Abstract
Background & objectives:
Infection with Salmonella enterica serovar Typhi (hereafter S. Typhi) is an important public health problem in India. There has been an increase in the number of reported clinical failures to ciprofloxacin treatment but the data on possible mechanism of failure are limited. One mechanism that has been widely reported and found associated with ciprofloxacin resistance, is the mutations in target genes in QRDR (quinolone resistance determining region). It is hypothesized that mutations in DNA gyrase or topoisomerase IV result in therapeutic failure under selective pressure of antibiotic while the patient is on treatment. We undertook in vitro sequential selection studies to expose the clinical isolates of S. Typhi to different concentration of ciprofloxacin to study the role of antibiotic selective pressure in the development of mutations in QRDR.
Methods:
Total 26 clinical isolates were divided in to two parts: part I included six isolates obtained from three patients with relapse of enteric fever and part II included 20 isolates with different ciprofloxacin MIC levels. For in vitro induction of mutation experiment, five S. Typhi isolates were selected which included three NAS (nalidixic acid sensitive) and 2 NAR (nalidixic acid resistant) S. Typhi. These isolates were grown under increasing concentrations of ciprofloxacin and mutations acquired in QRDR of DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) were investigated by sequencing.
Results:
For the isolates included in the part I of the study, it was found that the MIC to ciprofloxacin increased in the isolates obtained during the relapse of enteric fever as compare to the first isolate. All isolates had single mutation in gyrA gene at S83 without additional mutation in the second isolate. In the second part of the study, the nine isolates with varying MICs to ciprofloxacin also had single mutation in gyrA gene at S83 and another six had triple mutations, two mutations in gyrA gene (at S83 and D87) and one mutation in parC gene (at S80). In in vitro induction of mutation experiment, all mutated isolates showed triple mutation (two mutation in gyrA and one in parC gene) while no mutations were found in wild isolates.
Interpretation & conclusions:
Upon exposure to the step-wise increased concentration of ciprofloxacin, isolates become more tolerant to the ciprofloxacin and showed 2-4 fold higher MICs without new mutation after 8 μg/ml. So the accumulation of mutations under continuous ciprofloxacin pressure and tolerance of the mutant isolates led to the clinical failure. These results also suggested that there could be another mechanism responsible for resistance.
Keywords: fluoroquionolones, in vitro induction, Mutants, Quinolone resistance determining region, S. Typhi
The introduction of ciprofloxacin as the first line of treatment for enteric fever in the late 1980s followed by its wide use as the drug of choice led to the reports on decrease in the susceptibility of Salmonella enterica serovar Typhi (hereafter S. Typhi) to ciprofloxacin, with consequent therapeutic failure1,2,3,4,5,6. Two enzymes are the principal targets for the antibacterial activity of quinolones: DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE)7,8. One mechanism that has been widely reported and found associated with ciprofloxacin resistance, is mutations in target genes in QRDR9,10,11. However, these mutations are not able to impart in vitro resistance to ciprofloxacin.
With the initial isolates of S. Typhi from the blood culture of patient being susceptible to ciprofloxacin and still specific therapy resulting in clinical failure, it is hypothesized that mutations in DNA gyrase or topoisomerase IV result in therapeutic failure under selective pressure of antibiotic while the patient is still on treatment. This has been observed in animal isolates of Salmonella spp. by Giraud et al12 and in S. Typhi and S. Paratyphi A by Hirose et al13.
To study the role of antibiotic selective pressure in the development of mutations in QRDR (quinolone resistance determining region), we carried out in vitro sequential selection studies to expose the clinical isolates of S. Typhi from enteric fever patients with clinical failure to ciprofloxacin to different concentration of ciprofloxacin.
Material & Methods
Total 26 clinical isolates of S. Typhi recovered from children presenting with enteric fever and documented clinical failure to ciprofloxacin at the Paediatric Outpatients department, as also inpatients, All India Institute of Medical sciences (AIIMS), New Delhi, India, from 2005-2010 were included in the study. All the isolates were identified by standard biochemical tests and serotyped by using specific antisera (dH and O9) (Murex Diagnostics Ltd, UK), in the microbiology laboratory at AIIMS. The isolates were stored in glycerol at -70°c till the time of use. The study was divided in to 2 parts: - Part I and II.
Part I included characterization of six isolates, obtained from three patients with relapse of enteric fever who were culture positive both in first and second episode for S. Typhi (2 patients) and S. Paratyphi (1 patient). During the first episode, these patients were treated with ciprofloxacin for 10 days. No other antimicrobial was used initially because they all showed response to the treatment in terms of defervascence of fever in 5-6 days of initiating treatment.
However, these patients had recurrence of fever after 10-14 days and this second episode was defined as relapse because the fever reappeared after completion of first line treatment. All these patients were treated with intravenous ceftriaxone for 14 days during the second episode.
Part II of the study included 20 isolates, 3 NAS (nalidixic acid sensitive) and 17 NAR (nalidixic acid resistant) with different levels of MIC (minimum inhibitory concentration) to ciprofloxacin against S. Typhi.
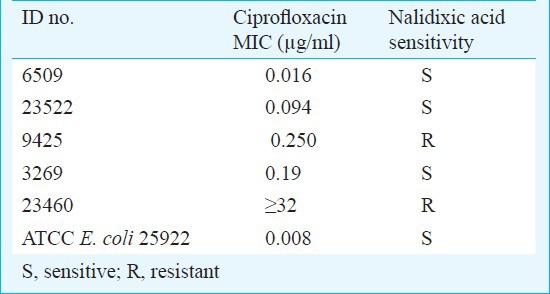
Susceptibility testing: Antimicrobial susceptibility was determined to chloramphenicol (30 μg), amoxicillin (10 μg), co-trimoxazole (1.25/23.75 μg), ceftriaxone (30 μg), cefexime (30 μg), nalidixic acid (30 μg) and ciprofloxacin (5 μg) (Hi-Media Labortories limited, Mumbai, India) by disk diffusion method according to the Clinical Laboratory Standards Institute (CLSI) guidelines, 201114. Escherichia coli ATCC 25922 was used as reference strains for quality control. MIC to ciprofloxacin was determined using the E-test (AB Bio disk, Solna, Sweden) following the manufacturer's instructions.
DNA isolation: DNA was isolated by boiling method as earlier described in our laboratory15. Briefly, DNA of each sample was isolated from one millilitre of overnight culture (OD600= 5) equivalent to approximately 109 bacterial cells. Culture was centrifuged at 14000 g for 5 min and DNA extraction yielded about 2 μg DNA (elution volume was 100 μl). So 100 ng of DNA (per 5μl) was used in the PCR as template.
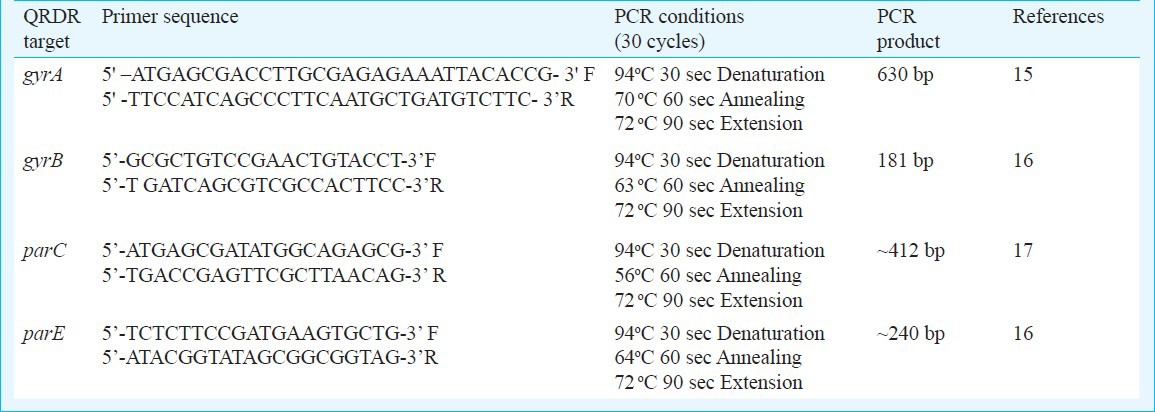
PCR amplification and DNA sequencing of the QRDR: PCR for QRDR of all subunits for DNA gyrase (gyrA and gyrB gene) and topoisomerase IV (parC and parE) was run using published primers15,16,17. DNA sequence analysis for mutation detection was done as described earlier15.
Briefly, PCR was performed on genomic DNA of each isolates in a total volume of 50 μl, which contained 5 μl of supernatant, 5 pmol of each primer, 200 μM deoxynucleoside triphosphates and 2.5 U of Taq polymerase. The details of primers and PCR reactions are given in Table I. Multiple DNA sequencing reactions were performed for each QRDR of individual isolates using the Applied Biosystems Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) for cycle sequencing reactions on ‘Gene Amp PCR system 2400’ thermal cycler (Applied Biosystems, USA) and ABI Prism 310 Genetic Analyzer (Perkin-Elmer, Applied Biosystems, USA). AmpliTaq Gold DNA polymerase (Applied Biosystems, USA) was used for PCR, which is a modified form of AmpliTaq DNA Polymerase.
Table I.
Details of primer and PCR conditions for the amplification of QRDRs
Sequence analysis: Reference sequences for all QRDRs (gyrA, gyrB, parC and parE) were obtained from http://www.ncbi.nlm.nih.gov/gene and compared with the sequence of S. Typhi strain CT18 (accession no. AL627274). Sequence chromatograph files were analyzed using BioEdit ver 5.0.918 to resolve nucleotide ambiguities. Global alignment of sequences was done using the software Clustal X ver 1.8 and Gene Doc version 2.1.0219.
In vitro induction of mutation
Of the 20 isolates included in the second part of the study, five (all the three NAS and two NAR) were selected based on their MIC to ciprofloxacin for the in vitro induction experiment12,13,20. These were designated as wild type (Table II). The isolates selected after ciprofloxacin exposure designated as mutants.
Table II.
Phenotypic characteristics of the five isolates selected for in vitro induction
Multistep induction and selection of resistant mutants: Wild isolates were grown in antibiotic-free brain heart infusion broth (Difco Laboratories, USA) at 37°C for 18 h12. To start with about 1×108 cfu/ml (0.5 McFarland standard) of each isolates (1 ml) was inoculated into 2 ml of cation-adjusted Muller Hinton broth (Difco Laboratories, USA) containing ciprofloxacin concentration equal to MIC of corresponding wild isolates. The ciprofloxacin concentrations used for mutant induction ranged from 2- to 4-fold higher the MIC for the wild isolates or sub-wild mutant isolates resulting from the prior induction step13,20. After an overnight incubation at 37°C, cells growing in the tubes with the highest ciprofloxacin exposure concentration were selected for mutants with increased MICs by plating onto ciprofloxacin containing cation-adjusted Muller Hinton agar plates at concentrations of 0, 2, 4, 8 and 16 mg/l, or higher when necessary. After each two subcultures the organism was confirmed by MIC and serotyping. MICs were determined in cation-adjusted Muller Hinton agar medium without drugs. Three colonies were randomly selected for MIC determination, and isolates with the highest MIC were subjected to sequence analysis and further induction. This was repeated when a higher exposure concentration was used for the next step of the induction/selection cycle until mutants with significantly high MICs were selected. When MICs of mutant isolates increased up to four-fold higher than wild type, and mutant isolate stop growing in a particular concentration of ciprofloxacin, it was considered as “significantly high MICs”. The stability of the selected resistant mutants was confirmed by sub-culture onto antibiotic-free sheep blood agar plates for 10 serial passages.
Susceptibility testing, MIC determination, DNA isolation, PCR amplification and sequencing of QRDR region were done as described above.
Results
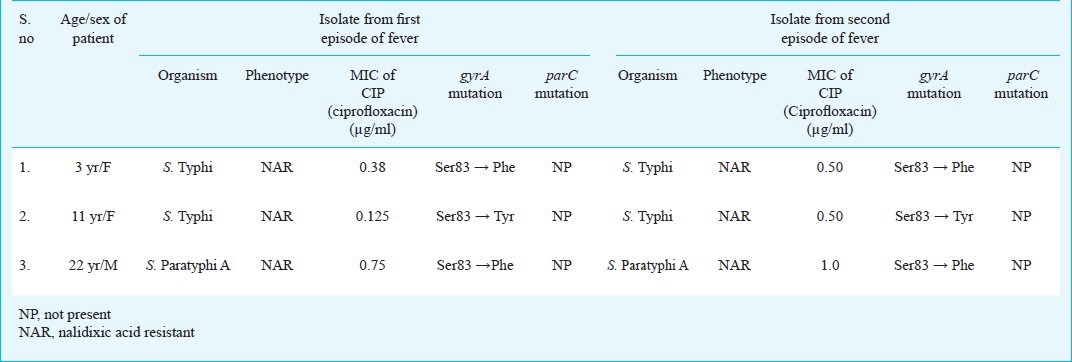
The results for part 1 of the study are shown in Table III. In S. Typhi MIC was increased from 0.38 and 0.125 to 0.50 in both the isolates, respectively. These isolates showed mutation at S83 to F/Y (serine to phenylalanine, or tyrosine), respectively in gyrA gene. The MIC in S. Paratyphi A was increased from 0.75 to 1 in subsequent isolate and the mutation was at S83 to F (serine to phenylalanine) in gyrA gene only and no mutations were found in gyrB, parC and parE genes in all the isolates.
Table III.
Phenotypic and genotypic characteristics of bacteria isolated from the patients with relapse of enteric fever (Part I)
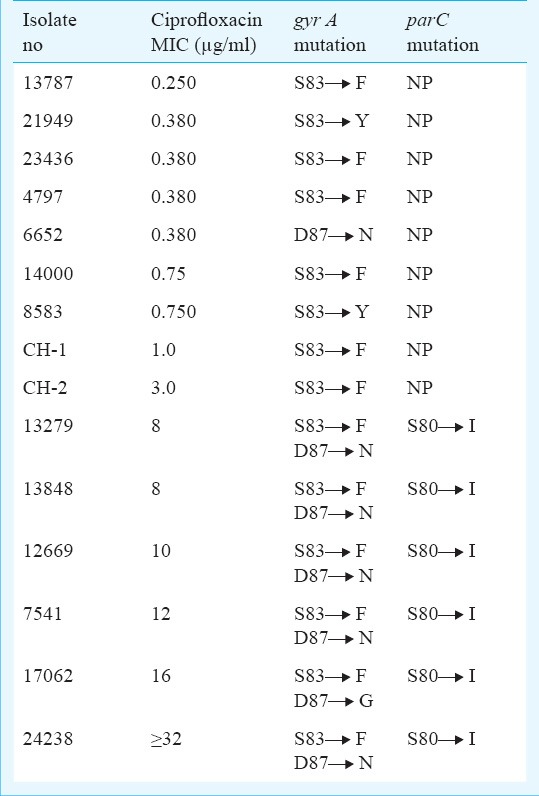
In part II of the study, nine isolates had single mutation in gyrA gene at S83 to F/Y while six had triple mutation (double mutation in gyrA gene at S83 to F/Y and D87 to N with single mutation in parC gene at S80 to I) (Figure). All these clinical isolates with triple mutations were ciprofloxacin resistant and showed higher MIC (>4 μg/ml) (Table IV). S83 to F/Y was the most common mutation except in one isolate where only single mutation was detected at D87 to N.
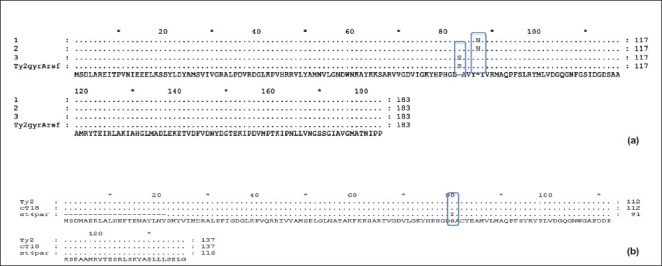
Fig. a. Double mutation in gyrA gene at S83 and D87. Fig. b. Mutation in parC gene at S80. Fig. a and b show mutations, found in a clinical isolate of S. Typhi.
Table IV.
Phenotypic and genotypic characteristics of isolates included in Part II study
In the in vitro induction of mutation experiment, MIC of wild and mutant isolates is shown in Table V. With each set of wild/mutant, the MIC of ciprofloxacin for mutants was increased following induction/selection cycles. All mutants reached MICs of ciprofloxacin >32 μg/l after six to eight cycles of induction/selection except in one strain that is reached up to 16 μg/l.
Table V.
Phenotypic and genotypic characteristics of wild and mutated isolates of S. Typhi obtained after selection by ciprofloxacin
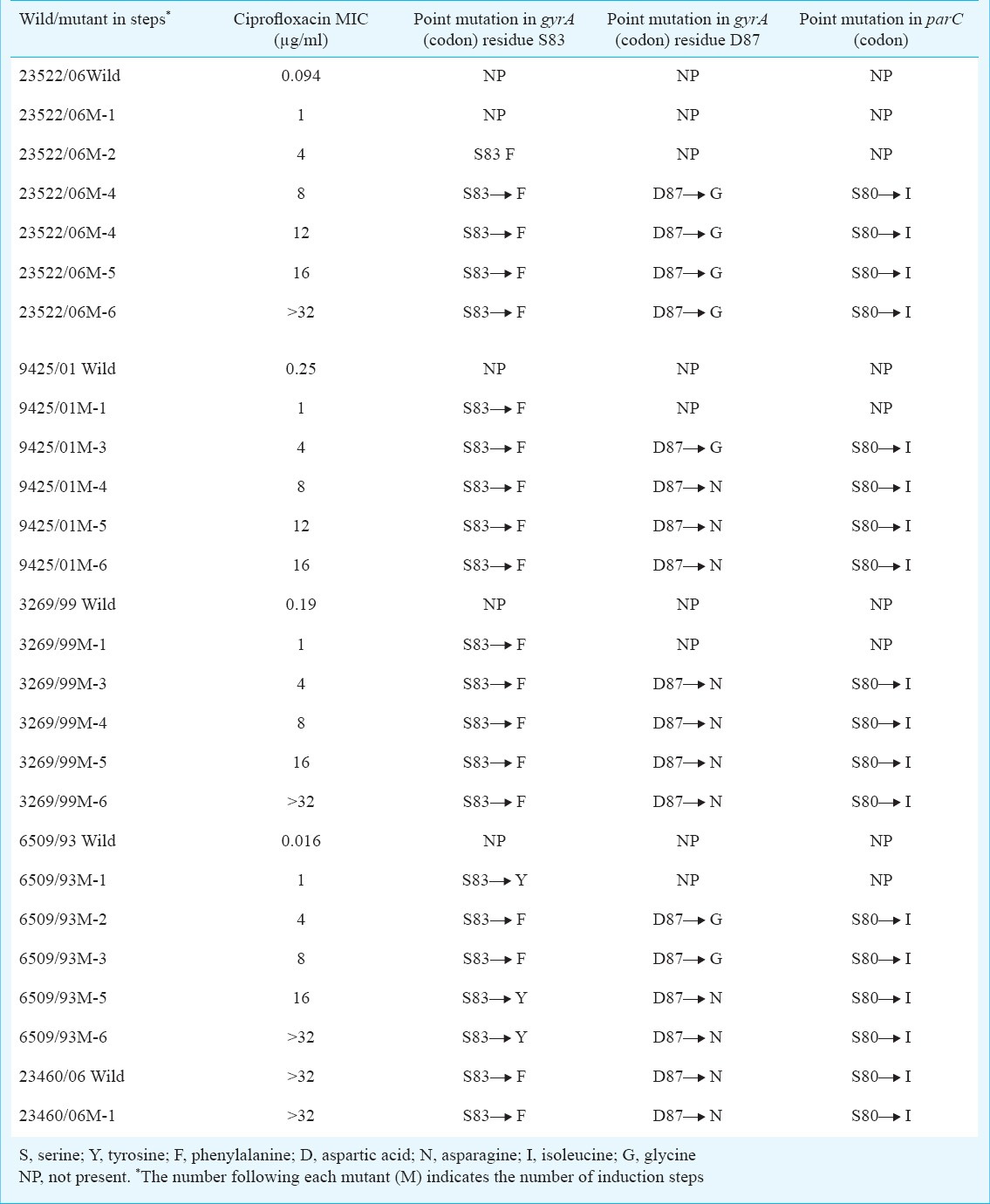
The isolates with single gyrA mutation were less resistant to fluoroquinolones than those with additional parC mutations. Bacterial isolates acquired mutations (triple mutations) thus exhibiting MICs (> 4μg/ml) except in one isolate. No mutations were detected in the gyrB and parE in our study.
Discussion
Enteric fever remains to be an important public health problem despite the availability of vaccine and effective antibiotics particularly in endemic countries and this has been compounded by the global emergence of multidrug resistant S. Typhi leading to treatment failures1,2,3,4,5,6. The data on the status of Salmonella strains in a patient on documented treatment failure are limited because the sensitivity of blood culture is very poor in a patient already on ciprofloxacin treatment, although there is no clinical response. Subsequent treatment with ceftriaxone or cefexime helps in curing the patient.
The isolates from the three patients who relapsed also showed an increased MIC to ciprofloxacin during the relapse. It is possible that the MIC increases while the patient is on treatment with ciprofloxacin, which results in clinical non-response. It may be hypothesized that during the first stages of antibiotic treatment, a subpopulation of S. Typhi with reduced susceptibility to ciprofloxacin may be able to survive, replicate and acquire substitutions conferring resistance to higher concentrations of drug, while the antimicrobial level is still inhibitory to the remaining population. In in vitro induction of mutation experiment, isolates acquired an increased MIC to ciprofloxacin under sequential increase of ciprofloxacin concentration. Further, single point mutation in gyrA gene from S83 to F/Y was associated with decreased susceptibility to ciprofloxacin, while accumulation of mutations in gyrA gene (mainly at S83 to F/Y and D87 to G/N/Y) and parC gene (at S80 to I) led to the increased MIC to ciprofloxacin and complete resistance. We found triple mutations in all isolates after 2-3 initial induction/selection cycles in gyrA and parC gene which resulted in increased MIC. While Hirose et al13 found that the isolates with high-level resistance to fluoroquinolones (MIC range, 4 to >32 μg/ml) induced by in vitro selection with ciprofloxacin had double mutation at both Ser83 and Asp87 in the gyrA gene, they did not report any mutation in parC gene. Another important factor noticeable in the mutation experiment was that there was 2-4 fold rise of MIC of the mutant isolates on sequential exposure to ciprofloxacin without any new mutation after 8 μg/ml. Thus, these mutated isolates become more tolerant to ciprofloxacin on exposure and could be another important factor for clinical failure. As the increase in MIC to ciprofloxacin did not directly lead to increase in mutations, there could be other mechanisms responsible for raised MICs such as efflux pumps associated with multi-antibiotic resistance (outer membrane proteins, MAR locus), up/downregulation of operon genes. Plasmid which are responsible for quinolone resistance are qnr (qnr A, qnr B), aminoglycoside acetyltransferase (AAC)13,20,21. The mixture of mutations in target enzymes and efflux pump perhaps builds up high-level resistance in enteric fever22.
Of the 20 isolates included in the second part of the study, triple mutations were found in six isolates, which means there is accumulation of mutations and clonal spread of highly resistant S. Typhi clone. This also has been reported by Gaind et al23 from India. Upon exposure to the selective pressure of the ciprofloxacin, the increase in the level of resistance could be due to an increase in the rate of mutations or clonal expansion of mutant with single mutation on to which the second mutation can add up24,25. This step-wise evolution is consistent with the conclusions that the acquisition of mutations during treatment is one of the most important factors contributing to therapy failure26.
There is no new antibiotic molecule developed for GNB (Gram-negative bacteria) in the recent years27, which again emphasizes the need for strict and rational use of ciprofloxacin in human and veterinary therapeutics. The judicious use by optimal dosage intervals taking Pharmacodynamics and pharmacokinetics parameters that prevent emergence of pre-existing or newly formed mutants should now be guiding the treatment in such infections28. Thus, the molecular analysis of fluoroquinolone resistance with continuous antimicrobial surveillance is warranted in future.
Acknowledgment
This work was partially financially supported by the Indian Council of Medical Research (ICMR), New Delhi.
References
- 1.Capoor MR, Nair D. Quinolone and cephalosporin resistance in enteric fever. J Glob Infect Dis. 2010;2:258–62. doi: 10.4103/0974-777X.68529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadhiravan T, Wig N, Kapil A, Kabra SK, Renuka K, Misra A. Clinical outcomes in typhoid fever: adverse impact of infection with nalidixic acid-resistant Salmonella Typhi. BMC Infect Dis. 2005;5:37. doi: 10.1186/1471-2334-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapil A, Ayyagari A, Garg RK, Agarwal KC. S. typhi with transferable chloramphenicol resistance isolated in Chandigarh during 1983-87. Indian J Pathol Microbiol. 1994;37:179–83. [PubMed] [Google Scholar]
- 4.Kapil A, Sood S, Dash NR, Das BK, Seth P. Ciprofloxacin in typhoid fever. Lancet. 1999;354:164. doi: 10.1016/s0140-6736(05)75300-9. [DOI] [PubMed] [Google Scholar]
- 5.Capoor MR, Nair D, Hasan AS, Aggarwal P, Gupta B. Typhoid fever: narrowing therapeutic options in India. Southeast Asian J Trop Med Public Health. 2006;37:1170–4. [PubMed] [Google Scholar]
- 6.Saha SK, Darmstadt GL, Baqui AH, Crook DW, Islam MN, Islam M, et al. Molecular basis of resistance displayed by highly ciprofloxacin-resistant Salmonella enterica serovar Typhi in Bangladesh. J Clin Microbiol. 2006;44:3811–3. doi: 10.1128/JCM.01197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khodursky AB, Cozzarelli NR. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J Biol Chem. 1998;273:27668–77. doi: 10.1074/jbc.273.42.27668. [DOI] [PubMed] [Google Scholar]
- 8.Hooper DC. Quinolone mode of action. Drugs. 1995;49(Suppl 2):10–15. doi: 10.2165/00003495-199500492-00004. [DOI] [PubMed] [Google Scholar]
- 9.Maxwell A. DNA gyrase as a drug target. Trends Microbiol. 1997;5:102–9. doi: 10.1016/S0966-842X(96)10085-8. [DOI] [PubMed] [Google Scholar]
- 10.Hirose K, Tamura K, Watanabe H. Screening method for Salmonella enterica serovar Typhi and serovar Paratyphi A with reduced susceptibility to fluoroquinolones by PCR-restriction fragment length polymorphism. Microbiol Immunol. 2003;47:161–5. doi: 10.1111/j.1348-0421.2003.tb02800.x. [DOI] [PubMed] [Google Scholar]
- 11.Renuka K, Sood S, Das BK, Kapil A. High-level ciprofloxacin resistance in Salmonella enterica serovar Typhi in India. J Med Microbiol. 2005;54:999–1000. doi: 10.1099/jmm.0.45966-0. [DOI] [PubMed] [Google Scholar]
- 12.Giraud E, Brisabois A, Martel JL, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in vitro and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob Agents Chemother. 1999;43:2131–7. doi: 10.1128/aac.43.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirose K, Hashimoto A, Tamura K, Kawamura Y, Ezaki T, Sagara H, et al. DNA sequence analysis of DNA gyrase and DNA topoisomerase IV quinolone resistance-determining regions of Salmonella enterica serovar Typhi and serovar Paratyphi A. Antimicrob Agents Chemother. 2002;46:3249–52. doi: 10.1128/AAC.46.10.3249-3252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute (CLSI) Wayne, PA, USA: CLSI; 2011. Performance standards for antimicrobial susceptibility testing; twenty first informational supplement. CLSI document M100-S21. [Google Scholar]
- 15.Renuka K, Kapil A, Kabra SK, Wig N, Das BK, Prasad VV, et al. Reduced susceptibility to ciprofloxacin and gyra gene mutation in North Indian strains of Salmonella enterica serotype Typhi and serotype Paratyphi A. Microb Drug Resist. 2004;10:146–53. doi: 10.1089/1076629041310028. [DOI] [PubMed] [Google Scholar]
- 16.Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White PA, et al. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob Agents Chemother. 2004;48:4012–5. doi: 10.1128/AAC.48.10.4012-4015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling JM, Chan EW, Lam AW, Cheng AF. Mutations in topoisomerase genes of fluoroquinolone-resistant salmonellae in Hong Kong. Antimicrob Agents Chemother. 2003;47:3567–73. doi: 10.1128/AAC.47.11.3567-3573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 19.Nicholas KB, Nicholas HB, Jr, Deerfield DW., II GeneDoc: analysis and Visualization of Genetic Variation. 1997. [accessed on November 16, 2011]. Available from: http://www.psc.edu/biomed/genedoc .
- 20.Levy DD, Sharma B, Cebula TA. Single nucleotide polymorphism mutation spectra and resistance to quinolones in Salmonella enterica serovar enteritidis with a mutator phenotype. Antimicrob Agents Chemother. 2004;48:2355–63. doi: 10.1128/AAC.48.7.2355-2363.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair S, Unnikrishnan M, Turner K, Parija SC, Churcher C, Wain J, et al. Molecular analysis of fluoroquinolone-resistant Salmonella Paratyphi A isolate, India. Emerg Infect Dis. 2006;12:489–91. doi: 10.3201/eid1203.050560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komp Lindgren P, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother. 2003;47:3222–32. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaind R, Paglietti B, Murgia M, Dawar R, Uzzau S, Cappuccinelli P, et al. Molecular characterization of ciprofloxacin- resistant Salmonella enterica serovar Typhi and Paratyphi A causing enteric fever in India. J Antimicrob Chemother. 2006;58:1139–44. doi: 10.1093/jac/dkl391. [DOI] [PubMed] [Google Scholar]
- 24.Blazquez J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin Infect Dis. 2003;37:1201–9. doi: 10.1086/378810. [DOI] [PubMed] [Google Scholar]
- 25.Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fish DN, Piscitelli SC, Danziger LH. Development of resistance during antimicrobial therapy: a review of antibiotic classes and patient characteristics in 173 studies. Pharmacotherapy. 1995;15:279–91. [PubMed] [Google Scholar]
- 27.Bax RP. Antibiotic resistance: a view from the pharmaceutical industry. Clin Infect Dis. 1997;24(Suppl 1):151–3. doi: 10.1093/clinids/24.supplement_1.s151. [DOI] [PubMed] [Google Scholar]
- 28.Thomas JK, Forrest A, Bhavnani SM, Hyatt JM, Cheng A, Ballow CH, et al. Pharmacodynamic evaluation of factors associated with the development of bacterial resistance in acutely ill patients during therapy. Antimicrob Agents Chemother. 1998;42:52–7. doi: 10.1128/aac.42.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]


