Abstract
Meiotic nodules are the sites of double-stranded DNA break repair. Rpa is a single-stranded DNA-binding protein, and Rad51 is a protein that assists in the repair of DNA double strand breaks. The localisation of Rad51 to meiotic nodules before the localisation of Rpa in mice introduces the issue of whether Rpa is involved in presynaptic filament formation during mammalian meiosis. Here, we show that a protein with unknown function, Spata22, colocalises with Rpa in meiotic nodules in rat spermatocytes. In spermatocytes of Spata22-deficient mutant rats, meiosis was arrested at the zygotene-like stage, and a normal number of Rpa foci was observed during leptotene- and zygotene-like stages. The number of Rad51 foci was initially normal but declined from the leptotene-like stage. These results suggest that both formation and maintenance of Rpa foci are independent of Spata22, and the maintenance, but not the formation, of Rad51 foci requires Spata22. We propose a possible model of presynaptic filament formation in mammalian meiosis, which involves Rpa and Spata22.
Spo11-induced double-stranded DNA breaks (DSBs) are repaired through homologous recombination during the first meiotic prophase1. During the early stage of homologous recombination after DSB induction, the processing of 5′ to 3′ DNA end resection produces molecules with 3′ single-stranded DNA (ssDNA) tails; ssDNA tails invade intact homologous duplex DNAs leading to the creation of joint molecule intermediates2,3. The eukaryotic RecA homolog Rad51 mediates DNA strand exchange in vitro by forming a helical protein filament on ssDNA, the so-called presynaptic filament4,5. The secondary structure of ssDNA inhibits assembly of a contiguous nucleoprotein filament. Heterotrimeric ssDNA binding factor replication protein A (Rpa) overcomes this inhibition through its ability to destabilise base pairs6,7. When Rpa is added after preincubation of Rad51 with the ssDNA substrate, Rpa stimulates DNA strand exchange and adenosine triphosphate (ATP) hydrolysis in vitro, which are mediated by Rad517,8. Since Rpa has a high affinity for ssDNA, the presence of Rpa interferes with the binding of Rad51 to ssDNA7,8. The availability of Rpa during the formation of presynaptic filaments in vivo suggests that a molecular mechanism underlies the filament formation in the presence of Rpa9. In fact, several proteins known as mediators have been shown to facilitate Rad51 assembly on Rpa-coated ssDNAs in vitro7,10,11. However, although double-stranded DNA (dsDNA) is a substrate in DNA strand exchange, it suppresses presynaptic filament formation because Rad51 protein binds not only to ssDNA but also to dsDNA12. Thus, efficient targeting of Rad51 to ssDNA is necessary for the formation of an active presynaptic filament. Recent biochemical studies suggest that Brca2 promotes the preferential binding of Rad51 to ssDNA3,11. Rad52 protein does not reverse the inhibitory effect of Rad51 protein binding to dsDNA9. In line with these biochemical findings, in vivo analysis demonstrated that mediators are required for the normal formation of Rad51 foci, but not Rpa foci, during meiotic recombination, as well as during mitotic DSB repair via homologous recombination. Mutations in Rad52, Rad55, and Rad57 in the budding yeast Saccharomyces cerevisiae, and Brca2 orthologs in the corn smut fungus Ustilago maydis, the nematode Caenorhabditis elegans, and the mouse Mus musculus induce a failure of Rad51 to appear at DSB sites, but do not have any obvious effects on the accumulation of Rpa at DSB sites13,14,15,16. Rpa localises at DSB repair sites prior to the localisation of Rad51 at those sites during both meiosis and mitosis in S. cerevisiae3. Thus, based on studies of homologous recombination in S. cerevisiae, assembly of presynaptic filaments can be thought to progress as follows: Rpa first unwinds secondary structures in ssDNA, and then Rad51 coats ssDNA by the displacement of Rpa with the aid of mediators2,3,4. However, in contrast to the case in S. cerevisiae, Rad51 localisation to meiotic nodules, which are the sites of DSB repair17,18,19, is known to precede Rpa localisation in mice20,21. This finding raises the issue of whether Rpa is involved in assembly of the presynaptic complex during mammalian meiosis21. No proteins that mediate the unwinding of secondary structures of ssDNA by substituting for Rpa have been identified.
Spata22, a vertebrate-specific gene, is responsible for sterility associated with gonadal hypoplasia, arrest of meiosis during prophase I, abnormal chromosome synapsis, and failure of DSB repair in Spata22-deficient mice and rats, and the Spata22 protein was shown to be expressed predominantly in primary spermatocytes22,23. Recent studies identified a novel protein, Meiob, which associates with Spata22 and Rpa, and showed that Meiob is required for meiotic progression, chromosome synapsis, and meiotic DSB repair24,25. However, the meiotic event(s) that involve Spata22 have not been elucidated.
In this study, we reveal that Spata22 is localised to meiotic nodules during prophase I in rat spermatocytes. Spata22 is observed as discrete foci along meiotic chromosomes by immunofluorescence analysis. Spata22 foci coexist almost completely with Rpa foci, and partially with Rad51 and Mlh1 foci, the latter being a marker of presumptive crossover sites26,27. Spata22 foci first appear in meiotic chromosomes after Rad51 foci appear. In spermatocytes of Spata22-deficient rats, meiosis is arrested at the zygotene-like stage. In these spermatocytes, the DSB marker phosphorylated H2AFX (γ-H2AFX)28,29 is distributed throughout the chromatin at the leptotene-like stage as well as in leptotene spermatocytes of the wild type. Moreover, a normal number of Rpa foci are observed during leptotene- and zygotene-like stages, but the number of Rad51 foci is reduced from the leptotene-like stage onwards. In mutant spermatocytes, γ-H2AFX staining and Rpa foci are not removed from meiotic chromosomes, and Mlh1 foci are undetectable. Our data are novel in that the careful quantification of the number of Rpa and Rad51 foci has not been reported in the Spata22 mutant, and the distribution of Spata22 has not been studied in rats. Based on our present findings, and on the biochemical functions of Rpa and Brca2 in the assembly of the presynaptic filament and the association of Spata22 with Meiob, we propose a possible model for presynaptic filament formation in mammalian meiosis. Further, we discuss the additional roles of Spata22 in meiotic recombination.
Results
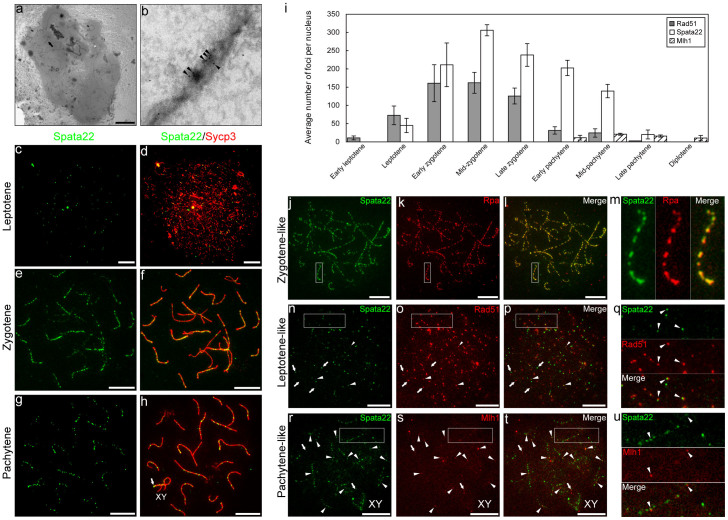
To investigate the function of Spata22 in meiosis, antibodies were raised against the protein, and immunochemical analysis was performed at the histological and chromosomal levels. Immunostaining of testis sections showed discrete staining in the nuclei of spermatocytes (Supplementary Fig. 1a, b). This suggested that Spata22 is expressed on meiotic chromosomes. To determine the subnuclear distribution of Spata22 in spermatocytes during prophase I, we performed electron microscopic observation of spread nuclei of rat spermatocytes immunolabelled with anti-Spata22 antibody. Spata22 localised to electron-dense bodies, namely meiotic nodules, along synaptonemal complexes (SCs)17,18,19 (Fig. 1a, b). We next performed immunofluorescent localisation analysis with anti-Spata22 antibody and antibodies to Sycp3, which is a protein component of both the axial elements and the lateral elements of the SCs19. Immunostaining revealed that Spata22 was distributed as punctate foci along axial and/or lateral elements during the leptotene, zygotene, and pachytene stages (Fig. 1c–h), but was not detected during the diplotene stage (data not shown). We observed intense immunofluorescent signals of Spata22 in the terminal segments of pseudoautosomal regions of the X and Y sex chromosomes in the pachytene nuclei (Fig. 1g, h). The pseudoautosomal regions of mammalian sex chromosomes undergo frequent reciprocal recombination30, and large protein complexes that include Rpa are formed in this region during prophase I20.
Figure 1. Spata22 colocalises with Rpa in meiotic nodules.
(a, b) Immunoelectron microscopy of a pachytene spermatocyte nucleus. Higher magnification of the SC indicated by an arrow in (a) (b). Spata22 localised to meiotic nodules (c–h). Distribution of Spata22 in spermatocyte nuclei during leptotene (c, d), zygotene (e, f), and pachytene (g, h). Punctate staining signals of Spata22 (green) were distributed along axial/lateral elements (red). Strong immunofluorescent signals were present in the terminal segments of the sex chromosomes (XY) in the pachytene nucleus ((g), (h), arrows). (i) Average number of Spata22, Rad51, and Mlh1 foci in each substage of meiotic prophase I. Bars, standard deviations. (j–u) Comparison of the distribution of Spata22 foci (green) with the distribution of Rpa, Rad51, and Mlh1 foci (red) in the spermatocyte nuclei. Overlap of the two immunofluorescent signals is indicated in yellow. Spata22 foci and Rpa foci coexisted almost completely in the zygotene-like nucleus (j–m). Spata22 only, Rpa only, and the overlap were 0.8%, 1.1%, and 98.1% of all the foci in the nucleus, respectively. Higher magnification of the white boxes in (j–l) shows that Spata22 foci overlapped almost completely with Rpa foci (m). Rad51 foci and Spata22 foci partially coexisted in the leptotene-like nucleus (n–q). Spata22 only, Rad51 only, and the overlap were 18.2%, 60.1%, and 21.7% of all the foci in the nucleus, respectively. Complete overlap, partial overlap, and non-overlap of the two proteins' foci are indicated by large arrowheads, small arrowheads, and arrows, respectively. Higher magnification of the white boxes in (n–p) shows that Rad51 foci partially overlapped with Spata22 foci ((q), arrowheads). Mlh1 foci and Spata22 foci partially coexisted in the pachytene-like nucleus (r–u). Spata22 only, Mlh1 only, and the overlap were 83.3%, 4.4%, and 12.3% of all the foci in the nucleus, respectively. Complete or partial overlap and non-overlap between the two proteins' foci are indicated by arrowheads and arrows, respectively (r–t). Higher magnification of the white boxes in (r–t) shows that Mlh1 foci overlapped partially with Spata22 foci ((u), arrowheads). Scale bars in all panels, 10 μm.
Time course experiments were carried out with co-immunostaining of samples to determine the relative times at which Rad51, Spata22, and Mlh1 foci appeared and disappeared. We used anti-Sycp3 antibody and antibodies to each of Rad51, Spata22, and Mlh1, and counted the number of foci of these meiotic nodule proteins per nucleus. Immunostaining revealed that both Rad51 and Spata22 localised to meiotic chromosomes during leptotene, zygotene, and pachytene stages. Rad51 foci were observed from the early leptotene stage until the mid-pachytene stage, whereas Spata22 foci were first observed during the leptotene stage and persisted until the late pachytene stage (Fig. 1i, Supplementary Table 1). Mlh1 foci were detected along SCs from early pachytene to diplotene (Fig. 1i, Supplementary Table 1). To examine whether Spata22 colocalises with other meiotic nodule proteins Rpa, Rad51, and Mlh1, we performed co-immunostaining of the spread nuclei of rat spermatocytes with anti-Spata22 antibody and antibodies to Rpa, Rad51, and Mlh1. Spata22 colocalised with Rpa in meiotic chromosomes during prophase I: Spata22 foci and Rpa foci coexisted in 60.3%, 96.7%, and 95.2% of all the foci at leptotene-, zygotene-, and pachytene-like stages, respectively (Fig. 1j–m, Supplementary Table 2, Supplementary Fig. 2 and 3). This suggests that Spata22 and Rpa might function at nearly the same location. As shown in previous studies that revealed the colocalisation of Rpa and Rad5120,31, Spata22 colocalised with Rad51 in meiotic chromosomes; this was indicated by the partial or complete overlap of two foci (Fig. 1n–q, Supplementary Fig. 2). The colocalisation occurred in only a subset of the two proteins' foci: Spata22 foci and Rad51 foci coexisted in 36.2%, 31.0%, and 30.4% of all the foci at leptotene-zygotene-, zygotene-, and early-mid-pachytene-like stages, respectively (Fig. 1n–q, Supplementary Table 3, Supplementary Fig. 3). As shown in a previous study that revealed the colocalisation of Rpa and Mlh1 in meiotic chromosomes31, most of the Mlh1 foci colocalised with Spata22 foci during the early-mid-pachytene-like stage (Fig. 1r–u, Supplementary Table 4); however, during the late pachytene-like stage, the colocalisation was observed in only a subset of the Mlh1 foci (Supplementary Table 4, Supplementary Fig. 2 and 3). Spata22 foci and Mlh1 foci coexisted in 11.9% and 4.8% of all the foci at early-mid-pachytene- and late pachytene-like stages, respectively (Supplementary Fig. 3).
To determine the meiotic function of Spata22, we examined the effect of Spata22 deficiency on the progression of meiotic recombination using the tremor rat. This rat is an autosomal recessive mutant that exhibits meiotic failure, as well as neurological disorders and abnormalities of the hair and whiskers22,32,33,34. The causative mutation, named tremor (tm), is a >200-kb genomic deletion located at rat Chr10q24 that contains at least 13 genes, including Spata2222. Forced expression of Spata22 rescued the meiotic phenotype of the tremor mutant, consistent with the possibility that deletion of Spata22 is responsible for the phenotype22. However, we cannot exclude the possibility that the absence of one or more of the 12 genes deleted in the tremor mutant modifies the meiotic phenotype. Generation of a Spata22−/− knockout will be required to definitively address this question. Examination of chromosome synapsis in spermatocytes of tm homozygous rats revealed that meiosis progressed normally at the onset of the zygotene-like stage; however, no nuclei with complete synapsis were found (Supplementary Fig. 4). There were mid-late zygotene-like spermatocytes that exhibited abnormal chromosome synapsis, including non-homologous pairing, in the mutant (Supplementary Fig. 4). Thus, we examined the distributions of molecular markers related to meiotic recombination during leptotene- and zygotene-like stages in the mutant spermatocytes. These markers were the DSB marker phosphorylated H2AFX (γ-H2AFX)28,29, Rpa, Rad51, and Mlh1. Here, the possibility that stages of mutant nuclei and wild-type controls do not correspond to each other in terms of cell-cycle progression should be noted. Our criteria for the classification of stages are described in detail in the Methods section.
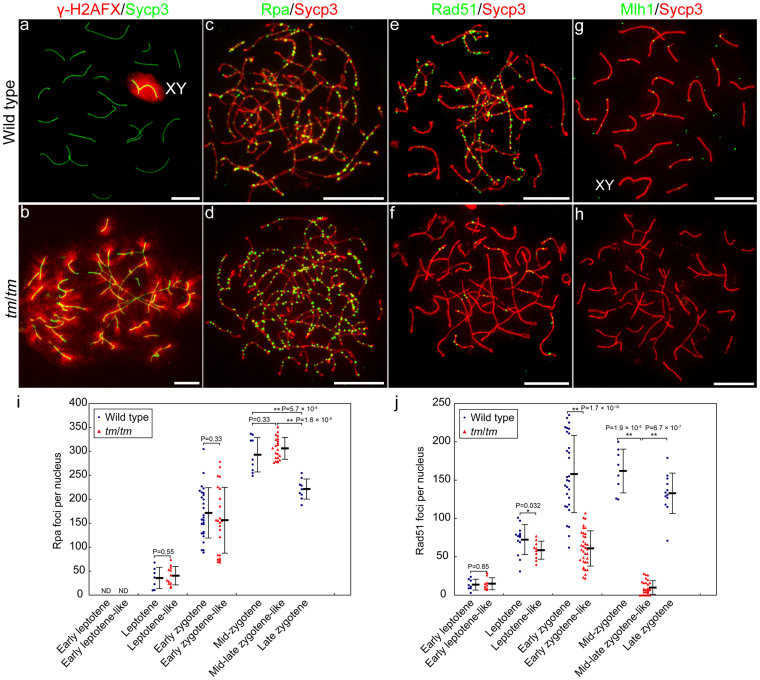
Immunolocalisation analysis revealed that γ-H2AFX was distributed throughout the chromatin in leptotene-like spermatocytes of the mutant and the wild-type controls (Supplementary Fig. 5). Whereas γ-H2AFX staining was distributed solely around the sex chromosomes in the wild-type pachytene spermatocytes, it was distributed extensively in the nuclei of the mutant mid-late-zygotene-like spermatocytes (Fig. 2a, b), as seen previously in the zygotene-like spermatocytes of the Spata22-deficient mouse23. These findings suggest that although DSBs are induced normally in the mutant, they remain unrepaired owing to the inadequacy of repair mechanisms28,35. The number of Rpa foci was comparable between the mutant spermatocytes and the wild-type controls during leptotene- and early zygotene-like stages (Mann–Whitney U-test, P = 0.55 during the leptotene-like stage; P = 0.33 during the early zygotene-like stage) (Fig. 2c, d, i, Supplementary Fig. 6, Supplementary Table 5). Moreover, a similar number of Rpa foci was observed between the mutant mid-late-zygotene-like and wild-type mid-zygotene spermatocytes (P = 0.33, adjusted 10% significance level is 0.067). In the wild-type spermatocytes, fewer Rpa foci were observed during late zygotene than during mid-zygotene (P = 5.7 × 10−4, adjusted 1% significance level is 0.0067); however, a larger number of Rpa foci were observed in the mutant mid-late-zygotene-like spermatocytes than in the wild-type late-zygotene spermatocytes (P = 1.6 × 10−5, adjusted 1% significance level is 0.0033) (Fig. 2i, Supplementary Fig. 6, Supplementary Table 5). These findings suggest that DSBs are induced normally in the mutant13,16,35, as shown by the γ-H2AFX staining, and that the DSBs remain unrepaired in the mid-late zygotene-like nuclei of the mutant. The number of Rad51 foci was similar between the mutant spermatocytes and the wild-type controls during the early leptotene-like stage (P = 0.85), but fewer Rad51 foci were observed in the mutant spermatocytes than in the wild-type controls from the leptotene-like stage to the mid-late zygotene-like stage (P = 0.032 during the leptotene-like stage; P = 1.7 × 10−10 during the early zygotene-like stage; P = 1.9 × 10−5 mutant mid-late-zygotene-like vs. wild-type mid-zygotene spermatocytes; and P = 6.7 × 10−7 mutant mid-late-zygotene-like vs. wild-type late-zygotene spermatocytes) (Fig. 2e, f, j, Supplementary Fig. 6, Supplementary Table 5). These findings suggest the possibility that Rad51-mediated repair processes, such as Rad51 filament formation, strand invasion, and joint molecule intermediate formation, are impaired in mutant spermatocytes. We note that Mlh1 is thought to function in chiasma formation26,27. Mlh1 foci were not detected in mutant spermatocytes (Fig. 2g, h). This suggests that the recombination-defective spermatocytes are eliminated before the normal formation of Mlh1 foci19.
Figure 2. Spata22 deficiency causes defective DSB repair with reduced Rad51 expression and persistent Rpa expression.
(a–h) Comparison of γ-H2AFX distribution and the number of Rpa, Rad51, and Mlh1 foci between wild-type (upper panels) and tm/tm (lower panels) spermatocytes. The γ-H2AFX labelling was distributed only around the sex chromosomes (XY) in the wild-type pachytene nucleus, but was observed throughout the chromatin in the mutant zygotene-like nucleus (a, b). A number of Rpa foci were distributed along axial and lateral elements in the mutant zygotene-like nucleus, as seen in the wild-type zygotene nucleus (c, d). Fewer Rad51 foci were observed in the mutant zygotene-like nucleus than in the wild-type zygotene nucleus (e, f). Mlh1 foci were distributed along SCs in the wild-type pachytene nucleus, but were not detected in the mutant mid-late-zygotene-like nucleus (g, h). The sex chromosomes, XY; scale bars in all panels, 10 μm. (i, j) Time course of the number of Rpa foci (i) and Rad51 foci (j) per nucleus in wild-type (blue circles) and tm/tm (red triangles) spermatocytes. Stages of examined nuclei are shown below dots. We should note the possibility that stages of mutant nuclei and wild-type controls are not corresponding. Bold horizontal lines, averages; bars, standard deviations. ND, not detected. The number of examined nuclei is shown in Supplementary Table. 5. Two-tailed P values are shown above dots. The number of Rpa foci per nucleus was comparable between the mutant and the wild-type controls during leptotene- and early zygotene-like stages (P > 0.05) (i). The number of Rpa foci was similar between the mutant mid-late-zygotene-like and wild-type mid-zygotene nuclei (P > 0.1, adjusted 10% significance level is 0.067), and was larger in the mutant mid-late-zygotene-like nuclei than in the wild-type late-zygotene nuclei (**P < 0.01, adjusted 1% significance level is 0.0033) (i). The number of Rad51 foci per nucleus was similar between the mutant and the wild-type controls during the early leptotene-like stage (P > 0.05), but was significantly lower in the mutant than in the wild-type controls from the leptotene-like stage to the mid-late zygotene-like stage (*P < 0.05, **P < 0.01) (j).
Discussion
We have shown that meiotic recombination was defective in the Spata22-deficient tremor rats. In the spermatocytes of the mutant, Rpa foci appeared normally in meiotic chromosomes from the leptotene-like stage to the zygotene-like stage, but seemed to persist without a reduction in their number during the mid-to-late zygotene-like stage. The number of Rad51 foci in the mutant was also normal during the early leptotene-like stage, but was significantly reduced from the leptotene-like stage to the mid-late zygotene-like stage, at which time meiosis was arrested, compared with the level in the wild type. These findings suggest that both formation and maintenance of Rpa foci are independent of Spata22, but Spata22 is required for the maintenance, but not the formation, of Rad51 foci in mammalian meiosis.
Recently, Meiob was identified as a protein that associates with Spata22 and with Rpa during prophase I24,25. Meiob and Spata22 are localised to meiotic chromosomes interdependently25. In spermatocytes of the Meiob−/− mouse, γ-H2AFX persists24,25, as seen in the Spata22-deficient rat (this study) and mouse23. In Meiob−/− mouse spermatocytes, there is a normal number of Rpa foci during the leptotene- and zygotene-like stages, but they are not removed from meiotic chromosomes during a pachytene-like stage, at which cells exhibit abnormal chromosome synapsis but contain condensed axial and/or lateral elements that are similar to those of wild-type cells at late zygotene and pachytene stages24,25, and the number of Rad51 foci decreases during the zygotene- and pachytene-like stages24. These phenotypes are similar to the meiotic phenotype of the Spata22-deficient rat. However, Luo et al. reported that Rad51 foci appear normally even during the leptotene- and zygotene-like stages in the Meiob−/− mouse, and the number of Rad51 foci is reduced only at the pachytene-like stage, which is similar to the meiotic phenotype of the Spata22-deficient mouse23,25. Mlh1 foci are undetectable in both Meiob−/−24,25 and Spata22-deficient rat spermatocytes. We cannot exclude the possibility that the loss of genes besides Spata22 in the tremor rat contribute to the differences in the time course of the number of Rad51 between the tremor rat and the mouse Meiob−/− mutants. However, the results of this study and previous studies are consistent in the overall conclusion that Rad51 foci disappear prematurely from meiotic chromatin in the absence of Spata22 function.
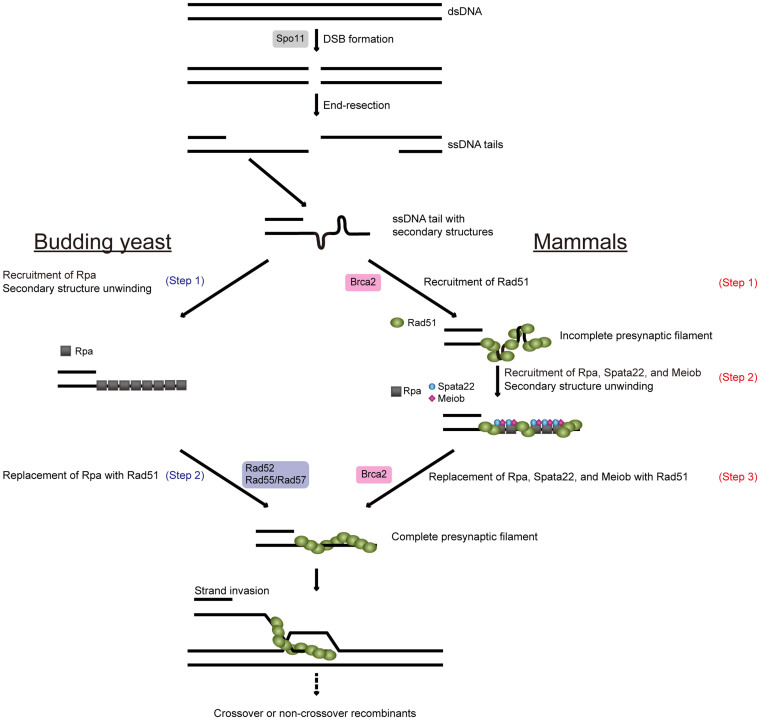
Human BRCA2 facilitates RAD51 nucleofilament formation in vitro by localising RAD51 to ssDNA, and also by performing a mediator function similar to S. cerevisiae Rad52 and Rad55-Rad577,10,11. One may speculate that Spata22 is involved in Brca2-mediated presynaptic filament formation. The premature disappearance of Rad51 foci in the tremor rat suggests that Spata22 deficiency does not affect the localisation of Rad51 to ssDNA tails but affects the stabilisation of Rad51-ssDNA filaments. Considering that Rpa unwinds the secondary structure of ssDNA in vitro, the localisation of Rad51 to meiotic nodules before the localisation of Rpa probably reflects the formation of incomplete presynaptic filaments that contain secondary structures of ssDNA. Thus, based on our present results, as well as the biochemical functions of Rpa and Brca2 in presynaptic filament formation3,8,10,11 and the association of Meiob with Rpa and with Spata2224,25, we propose a possible model for Rad51 filament formation in mammalian meiosis, as follows. Rad51 proteins are loaded onto a 3′ ssDNA tail with the aid of the mediator Brca2 and form an incomplete Rad51 filament; Rpa, Spata22, and Meiob proteins are subsequently loaded onto the ssDNA and circumvent the secondary structures in the substrate; thereafter, Rad51 proteins replace Rpa, Spata22, and Meiob proteins with the aid of Brca2, after which a complete presynaptic complex is eventually formed (Fig. 3). We name this model the three-step model because the formation of the Rad51-ssDNA complex is added as an initial step that is not part of the present model of Rad51 filament formation2,3,4. This hypothesis should be inspected experimentally. Our model can explain the difference in the order of localisation of Rad51 and Rpa proteins between yeast and mammals, although the molecular mechanism that underlies this difference should be elucidated in future studies. Rpa competes with Rad51 for the same binding sites on ssDNA in vitro8. When there is insufficient Rad51 for complete coating of the ssDNA in vitro, the Rad51 that partially coats the ssDNA substrate is displaced by subsequently loaded Rpa, if there is enough of it to coat the entire substrate8. This result indicates that when the amount of Rad51 is not in excess in vivo, Rad51 is displaced from the incomplete Rad51-ssDNA filament by subsequent loading of Rpa in the absence of normal mediators. This appears as premature disappearance of Rad51 foci and persistence of Rpa foci, as seen in the tremor rat. Therefore, we propose that Spata22 may function in the removal of Rpa from an ssDNA end by associating with Meiob during the assembly of presynaptic filament; Rad51 is displaced from an ssDNA end by Rpa in the absence of Spata22. Based on the ssDNA binding ability of both Rpa and Meiob, Souquet et al. suggested a similar possibility that Meiob may function in the proper assembly of presynaptic filament, as a Rad51/Dmc1 mediator24. In addition, the authors proposed a later role of Meiob in the activity of recombinases: Meiob may influence the stability of the presynaptic filament24. Luo et al. suggested that Meiob functions after strand invasion25. The authors proposed that Meiob may mediate second-end capture through interacting with Rpa that remains on the displaced strand of the D-loop and the ssDNA of the second end. In addition, they proposed another hypothesis that Meiob may function in removing 3′-flaps that result from second-end capture in the dHJ pathway and from annealing of the newly synthesized strand with the second resected end in the synthesis-dependent strand annealing (SDSA) pathway, which is the predominant pathway used for meiotic recombination36. These hypotheses should be taken into consideration in future studies.
Figure 3. Schematic representation of models for Rad51 filament formation during meiosis in the budding yeast S. cerevisiae and mammals.
After Spo11-mediated DSB induction, 3′ ssDNA tails that contain secondary structures are generated by 5′ to 3′ DNA end resection. During meiosis in budding yeast (left), Rpa proteins are recruited into an ssDNA tail prior to Rad51 recruitment and then unwind secondary structures in ssDNA (Step 1). The Rad51 nucleofilament is formed by the displacement of Rpa proteins from the ssDNA with the aid of the mediators Rad52 and Rad55-Rad57 heterodimer (Step 2). In mammalian meiosis (right), Rad51 proteins are recruited into a 3′ ssDNA tail with the aid of the mediator Brca2 prior to the recruitment of Rpa, Spata22 and Meiob, and form an incomplete Rad51 nucleofilament on the 3′ ssDNA tail that contains secondary structures (Step 1). Rpa, Spata22, and Meiob proteins then bind to the 3′ ssDNA tail and thereby unwind secondary structures in the ssDNA (Step 2), and Rad51 proteins form a complete presynaptic filament by the displacement of the Rpa, Spata22, and Meiob proteins from the ssDNA with the aid of Brca2 (Step 3). The complete presynaptic filament mediates strand invasion and joint molecule intermediate formation.
The premature disappearance of Rad51 foci in the tremor rat raised another possibility that Spata22 may function at a later stage of meiotic recombination. Rad54 family proteins (Rad54 and Rad54B in mammals) are ATP-dependent translocases21. The ability of yeast Rad54 to remove Rad51 from DNA in vitro suggests that Rad54 and Rad54B are involved in the formation of heteroduplex DNA (hDNA) by removing Rad51 from an invading end after strand invasion37. Rad54 has also been implicated in branch migration for D-loop expansion37. One may speculate that excessive translocase activity of Rad54 causes rapid removal of the Rad51 remaining on the invading end after the strand invasion, and causes an abnormal expansion of the D-loop by abnormal branch migration. Therefore, misregulation of Rad54 activity may block SDSA by the prevention of D-loop disruption, which is the dissociation of the invading end from the targeted strand. The failure in D-loop disruption may be associated with the persistence of Rpa on the displaced strand, as suggested by the results of biochemical studies, which showed that Rpa promotes strand invasion by binding to the displaced strand and preventing it from reannealing2. Accordingly, we propose that Spata22 may function in Rad54- and Rad54B-mediated hDNA formation and D-loop disruption through interacting with Rpa that binds to the displaced strand. This hypothesis should be tested experimentally.
In normal meiotic recombination, Rpa and Spata22 seem to remain in DSB sites after Rad51 is removed19,20,31. Therefore, Spata22 may function in recombination processes during the postsynaptic stage, such as post-invasion DNA synthesis, second-end capture, resolution of recombination intermediates, and crossover control. In mammals, DNA polymerases (Pols) δ, η, and ζ with their cofactor proliferating cell nuclear antigen/replication factor C (PCNA/RFC) appear to be primary polymerases for post-invasion DNA synthesis38. A recent biochemical study proposed a model of the interaction of Rpa, RFC, and PCNA in this process38. We speculate that Spata22 may function in regulating the post-invasion DNA synthesis through interacting with Rpa on the displaced strand.
Methods
Animals
The tremor rats TRM/Kyo, which were bred in the Genome Dynamics Research Center at Hokkaido University, were supplied by the Institute of Laboratory Animals, Graduate School of Medicine, Kyoto University (Kyoto, Japan). To generate the homozygous mutant (tm/tm) and wild-type control (+/+) rats used in this study, heterozygous female rats (tm/+) were mated with heterozygous male rats (tm/+). To discriminate between heterozygous carriers and the wild type, we determined the genotype of the tm locus in unaffected (+/+ or tm/+) offspring by polymerase chain reaction (PCR) amplification using two primer sets. For detection of the deletion mutation, one primer set was designed to be located at both ends of the breakage site22. In the other primer set, for detection of the wild-type allele, one primer was designed to be located in the centromere-proximal end of the breakage site, and the other was designed to be located in the centromere-proximal end of the deleted region22. All of the rats were maintained in a room with controlled temperature and a 12-h light/dark cycle. Food and water were provided ad libitum. All the animal experiments in this study were approved by the Institutional Animal Care and Use Committee, Hokkaido University, and were performed in accordance with National University Corporation Hokkaido University Regulations on Animal Experimentation.
Antibodies
To generate polyclonal antibodies to Sycp3, the coding sequence of Sycp3 was obtained by PCR from a rat testis cDNA library (Takara Bio). The full coding sequence of Sycp3 and the sequence encoding the N-terminal 114-aa section of Sycp3 were inserted into the pET102/D-TOPO vector (Life Technologies). Sequences that encode amino acids 1–195 and 89–195 of rat Spata22 were inserted into the pGEX-4T-1 vector (GE Healthcare). The recombinant proteins were expressed in E. coli and purified using Glutathione Sepharose 4B or Ni Sepharose 6 Fast Flow (GE Healthcare) columns. Mouse polyclonal antibodies to Sycp3 and Spata22 were generated by the immunisation of mice with recombination proteins of the full-length Sycp3 and the N-terminal 195 aa of Spata22. Rabbit polyclonal antibodies to Sycp3 and Spata22 were generated by the immunisation of rabbits with recombination proteins of the partial Sycp3 and 89–195 aa of Spata22, respectively. Anti-Rpa2 mouse antibody (Ab-3, NA19L), anti-Rad51 rabbit antibody (Ab-1, PC130), and anti-Mlh1 rabbit antibody (Ab-2, PC56T) were purchased from Merck Millipore. Anti-γ-H2AFX rabbit antibody was also purchased (4411-PC-020, Trevigen). The primary antibodies were used in immunocytological analysis by dilution in antibody dilution buffer (adb) [3% bovine serum albumin (BSA) and 0.05% Nonidet P-40 (NP-40) in phosphate buffered saline (PBS)] as follows: 1:250 for anti-Spata22, anti-Sycp3, and anti-Rad51; 1:100 for anti-γ-H2AFX; 1:50 for anti-Rpa2; and 1:25 for anti-Mlh1. Goat anti-mouse immunoglobulin fluorescein isothiocyanate (FITC) conjugate (F0479) and swine anti-rabbit immunoglobulin TRITC conjugate (R0156) were purchased from Dako and used at dilutions of 1:333 and 1:500, respectively. For electron microscopy analyses, 12-nm colloidal gold-goat anti-rabbit IgG was also purchased (111-205-144, Jackson ImmunoResearch) and used at 1:100 dilution.
Immunocytological analysis
Preparations of meiotic nuclei for immunofluorescent analysis were made by the dry-down method, as described previously39. For electron microscopy, spermatocytes were prepared as described previously40, with some modifications. Testicular cells collected in DMEM (Life Technologies) were surface-spread on a hypotonic salt solution, picked up on plastic-coated glass slides, and fixed in 1% paraformaldehyde, pH 8.2, with 0.05% sodium dodecyl sulphate (SDS) for 3 min, and 1% paraformaldehyde without SDS for an additional 3 min. After three 1-min rinses in 0.4% DRIWEL (Fujifilm), the slides were air-dried. The slides were stained with 1% ethanolic phosphotungstic acid for 30 min, rinsed with 95% ethanol, and dried. The plastic film with cells was floated on the surface of water and grids were placed on the floating film. Film and grids were picked up with Parafilm and air-dried. The cells on the grids were examined by electron microscopy. Immunostaining of preparations of meiotic nuclei was performed as described previously40. Antibodies used for immunostaining are described above. Preparations were blocked at room temperature as follows: 10 min in 10% adb in PBS (10% adb/PBS), 10 min in 10% adb/PBS containing 0.05% NP-40, and 10 min in 10% adb/PBS. After blocking, preparations were incubated with adb containing primary antibodies for 14–16 h at 4°C. Washing was performed on ice as follows: 10 min in 10% adb/PBS containing 0.2% DRIWEL, 10 min in 10% adb/PBS, 10 min in 10% adb/PBS containing 0.05% NP-40, and 10 min in 10% adb/PBS. After incubation with adb containing secondary antibodies for 3–4 h at 4°C, slides were washed on ice as follows: 10 min in 10% adb/PBS containing 0.2% DRIWEL, 10 min in 10% adb/PBS containing 0.05% NP-40, and two washes (10 min each) in 0.2% DRIWEL in distilled water. Slides were mounted in a drop of mounting solution [90% glycerol in PBS containing 1,4-diazabicyclo(2,2,2)octane (DABCO), p-phenylenediamine (PPD), and 4′,6-diamidino-2-phenylindole (DAPI)]. For comparison of the expressions of recombination-related proteins between mutants and the wild type, spermatocytes were isolated from 17–25-day-old wild-type and mutant rats. After mutant and wild-type control nuclei were co-immunostained under identical conditions, the numbers of foci detected per nucleus were counted. For co-immunostaining with anti-Spata22 and anti-Sycp3 antibodies and co-immunostaining with anti-Spata22 and antibodies to recombination nodule proteins, spermatocytes were isolated from 25–35-day-old wild-type male rats. For co-immunostaining with anti-Spata22 and anti-Rpa antibodies, 5–10 nuclei at each of the leptotene-, zygotene-, and pachytene-like stages were observed. Nuclei were staged based on anti-Sycp3 staining or by the nuclear distribution pattern and number of Spata22 foci when anti-Sycp3 staining was not performed.
Analysis of chromosome synapsis
For the analysis of chromosome synapsis, we isolated spermatocytes from each of the two 30-day-old and 6-month-old mutants as well as each two of their wild-type littermates. We immunostained preparations of spermatocyte nuclei with anti-Sycp3 rabbit antibody.
Classification of stages of spermatocytes
Stages in prophase I were classified based on the morphologies of axial elements (AEs) and/or lateral elements (LEs). The criteria used for the classification are as follows. In the early leptotene stage, AEs appeared mainly as dot-like foci. In the leptotene stage, AEs were observed as a number of short fragments. In the early zygotene stage, AEs had developed as thin elongated filamentous structures, and synapsed regions were not apparent or were observed mainly as short segments of synapsed LEs. In the mid-zygotene stage, AEs and LEs became thicker and shorter than in the early zygotene stage, and synapsed regions had extended. In the late zygotene stage, almost all AEs were synapsed extensively with their homologs, and AEs and LEs became thick and short. In the early pachytene stage, all chromosomes were synapsed; LEs were well condensed but longer than in the mid-pachytene stage. In the mid-pachytene stage, highly condensed LEs were observed. In the late pachytene stage, all chromosomes remained synapsed, but LEs became elongated and loose. In the diplotene stage, synapsed LEs began to separate, and LEs became looser.
Mutant cells that resembled the early leptotene, leptotene, and early zygotene wild-type cells were classified as early leptotene-, leptotene-, and early zygotene-like cells, respectively. Mutant cells that appeared to progress beyond the early zygotene-like stage were classified as mid-late zygotene-like cells. In these cells, AEs and LEs were condensed as seen in mid-zygotene, late zygotene, and pachytene cells of the wild-type. We should note the possibility that stages of mutant spermatocytes and wild-type controls do not correspond to each other in terms of cell-cycle-progression. For example, although the mutant cells resemble the early zygotene wild-type cells, they may have reached the pachytene stage in terms of cell-cycle-progression.
Imaging
Immunolabelled chromosome spreads were imaged using a cooled charge-coupled device (CCD) camera (MicroMAX 782Y, Princeton Instruments) mounted on a Leica DMRA microscope and were analysed with the 550CW-QFISH application program of Leica Microsystems Imaging Solution. For electron microscopy, we used a transmission electron microscope (JEM-1220, JEOL).
Statistical analysis
To compare levels of expression of Rpa and Rad51 between tm homozygous spermatocytes and the wild type, we counted the number of foci of these proteins per nucleus at each substage of prophase I, and then pooled data obtained from two tm homozygous mutant males (one at 17 days after birth and the other at 25 days), pooled data obtained from two of their wild-type male littermates (one at 17 days after birth and the other at 25 days), and used the Mann–Whitney U-test (two-tailed), which is a non-parametric test, for statistical analysis. To compare numbers of Rpa foci per nucleus between mutant mid-late-zygotene-like and wild-type mid-zygotene spermatocytes as well as between mutant mid-late-zygotene-like and wild-type late-zygotene spermatocytes, we adjusted significance levels for multiple comparisons by following the Ryan procedure.
Author Contributions
S.I. designed, performed, and analysed all of the experiments in this study; Y.M. provided technical expertise required for the chromosome analysis; S.I. and K.K. wrote the manuscript with comments from Y.M.; and K.K. conceived and designed the study.
Supplementary Material
Supplementary information
Acknowledgments
We thank T. Serikawa for providing essential materials, K. Yamada, T. Isobe, and T. Ijiri for instructions on the chromosome analysis, and Y. Deguchi for help with animal breeding. This work was partially supported by a grant from the Uehara Memorial Foundation.
References
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1–53 (2001). [DOI] [PubMed] [Google Scholar]
- San Filippo J., Sung P. & Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008). [DOI] [PubMed] [Google Scholar]
- Krejci L., Altmannova V., Spirek M. & Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795–5818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A. & Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet. Genome Res. 107, 201–207 (2004). [DOI] [PubMed] [Google Scholar]
- Baumann P., Benson F. E. & West S. C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87, 757–766 (1996). [DOI] [PubMed] [Google Scholar]
- Wold M. S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S. & Sehorn M. G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278, 42729–42732 (2003). [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Zaitseva E. M. & Kowalczykowski S. C. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 272, 7940–7945 (1997). [DOI] [PubMed] [Google Scholar]
- New J. H., Sugiyama T., Zaitseva E. & Kowalczykowski S. C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 (1998). [DOI] [PubMed] [Google Scholar]
- Thorslund T. & West S. C. BRCA2: a universal recombinase regulator. Oncogene 26, 7720–7730 (2007). [DOI] [PubMed] [Google Scholar]
- Holloman W. K. Unraveling the mechanism of BRCA2 in homologous recombination. Nat. Struct. Mol. Biol. 18, 748–754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. & Robberson D. L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82, 453–461 (1995). [DOI] [PubMed] [Google Scholar]
- Gasior S. L., Wong A. K., Kora Y., Shinohara A. & Bishop D. K. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 12, 2208–2221 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman W. K., Schirawski J. & Holliday R. The homologous recombination system of Ustilago maydis. Fungal Genet. Biol. 45, S31–S39 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. S., Winkelmann N., Petalcorin M. I. R., McIlwraith M. J. & Boulton S. J. RAD-51-Dependent and -Independent Roles of a Caenorhabditis elegans BRCA2-Related Protein during DNA Double-Strand Break Repair. Mol. Cell. Biol. 25, 3127–3139 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S. K. et al. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131, 131–142 (2004). [DOI] [PubMed] [Google Scholar]
- Roeder G. S. Meiotic chromosomes: it takes two to tango. Genes Dev. 11, 2600–2621 (1997). [DOI] [PubMed] [Google Scholar]
- Cohen P. E. & Pollard J. W. Regulation of meiotic recombination and prophase I progression in mammals. BioEssays 23, 996–1009 (2001). [DOI] [PubMed] [Google Scholar]
- Cohen P. E., Pollack S. E. & Pollard J. W. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr. Rev. 27, 398–426 (2006). [DOI] [PubMed] [Google Scholar]
- Moens P. B. et al. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA−DNA interactions without reciprocal recombination. J. Cell Sci. 115, 1611–1622 (2002). [DOI] [PubMed] [Google Scholar]
- Bannister L. A. & Schimenti J. C. Homologous recombinational repair proteins in mouse meiosis. Cytogenet. Genome Res. 107, 191–200 (2004). [DOI] [PubMed] [Google Scholar]
- Ishishita S., Inui T., Matsuda Y., Serikawa T. & Kitada K. Infertility Associated with Meiotic Failure in the tremor Rat (tm/tm) is Caused by the Deletion of Spermatogenesis associated 22. Exp. Anim. 62, 219–227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S. et al. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol. Reprod. 86, 45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquet B. et al. MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination. PLoS Genet 9, e1003784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M. et al. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat. Commun. 4, 2788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N. K. & Cohen P. E. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet. Genome Res. 107, 216–231 (2004). [DOI] [PubMed] [Google Scholar]
- Reynolds A. et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah S. K. et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271–276 (2001). [DOI] [PubMed] [Google Scholar]
- Bellani M. A., Romanienko P. J., Cairatti D. A. & Camerini-Otero R. D. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J. Cell. Sci. 118, 3233–3245 (2005). [DOI] [PubMed] [Google Scholar]
- Otto S. P. et al. About PAR: The distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 27, 358–367 (2011). [DOI] [PubMed] [Google Scholar]
- Plug A. W. et al. Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell Sci. 111, 413–423 (1998). [DOI] [PubMed] [Google Scholar]
- Yamada J. et al. Rats with congenital tremor and curled whiskers and hair. Jikken Dobutsu 34, 183–188 (1985). [DOI] [PubMed] [Google Scholar]
- Serikawa T., Ohno Y., Sasa M., Yamada J. & Takaori S. A new model of petit mal epilepsy: spontaneous spike and wave discharges in tremor rats. Lab. Anim. 21, 68–71 (1987). [DOI] [PubMed] [Google Scholar]
- Kitada K. et al. Accumulation of N-acetyl-L-aspartate in the brain of the tremor rat, a mutant exhibiting absence-like seizure and spongiform degeneration in the central nervous system. J. Neurochem. 74, 2512–2519 (2000). [DOI] [PubMed] [Google Scholar]
- Reinholdt L. G. & Schimenti J. C. Mei1 is epistatic to Dmc1 during mouse meiosis. Chromosoma 114, 127–134 (2005). [DOI] [PubMed] [Google Scholar]
- McMahill M. S., Sham C. W. & Bishop D. K. Synthesis-Dependent Strand Annealing in Meiosis. PLoS Biol 5, e299 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos S. J. & Heyer W. D. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim Biophys Acta 1809, 509–523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Holzschu D. L. & Sugiyama T. PCNA is efficiently loaded on the DNA recombination intermediate to modulate polymerase δ, η, and ζ activities. PNAS 110, 7672–7677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. H., Plug A. W., Van Vugt M. J. & De Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997). [DOI] [PubMed] [Google Scholar]
- Matsuda Y., Moens P. B. & Chapman V. M. Deficiency of X and Y chromosomal pairing at meiotic prophase in spermatocytes of sterile interspecific hybrids between laboratory mice (Mus domesticus) and Mus spretus. Chromosoma 101, 483–492 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information