The discovery of somatic mutations within genes primarily involved in epigenetic regulation has been one of the major success stories of the genomics era. Within the past 5 years, this novel class of recurrent mutations has been elucidated, which consists of critical regulators of post-translational events including DNA methylation and histone modification. The updated list of mutated epigenetic modifiers in myeloid malignancies now includes the genes isocitrate dehydrogenase 1 (IDH1)(1), IDH2(2), tet methylcytosine dioxygenase 2 (TET2)(3), enhancer of zeste homologue 2 (EZH2)(4), mixed lineage leukemia (MLL)(5), additional sex combs-like 1 (ASXL1)(6) and DNA methyltransferase 3A (DNMT3A).(7, 8)
Importantly, many of these epigenetic mutations have subsequently been identified to have prognostic importance for AML patients, particularly within the sizeable cohort of patients with diploid karyotype or otherwise intermediate-risk cytogenetics, and specifically within those intermediate-risk patients with wild-type FMS-related tyrosine kinase 3 (FLT3) status. DNMT3A mutations, for instance, identified in approximately 20% of adult AML patients, have been shown to lead to both an increased risk of relapse and decreased overall survival (OS), correlate with advanced age and a higher white blood cell count at diagnosis, and are associated with mutations in FLT3, nucleophosmin 1 (NPM1) and IDH1.(7–10) TET2 mutations, with a frequency of 7 to 20% in adult AML, also appear to confer an adverse prognosis in intermediate-risk AML.(11, 12) IDH1 and IDH2 mutations, taken together, occur in AML with an estimated prevalence of 10–30%, also occurring at an increased frequency in patients with normal diploid karyotype (CN-AML), older age and NPM1 mutations.(1, 2, 13) The prognostic significance of IDH mutations has been less apparent, perhaps related to a difference in prognosis attributed to the three different allelic mutations observed in AML; namely IDH1-R132, IDH2-R140 and IDH2-R172. While IDH1-R132 and IDH2-R172 mutations appear to have a negative or null clinical impact within the CN-AML patient cohort,(14) IDH2-R140 mutations or any IDH mutation in conjunction with an NPM1 mutation rather unexpectedly confers a favorable outcome in otherwise intermediate-risk AML patients.(10, 15)
Despite the clinically significant prognostic information gained through identification of epigenetic mutations, less insight is currently available to determine how these mutations should affect therapeutic decisions. Recent retrospective data in adults aged 18–60 with de novo AML has notably suggested that the dose-intensity of induction therapy may be especially important for AML patients with DNMT3A, NPM1 or 11q23 abnormalities involving MLL.(10) Nonetheless, AML is primarily a disease of the elderly, with a median age at diagnosis above 65 and with a median survival of less than one year in elderly patients receiving treatment.(16) Given the poor overall response and often increased toxicity associated with intensive chemotherapy in elderly AML, much attention has been paid to the potential role of the hypomethylating agents (HMAs), 5-azacitadine and decitabine, as front-line therapy for elderly patients with AML. With this approach, an improved median survival of 24.5 (versus 16 months with conventional care) was seen in the subgroup of 113 elderly patients with AML receiving azacitidine in the AZA-001 trial.(17) An improved remission rate (CR or CRp) of 18% and median survival of 8 months was seen in a multicenter Phase III study of front-line decitabine versus standard of care (supportive care or low-dose cytarabine) for older patients with AML,(18) suggesting that therapy with HMAs may be beneficial for elderly patients with AML.
Whether there is a relationship between patient response to HMA therapy based on the presence or absence of mutations with epigenetic impact is currently unknown. In one retrospective review by the Group Francophone des Myelodysplasies, the presence of TET2 mutations in 13 of 86 patients (15%) with MDS or AML with 20–30% marrow blasts treated with azacitidine correlated with a higher overall response rate (ORR).(19) This improved ORR, however, did not translate into a benefit in either response duration or overall survival. As most epigenetic mutations are rare in pediatric AML but occur with increased frequency with increased age, this relationship is particularly important to define among the HMA-treated elderly patients.
To investigate the utility of epigenetic mutations as a marker for the effectiveness of epigenetically-targeted therapy in elderly AML, we herein evaluate the association of IDH1, IDH2, and DNMT3A mutations with clinical outcome, in AML patients over 60 years of age who were treated with front-line HMA therapy.
From 2000 to 2010, 68 patients aged sixty years or older with a diagnosis of AML treated with front-line hypomethylating agent therapy out of an eligible cohort of 110 patients were included. Patients were chosen based on the availability of remaining research samples for epigenetic mutation analysis. Patients received therapy on one of seven clinical protocols of HMA-therapy, including decitabine alone (n=24), decitabine + valproic acid (n=14), azacitidine + ATRA + valproic acid (n=20), azacitidine + vorinostat (n=4), azacitidine + valproic acid (n=3), azacitidine + low-dose cytarabine (n=2) and decitabine + vorinostat (n=1). All patients signed informed consent following institutional guidelines and in accordance with the Declaration of Helsinki. This study was approved by the University of Texas MD Anderson Cancer Center Institutional Review Board. Clinical information of all patients is provided in Table 1, including patient age, laboratory parameters at diagnosis, cytogenetic analysis and presence of FLT3, NPM1 and RAS mutations at AML diagnosis.
Table 1.
Clinicopathologic characteristics of patient cohort (n=68)
| Patient Characteristics | Total | IDH1/IDH2 WT | IDH1/IDH2 Mutated | P value | DNMT3A WT | DNMT3A Mutated | P value |
|---|---|---|---|---|---|---|---|
| Number | 68 | 57 | 11 | 58 | 10 | ||
|
| |||||||
| Median age, yrs [Range] | 72 [60–83] | 72 [60–83] | 74 [63–79] | NS | 72 [60–83] | 73 [63–77] | NS |
|
| |||||||
| Median WBC x 109/L [Range] | 7.2 [1.2–79.1] | 6.4 [1.2–79.1] | 8.1 [1.2–30.5] | NS | 7.2 [1.2–79.1] | 6.2 [1.9–37.0] | NS |
|
| |||||||
| Median Plt x 109/L [Range] | 52 [2–470] | 61 [3–470] | 26 [2–61] | 0.004 | 59 [2–470] | 36 [19–113] | NS |
|
| |||||||
| AML type | NS | NS | |||||
| De Novo | 36 | 30 | 6 | 32 | 4 | ||
| Secondary | 17 | 15 | 2 | 15 | 2 | ||
| Therapy-related | 15 | 12 | 3 | 11 | 4 | ||
|
| |||||||
| Cytogenetics | 0.026 | NS | |||||
| +8 | 5 | 5 | 0 | 5 | |||
| −5/5q- | 5 | 4 | 1 | 5 | |||
| −5 and −7 | 12 | 11 | 1 | 12 | |||
| −7/7q- | 10 | 8 | 2 | 7 | 3 | ||
| Diploid | 19 | 13 | 6 | 14 | 5 | ||
| IM | 3 | 2 | 1 | 3 | |||
| Miscellaneous | 14 | 14 | 12 | 2 | |||
|
| |||||||
| FLT3 | NS | NS | |||||
| ITD | 7 | 1 | 2 | 6 | 1 | ||
| D835 | 1 | 5 | 1 | ||||
| Wild-type | 56 | 47 | 9 | 47 | 9 | ||
| Not done | 4 | 4 | 4 | ||||
|
| |||||||
| NPM1 | 0.023 | 0.006 | |||||
| Mutated | 9 | 5 | 4 | 5 | 4 | ||
| Wild-type | 53 | 46 | 7 | 48 | 5 | ||
| Not done | 6 | 6 | 5 | 1 | |||
|
| |||||||
| RAS | NS | NS | |||||
| Mutated | 4 | 1 | 3 | 3 | 1 | ||
| Wild-type | 52 | 7 | 45 | 44 | 8 | ||
| Not done | 12 | 3 | 9 | 11 | 1 | ||
|
| |||||||
| Cytogenetics | 0.007 | 0.041 | |||||
| ELN Favorable | 1 | 0 | 1 | 0 | 1 | ||
| ELN Int-1 | 8 | 4 | 4 | 6 | 2 | ||
| ELN Int-2 | 12 | 11 | 1 | 9 | 3 | ||
| ELN Adverse | 29 | 26 | 3 | 27 | 2 | ||
| Unknown | 18 | 16 | 2 | 16 | 2 | ||
IDH1, IDH2, DNMT3A, NPM1, FLT3 and RAS mutation analysis: Exon 4 mutations of codon R132 of IDH1 and codon R172 and R140 of IDH2 were detected using polymerase chain reaction (PCR) amplification followed by Sanger sequencing using previously described methodology and PCR primers from Integrated DNA Technologies, Coralville, IA.(20) Mutations in exon 12 of NPM1 were detected using PCR amplification followed by capillary gel electrophoresis using previously published methodology.(20) Mutation testing of codons 12, 13 and 61 of KRAS and NRAS were performed using pyrosequencing and FLT3 by PCR followed by capillary gel electrophoresis as previously described.(20, 21) DNMT3A mutations were evaluated by high-resolution melting (HRM) analysis of exons 9, 10, 15 to 19, and 21 to 23, and positive results were confirmed by Sanger sequencing.
Patient characteristics are summarized using median (range) for continuous variables and frequency (percentage) for categorical variables. Categorical variables were compared using the χ2 or Fisher’s exact test, and continuous variables using the Wilcoxon Rank-Sum, and statistical significance was defined as a p value of < 0.05. Event-free survival (EFS) and overall survival (OS) were based on the Kaplan-Meier method, with differences compared between groups by the long-rank test. OS was measured as the time from date of treatment initiation to death or date of last follow-up (censored). EFS was defined as the time interval between date of initiation of treatment and date of treatment failure, relapse, death, or date of last follow-up (censored). The predictive effects of mutation status and other patient characteristics on OS and EFS were examined using univariate and multivariate Cox proportional hazards models. All statistical analyses were conducted in SAS 9.0.
Clinical and disease-specific characteristics of the patients are detailed in Table 1. The median age of the treated patients was 72 years of age, with a range of 60 to 83 years. 36 patients (53%) had a diagnosis of de novo AML; the remainder had secondary or therapy-related AML. By European LeukemiaNet (ELN) cytogenetic and molecular classification system(22), 29 patients (43%) had unfavorable risk, 20 patients (29%) were of intermediate risk, one had ELN favorable risk disease, and 18 (26%) were unknown. Of note, nineteen patients (28%) had diploid cytogenetics (CN-AML).
Overall, 11 patients (16%) had IDH1 or IDH2 mutations; no patient had both an IDH1 and IDH2 mutation present. Of the 11 IDH mutant patients, 3 were IDH1-R132 mutated, 7 were IDH2-R140 mutated, and one patient had an IDH2-R172 mutation. Ten patients (15%) had DNMT3A mutations, and seven of these 10 patients had mutations affecting arginine codon 882 (R882-DNMT3A). Five patients (7%) had both an IDH1/2 and DNMT3A mutation present. The presence of IDH1/2 mutations were associated with a diploid karyotype (p=0.03) and the presence of NPM1 mutations (p=0.02). DNMT3A mutations were not associated with specific cytogenetics, FLT3, or RAS mutations, but were also significantly correlated with NPM1 mutations (p=0.006). Overall, eight patients had FLT3 mutations (seven with FLT3-ITD and one with FLT3-D835) and four patients had RAS mutations.
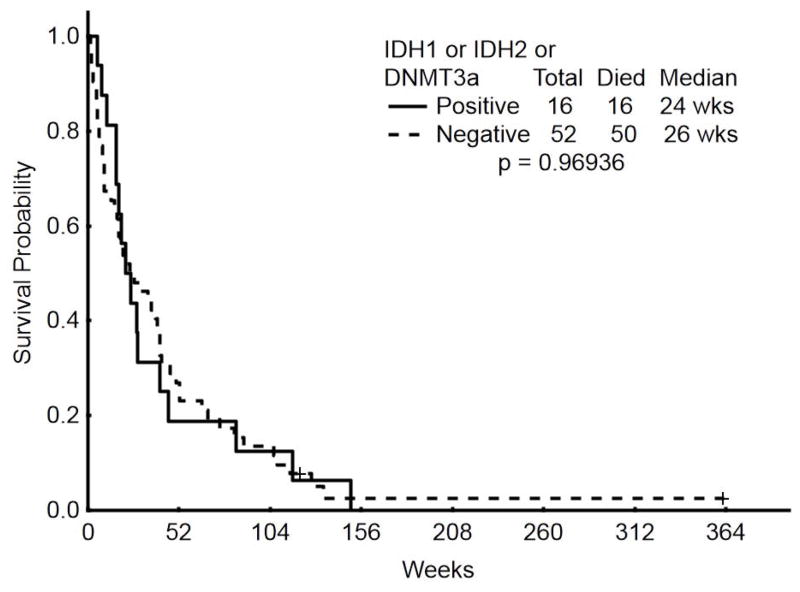
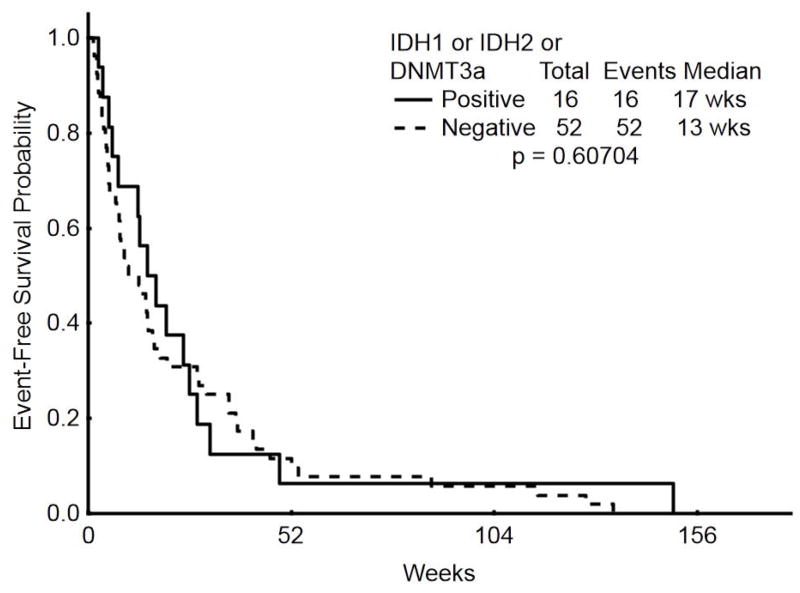
All patients were treated with hypomethylating agent therapy, decitabine in 39 (57%) and azacitidine in 29 (43%). 42 patients (62%) also received concomitant histone deacetylase inhibitor (HDACI) therapy with either vorinostat (n=5) or valproic acid (n=37). Overall, 17 patients (25%) achieved a CR; neither the presence of IDH1/2 mutations nor of DNMT3A mutations (or both) were associated with the achievement of CR (Table 2). With a median duration of follow-up of 60 months, median EFS was 3 months for all patients and median OS was 7 months. IDH1/2 mutations were not associated with an impact on either EFS (p=0.29) or OS (p=0.14) [Supplemental Figure 1]. Similarly, DNMT3A mutations were not associated with an effect on EFS (p=0.21) or OS (p=0.58) [Supplemental Figure 2]. When any epigenetic mutation (IDH1, IDH2 or DNMT3A mutation, n=16) was compared to the lack of an epigenetic mutation (n=52), there was likewise no association with EFS (p=0.6) or OS (p=0.97) [Figure 1].
Table 2.
Response by mutational status in patient cohort (n=68)
| Response to therapy | Total | IDH WT | IDH Mutated | P value | DNMT3A WT | DNMT3A Mutated | P value |
|---|---|---|---|---|---|---|---|
| N (%) | N (%) | ||||||
| CR | 17 (25) | 16 (28) | 1 (9) | NS | 13 (22) | 4 (40) | NS |
| CRp | 1 (1.5) | 1 (2) | 1 (2) | ||||
| PR | 1 (1.5) | 1 (2) | 1 (2) | ||||
| ID | 16 (24) | 12 (21) | 4 (36) | 14 (24) | 2 (20) | ||
| Resistant | 33 (49) | 27 (47) | 6 (55) | 29 (50) | 4 (40) |
CR indicates complete remission; CRp indicates CR with incomplete platelet recovery; PR indicates partial response; ID indicates induction death; WT indicates wild-type; NS indicates not significant.
Figure 1.
Overall, 16 patients (24%) died during induction, which was defined as a death before the protocol-defined response assessment (induction death, Table 2). Since increased toxicity from histone deacetylase therapy has been described in the clinical setting, we additionally examined patient outcome based on whether a patient did or did not receive HDACI during HMA-therapy. There was no difference in EFS (p=0.51) or OS (p=0.96) in our cohort based on the addition of HDACI therapy, with a median EFS of 13 weeks in patients receiving HDACI and 15 weeks in those receiving HMA-therapy alone [Supplemental Figure 3].
Treatment of the older AML patient requires a personalized and risk-adapted approach.(23) Age, comorbidities, cytogenetics, and emerging molecular data need to be analyzed and incorporated into a therapeutic decision that is most appropriate for each individual patient, especially as there is no standard of care for this population. In some patients, intensive therapy with a cytarabine and anthracycline-based regimen is appropriate, and as reported by Juliusson et al., in a selected population of elderly patients, intensive chemotherapy can improve both short and long-term survival compared with palliative approaches.(24) HMA therapy with 5-azacitidine and decitabine is frequently used for older patients with AML who are deemed unfit to receive standard chemotherapy, and decitabine has recently been approved by the European Medicines Agency (EMA) for this front-line indication. As therapy with HMAs is generally well-tolerated with predominantly myelosuppressive adverse events, HMAs are an important therapeutic alternative for elderly patients with AML who are unable to tolerate standard intensive chemotherapy regimens.
We evaluated the utility of front-line HMA therapy in older patients with AML, based on the presence or absence of relevant mutations with known epigenetic impact. This is a fundamental research question, for it follows that if these mutations were found to confer a particularly favorable response to HMA-therapy, epigenetic therapy could be offered to patients using a personalized therapeutic approach to maximize patient response. Unfortunately, our study failed to identify a direct correlation between the presence of various epigenetic mutations and response, event-free or overall survival in this cohort of older patients treated with front-line HMA therapy.
Certain limitations should be taken into account during interpretation of this study. First, this was a retrospective review including several different treatment regimens, several of which additionally incorporated the use of histone deacetylase inhibitors (HDACIs). While HDACIs are thought to have a similar epigenetic mechanism of action that may also be beneficial to patients with epigenetically active mutations, their clinical utility has been limited by their significant toxicity, particularly the first generation HDACIs. Valproic acid, one such first generation HDACI, was received by the majority (37/68, or 54%) of patients in this analysis. However, there was no evidence that patients receiving HDACI therapy had increased toxicity or worse outcomes [Figure 4]. Death during induction was observed in 16 of 68 patients (24%), of whom 10 (62.5%) received HDACI therapy. This corresponds with the 62% of patients receiving HDACI therapy overall. Thus, we believe it unlikely that increased therapy-related toxicity from the combination of HMAs and HDACIs was responsible for the limited benefit seen in our cohort.
Complete molecular characterization using the currently updated list of mutations with epigenetic importance was not possible in this retrospective study, which most conspicuously lacks TET2 mutation analysis. While this is a bona fide limitation to our analysis, our study includes the largest cohort of elderly AML patients treated with HMA-therapy described to date, and provides important information to clinician and researcher alike with regards to epigenetic therapy.
Furthermore, several important conclusions and topics for future research are raised. The lack of an inferior overall survival in patients with a DNMT3A mutation treated with HMAs is noteworthy. This is counter-intuitive as it could be hypothesized that such mutations may interfere with response to HMA therapy. However, our data may provide support to the notion that DNMT3A mutations can mechanistically lead to impaired DNA methyltransferase activity, and therefore lead to an improved response to HMA therapy.(25) Additional prospective analysis of DNMT3A mutations and HMA-therapy in larger cohorts is both warranted and ongoing at our institution.
As AML is fundamentally a heterogeneous disease arising from the acquisition of diverse clonal alterations leading to a malignant phenotype, and somatic mutations in genes essential to epigenetic regulation have been recurrently identified in AML patients, it follows that aberrant epigenetic patterns are likely an important mechanism of leukemic transformation. Ultimately, however, our study supports the increasingly evident principle that epigenetic mutations, methylation profiles and hypomethylating agents in AML are not related by any simple algorithm. There appears to be a particular link between methylation signatures and certain mutations, such as the hyper-methylated phenotype reported with IDH1/2 and TET2 mutations, which subsequently leads to decreased gene expression of genes critical for myeloid development.(26) Other aberrations such as DNMT3A mutations are not obviously correlated with any robust changes in gene expression or specific methylation profiles.(7, 27) Whether effective HMA therapy is ultimately related to re-expression of critically silenced genes, and whether this response is conditional upon the presence of specific epigenetic mutations, remains a clinically and scientifically crucial question that will require larger prospective studies for a definitive answer.
Supplementary Material
Acknowledgments
This work was supported in part by the MD Anderson Cancer Center Support Grant (CCSG) CA016672 and the Leukemia Specialized Program on Research and Education (SPORE) grant 5 P50 CA100632.
Many thanks to Hamed Rahimi and R. Craig Cason for their help with molecular analyses.
Footnotes
Conflict of Interest Disclosure: The authors declare no competing financial interest
This work represents original research not previously published and has not been submitted for publication elsewhere. It was presented in abstract form at the American Society of Hematology Annual Meeting in Atlanta, December 2012.
Author Contributions:
CD reviewed and analyzed data and wrote the manuscript.
KP and RL performed the mutational analysis.
SP collected and analyzed the data.
GGM, MK, EJ, GB, TK, NP, SF, JC, HK designed and treated patients on the clinical trials and critically reviewed the manuscript.
FR designed the study, coordinated the analysis, treated the patients, analyzed the data and critically reviewed the manuscript.
References
- 1.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. The New England journal of medicine. 2009;361(11):1058–66. doi: 10.1056/NEJMoa0903840. Epub 2009/08/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(14):2348–55. doi: 10.1200/JCO.2009.27.3730. Epub 2010/04/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. The New England journal of medicine. 2009;360(22):2289–301. doi: 10.1056/NEJMoa0810069. Epub 2009/05/29. [DOI] [PubMed] [Google Scholar]
- 4.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature genetics. 2010;42(8):722–6. doi: 10.1038/ng.621. Epub 2010/07/06. [DOI] [PubMed] [Google Scholar]
- 5.Ziemin-van der Poel S, McCabe NR, Gill HJ, Espinosa R, 3rd, Patel Y, Harden A, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(23):10735–9. doi: 10.1073/pnas.88.23.10735. Epub 1991/12/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metzeler KH, Becker H, Maharry K, Radmacher MD, Kohlschmidt J, Mrozek K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118(26):6920–9. doi: 10.1182/blood-2011-08-368225. Epub 2011/10/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363(25):2424–33. doi: 10.1056/NEJMoa1005143. Epub 2010/11/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thol F, Damm F, Ludeking A, Winschel C, Wagner K, Morgan M, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(21):2889–96. doi: 10.1200/JCO.2011.35.4894. Epub 2011/06/15. [DOI] [PubMed] [Google Scholar]
- 9.Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrozek K, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30(7):742–50. doi: 10.1200/JCO.2011.39.2092. Epub 2012/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. The New England journal of medicine. 2012;366(12):1079–89. doi: 10.1056/NEJMoa1112304. Epub 2012/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–7. doi: 10.1182/blood-2009-03-210039. Epub 2009/05/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118(14):3803–10. doi: 10.1182/blood-2011-02-339747. Epub 2011/08/11. [DOI] [PubMed] [Google Scholar]
- 13.Ravandi F, Patel K, Luthra R, Faderl S, Konopleva M, Kadia T, et al. Prognostic significance of alterations in IDH enzyme isoforms in patients with AML treated with high-dose cytarabine and idarubicin. Cancer. 2012;118(10):2665–73. doi: 10.1002/cncr.26580. Epub 2011/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(22):3636–43. doi: 10.1200/JCO.2010.28.3762. Epub 2010/06/23. [DOI] [PubMed] [Google Scholar]
- 15.Green CL, Evans CM, Zhao L, Hills RK, Burnett AK, Linch DC, et al. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood. 2011;118(2):409–12. doi: 10.1182/blood-2010-12-322479. Epub 2011/05/21. [DOI] [PubMed] [Google Scholar]
- 16.Estey E. Acute myeloid leukemia and myelodysplastic syndromes in older patients. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25(14):1908–15. doi: 10.1200/JCO.2006.10.2731. Epub 2007/05/10. [DOI] [PubMed] [Google Scholar]
- 17.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Gattermann N, Germing U, et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(4):562–9. doi: 10.1200/JCO.2009.23.8329. Epub 2009/12/23. [DOI] [PubMed] [Google Scholar]
- 18.Kantarjian HM, Thomas XG, Dmoszynska A, Wierzbowska A, Mazur G, Mayer J, et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30(21):2670–7. doi: 10.1200/JCO.2011.38.9429. Epub 2012/06/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2011;25(7):1147–52. doi: 10.1038/leu.2011.71. Epub 2011/04/16. [DOI] [PubMed] [Google Scholar]
- 20.Patel KP, Ravandi F, Ma D, Paladugu A, Barkoh BA, Medeiros LJ, et al. Acute myeloid leukemia with IDH1 or IDH2 mutation: frequency and clinicopathologic features. American journal of clinical pathology. 2011;135(1):35–45. doi: 10.1309/AJCPD7NR2RMNQDVF. Epub 2010/12/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin P, Jones D, Medeiros LJ, Chen W, Vega-Vazquez F, Luthra R. Activating FLT3 mutations are detectable in chronic and blast phase of chronic myeloproliferative disorders other than chronic myeloid leukemia. American journal of clinical pathology. 2006;126(4):530–3. doi: 10.1309/JT5BE2L1FGG8P8Y6. Epub 2006/08/30. [DOI] [PubMed] [Google Scholar]
- 22.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–74. doi: 10.1182/blood-2009-07-235358. Epub 2009/11/03. [DOI] [PubMed] [Google Scholar]
- 23.Ravandi F. Acute Myeloid Leukemia in the Elderly: Who Should Be Treated and How? ASCO Educational Book. 2010:257–61. [Google Scholar]
- 24.Juliusson G, Antunovic P, Derolf A, Lehmann S, Mollgard L, Stockelberg D, et al. Age and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish Acute Leukemia Registry. Blood. 2009;113(18):4179–87. doi: 10.1182/blood-2008-07-172007. Epub 2008/11/15. [DOI] [PubMed] [Google Scholar]
- 25.Metzeler KH, Walker A, Geyer S, Garzon R, Klisovic RB, Bloomfield CD, et al. DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012;26(5):1106–7. doi: 10.1038/leu.2011.342. Epub 2011/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. Epub 2010/12/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, Rockova V, Sanders M, Abbas S, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119(24):5824–31. doi: 10.1182/blood-2011-07-367961. Epub 2012/04/12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.