SUMMARY
The CRISPR-associated endonuclease Cas9 can be targeted to specific genomic loci by single guide RNAs (sgRNAs). Here, we report the crystal structure of Streptococcus pyogenes Cas9 in complex with sgRNA and its target DNA, at 2.5 Å resolution. The structure revealed a bilobed architecture composed of target recognition and nuclease lobes, accommodating the sgRNA:DNA heteroduplex in a positively-charged groove at their interface. Whereas the recognition lobe is essential for binding sgRNA and DNA, the nuclease lobe contains the HNH and RuvC nuclease domains, which are properly positioned for cleavage of the complementary and non-complementary strands of the target DNA, respectively. The nuclease lobe also contains a carboxyl-terminal domain responsible for the interaction with the protospacer adjacent motif (PAM). This high-resolution structure and accompanying functional analyses have revealed the molecular mechanism of RNA-guided DNA targeting by Cas9, thus paving the way for the rational design of new, versatile genome-editing technologies.
INTRODUCTION
The CRISPR (clustered regularly interspaced palindromic repeat)-Cas system is a naturally occurring, adaptive microbial immune system for defense against invading phages and other mobile genetic elements (Deveau et al., 2010; Horvath and Barrangou, 2010; Marraffini and Sontheimer, 2010; Terns and Terns, 2011). Three types (I–III) of CRISPR-Cas systems have been functionally identified across a wide range of microbial species (Barrangou et al., 2007; Brouns et al., 2008; Marraffini and Sontheimer, 2008), and each contains a cluster of CRISPR-associated (Cas) genes and its corresponding CRISPR array. These characteristic CRISPR arrays consist of repetitive sequences (direct repeats, referred to as repeats) interspaced by short stretches of non-repetitive sequences (spacers) derived from short segments of foreign genetic material (protospacers). The CRISPR array is transcribed and processed into short CRISPR RNAs (crRNAs), which direct the Cas proteins to the target nucleic acids via Watson-Crick base pairing to facilitate nucleic acid destruction.
The Type I and III CRISPR systems utilize ensembles of Cas proteins complexed with crRNAs to mediate the recognition and subsequent degradation of target nucleic acids (Spilman et al., 2013; Wiedenheft et al., 2011). In contrast, the Type II CRISPR system recognizes and cleaves the target DNA (Garneau et al., 2010) via the RNA-guided endonuclease Cas9 (Sapranauskas et al., 2011) along with two non-coding RNAs, the crRNA and the trans-activating crRNA (tracrRNA) (Deltcheva et al., 2011). The crRNA hybridizes with the tracrRNA to form a crRNA:tracrRNA duplex, which is loaded onto Cas9 to direct the cleavage of cognate DNA sequences bearing appropriate protospacer adjacent motifs (PAM) (Mojica et al., 2009). Cas9 contains two nuclease domains, HNH and RuvC, which cleave the DNA strands that are complementary and non-complementary to the 20-nucleotide (nt) guide sequence in crRNAs, respectively (Gasiunas et al., 2012; Jinek et al., 2012).
The Type II CRISPR system was the first to be adapted for facilitating genome editing in eukaryotic cells (Cong et al., 2013; Mali et al., 2013b). The Cas9 protein from Streptococcus pyogenes, along with a single guide RNA (sgRNA), a synthetic fusion of crRNA and tracrRNA (Jinek et al., 2012), could be programmed to cleave virtually any sequence preceding a 5′-NGG-3′ PAM sequence in mammalian cells (Cong et al., 2013; Mali et al., 2013b). This unprecedented flexibility has enabled a broad range of applications, including rapid generation of genetically modified cells and animal models (Gratz et al., 2013; Hwang et al., 2013; Wang et al., 2013; Yang et al., 2013), and genome-scale genetic screening (Shalem et al., 2014; Wang et al., 2014).
However, despite the brisk progress in the development of Cas9 technology, the mechanism by which the Cas9–sgRNA complex recognizes and cleaves its target DNA remains to be elucidated. Biochemical analyses at the domain levels have enabled site-specific engineering to convert the native Cas9 into a DNA nicking enzyme (Gasiunas et al., 2012; Jinek et al., 2012; Sapranauskas et al., 2011) that facilitates homology-directed repair in eukaryotic cells (Cong et al., 2013; Mali et al., 2013b) and cleaves DNA with improved specificity, given appropriately paired sgRNAs (Mali et al., 2013a; Ran et al., 2013). Moreover, a catalytically inactive Cas9 can serve as an RNA-guided DNA-binding platform to target effector domains and modulate endogenous transcription (Gilbert et al., 2013; Konermann et al., 2013; Maeder et al., 2013; Perez-Pinera et al., 2013; Qi et al., 2013). These advances in Cas9 engineering represent just the first steps toward fully realizing the potential of this flexible RNA-guided genome positioning system. Precise structural information about Cas9 will thus not only enhance our understanding of how this elegant RNA-guided, adaptive microbial immune system functions, but also facilitate further improvements in the Cas9 targeting specificity, the in vitro and in vivo delivery, and the engineering of Cas9 for novel functions and optimized features.
Here, we report the crystal structure of S. pyogenes Cas9 in complex with sgRNA and its target DNA at 2.5 Å resolution. This high-resolution structure, along with functional analyses, reveals the key functional interactions that integrate the guide RNA, the target DNA, and the Cas9 protein, thus paving the way towards enhancing Cas9 function as well as engineering novel applications.
RESULTS
Overall structure of the Cas9–sgRNA–DNA ternary complex
We solved the crystal structure of full-length S. pyogenes Cas9 (residues 1–1368; D10A/C80L/C574E/H840A), in complex with a 98-nt sgRNA and a 23-nt target DNA, at 2.5 Å resolution by the SAD (single-wavelength anomalous dispersion) method, using a SeMet-labeled protein (Figure 1, Figure S1 and Table S1). To improve the solution behavior of Cas9, we replaced two less conserved cysteine residues (Cys80 and Cys574) with leucine and glutamic acid, respectively. This C80L/C574E mutant retained the ability to efficiently cleave genomic DNA in human embryonic kidney 293FT (HEK293FT) cells, confirming that these mutations have no effects on the Cas9 nuclease function (Figure S2). Additionally, to prevent target DNA cleavage during crystallization, we replaced two catalytic residues, Asp10 from the RuvC domain and His840 from the HNH domain, with alanines.
Figure 1. Overall structure of the Cas9–sgRNA–DNA ternary complex.
(A) Domain organization of S. pyogenes Cas9. BH, Bridge helix.
(B) Schematic representation of the sgRNA:target DNA complex.
(C) Ribbon representation of the Cas9–sgRNA–DNA complex. Disordered linkers are shown as red dotted lines.
(D) Surface representation of the Cas9–sgRNA–DNA complex. The active sites of the RuvC (D10A) and HNH (H840A) domains are indicated by dashed yellow circles.
(E) Electrostatic surface potential of Cas9. The HNH domain is omitted for clarity. Molecular graphic images were prepared using CueMol (http://www.cuemol.org).
See also Figures S1, S2 and Table S1.
The crystallographic asymmetric unit contained two Cas9–sgRNA–DNA ternary complexes (Mol A and Mol B). Although there are conformational differences between the two complexes, the sgRNA and the DNA are recognized by Cas9 in similar manners. Most notably, while the HNH domain in Mol A is connected to the RuvC domain by a disordered linker, the HNH domain in Mol B is not visible in the electron density map, indicating the flexible nature of the HNH domain. Thus, we will first describe the structural features of Mol A unless otherwise stated, and then discuss the structural differences between the two complexes, which suggest the conformational flexibility of Cas9.
The crystal structure revealed that Cas9 consists of two lobes, a recognition (REC) lobe and a nuclease (NUC) lobe (Figures 1A–D). The REC lobe can be divided into three regions, a long α-helix referred to as the Bridge helix (residues 60–93), the REC1 (residues 94–179 and 308–713) domain, and the REC2 (residues 180–307) domain (Figures 1A–D). The NUC lobe consists of the RuvC (residues 1–59, 718–769 and 909–1098), HNH (residues 775–908), and PAM-interacting (PI) (residues 1099–1368) domains (Figures 1A–D). The negatively-charged sgRNA:target DNA heteroduplex is accommodated in a positively-charged groove at the interface between the REC and NUC lobes (Figure 1E). In the NUC lobe, the RuvC domain is assembled from the three split RuvC motifs (RuvC I–III), and interfaces with the PI domain to form a positively-charged surface that interacts with the 3′ tail of the sgRNA (Figure 1E). The HNH domain lies in between the RuvC II–III motifs and forms only a few contacts with the rest of the protein.
The REC lobe interacts with the repeat:anti-repeat duplex
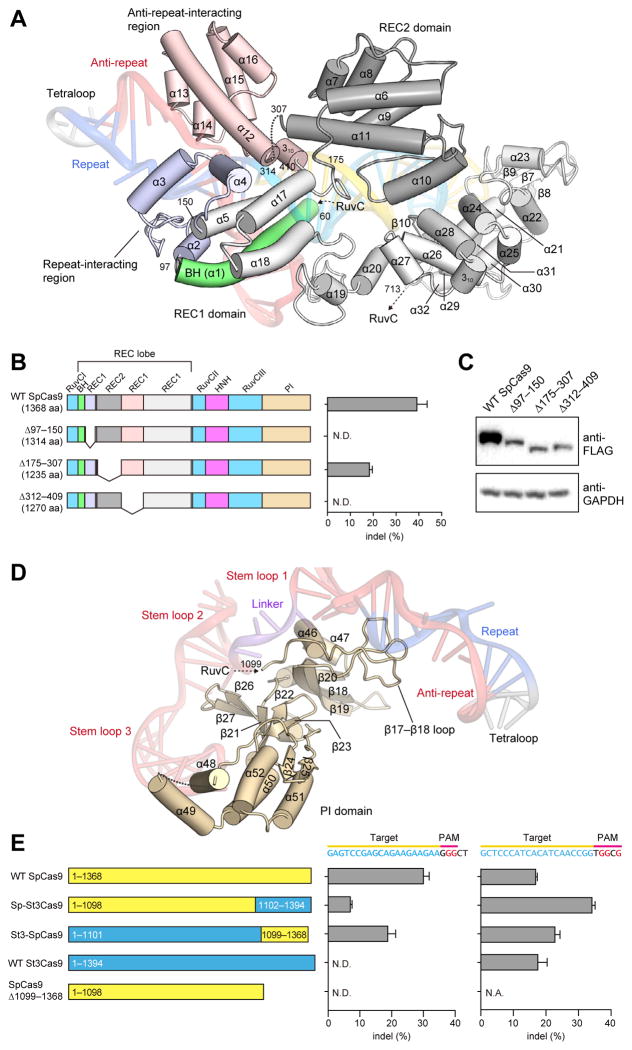
The REC lobe includes the REC1 and REC2 domains. REC1 adopts an elongated, α-helical structure comprising 25 α-helices (α2–α5 and α12–α32) and two β-sheets (β6 and β10, and β7–β9), whereas REC2 adopts a six-helix bundle structure (α6–α11) (Figure 2A and Figure S3). A Dali search (Holm and Rosenstrom, 2010) revealed that the REC lobe does not share structural similarity with other known proteins, indicating that it is a Cas9-specific functional domain.
Figure 2. REC lobe and PI domain.
(A) Structure of the REC lobe. The REC2 domain and the Bridge helix are colored dark gray and green, respectively. The REC1 domain is colored gray, with the repeat-interacting and anti-repeat-interacting regions colored pale blue and pink, respectively. The bound sgRNA:DNA is shown as a semi-transparent ribbon representation.
(B) Mutational analysis of the REC lobe. Schematics show the truncation mutants. The bar graph shows indel mutations generated by the truncation mutants, measured by the SURVEYOR assay (n = 3, error bars show mean ± S.E.M., N.D., not detectable).
(C) Western blot showing the expression of the truncation mutants in HEK 293FT cells.
(D) Structure of the PI domain. The bound sgRNA is shown as a semi-transparent ribbon representation.
(E) Mutational analysis of the PI domain. Schematics show wild-type SpCas9 and St3Cas9, chimeric Sp-St3Cas9 and St3-SpCas9, and the SpCas9 PI domain truncation mutant. Cas9s were assayed for indel generation at target sites upstream of either NGG (left bar graph) or NGGNG (right bar graph) PAMs (n = 3, error bars show mean ± S.E.M., N.D., not detectable).
See also Figures S3–S5.
The REC lobe is one of the least conserved regions across the three Cas9 families within the Type II CRISPR system (IIA, IIB and IIC), and many Cas9 orthologs contain significantly shorter REC lobes (Figures S4, S5). In the present structure, the REC2 domain does not contact the bound guide:target heteroduplex. We thus hypothesized that truncations in the REC lobe could be tolerated. As expected, a Cas9 mutant lacking the REC2 domain (Δ175–307) retained ~50% of the wild-type Cas9 activity (Figure 2B), indicating that the REC2 domain is not critical for DNA cleavage. The lower cleavage efficiency may be attributed in part to the reduced expression levels of the Δ175–307 mutant relative to that of the wild-type protein (Figure 2C). In striking contrast, the deletion of either the repeat-interacting region (Δ97–150) or the anti-repeat-interacting region (Δ312–409) of the REC1 domain abolished the DNA cleavage activity (Figure 2B), indicating that the recognition of the repeat:anti-repeat duplex by the REC1 domain is critical for the Cas9 function.
The PAM-interacting (PI) domain confers PAM specificity
The NUC lobe contains the PI domain, which forms an elongated structure comprising seven α-helices (α46–α52), a three-stranded antiparallel β-sheet (β18–β20), a five-stranded antiparallel β-sheet (β21–β23, β26 and β27), and a two-stranded antiparallel β-sheet (β24 and β25) (Figure 2D and Figure S3). Similar to the REC lobe, the PI domain also adopts a novel protein fold unique to the Cas9 family.
The locations of the bound complementary DNA strand and the active site of the RuvC domain in the present structure suggested that the PI domain is positioned to recognize the PAM sequence on the non-complementary DNA strand. We tested whether the replacement of the PI domain of S. pyogenes Cas9 (SpCas9; Cas9 in this study) with that of an orthologous Cas9 protein, recognizing a different PAM, would be sufficient to alter their PAM specificities. The Streptococcus thermophilus CRISPR-3 Cas9 (St3Cas9) shares ~60% sequence identity with SpCas9. While their crRNA repeats and tracrRNAs are interchangeable (Fonfara et al., 2013), SpCas9 and St3Cas9 require different PAM sequences (5′-NGG-3′ for SpCas9 and 5′-NGGNG-3′ for St3Cas9) for target DNA cleavage (Fonfara et al., 2013).
We swapped their PI domains to generate two chimeras, Sp-St3Cas9 (SpCas9 with the PI domain of St3Cas9) and St3-SpCas9 (St3Cas9 with the PI domain of SpCas9), and examined their cleavage activities for target DNA sequences bearing 5′-NGG-3′ PAM (5′-GGGCT-3′) or 5′-NGGNG-3′ PAM (5′-GGGCG-3′) (Figure 2E). SpCas9 and St3-SpCas9, but not St3Cas9, cleaved the target DNA with 5′-NGG-3′ PAM (Figure 2E), indicating that the PI domain of SpCas9 is required for the recognition of 5′-NGG-3′ PAM and is sufficient to alter the PAM specificity of St3Cas9. Sp-St3Cas9 retained the cleavage activity for the target DNA with 5′-NGG-3′ PAM, albeit at a lower level than that of SpCas9 (Figure 2E). Additionally, the deletion of the PI domain (Δ1099–1368) abolished the cleavage activity (Figure 2E), indicating that the PI domain is critical for the Cas9 function. These results revealed that the PI domain is a major determinant of the PAM specificity.
The RuvC domain has an RNase H fold
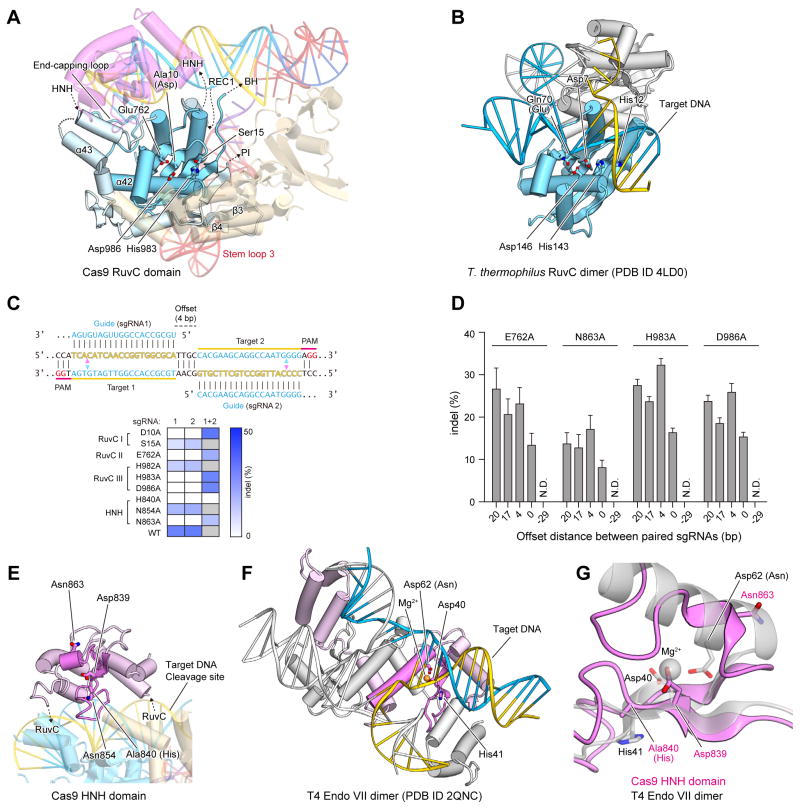
The RuvC domain consists of a six-stranded mixed β-sheet (β1, β2, β5, β11, β14 and β17) flanked by α-helices (α33, α34 and α39–α45) and two additional two-stranded antiparallel β-sheets (β3/β4 and β15/β16) (Figure 3A and Figure S3). It shares structural similarity with the retroviral integrase superfamily members characterized by an RNase H fold, such as Escherichia coli RuvC (Ariyoshi et al., 1994) (PDB code 1HJR, 14% identity, root-mean-square deviation (rmsd) of 3.6 Å for 126 equivalent Cα atoms) and Thermus thermophilus RuvC (Gorecka et al., 2013) (PDB code 4LD0, 12% identity, rmsd of 3.4 Å for 131 equivalent Cα atoms) (Figure 3B). The RuvC nucleases have four catalytic residues (e.g., Asp7, Glu70, His143 and Asp146 in T. thermophilus RuvC), and cleave Holliday junctions through a two-metal mechanism (Ariyoshi et al., 1994; Chen et al., 2013; Gorecka et al., 2013). Asp10 (Ala), Glu762, His983 and Asp986 of the Cas9 RuvC domain are located at positions similar to those of the catalytic residues of T. thermophilus RuvC (Figures 3A, B), consistent with previous results showing that Asp10 is critical for the cleavage of the non-complementary DNA strand, and that Cas9 requires Mg2+ ions for the cleavage activity (Gasiunas et al., 2012; Jinek et al., 2012). Moreover, the alanine substitution of Glu762, His983 or Asp986 also converted Cas9 into a nickase (Figures 3C, D). Each nickase mutant was able to facilitate targeted double strand breaks using a pair of juxtaposed sgRNAs (Figures 3C, D, Table S2), as previously demonstrated with the D10A nickase (Ran et al., 2013). This combination of structural observations and mutational analyses suggested that the Cas9 RuvC domain cleaves the non-complementary strand of the target DNA through the two-metal mechanism, as previously observed for other retroviral integrase superfamily nucleases.
Figure 3. NUC lobe.
(A) Structure of the RuvC domain. The core structure of the RNase H fold is highlighted in cyan. The active-site residues are shown as stick models.
(B) Structure of the T. thermophilus RuvC dimer in complex with a Holliday junction (PDB ID 4LD0). The two protomers are colored cyan and gray, respectively.
(C) Mutational analysis of the RuvC and HNH domains. The sequences (top) illustrate Cas9 nicking targets on opposite strands of DNA. Targets 1 and 2 are offset by a distance of 4-bp in between. The cleavage sites by the HNH and RuvC domains are indicated by pink and cyan triangles, respectively. The heatmap (bottom) shows the ability of each catalytic mutant to induce double- (with either sgRNA 1 or 2) or single-stranded breaks (only with both sgRNAs together). Gray boxes, not assayed.
(D) Indel formation by Cas9 nickases depends on the off-set distance between sgRNA pairs. The off-set distance is defined as the number of base pairs between the PAM-distal (5′) ends of the guide sequence of a given sgRNA pair (n = 3, error bars show mean ± S.E.M., N.D., not detectable).
(E) Structure of the HNH domain. The core structure of the ββα-metal fold is highlighted in magenta. The active-site residues are shown as stick models.
(F) Structure of the T4 Endo VII dimer in complex with a Holliday junction (PDB ID 2QNC). The two protomers are colored pink and gray, respectively, with the ββα-metal fold core highlighted in magenta. The bound Mg2+ ion is shown as an orange sphere.
(G) Superimposition of the Cas9 HNH domain and T4 Endo VII (PDB ID 2QNC).
See also Table S2.
It is important to note that there are key structural dissimilarities between the Cas9 RuvC domain and the RuvC nucleases, which explain their functional differences. Unlike the Cas9 RuvC domain, the RuvC nucleases form dimers and recognize Holliday junctions (Gorecka et al., 2013) (Figure 3B). In addition to the conserved RNase H fold, the Cas9 RuvC domain has other structural elements involved in interactions with the guide:target heteroduplex (an end-capping loop between α42 and α43) and the PI domain/stem loop 3 (β-hairpin formed by β3 and β4) (Figure 3A).
The HNH domain has a ββα-metal fold
The HNH domain comprises a two-stranded antiparallel β-sheet (β12 and β13) flanked by four α-helices (α35–α38) (Figure 3E). It shares structural similarity with the HNH endonucleases characterized by a ββα-metal fold, such as phage T4 endonuclease VII (Endo VII) (Biertümpfel et al., 2007) (PDB code 2QNC, 20% identity, rmsd of 2.7 Å for 61 equivalent Cα atoms) and Vibrio vulnificus nuclease (Li et al., 2003) (PDB code 1OUP, 8% identity, rmsd of 2.7 Å for 77 equivalent Cα atoms) (Figure 3F). HNH nucleases have three catalytic residues (e.g., Asp40, His41, and Asn62 in Endo VII), and cleave nucleic acid substrates through a single-metal mechanism (Biertümpfel et al., 2007; Li et al., 2003). In the structure of the Endo VII N62D mutant in complex with a Holliday junction, a Mg2+ ion is coordinated by Asp40, Asp62, and the oxygen atoms of the scissile phosphate group of the substrate, while His41 acts as a general base to activate a water molecule for catalysis (Figure 3F). Asp839, His840, and Asn863 of the Cas9 HNH domain correspond to Asp40, His41, and Asn62 of Endo VII, respectively (Figure 3E), consistent with the observation that His840 is critical for the cleavage of the complementary DNA strand (Gasiunas et al., 2012; Jinek et al., 2012). The N863A mutant functions as a nickase (Figure 3C, D), indicating that Asn863 participates in catalysis. These observations suggested that the Cas9 HNH domain may cleave the complementary strand of the target DNA through a single-metal mechanism, as observed for other HNH superfamily nucleases. However, in the present structure, Asn863 of Cas9 is located at a different position from that of Asn62 in Endo VII, whereas Asp839 and His840 (Ala) of Cas9 are located at positions similar to those of Asp40 and His41 in Endo VII, respectively (Figures 3G). This might be due to the absence of divalent ions, such as Mg2+, in our crystallization solution, and Asn863 may point towards the active site and participate in catalysis. Although the HNH domain shares a ββα-metal fold with other HNN endonucleases, their overall structures are distinct (Figures 3E, F), consistent with the differences in their substrate specificities.
The sgRNA:DNA complex adopts a T-shaped architecture
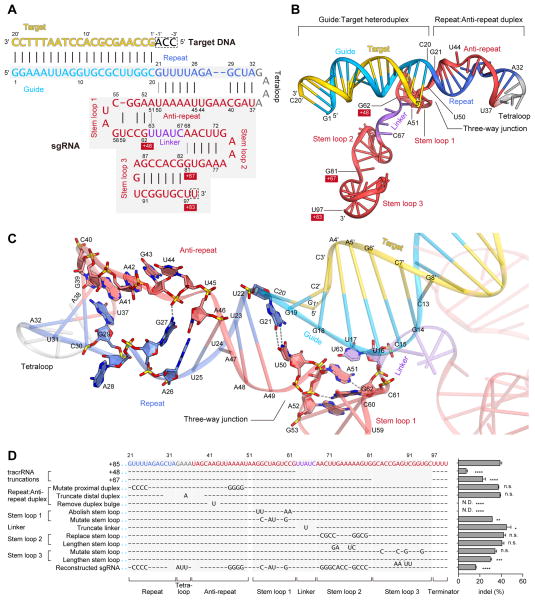
The sgRNA consists of crRNA- and tracrRNA-derived sequences connected by an artificial tetraloop (Figure 4A). The crRNA sequence can be divided into guide (20-nt) and repeat (12-nt) regions, while the tracrRNA sequence can be divided into anti-repeat (14-nt) and three tracrRNA stem loops (Figure 4A). The crystal structure revealed that the sgRNA binds the target DNA to form a T-shaped architecture comprising a guide:target heteroduplex, a repeat:anti-repeat duplex, and stem loops 1–3 (Figures 4A, B). The repeat:anti-repeat duplex and stem loop 1 are connected by a single nucleotide (A51), while stem loops 1 and 2 are connected by a 5-nt single-stranded linker (nucleotides 63–67).
Figure 4. sgRNA and its target DNA.
(A) Schematic representation of the sgRNA:target DNA complex. The guide and repeat regions of the crRNA sequence are colored sky blue and blue, respectively. The tracrRNA sequence is colored red, with the linker region colored violet. The target DNA and the tetraloop are colored yellow and gray, respectively. The numbering of the 3′ tails of the tracrRNA is shown on a red background. Watson-Crick and non-Watson-Crick base pairs are indicated by black and gray lines, respectively. Disordered nucleotides are boxed by dashed lines.
(B) Structure of the sgRNA:target DNA complex.
(C) Close-up view of the repeat:anti-repeat duplex and the three-way junction. Key interactions are shown with gray dashed lines.
(D) Effects of sgRNA mutations on the ability to induce indels. Base changes from the sgRNA(+85) scaffold are shown at the respective positions, with dashes indicating unaltered bases (n = 3, error bars show mean ± S.E.M., p values based on unpaired Student’s t-test, N.D., not detectable).
See also Figure S6.
The guide (nucleotides 1–20) and target DNA (nucleotides 1′–20′) form the guide:target heteroduplex via 20 Watson-Crick base pairs (Figures 4A, B). The conformation of the heteroduplex is distorted from that of the canonical A-form RNA duplex (Figure S6). The repeat (nucleotides 21–32) and the anti-repeat (nucleotides 37–50) form the repeat:anti-repeat duplex via nine Watson-Crick base pairs (U22:A49–A26:U45 and G29:C40–A32:U37) (Figures 4A, B). Within this region, G27, A28, A41, A42, G43 and U44 are unpaired, with A28 and U44 flipped out from the duplex (Figure 4C). The G27 and A41 nucleobases stack with the A26:U45 and G29:C40 pairs, respectively, and the 2-amino group of G27 interacts with the backbone phosphate group between G43 and U44, stabilizing the duplex structure (Figure 4C). G21 and U50 form a wobble base pair at the three-way junction between the guide:target heteroduplex/repeat:anti-repeat duplexes and stem loop 1, contributing to the formation of the T-shaped architecture (Figure 4C).
As expected from the RNA-fold predictions based on the nucleotide sequence, the tracrRNA 3′ tail (nucleotides 68–81 and 82–96) forms stem loops 2 and 3 via four and six Watson-Crick base pairs (A69:U80–U72:A77 and G82:C96–G87:C91), respectively (Figures 4A, B). In addition, nucleotides 52–62 form the newly detected stem loop (stem loop 1) via three Watson-Crick base pairs (G53:C61, G54:C60 and C55:G58), with U59 flipped out from the stem (Figures 4A, B). Stem loop 1 is stabilized by the G62–G53:C61 stacking interaction and the G62–A51/A52 polar interactions (Figure 4C).
The guide:target and repeat:anti-repeat duplexes are deeply buried in a positively-charged groove at the interface of the two lobes, while the rest of the sgRNA extensively interacts with the positively-charged surface on the back side of the protein (Figure 1E). In Mol A, the three nucleotides at the 5′ end of the target DNA (3′-ACC-5′, complementary to the PAM) are not visible in the electron density map. Although the two adjacent nucleotides (3′-AC-5′) in Mol B are structurally ordered due to the crystal packing interactions, and are visible in the electron density map, these nucleotides are not recognized by Cas9 (data not shown). These observations suggested that the 3′-NCC-5′ sequence complementary to the 5′-NGG-3′ PAM is not recognized by Cas9, and are consistent with previous biochemical data showing that Cas9-catalyzed DNA cleavage requires the 5′-NGG-3′ PAM on the non-complementary strand but not the 3′-NCC-5′ sequence on the complementary strand (Jinek et al., 2012).
Previous studies showed that, although sgRNA with a 48-nt tracrRNA tail [referred to as sgRNA(+48)] is the minimal region for the Cas9-catalyzed DNA cleavage in vitro (Jinek et al., 2012), sgRNAs with extended tracrRNA tails, sgRNA(+67) and sgRNA(+85), dramatically improved the Cas9 cleavage activity in vivo (Hsu et al., 2013). The present structure revealed that sgRNA(+48), sgRNA(+67) and sgRNA(+85) contain stem loop 1, stem loops 1–2 and stem loops 1–3, respectively (Figures 4A, B). These observations indicated that, whereas stem loop 1 is essential for the formation of the functional Cas9–sgRNA complex, stem loops 2 and 3 further support the stable complex formation and enhance the stability of the sgRNA, thus improving the in vivo activity.
To determine the significance of each sgRNA structural component on the Cas9 function, we tested a number of sgRNAs with mutations in the repeat:anti-repeat duplex, stem loops 1–3, and the linker between stem loops 1 and 2. Our results revealed that, whereas stem loops 2 and 3 as well as the linker region can tolerate a large number of mutations, the repeat:anti-repeat duplex and stem loop 1 are critical for the Cas9 function (Figure 4D). Moreover, the sgRNA sequence can tolerate a large number of mutations (Figure 4D, reconstructed sgRNA). These results highlight the functional significance of the structure-dependent recognition of the repeat:anti-repeat duplex by Cas9.
The conserved arginine cluster on the Bridge helix is critical for sgRNA:DNA recognition
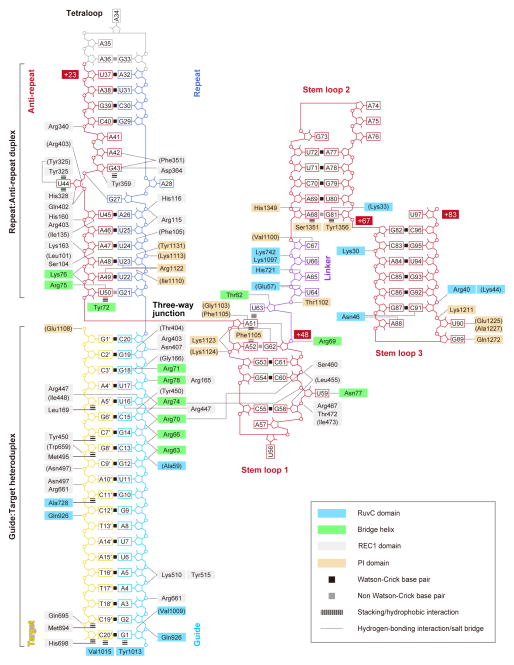
The sgRNA guide region is primarily recognized by the REC lobe (Figure 5). The backbone phosphate groups of the guide region (nucleotides 2, 4–6 and 13–20) interact with the REC1 domain (Arg165, Gly166, Arg403, Asn407, Lys510, Tyr515 and Arg661) and the Bridge helix (Arg63, Arg66, Arg70, Arg71, Arg74 and Arg78) (Figure 6A). The 2′-hydroxyl groups of G1, C15, U16 and G19 hydrogen bond with Val1009, Tyr450, Arg447/Ile448 and Thr404, respectively (Figure 6A). These observations suggested that the Watson-Crick faces of eight PAM-proximal nucleotides in the Cas9-bound sgRNA are exposed to the solvent, thus serving as a nucleation site for pairing with the complementary strand of the target DNA. This is consistent with previous reports that the 10–12 bp PAM-proximal “seed” region is critical for the Cas9-catalyzed DNA cleavage (Cong et al., 2013; Fu et al., 2013; Hsu et al., 2013; Jinek et al., 2012; Mali et al., 2013a; Pattanayak et al., 2013).
Figure 5. Schematic representation of sgRNA:target DNA recognition by Cas9.
Residues that interact with the sgRNA:DNA via their main chain are shown in parentheses. Note that water-mediated hydrogen-bonding interactions are not shown, for clarity.
Figure 6. sgRNA:target DNA recognition by Cas9.
(A and C–J) Recognition of the guide (A), the guide:target heteroduplex (C), the repeat (D), the anti-repeat (E), the three-way junction (F), stem loop 1 (G), the linker (H), stem loop 2 (I) and stem loop 3 (J). Hydrogen bonds and salt bridges are shown as dashed lines. In (A), the target DNA is omitted, for clarity.
(B) Effects of Cas9 (top) and sgRNA (bottom) mutations on the ability to induce indels (n = 3, error bars show mean ± S.E.M., p values based on unpaired Student’s t-test. N.D., not detectable).
A mutational analysis demonstrated that the R66A, R70A and R74A mutations on the Bridge helix markedly reduced the DNA cleavage activities (Figure 6B), highlighting the functional significance of the recognition of the sgRNA “seed” region by the Bridge helix. Although Arg78 and Arg165 also interact with the “seed” region, the R78A and R165A mutants showed only moderately decreased activities (Figure 6B). These results are consistent with the fact that Arg66, Arg70 and Arg74 form multiple salt bridges with the sgRNA backbone, while Arg78 and Arg165 form a single salt bridge with the sgRNA backbone (Figure 6A). The cluster of arginine residues on the Bridge helix is highly conserved among the Cas9 proteins in the Type II-A–C systems (Figures S4, S5), suggesting that the Bridge helix is a universal structural feature of the Cas9 proteins. This notion is supported by the previous observation that a strictly conserved arginine residue, equivalent to Arg70 of S. pyogenes Cas9, is essential for the function of Francisella novicida Cas9 in the Type II-B system (Sampson et al., 2013). Moreover, the alanine mutations of the repeat:anti-repeat duplex-interacting residues (Arg75 and Lys163) and the stem loop 1-interacting residue (Arg69) resulted in decreased DNA cleavage activity (Figure 6B), confirming the functional importance of the recognition of the repeat:anti-repeat duplex and stem loop 1 by Cas9.
The sgRNA guide region is recognized by Cas9 in a sequence-independent manner, except for the U16–Arg447 and G18–Arg71 interactions (Figures 5 and 6A). This base-specific G18–Arg71 interaction may partly explain the observed preference of Cas9 for sgRNAs with guanines in the four PAM-proximal guide region (Wang et al., 2014).
The REC1 and RuvC domains facilitate RNA-guided DNA targeting
Cas9 recognizes the 20-bp guide:target heteroduplex in a sequence-independent manner (Figure 5). The backbone phosphate groups of the target DNA (nucleotides 1′, 9′–11′, 13′ and 20′) interact with the REC1 (Asn497, Trp659, Arg661 and Gln695), RuvC (Gln926), and PI (Glu1108) domains. The C2′ atoms of the target DNA (nucleotides 5′, 7′, 8′, 11′, 19′ and 20′) form van der Waals interactions with the REC1 domain (Leu169, Tyr450, Met495, Met694 and His698) and the RuvC domain (Ala728) (Figure 5). These interactions are likely to contribute toward the ability of Cas9 to discriminate between DNA and RNA targets. The terminal base pair of the guide:target heteroduplex (G1:C20′) is recognized by the RuvC domain via end-capping interactions (Figure 6C); the sgRNA G1 and target DNA C20′ nucleobases interact with the Tyr1013 and Val1015 side chains, respectively, whereas the 2′-hydroxyl and phosphate groups of sgRNA G1 interact with Val1009 and Gln926, respectively. These end-capping interactions are consistent with the previous observation that Cas9 recognizes a 17–20-bp guide:target heteroduplex, and that extended guide sequences are degraded in cells and do not contribute to improving sequence specificity (Ran et al., 2013). Taken together, these structural findings explain the RNA-guided DNA targeting mechanism of Cas9.
The repeat:anti-repeat duplex is recognized by the REC and NUC lobes in a sequence-dependent manner
In contrast to the sequence-independent recognition of the sgRNA guide region, sequence-dependent interactions exist between Cas9 and the repeat:anti-repeat duplex (Figures 5, 6D and 6E). The nucleobases of U23/A49 and A42/G43 hydrogen bond with the side chain of Arg1122 and the main-chain carbonyl group of Phe351, respectively (Figure 6D). The nucleobase of the flipped U44 is sandwiched between Tyr325 and His328, with its N3 atom hydrogen bonded with Tyr325, while the nucleobase of the unpaired G43 stacks with Tyr359 and hydrogen bonds with Asp364 (Figure 6E).
The present structure revealed that the repeat:anti-repeat duplex is recognized by the REC lobe, which is divergent in both sequence and length among the Cas9 orthologs within the Type II-A–C systems (Figures S4, S5). This structural finding explains the previous observation that Cas9 and sgRNA are interchangeable only between closely related Type II systems (Fonfara et al., 2013). The three PAM-distal base pairs (C30:G39–A32:U37) are not recognized by Cas9 and protrude from the complex (Figure 5), consistent with a proposed model in which the Cas9-bound repeat:anti-repeat duplex is processed by the host RNase III enzyme (Deltcheva et al., 2011).
The nucleobases of G21 and U50, in the G21:U50 wobble pair, stack with the terminal C20:G1′ pair in the guide:target heteroduplex and Tyr72 on the Bridge helix, respectively, with the U50 O4 atom hydrogen bonded with Arg75 (Figure 6D). Notably, A51 adopts the syn-conformation, and is oriented in the direction opposite to U50 (Figures 4C and 6F). The nucleobase of A51 is sandwiched between Phe1105 and U63, with its N1, N6 and N7 atoms hydrogen bonded with G62, Gly1103 and Phe1105, respectively (Figure 6F). Whereas the repeat:anti-repeat duplexes have diverse sequences and lengths among the Type II-A–C systems, the G:U base pair at the three-way junction is highly conserved among the repeat:anti-repeat duplexes in these three systems (Fonfara et al., 2013), suggesting that this wobble pairing is a universal structural feature involved in the three-way junction formation.
To verify the importance of the sequence-dependent recognition of the repeat:anti-repeat duplex by Cas9, we evaluated the effects of repeat:anti-repeat mutations on the Cas9-mediated DNA cleavage (Figure 6B). The replacement of G43, which forms base-specific hydrogen bonds with Phe351 and Asp364, with cytosine reduced the Cas9 activity by over 3-fold. In addition, the replacement of the flipped U44 with guanine resulted in an over 5-fold drop in the cleavage activity, whereas the replacement of U44 with another pyrimidine base (cytosine) did not significantly affect the cleavage activity (Figure 6B). These results confirmed the functional importance of the base-specific recognition of G43 and U44 by Cas9.
Stem loops 1–3 reinforce the interaction between Cas9 and sgRNA
Stem loop 1 is primarily recognized by the REC lobe, together with the PI domain (Figure 5). The backbone phosphate groups of stem loop 1 (nucleotides 52, 53 and 59–61) interact with the REC1 domain (Leu455, Ser460, Arg467, Thr472 and Ile473), the PI domain (Lys1123 and Lys1124), and the Bridge helix (Arg70 and Arg74), with the 2′-hydroxyl group of G58 hydrogen bonded with Leu455 (Figure 6G). A52 interacts with Phe1105 through a face-to-edge π–π stacking interaction (Figure 6F), and the flipped U59 nucleobase hydrogen bonds with Asn77 (Figure 6G).
The single-stranded linker and stem loops 2 and 3 are primarily recognized by the NUC lobe (Figure 5); in contrast, stem loop 1 and the guide:target/repeat:anti-repeat duplexes are recognized by both the REC and NUC lobes. The backbone phosphate groups of the linker (nucleotides 63–65 and 67) interact with the RuvC domain (Glu57, Lys742 and Lys1097), the PI domain (Thr1102), and the Bridge helix (Arg69), with the 2′-hydroxyl groups of U64 and A65 hydrogen bonded with Glu57 and His721, respectively (Figure 6H). The C67 nucleobase forms two hydrogen bonds with Val1100 (Figure 6H).
Stem loop 2 is recognized by Cas9 via the interactions between the NUC lobe and the non-Watson-Crick A68:G81 pair, which is formed by direct (between the A68 N6 and G81 O6 atoms) and water-mediated (between the A68 N1 and G81 N1 atoms) hydrogen-bonding interactions (Figure 6I). The A68 and G81 nucleobases contact Ser1351 and Tyr1356, respectively, while the A68:G81 pair interacts with Thr1358 via a water-mediated hydrogen bond (Figure 6I). The 2′-hydroxyl group of A68 hydrogen bonds with His1349, while the G81 nucleobase hydrogen bonds with Lys33 (Figure 6I).
Stem loop 3 interacts with the NUC lobe more extensively, as compared to stem loop 2 (Figure 5). The backbone phosphate groups of C91 and G92 interact with the RuvC domain (Arg40 and Lys44), while the G89 and U90 nucleobases hydrogen bond with Gln1272 and Glu1225/Ala1227, respectively (Figure 6J). The A88 and C91 nucleobases are recognized by Asn46 via multiple hydrogen-bonding interactions (Figure 6J).
Structural flexibility of Cas9 and sgRNA
Although the HNH domain cleaves the complementary strand of the target DNA at a position three nucleotides upstream of the PAM sequence (Gasiunas et al., 2012; Jinek et al., 2012), in the present structure, the HNH domain is located away from the scissile phosphate group of the bound complementary strand (Figure 7A). A structural comparison of Mol A and Mol B provided mechanistic insights into complementary strand cleavage by the HNH domain. In Mol A, the HNH domain is followed by the α39 helix of the RuvC domain, which is connected to the α40 helix by the α39–α40 linker (residues 919–925) (Figure 7A). In Mol A, residues 913–925 form the C-terminal portion of the α39 helix and the α39–α40 linker, while in Mol B, these residues form an extended α-helix, which is directed toward the cleavage site of the complementary strand (Figure 7A). These observations suggested that the HNH domain can approach and cleave the target DNA through conformational changes in the segment connecting the HNH and RuvC domains.
Figure 7. Structural flexibility of the complex and a model for RNA-guided DNA cleavage by Cas9.
(A) Structural comparison of Mol A and Mol B. In Mol A (left), the disordered linker between the RuvC and HNH domains is indicated by a dotted line. In Mol B (right), the disordered HNH domain is shown as a dashed circle. The flexible connecting segment (α39 and α40) in the RuvC domain is colored orange.
(B) Superimposition of the Cas9 proteins in Mol A and Mol B. The two complexes are superimposed based on the core β-sheet of the two RuvC domains. The HNH domain and the bound sgRNA:target DNA complex were omitted, for clarity.
(C) Superimposition of the sgRNA:target DNA complex in Mol A and Mol B. After superimposition of the two complexes as in (B), the Cas9 proteins were omitted to show the sgRNA:target DNA complex.
(D) Molecular surface of Cas9. The HNH domain and the sgRNA:target DNA complex were omitted, for clarity.
(E) Model of RNA-guided DNA cleavage by Cas9.
The structural comparison further revealed the conformational flexibility between the REC and NUC lobes (Figure 7B). As compared to Mol A, Mol B adopts a more open conformation, in which the two lobes are rotated by 15° at a hinge loop between the Bridge helix and strand β5 in the RuvC domain (Figure 7B). The bound sgRNA also undergoes an accompanying conformational change at the linker, which interacts with the hinge loop (Figure 7C). We also observed the concomitant displacement of the β17–β18 loop of the PI domain, which interacts with the repeat:anti-repeat duplex and the α2–α3 loop of the REC1 domain (Figure 7B). Notably, there is no direct contact between the two lobes in the present structure, except for the interactions between the α2–α3 and β17–β18 loops (Figure 7D), suggesting that Cas9 is highly flexible in the absence of the sgRNA. The flexible nature of Cas9 is likely to play a role in the assembly of the Cas9–sgRNA–DNA ternary complex.
DISCUSSION
The present structure revealed that the 20-bp heteroduplex, formed by the sgRNA guide region and the complementary strand of the target DNA, is accommodated in the positively-charged groove at the interface between the REC and NUC lobes, with the scissile phosphate group of the target DNA properly positioned for cleavage by the HNH domain. Although the present structure does not contain the non-complementary DNA strand, the position of the bound complementary strand suggested that the scissile phosphate of the non-complementary strand is located in the vicinity of the active site of the RuvC domain, consistent with previous biochemical data (Gasiunas et al., 2012; Jinek et al., 2012). Furthermore, our structural and functional analyses indicated that the PI domain participates in the PAM recognition.
Based on these observations, we propose a model for the Cas9-catalyzed RNA-guided DNA cleavage (Figure 7E). Cas9 recognizes the PAM-proximal guide region and the repeat:anti-repeat duplex of sgRNA, to form the Cas9–sgRNA binary complex. The binary complex subsequently recognizes the DNA sequence complementary to the 20-nt guide region of the bound sgRNA, to form the final Cas9–sgRNA–target DNA ternary complex. Prior to the ternary complex formation, the PI domain recognizes the PAM sequence on the non-complementary strand, thereby triggering the R-loop formation. Upon the assembly of the ternary complex, the mobile HNH domain approaches and cleaves the complementary strand in the guide:target heteroduplex, whereas the RuvC domain cleaves the single-stranded, non-complementary strand. Biochemical studies indicated that PAM recognition by Cas9 is important for both the binding and cleavage of the target DNA, suggesting that the Cas9–sgRNA complex may indeed undergo an inactive-to-active conformational transition upon PAM recognition (Gasiunas et al., 2012; Sternberg et al., 2014). This notion is consistent with the fact that the present structure is likely to represent an inactive state, where the HNH domain is located away from the complementary strand.
The present crystal structure provides a critical step towards understanding the molecular mechanism of RNA-guided DNA targeting by Cas9. Further structural and functional studies with S. pyogenes Cas9 or related orthologs, including the structural elucidation of the Cas9–sgRNA–DNA ternary complex containing the non-complementary strand, will be important for illuminating the mechanisms of PAM recognition, the conformational changes occurring upon PAM recognition, and the mismatch tolerance between the guide:target heteroduplex. However, this study has provided a useful scaffold for the rational engineering of Cas9-based genome modulating technologies. For example, we created an S. pyogenes Cas9 truncation mutant (Figure 2B) that will facilitate the packaging of Cas9 into size-constrained viral vectors for in vivo and therapeutic applications. Moreover, future engineering of the PI domain may allow us to program the PAM specificity, improve the target site recognition fidelity, and increase the versatility of the Cas9 genome engineering platform.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are described in the Supplemental Experimental Procedures.
The S. pyogenes Cas9 D10A/C80L/C574E/H840A mutant (residues 1–1368) was expressed in Escherichia coli Rosetta 2 (DE3) (Novagen) and purified by chromatography on Ni-NTA Superflow (QIAGEN), Mono S (GE Healthcare) and HiLoad Superdex 200 16/60 (GE Healthcare) columns. The SeMet-labeled Cas9 protein was expressed in E. coli B834 (DE3), and was purified using a similar protocol as for the native protein. The 98-nt sgRNA was in vitro transcribed with T7 polymerase and purified by 10% denaturing polyacrylamide gel electrophoresis. The 23-nt target DNA was purchased from Sigma-Aldrich. The purified Cas9 protein was mixed with sgRNA and DNA (molar ratio, 1:1.5:2.3), and the complex was purified by gel filtration chromatography on a Superdex 200 Increase column (GE Healthcare).
The purified complex was crystallized at 20°C by the hanging-drop vapor diffusion method. Crystals were obtained by mixing 1 μl of complex solution (A260 nm, 15) and 1 μl of reservoir solution (13% PEG 3,350, 100 mM Tris-HCl, pH 8.0, 200 mM ammonium acetate and 100 mM NDSB-256). The SeMet-labeled protein was crystallized under conditions similar to those for the native protein. X-ray diffraction data were collected at 100 K on the beamlines BL32XU and BL41XU at SPring-8 (Hyogo, Japan). The structure was determined by the SAD method, using the 2.6 Å resolution data set from the SeMet-labeled crystals. The final model was refined using the 2.5 Å resolution native data set.
The human embryonic kidney (HEK) cell line 293FT (Life Technologies) was seeded into 24-well plates (Corning) at a density of 120,000 cells/well, 24 h prior to transfection. Cells at 70–80% confluency were transfected using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s recommended protocol. The Cas9 plasmid (400 ng) and the U6::sgRNA PCR product (100 ng) were transfected into each well. Genomic modifications were evaluated using the SURVEYOR nuclease assay, as described previously (Cong et al., 2013).
Supplementary Material
Acknowledgments
We thank Arisa Kurabayashi for assistance with vector construction, Tomohiro Nishizawa and Kazuki Kato for helpful comments on the manuscript, Ryohei Ishii and Motoyuki Hattori for assistance in data collection, and Chie-yu Lin for helpful discussions regarding the sgRNA mutagenesis. We thank the beam-line staffs at BL32XU and BL41XU of SPring-8, Japan, and at IO3 of Diamond Light Source Ltd., UK, for assistance with data collection.
This work was supported by PRESTO from the Japan Science and Technology (JST) Agency and Platform for Drug Discovery and Informatics (to H.N.), by a grant from the Japan Society for the Promotion of Science (JSPS), through its “Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST program)” (to O.N.), and by the Core Research for Evolutional Science and Technology (CREST) Program ‘The Creation of Basic Medical Technologies to Clarify and Control the Mechanisms Underlying Chronic Inflammation’ of Japan Science and Technology Agency (JST) (to O.N.). F.Z. is supported by an NIH Director’s Pioneer Award (1DP1-MH100706), the Keck, McKnight, Poitras, Merkin, Vallee, Damon Runyon, Searle Scholars, Klingenstein, and Simons Foundations, Bob Metcalfe, and Jane Pauley.
Footnotes
ACCESSION NUMBER
The atomic coordinates of the Cas9–sgRNA–DNA ternary complex have been deposited in the Protein Data Bank, with the PDB code 4OO8.
Supplemental Information includes Extended Experimental Procedures, DNA sequences of constructs, sgRNAs, and PCR primers used in the study, six figures, and two table, and can be found with this article online.
AUTHOR CONTRIBUTIONS
H.N. performed the crystallization and structural analysis; F.A.R. designed and performed the Cas9 truncation, sgRNA mutation, and nickase analyses; P.D.H., S.K., and S.S. designed and performed the Cas9 domain switching and point mutation analyses; N.D. performed the mass spectrometric analysis; R.I. performed the structural analysis; and H.N., F.A.R., P.D.H., F.Z., and O.N. wrote the manuscript with help from all authors. H.N., F.Z. and O.N. directed and supervised all of the research.
The authors have no conflicting financial interests. A patent application has been filed related to this work, and the authors plan to make the reagents widely available to the academic community through Addgene and to provide software tools via the Zhang lab Web site (www.genome-engineering.org).
References
- Ariyoshi M, Vassylyev DG, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Biertümpfel C, Yang W, Suck D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007;449:616–620. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Shi K, Yin Z, Aihara H. Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution. Nucleic Acids Res. 2013;41:648–656. doi: 10.1093/nar/gks1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Ann Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecka KM, Komorowska W, Nowotny M. Crystal structure of RuvC resolvase in complex with Holliday junction substrate. Nucleic Acids Res. 2013;41:9945–9955. doi: 10.1093/nar/gkt769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Hor LI, Chang ZF, Tsai LC, Yang WZ, Yuan HS. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 2003;22:4014–4025. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014 doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiology. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.