cancer is poorly understood. In 2011, Paul et al. first detected no significant association of rs10505477 with upper gastrointestinal cancer in Caucasians [24]. However, there was no research conducted to investigate if the mutation of POU5F1P1 rs10505477 is associated with gastric cancer or not in Asians. Thus we performed this study to investigate the correlation of POU5F1P1 rs10505477 with the survival of gastric cancer patients in a Chinese Han population. We found that the patients with the A allele receiving CDDP-based chemotherapy after gastrectomy had worse prognosis.
The REAL-2 and some other studies [34,35] similarly demonstrated the same result that there was no significant difference between oxaliplatin-containing and cisplatin-containing regimens and Arbeitsgemeinschaft Internistische Onkologie (AIO) trial revealed that oxaliplatin could be substituted for cisplatin [36]. Thus, in this study, patients were stratified by chemotherapy regimens (oxaliplatin-based and cisplatin-based), then the prognosis was analyzed for the different genetic models. The result showed that the A allele was a risk factor for the prognosis of these patients having chemotherapy based on cisplatin. But there was no relationship with the prognosis for rs10505477 and gastric cancer patients receving oxaliplatin-based chemotherapy. Thus, we propose that the rs10505477 genotypes can be a potential predictive biomarker of response to cisplatin-based chemotherapy. As far as we are aware, cisplatin and oxaliplatin are the standard platinum drugs. They share the same basic mechanism of anti-tumor action by influencing their common pharmacological target namely DNA. But their mechanism of antitumor action and drug resistance are not exactly the same. This may be the reason why rs10505477 can predict response to cisplatin but not be related to oxaliplatin. The underlying mechanisms of cisplatin resistance are complicated, including reduced concentration of the drug via efflux pumps and detoxification enzymes, or enhanced DNA repair activity, and so on [37,38]. Previous studies have given evidence that over-expression of AKT [39,40,41,42], activation of the STAT3 [43,44] and Wnt signaling pathways [45,46] and down-regulation of c-Myc expression [47] all contribute to cisplatin resistance. In our study, we found that the polymorphism of rs10505477 was significantly associated with the outcome of GC patients treated with cisplatin. We suspect that rs10505477 variants may lead to cisplatin resistance. If this is the case, the underlying mechanisms not fully explored here need further investigation.
A number of limitations should be addressed in this study. First, we only have data for overall survival of the gastric cancer patients, and lack information on disease-specific survival and relapse-free survival. We estimate that most of the patients died of gastric cancer, but lack definitive data on this outcome; Second, the study samples were 909 Chinese GC patients without matched group, and this may lead to bias. Larger sample sizes studies and case-control studies in different populations are needed in the future; Third, in chemotherapy regimen based on CDDP, we found that compared with GG genotype, overall survival of the patients with GA/AA genotypes is decreased significantly. But we cannot make a conclusion that this mutation induces chemo-resistance for lack of large multicenter clinical trials, thus more studies are needed to be carried out to validate our hypothesis.
In conclusion, our results show that POU5F1P1 rs10505477 polymorphisms have no overall significant association with the survival time of gastric cancer patients; the A allele is a risk factor of the prognosis for these patients only in the subgroup regimen based on cisplatin. Further investigations are required to confirm these findings.
4. Material and Methods
4.1. Ethics Statement
All participants included in this study had provided written informed consent and the entire procedure was approved by the Institutional Review Board of Nanjing Medical University (Nanjing, China; register ID number: 201203121; 2 March 2012).
4.2. Study Subjects
Study subjects were patients with histopathologically confirmed gastric cancer who had received gastrectomy between January 1999 and December 2006 recruited from Yixing People’s Hospital (Yixing, China). None had received chemotherapy or radiotherapy at any point prior to surgery. Nine-hundred and forty-four formalin-fixed and paraffin-embedded samples were obtained. The end point was overall survival (OS). The survival time was calculated from the date of surgery until death or the end of follow-up in March 2009. Death dates were confirmed by review of death certificates of inpatient and outpatient records or obtained through follow-up telephone calls. Patients alive on the last follow-up date were censored. Clinical and pathological variables including age, gender, tumor size, tumor site, histological type, depth of invasion, lymph node metastasis, distant metastasis, TNM stage and chemotherapy regimens were obtained. The TNM stage classification was evaluated according to the criteria of the American Joint Committee on Cancer (AJCC) in 2010. Lauren’s criteria were used to classify the tumors into intestinal and diffuse type.
4.3. Genotyping
Genomic DNA of patients was extracted from paraffin sections of tissues by proteinase K digestion, isopropanol extraction and ethanol precipitation. Genotyping was performed with the SNaPshot method using an ABI fluorescence-based assay allelic discrimination method (Applied Biosystems, Foster City, CA, USA) as described previously [25,26,27]. The sequences of the primers used for multiplexed PCR are F-primer (5'-TGTCAATACTGACTTTGCCCCTTTTC-3') and R-primer (5'-TCACCACTTGTCTATCAAACAGGAAGC-3'). The SNaPshot products were analyzed by using ABI 3130xl genetic analyzer (Applied Biosystems) and the genotypes were determined by GeneMapper Analysis Software version 4 (Applied Biosystems). Genotyping assays were performed by two people independently in a blind fashion. More than 10% of the samples were randomly selected for confirmation, and the results were 100% concordant. Nevertheless, 35 samples failed to be genotyped because of poor DNA quality, which were excluded in further analysis. As a result, 909 gastric cancer patients were included in the final analysis.
4.4. Statistical Method
Statistical analyses were carried out by using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA) with a two-sided test. The correlations between rs10505477 SNP and clinicopathologic parameters were estimated by using the Pearson chi-square test for categorical variables and the Student t test for continuous data. Kaplan–Meier survival curves and the log-rank test were used to evaluate the associations of clinicopathologic variables or rs10505477 SNP with the prognosis of GC. Unvaried or multivariate Cox proportional hazard models, adjuseded for sex, age and TNM stage, were adopted to estimate the crude hazard ratios (HRs), adjusted HRs and their 95% confidence intervals (CIs). Moreover, Cox stepwise regression analysis was performed to assess the independent impacts of SNP or clinicopathologic features on the overall survival (OS) after adjusting for other covariates, with a significance level of p < 0.05 for entering and p > 0.10 for removing the respective explanatory variables. All tests were two-sided and p < 0.05 was considered statistically significant.
5. Conclusions
Our preliminary study indicates, for the first time, that POU5F1P1 rs10505477 polymorphism has no significant association with the survival of gastric cancer patients. However, the A allele is a risk factor for the prognosis of gastric cancer patients receiving cisplatin-based chemotherapy. Further studies are warranted to investigate the mechanism and to verify our results in different populations.
Acknowledgments
The work was partly supported by grants from the National Natural Science Foundation of China (Grant No. 81272469), the National 973 Basic Research Program of China (Grant No. 2013CB911300) and the clinical special project for Natural Science Foundation of Jiangsu Province (Grant No. BL2012016), and the grant from Nanjing 12th Five-Year key Scientific Project of medicine to Jinfei Chen.
Author Contributions
L.S., M.D., C.W. and M.W. designed the study and applied for Research Ethics Board approval. D.G., Q.Z., T.Z., X.Z. and X.H. recruited the patients and collected the data. Y.T., W.G. and Z.X. analyzed the data and prepared draft figures and tables. L.S. and M.D. prepared the manuscript draft. All authors approved the final article. Z.Z. and J.C. had complete access to the study data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics. CA Cancer J. Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Santoro R., Carboni F., Lepiane P., Ettorre G.M., Santoro E. Clinicopathological features and prognosis of gastric cancer in young European adults. Br. J. Surg. 2007;94:737–742. doi: 10.1002/bjs.5600. [DOI] [PubMed] [Google Scholar]
- 4.Takaishi S., Okumura T., Wang T.C. Gastric cancer stem cells. J. Clin. Oncol. 2008;26:2876–2882. doi: 10.1200/JCO.2007.15.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takaishi S., Okumura T., Tu S., Wang S.S., Shibata W., Vigneshwaran R., Gordon S.A., Shimada Y., Wang T.C. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han M.E., Jeon T.Y., Hwang S.H., Lee Y.S., Kim H.J., Shim H.E., Yoon S., Baek S.Y., Kin B.S., Kang C.D., et al. Cancer spheres from gastric cancer patients provide an ideal model system for cancer stem cell research. Cell. Mol. Life Sci. 2011;68:3589–3605. doi: 10.1007/s00018-011-0672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatematsu M., Tsukamoto T., Inada K. Stem cells and gastric cancer: Role of gastric and intestinal mixed intestinal metaplasia. Cancer Sci. 2003;94:135–141. doi: 10.1111/j.1349-7006.2003.tb01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C., Li C., He F., Cai Y., Yang H. Identification of CD44+CD24+ gastric cancer stem cells. J. Cancer Res. Clin. Oncol. 2011;137:1679–1686. doi: 10.1007/s00432-011-1038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alison M.R., Poulsom R., Forbes S., Wright N.A. An introduction to stem cells. J. Pathol. 2002;197:419–423. doi: 10.1002/path.1187. [DOI] [PubMed] [Google Scholar]
- 10.Pan G.J., Chang Z.Y., Scholer H.R., Pei D. Stem cell pluripotency and transcription factor OCT4. Cell Res. 2002;12:321–329. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- 11.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Boyer, LAZucker J.P., Guenther M.G., Kumar R.M., Murray H.L., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng C.J., Wu Y.C., Shu J.A., Ling T.Y., Kuo H.C., Wu J.Y., Chang E.E., Chang S.C., Huang Y.H. Aberrant expression and distribution of the OCT-4 transcription factor in seminomas. J. Biomed. Sci. 2007;14:797–807. doi: 10.1007/s11373-007-9198-7. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka J., Yashiro M., Sakurai K., Kubo N., Tanaka H., Muguruma K., Sawada T., Ohira M., Hirakawa K. Role of the stemness factors Sox2, OCT3/4 and nanog in gastric carcinoma. J. Surg. Res. 2012;174:130–135. doi: 10.1016/j.jss.2010.11.903. [DOI] [PubMed] [Google Scholar]
- 14.Suo G., Han J., Wang X., Zhang J., Zhao Y., Zhao Y., Dai J. OCT4 pseudogenes are transcribed in cancers. Biochem. Biophys. Res. Commun. 2005;337:1047–1051. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- 15.Panagopoulos I., Moller E., Collin A., Mertens F. The POU5F1P1 pseudogene encodes a putative protein similar to POU5F1 isoform 1. Oncol. Rep. 2008;20:1029–1033. [PubMed] [Google Scholar]
- 16.Pal P., Xi H., Guha S., Sun G., Helfand B.T., Meeks J.J., Suarez B.K., Catalona W.J., Deka R. Common variants in 8q24 are associated with risk for prostate cancer and tumor aggressiveness in men of European ancestry. Prostate. 2009;69:1548–1556. doi: 10.1002/pros.20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei W., Jiang M., Luo L., Li Z., Wang P., Dong W.Q. Colorectal cancer susceptibility variants alter risk of breast cancer in a Chinese Han population. Genet. Mol. Res. 2013;12:6268–6274. doi: 10.4238/2013.December.4.14. [DOI] [PubMed] [Google Scholar]
- 18.Gruber S.B., Moreno V., Rozek L.S., Rennerts H.S., Lejbkowicz F., Bonner J.D., Greenson J.K., Giordano T.J., Fearson E.R., Rennert G. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol. Ther. 2007;6:1143–1147. doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 19.Poynter J.N., Figueiredo J.C., Conti D.V., Kennedy K., Gallinger S., Siegmund K.D., Casey G., Thibodeau S.N., Jenkins M.A., Hopper J.L., et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: Results from the colon cancer family registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 20.Schafmayer C., Buch S., Volzke H., von Schonfels W., Egberts J.H., Schniewind B., Brosch M., Ruether A., Franke A., Mathiak M., et al. Investigation of the colorectal cancer susceptibility region on chromosome 8q24.21 in a large German case-control sample. Int. J. Cancer. 2009;124:75–80. doi: 10.1002/ijc.23872. [DOI] [PubMed] [Google Scholar]
- 21.Dai J., Gu J., Huang M., Eng C., Kopetz E.S., Ellis L.M., Hawk E., Wu X. GWAS-identified colorectal cancer susceptibility loci associated with clinical outcomes. Carcinogenesis. 2012;33:1327–1331. doi: 10.1093/carcin/bgs147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutter C.M., Slattery M.L., Duggan D.J., Muehling J., Curtin K., Beresford S.A., Rajkovic A., Sarto G.E., Marshall J.R., et al. Characterization of the association between 8q24 and colon cancer: Gene-environment exploration and meta-analysis. BMC Cancer. 2010;10:670. doi: 10.1186/1471-2407-10-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White K.L., Sellers T.A., Fridley B.L., Vierkant R.A., Phelan C.M., Tsai Y.Y., Kalli K.R., Berchuck A., Iversen E.S., Hartmann L.C., et al. Variation at 8q24 and 9p24 and risk of epithelial ovarian cancer. Twin. Res. Hum. Genet. 2010;13:43–56. doi: 10.1375/twin.13.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lochhead P., Ng M.T.H., Hold G.L., Rabkin C.S., Vaughan T.L., Gammon M.D., Risch H.A., Lissowska J., Mukhopadhya I., Chow W.H., et al. Possible association between a genetic polymorphism at 8q24 and risk of upper gastrointestinal cancer. Eur. J. Cancer. Prev. 2011;20:54–57. doi: 10.1097/CEJ.0b013e328341e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Wang M., Gu D., Wu D., Zhang X., Gong W., Tan Y., Zhou J., Wu X., Tang C, et al. Association of XRCC1 gene polymorphisms with the survival and clinicopathological characteristics of gastric cancer. DNA Cell Biol. 2013;32:111–118. doi: 10.1089/dna.2012.1840. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Zhu H., Wu X., Wang M., Gu D., Gu D., Gong W., Xu Z., Tan Y., Gong Y., et al. A genetic polymorphism in TOX3 is associated with survival of gastric cancer in a Chinese population. PLoS One. 2013;8:e72186. doi: 10.1371/journal.pone.0072186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Zhu H., Zhang X., Gu D., Zhou X., Wang M., Cao C., Zhang X., Wu X., Gong W., et al. linical significance of MYT1L gene polymorphisms in Chinese patients with gastric cancer. PLoS One. 2013;8:e71979. doi: 10.1371/journal.pone.0071979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spradling A., Drummond-Barbosa D., Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 29.Ezeh U.I., Turek P.J., Reijo R.A., Clark A.T. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104:2255–2265. doi: 10.1002/cncr.21432. [DOI] [PubMed] [Google Scholar]
- 30.Wen K., Fu Z., Wu X., Feng J., Chen W., Qian J. OCT-4 is required for an antiapoptotic behavior of chemoresistant colorectal cancer cells enriched for cancer stem cells: Effects associated with STAT3/Survivin. Cancer Lett. 2013;333:56–65. doi: 10.1016/j.canlet.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Davidson K.C., Adams A.M., Goodson J.M., McDonald C.E., Potter J.C., Berndt J.D., Biechele T.L., Taylor R.J., Moon R.T. Wnt/β-catenin signaling promotes differentiation not self-renewal of human embryonic stem cells and is repressed by OCT4. Proc. Natl. Acad. Sci. USA. 2012;109:4485–4490. doi: 10.1073/pnas.1118777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X.Q., Ongkeko W.M., Chen L., Yang Z.F., Lu P., Chen K.K., Lopez J.P., Poon R.T., Fan S.T. Octamer 4 (Oct4) mediates chemotherapeutic drug resistance in liver cancer cells through a potential Oct4–AKT–ATP-binding cassette G2 pathway. Hepatology. 2010;52:528–539. doi: 10.1002/hep.23692. [DOI] [PubMed] [Google Scholar]
- 33.Kastler S., Honold L., Luedeke M., Kuefer R., Moller P., Hoegel J., Vogel W., Maier C., Assum G. POU5F1P1, a putative cancer susceptibility gene, is overexpressed in prostatic carcinoma. Prostate. 2010;7:666–674. doi: 10.1002/pros.21100. [DOI] [PubMed] [Google Scholar]
- 34.Cunningham D., Rao S., Starling N., Iveson T., Nicolson M., Coxon F., Middleton G., Daniel F., Oates J., Norman A.R. Randomised multicentre phase III study comparingcapecitabine with fluorouracil and oxaliplatin with cisplatin in patients with advanced oesophagogastric (OG) cancer: The REAL 2 trial. J. Clin. Oncol. 2006;24:18S. doi: 10.1200/JCO.2006.06.1143. [DOI] [Google Scholar]
- 35.Kim Y.S., Sym S.J., Park S.H., Park I., Hong J., Ahn H.K., Park J., Cho E.K., Lee W.K., Chung M., et al. A randomized phase II study of weekly docetaxel/cisplatin vs. weekly docetaxel/oxaliplatin as first-line therapy for patients with advanced gastric cancer. Cancer Chemother. Pharmacol. 2014;73:163–169. doi: 10.1007/s00280-013-2334-3. [DOI] [PubMed] [Google Scholar]
- 36.Al-Batran S.E., Hartmann J.T., Probst S., Schmalenberg H., Hollerbach S., Hofheinz R., Rethwisch V., Seipelt G., Homann N., Wilhelm G., et al. Phase III trial in metastatic gastroesophageal adenocarcinoma with fluorouracil, leucovorin plus either oxaliplatin or cisplatin: A study of the arbeitsgemeinschaft internistische onkologie. J. Clin. Oncol. 2008;26:1435–1442. doi: 10.1200/JCO.2007.13.9378. [DOI] [PubMed] [Google Scholar]
- 37.Gottesman M.M., Fojo T., Bates S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 38.Stewart D.J. Mechanisms of resistance to cisplatin and carboplatin. Crit. Rev. Oncol. Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Chu W., Pak B.J., Bani M.R., Kapoor M., Lu S.J., Tamir A., Kerbel R.S., Ben-David Y. Tyrosinase-related protein 2 as a mediator of melanoma specific resistance to cis-diamminedichloroplatinum(II): Therapeutic implications. Oncogene. 2000;19:395–402. doi: 10.1038/sj.onc.1203315. [DOI] [PubMed] [Google Scholar]
- 40.Mizutani Y., Bonavida B. Overcoming cis-diamminedichloroplatinum(II) resistance of human ovarian tumor cells by combination treatment with cis-diamminedichloroplatinum(II) and tumor necrosis factor-α. Cancer. 1993;72:809–818. doi: 10.1002/1097-0142(19930801)72:3<809::AID-CNCR2820720329>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 41.Liu L.Z., Zhou X.D., Qian G., Shi X., Fang J., Jiang B.H. AKT1 amplification regulates cisplatin resistance in human lung cancer cells through the mammalian target of rapamycin/p70S6K1 pathway. Cancer Res. 2007;67:6325–6332. doi: 10.1158/0008-5472.CAN-06-4261. [DOI] [PubMed] [Google Scholar]
- 42.Meijer C., Mulder N.H., Timmer-Bosscha H., Sluiter W.J., Meersma G.J., de Vries E.G. Relationship of cellular glutathione to the cytotoxicity and resistance of seven platinum compounds. Cancer Res. 1992;52:6885–6889. [PubMed] [Google Scholar]
- 43.Zhang L.L., Zhang J., Shen L., Xu X.M., Yu H.G. Overexpression of AKT decreases the chemosensitivity of gastric cancer cells to cisplatin in vitro and in vivo. Mol. Med. Rep. 2013;7:1387–1390. doi: 10.3892/mmr.2013.1400. [DOI] [PubMed] [Google Scholar]
- 44.Huang S., Chen M., Shen Y., Shen W., Guo H., Gao Q., Zuo X. Inhibition of activated Stat3 reverses drug resistance to chemotherapeutic agents in gastric cancer cells. Cancer Lett. 2012;315:198–205. doi: 10.1016/j.canlet.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Xu N., Shen C., Luo Y., Xia L., Xue F., Xia Q., Zhang J. Upregulated miR-130a increases drug resistance by regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell. Biochem. Biophys. Res. Commun. 2012;425:468–472. doi: 10.1016/j.bbrc.2012.07.127. [DOI] [PubMed] [Google Scholar]
- 46.Gosepath E.M., Eckstein N., Hamacher A., Servan K., von Jonquieres G., Lage H., Györffy B., Royer H.D., Kassack M.U. Acquired cisplatin resistance in the head-neck cancer cell line Cal27 is associated with decreased DKK1 expression and can partially be reversed by overexpression of DKK1. Int. J. Cancer. 2008;123:2013–2019. doi: 10.1002/ijc.23721. [DOI] [PubMed] [Google Scholar]
- 47.Biroccio A., Benassi B., Fiorentino F., Zupi G. Glutathione depletion induced by c-Myc downregulation triggers apoptosis on treatment with alkylating agents. Neoplasia. 2004;6:195–206. doi: 10.1593/neo.03370. [DOI] [PMC free article] [PubMed] [Google Scholar]