Abstract
The three-dimensional structure of bacteriophage ϕXl74 (ϕX174) was determined to ~2.6 nm resolution from images of frozen-hydrated 114 S particles. The outer surface of ϕX174 is characterized by several prominent features: (i) 12 mushroom-shaped caps (~7.1 nm wide × 3.8 nm high) are situated at each of the vertices of the icosahedral virion and extend to a maximum radius of 16.8 nm; (ii) a “collar” of density surrounds the base of each apical cap; and (iii) 20 conical protrusions (~2.3 nm high) lie along the three-fold symmetry axes. The caps have a pentagonal morphology composed of five globular “subunits” and appear to be loosely connected to the underlying capsid. The distribution of the four gene products present in virions (60 copies each of gpF, gpG, and gpJ, and 12 copies of gpH), and the single-stranded DNA (ssDNA) genome cannot be directly discerned in the reconstructed density map, although plausible assignments can be made on the basis of solvent-excluded volume estimates and previous biochemical data. Thus, gpG accounts for most of the mass in the caps; gpH, a presumed cap protein, cannot be identified in part due to the symmetry-averaging procedures, but may be partially located within the interior of the capsid; and gpF and gpJ make up the remainder of the capsid. The genome appears to be less densely packaged inside the capsid compared to many dsDNA viruses whose nucleic acid is arranged in a liquid-crystalline state.
INTRODUCTION
Bacteriophage ϕX174 (ϕX174) is a small icosahedral virus that infects Escherichia coli. This relatively simple virus contains one copy of a closed, circular, single-stranded DNA (ssDNA) molecule encapsidated within a protein coat. Interest in the structure of ϕX174 extends over a period of at least 30 years beginning with Sinsheimer's (1959a) demonstration that the phage DNA was “unusual” because it reacted with formaldehyde, thus indicating that it probably did not adopt the classical Watson-Crick, hydrogen-bonded, double-helical structure. The DNA was later shown to be single-stranded (Sinsheimer, 1959b).
Sinsheimer (1959a) and Hall et al. (1959) were the first to record electron micrographs of ϕX174. Hall et al. (1959) recorded images of purified ϕX174 samples prepared both by routine metal-shadowing methods and by the relatively new (at that time) negative-stain technique (Brenner and Horne, 1959). The virus appeared to be polyhedral, ~24 nm in diameter, and the platinum-shadowed particles often showed a central “knob” or “spike.” On the basis of these images, ϕX174 was proposed to be icosahedral and composed of 12 knobs regularly arranged on the faces of a regular dodecahedron (Hall et al., 1959). Tromans and Horne (1961) subsequently confirmed this interpretation and suggested that ϕX174 was composed of 12 morphological subunits with “rod-shaped projections” at the icosahedral vertices. Maclean and Hall (1962) proposed that the ssDNA adheres to the morphological units of the coat and is packaged as a ball-like structure with .protein “knobs” partially covering the surface of the virion. They also recognized that each of the knobs must have five-fold symmetry if the virion coat is icosahedral. Edgell et al. (1969) later showed that urea treatment removes the spikes from virions and produces nearly spherical particles.
Among the most detailed images of ϕX174 published to date were those recorded by Yazaki (1981) who utilized an ingenious stain and rotary-shadow preparation procedure. Phage prepared in this way clearly exhibited 12 “pentagonal, frustum-shaped spikes” that were ~6 nm high with a base of ~9 nm diameter. Virions bound to a support film and treated with 0.1 M Ca2 + were found to extrude their DNA, apparently through the apex of one or more of the spikes, suggesting that all spikes are equivalent and therefore any are capable of extruding the genome into cells (Yazaki, 1981).
The entire DNA genome of ϕX174 was also the first to be completely sequenced (Sanger et al., 1977, 1978; Air et al., 1978). The ssDNA contains 5386 bases and encodes 10 genes which produce 11 gene products (for review see Hayashi et al., 1988). Virions contain 60 copies of the major capsid protein, gene product F (gpF, Mr 48 440); 60 copies of an internal protein, gpJ (Mr 4220), which is required for DNA packaging; 60 copies of the major spike protein, gpG (Mr 19 020); and 12 copies of gpH (Mr 34 370), which is believed to be a minor spike protein. Each of the 12 spikes at the icosahedral vertices appear to consist of five copies of gpG and a single copy of gpH, although there is no direct evidence that gpH is actually distributed in this manner. Both spike proteins are thought to function in host receptor recognition and binding, and gpH (also called the “pilot” protein) is injected into the host along with the viral DNA during the eclipse process (for reviews see Incardona 1978; Hayashi et al., 1988).
We have examined the unstained, frozen-hydra-ted structure of ϕX174 at ~2.6 nm resolution using cryoelectron microscopy and three-dimensional image reconstruction techniques. The shapes of the capsid shell and the prominent spikes are clearly revealed. The spikes, which actually have a flattened, pentagonal mushroom shape, could more accurately be referred to as apical caps. The association of these caps with the underlying capsid appears to be rather tenuous. Our results agree remarkably well with a recent three-dimensional electron density map that has been calculated from X-ray crystallographic data of partially empty (devoid of DNA) ϕX174 particles (McKenna et al., 1992). The close correspondence between features of the virion surface depicted in the two density maps emphasizes the quality and reliability of the reconstruction presented here.
MATERIALS AND METHODS
Infection of E. coli and purification of ϕX174 were carried out by established procedures (Willingmann et al., 1990L The final two steps of purification involved successive sucrose density gradient centrifugations which produced clearly separated fast (114S) and slow (70S) bands. The A260/280 uv ratio indicated that the faster, infectious band had a higher DNA content than the slower, noninfectious band; consequently, samples from the fast band were utilized for cryoelectron microscopy.
Samples were quickly frozen in a thin layer of vitreous ice without added stains or fixatives according to established procedures (Dubochet et al., 1988; Olson et al., 1990). Images were recorded under minimal dose conditions (~ 1500–2000 e−/nm2) and at a nominal magnification of × 49 000 and ~1.3 μm under-focus on a Philips EM420 electron microscope (Philips Electronics Instruments, Mahwah, NJ) equipped with a Gatan anticontaminator and a Gatan 626 cryotransfer holder (Gatan Inc., Warren-dale, PA). Polyoma virus was mixed with the sample and used as an internal magnification standard (Olson and Baker, 1989; Belnap et al., manuscript in preparation).
Selection of the micrograph to be processed, digitization, and image analysis procedures were performed essentially as described (Olson et al., 1990). Of 57 particle images that were boxed out of the selected micrograph, a final data set of 25 images was chosen to refine particle orientations and origins and to compute a three-dimensional reconstruction to a resolution limit of 2.1 nm. The absolute hand of the density map was established by comparison with the X-ray crystallographic structure (McKenna et al., 1992). Self-consistency of the 25 images with respect to handedness was established by use of cross-common lines procedures (Fuller, 1987). We assessed the reliable resolution of the reconstruction by splitting the data set into two nearly equal groups, separately refining the particle images, and calculating independent reconstructions and structure factor data sets (Baker et al., 1990; Yeager et al., 1990). From these data sets we calculated a reliability index, RAB-factor (Baker et al., 1991; Winkelmann et al., 1991), which compares the amplitudes and phases of common structure factors of the two data sets. The data showed excellent agreement at spatial frequencies <(1/2.6) nm−1 RAB ~0.5) but indicated progressively poorer correlation at higher spatial frequencies.
Computations were performed on a VAX/VMS 8550 computer (Digital Equipment Corp., Maynard, MAl with FORTRAN programs (Fuller, 1987; Baker et al., 1988, 1989). Various images and the shaded-surface and projected spherical-shell representations were displayed on a raster graphics device (Model 3400; Lexidata Corp., Billerica, MA) and photographed with a graphics recorder (Model 3000; Matrix Instruments, Orangeburg, NY) as previously described (Baker et al., 1991). Shaded-surface representations of the reconstruction were computed at a threshold density level chosen according to criteria described by Yeager et al. (1990). This produced a molecular envelope which closely matched the structure determined by crystallography (McKenna et al., 1992).
RESULTS
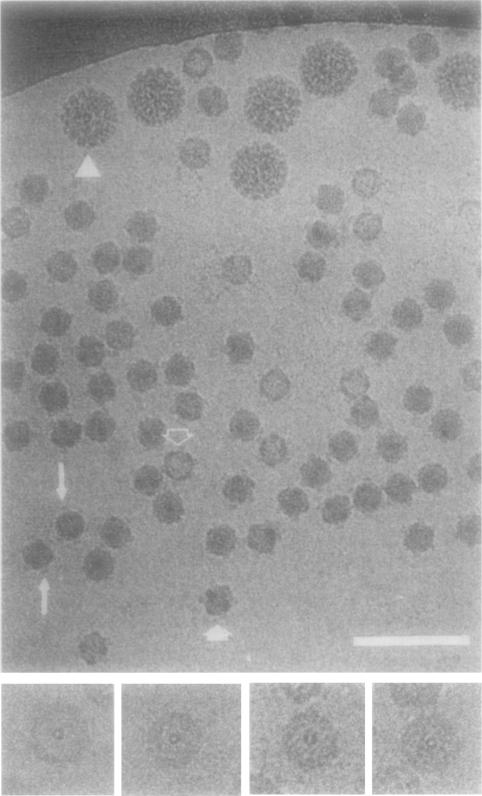
The capsid portion3 of frozen-hydrated ϕX174 virions appears to have a relatively featureless, spherical outer surface (Fig. 1). Faint spikes project radially from the virion surface and these distally flattened structures often appear to float as if disconnected from the capsid surface (Fig. 1, long arrows). This apparent anomaly is evidently a consequence of phase-contrast imaging, which results in strong, defocus-dependent Fresnel fringes that outline high density features in contact with the lower density solvent. Virus images from micrographs recorded much closer to focus do not exhibit this anomaly. Such micrographs, however, were not used because they exhibited very low contrast and consequently would have been more difficult to process. Particles oriented in a two-fold view display a six-sided, star-shaped profile (Fig. 1, short, filled arrow; for comparision see Fig. 2, left panel). In these images 4 of the 12 spikes are viewed edge-on and this produces the characteristic ϕX174 shape. Some particles appear less dense in the center, presumably because they have lost some or all of their DNA component (Fig. 1, open arrow). The wall of the capsid shell is clearly visible in these particles and, when aligned near a five-fold orientation, a small ring appears in the center presumably because two spikes superimpose in the projected view (Fig. 1, insets).
Fig. 1.
Frozen-hydrated ϕX174 and polyoma virus (triangle). Apical caps on some particles appear to float as if disconnected from the capsid surface (long arrows) presumably because of defocus-induced Fresnel fringes around the periphery of the particles. Some particles are viewed close to an icosahedral two-fold axis (filled arrow) or have lost most of the ssDNA component (unfilled arrow). Insets at bottom show “empty” particles viewed close to an icosahedral five-fold axis where apical caps on opposite sides (front and back) of the particle nearly superimpose and appear as a ring in the center of the particle. Bar, 100 nm (main panel) and 50 nm (bottom insets).
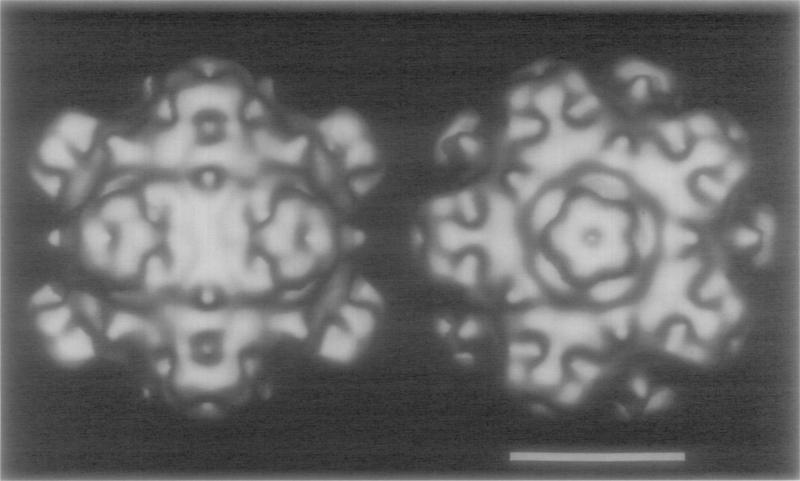
Fig. 2.
Shaded-surface representations of the ϕX17 4 reconstruction viewed along a two-fold axis (left) and along a five-fold axis (right). Bar, 15 nm.
Despite a relative lack of observable detail in the noisy images of frozen-hydrated <flX174 virions recorded with minimal irradiation conditions, the three-dimensional reconstruction that was computed from 25 independent virion images clearly reveals a detailed and highly sculpted virion surface (Fig. 2). The most prominent features of the particle are the 12 spikes at the icosahedral vertices. Despite a strong historical bias for use of the term spike to identify these features, the reconstruction presented here clearly indicates that they are not spike-like (i.e., “sharp, pointed projection[s)”; American Heritage Dictionary, 1985) and are more appropriately termed “apical caps.” A raised collar of density surrounds the base of each cap (Fig. 2, right panel) and small but prominent protrusions of density are situated at the icosahedral three-fold axes.
The apical caps are pentagonal, star-shaped structures (7.1 nm wide × 3.8 nm high) with a somewhat flattened distal surface. An axial dimple extends inward from the surface ~1.0 nm along the five-fold axis. The outlines of five globular morphological subunits are just discernable at the resolution of the reconstruction and the rounded tips of each of the subunits make up the blunt points of the star-shaped cap.
A raised, five-fold-modulated collar of density, of mean radius 4.6 nm, encircles each of the apical caps at its base. Density within the collar extends outward from the capsid surface and contacts the five points of the cap. The lowest regions of the collar coincide with the lobe regions between the cap points. This creates deep cavities outside and below the caps. The surface topology drops to lower radius just outside the collar and reaches its lowest point (12.4 nm) in a “valley” at the two-fold axis, but then raises up at the three-fold axis (14. 7 nm) in the form of a small, conical (0.9 nm diameter) protrusion at its base.
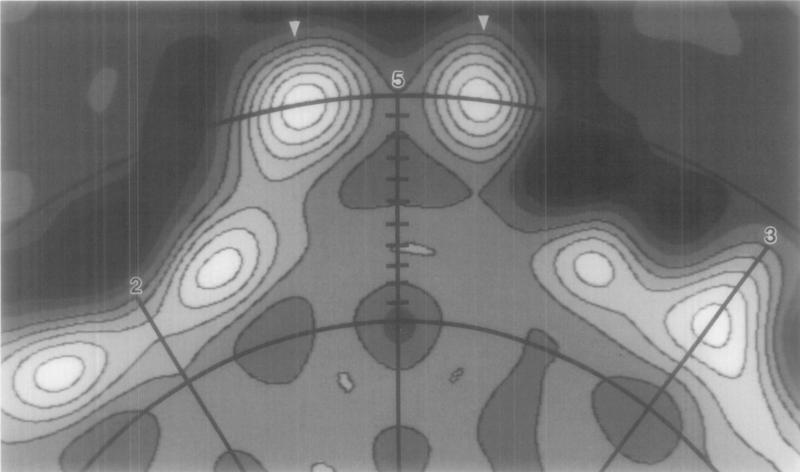
An equatorial section of the reconstruction helps clarify the relation of the apical cap to the underlying capsid (Fig. 3). The highest radius feature ofthe virion occurs at 16.8 nm at the distal portion of the cap ~8° off the five-fold axis (Fig. 3, arrowheads). Each cap appears to have a rather tenuous connection with the capsid because the density in the constricted region at the base of the cap is much lower than the other prominent features in the capsid. A “channel” of low density occurs along the axis of the cap, coincident with the icosahedral five-fold axis. This funnel-shaped channel is constricted at ~ 15.2 nm radius but then opens at lower radii to form a hollow cavity under the cap. Regions of high density occur along the three-fold axis (between radii of 10.5 and 14.7 nm) and on either side of the two-fold axis (the collar).
Fig. 3.
Upper portion of an equatorial slice of the reconstruction viewed along a two-fold axis. Arcs (at 10.0 and 15.5 nm radius) and tick marks (at 0.5-nm intervals) identify the radii corresponding to the spherical density projections in Fig. 4. Radial lines identify the positions of two-fold (2), three-fold (3), and five-fold (5) axes in the plane. Density at the largest radius in ϕX174 (16.8 nm) is indicated (arrowheads).
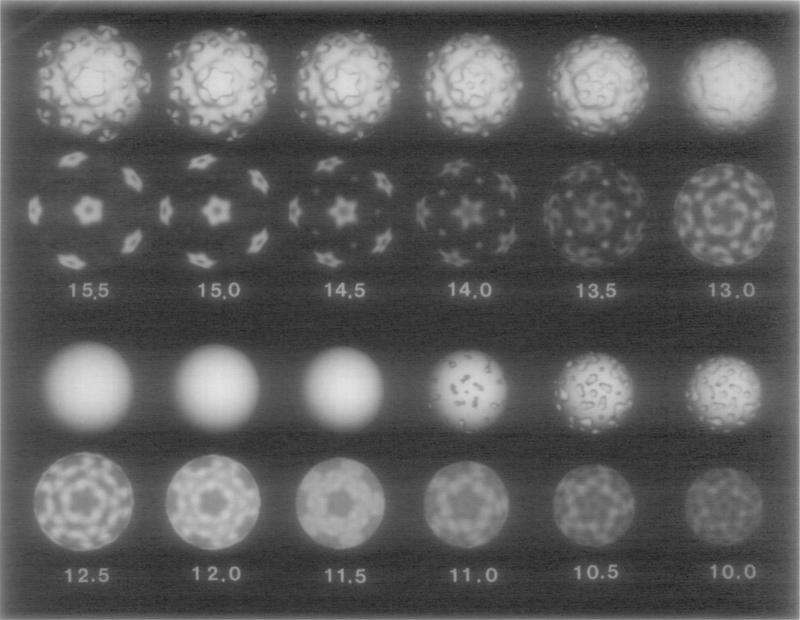
The distribution of density in <flX174 is visualized in a series of shaded-surface representations and concentric, spherical shells of decreasing radii (Fig. 4; radial positions of the shells correspond to those shown in Fig. 3). Individual morphological subunits that make up the rounded tips of the star-shaped cap and the axial depression are apparent at a radius of 15.5 nm (Fig. 4). Between radii of 16.0 and 14.0 nm the orientation of the main density features in the caps remains unchanged, indicating that the sub-units within the cap are mainly aligned in a radial direction with respect to the capsid sphere. Weak density that is present between the points of the cap at high radii (> 15 nm) is not present at lower radii. This gives the cap its mushroom shape. At radii between 14.0 and 12.0 nm the high density in the cap broadens tangentially away from the five-fold axis, first forming a right-handed “pinwheel” feature at 13.5 nm radius, and eventually forming the annular collar at 12.0–12.5 nm radius. The pinwheel feature is the highest radius density with a clearly defined handedness. We determined the absolute hand of the structure by comparing the image reconstruction to the high-resolution, X-ray crystallographic map of ϕX174 particles (McKenna et al., 1992).
Fig. 4.
Shaded-surface representations (first and third rows) and density projections (second and fourth rows) of the reconstruction truncated at successively lower radii (indicated in nanometers). The choice of a single threshold level for the surface representations at r = 12.5, 12.0, and 11.5 nm results in smooth spheres and gives a false impression of contiguous uniform density. The density projections at these radii give a more realistic rendering of the density distributions within the reconstruction.
The axial channel in the cap, which is narrowest at a radius of ~ 15.2 nm, opens out into a pentagonal cavity beneath the cap. At radii lower than 11.5 nm, density from the collar is directed toward the twofold axes. At ~ 10.0 nm radius the density becomes more diffuse. This radius corresponds approximately to the position of the inner surface of the protein shell (confirmed by preliminary analysis of empty ϕX174 particles; N. H. Olson, unpublished data). Densities at radii lower than 10.0 nm do not show features correlated with the icosahedral symmetry of the capsid and they are mainly expected to represent the DNA genome. Examination of near-focus, high-magnification (~60 000 ×) images of ϕX174 virions (data not shown) gives no indication of ordered or liquid-crystalline packing of the DNA as has been observed for double-stranded DNA (dsDNA) in other phages like T4 (Lepault and Leonard, 1985; Lepault et al., 1987) and λ(Lepault et al., 1987) and in the mammalian herpes simplex virus (Booy et al., 1991).
DISCUSSION
The three-dimensional reconstruction of ϕX174 clearly establishes the icosahedral structure of the bacteriophage. The atomic structure of slow band (70 S) particles ofϕX174 has recently been solved by X-ray crystallographic methods (McKenna et al., 1992). The size, shape, and location of all major surface features (apical cap with an axial dimple; tenuous contact of the cap with the capsid; the collar; the depression at the two-fold axis; and the protrusion at the three-fold axis) agree remarkably well in the two independently determined structures. Despite the limited resolution of the reconstruction (~2.6 nm), small features such as the protrusions at the three-fold axes (0.9 nm at the base and 2.3 nm high) are reliably represented in the reconstructed density map. This provides compelling proof of the high fidelity with which structural features are revealed in frozen-hydrated specimens examined with low-irradiation cryomicroscopy and image analysis techniques. Comparisons of the two structures are not as definitive at low radii. This could mainly be due to the different DNA compositions of the two structures examined and to the effects introduced by the contrast transfer function of the electron microscope (e.g., Erickson and Klug, 1971; Toyoshima and Unwin, 1988).
Tentative assignment of the locations and distribution of the protein and DNA components of ϕX174 can be made by examining the three-dimensional reconstruction and comparing it with existing biochemical data. Corroboration of the assignments can also be made by a comparison with the atomic structure (McKenna et al., 1992).
Burgess (1969) showed that ϕX174 contains 60 copies of gpG organized as pentameric aggregates in the 12 apical caps at the icosahedral vertices of the virion. Thus, each of the five morphological subunits in each cap (Fig. 2) most likely represents a single, globular gpG polypeptide (Mr 19 020). The volume within each cap at radii > ~14 nm and for densities greater than a threshold determined on the basis of fitting the reconstruction envelope to the X-ray structure (McKenna et al., 1992) is 118 nm3. Five copies of gpG, with an assumed partial specific volume of 0.74 g/cm3 which is typical for globular rroteins (Creighton, 1984), would occupy 117 nm . A spherical protein one-fifth this volume would have a calculated diameter of ~35 nm, which is consistent with the observed morphology and size of the cap subunits in the reconstruction (Fig. 2).
The location of gpH, which is also thought to be a cap protein (Burgess, 1969; Shank et al., 1977), is not clearly delineated in the reconstruction. The H or pilot protein recognizes a lipopolysaccharide receptor on the bacterium surface and is subsequently injected into the bacterium along with the ssDNA in a process defined as the “eclipse” reaction (Jazwinski et al., 1975a,b; Hayashi et al., 1988). Because there are only 12 copies of gpH per virion (Burgess, 1969), various models have been proposed that place a single copy of gpH in each apical cap, presumably at or exposed to the cap outer surface (Hayashi et al., 1988; Willingmann et al., 1990). If our volume estimate for the apical cap is correct, then there is insufficient space for all of the gpG and gpH. Some of gpH could be contained within the apical cap density if some of gpG extends to radii below 14 nm. Thus, portions of gpH might lie inside the virion associated with the other viral proteins (gpF or gpJ) and the ssDNA, and also in an extended and perhaps disordered conformation on the outside. In either case, gpH density is expected to be smeared out in the three-dimensional map as a consequence of symmetry averaging. Examination of the X-ray structure shows that all of gpG is ordered and most of it is located at radii > ~ 14 nm within the cap, but gpH cannot be directly identified (M. Rossmann, personal communication; McKenna et al., 1992). Our interpretation of the reconstruction based on knowledge of the X-ray structure thus indicates that little, if any, ordered gpH is revealed.
The remainder of the virion is composed of 60 copies each of gpF (Mr 48 440) and gpJ (Mr 4220) (Burgess, 1969) and a single copy ofthe ssDNA (Mr 1.7 × 106) (Sinsheimer, 1959b). There is no unambiguous way to discriminate the gpF, gpJ, and DNA densities in the reconstruction. However, gpF must account for most of the icosahedrally organized features within the capsid, especially those at the external surface, because gpJ constitutes a minor fraction of the capsid protein and it is known to be an internal protein which functions in DNA packaging (Aoyama et al., 1981, 1983; Hamatake et al., 1985; Hayashi et al., 1988).
There is ample space inside the virion to accommodate the entire DNA genome and also possibly a portion of gpH. The minimum radius of the protein shell (as determined from a reconstruction of empty ϕX174 particles, data not shown) is at ~9.8 nm which encompasses a spherical volume of 3900 nm3 . At a density of 1. 72 g/cm3 (Sinsheimer, 1959b; Eigner et al., 1963), the ϕX174 ssDNA would only occupy 1640 nm3 or about 42% of the available volume. Thus, if the DNA is uniformly distributed along with solvent, the overall density inside the protein shell of the virion would only be 1.30 g/cm3. This corresponds to a DNA packing density of ~0.7 g/ml which is ~12.5% lower than that found in bacteriophages such as T4 (Kellenberger et al., 1986). This is also consistent with studies that show that ϕX174 is capable of encapsidating a genome which is larger than the 5386 bases normally found within wild-type virions (Müller and Wells, 1980; Russell and Muller, 1984). For example, an additional 163 bases (3% larger) can be inserted into the J-F inter-cistronic region of the genome and be packaged into genetically and physically stable infectious particles that maintain this genome size through successive growth cycles (Müller and Wells, 1980). Up to 704 extra bases ( + 13%) have been packaged, but deletions occur during initial growth cycles (Russell and Müller, 1984). These data are consistent with a model in which the DNA is not as highly organized as it is in bacteriophages T4 and λ (Lepault and Leonard, 1985; Lepault et al., 1987) and the mammalian herpes simplex virus (Booy et al., 1991). This and the relatively small size of ϕX174 might also explain why we have been unsuccessful at visualizing ordered DNA structure in the central regions of images that were recorded at a low level of defocus ( <0.5 μm underfocus).
Models of the ϕ X174 structure obtained by cryo-microscopy and X-ray crystallography provide a firm basis for studying several additional aspects of bacteriophage morphology and morphogenesis. Preliminary reconstruction studies of empty particles found in fast band preparations (data not shown) reveal a nearly identical structure, with the exception of an empty center, compared to full virions. Difference maps may help identify how the DNA is organized and whether it influences the capsid structure as has been shown for mammalian herpes simplex virus (e.g., Booy et al., 1991). Reconstructions of slow-band particles would complement this study. Cryomicroscopy procedures will be especially powerful in studying dynamic events or structures that prove too difficult to crystallize for X-ray studies. For example, the location of gpH remains a puzzle. The existence of monoclonal antibodies to gpH (N. L. Incardona, unpublished results) and the recent development of cryomicroscopy methods capable of visualizing virus–Fab interactions (Prasad et al., 1990; Wang et al., 1992) may provide a way to definitively locate gpH. Cryomicroscopy and image analysis procedures are also well adapted to study a variety of ϕX174-related structures such as (i) intact capsids which have the apical caps (gpG, gpH) stripped away by urea (Edgell et al., 1969), (ii) pro-head intermediates, which also contain gpB and the scaffold phage protein, gpD, but are devoid of gpJ and the ssDNA (Hayashi et al., 1988), and (iii) 1328 intermediates, which contain gpD in addition to all the normal viral components (Hayashi et al., 1988). Finally, quick-freezing cryotechniques provide a way to trap and explore different, time-resolved functional states (e.g., Mandelkow and Milligan, 1990; Talmon et al., 1990; Lepault et al., 1991). For example, the process by which ϕX174 DNA is injected could be studied by inducing eclipse with isolated lipopolysaccharides (Incardona and Selvidge, 1973) or calcium (Incardona et al., 1972; Yazaki, 1981).
Acknowledgments
We gratefully acknowledge W. Murakami for providing the polyoma virus; L. Ilag and D. Xia for recent samples of “fast-band” ϕX174; R. McKenna, D. Xia, S. Krishnaswamy, and M. Rossmann for the map of ϕX174 derived from X-ray crystallographic data; R. Cheng and L. Tong for assistance in displaying the X-ray data; C. Music for photographic assistance; and M. Rossmann, L. Ilag, J. Johnson, S. Krishnaswamy, R. McKenna, and D. Xia for stimulating discussions. This work was supported by NIH Grant GM33050 (T.S.BJ, NSF Grant DMB84-16573 (N.L.L), NSF Grant DMB86-02753 (M. Rossmann), fellowship Wi873/1.1 from the Deutsche Forschungsgemeinschaft (P.W.), and a grant from The Lucille P. Markey Trust to the Purdue University Center for Macromolecular Structure Research.
Footnotes
For ϕX174, the term capsid strictly refers to the spherical portion of the virion coat structure that contains proteins gpF and gpJ.
REFERENCES
- Air GM, Coulson AR, Fiddes JC, Friedmann T, Hutchison CA,, III, Sanger F, Slocombe PM, Smith AJH. J. Mol. Biol. 1978;125:247–254. doi: 10.1016/0022-2836(78)90347-9. [DOI] [PubMed] [Google Scholar]
- American Heritage Dictionary, Second College Ed . Houghton Mifflin Co.; Boston, MA.: 1985. [Google Scholar]
- Aoyama A, Hamatake RK, Hayashi M. Proc. Natl. Acad. Sci. USA. 1981;78:7285–7289. doi: 10.1073/pnas.78.12.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama A, Hamatake RK, Hayashi M. Proc. Natl. Acad. Sci. USA. 1983;80:4195–4199. doi: 10.1073/pnas.80.14.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. Proc. Natl. Acad. Sei. USA. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. Biophys. J. 1989;55:243–253. doi: 10.1016/S0006-3495(89)82799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Newcomb WW, Booy FP, Brown JC, Steven AC. J. Virol. 1990;64:563–573. doi: 10.1128/jvi.64.2.563-573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Newcomb WW, Olson NH, Cowsert LM, Brown JC, Olson C. Biophys. J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Horne RW. Biochim. Biophys. Acta. 1959;34:103–110. doi: 10.1016/0006-3002(59)90237-9. [DOI] [PubMed] [Google Scholar]
- Burgess AB. Proc. Natl. Acad. Sci. USA. 1969;64:613–617. doi: 10.1073/pnas.64.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton TE. Proteins, Structures and Molecular Properties. Freeman; New York: 1984. [Google Scholar]
- Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall AW, Schultz P. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Edgell MH, Hutchison CA, III, Sinsheimer RL. J. Mol. Biol. 1969;42:547–557. doi: 10.1016/0022-2836(69)90242-3. [DOI] [PubMed] [Google Scholar]
- Eigner J, Stouthamer AH, Van der Sluys I, Cohen JA. J. Mol. Biol. 1963;6:61–84. [Google Scholar]
- Erickson HP, Klug A. Philos. Trans. R. Soc. London Ser. B. 1971;261:105–118. [Google Scholar]
- Fuller SD. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- Hall CE, Maclean EC, Tessman I. J. Mol. Biol. 1959;1:192–194. [Google Scholar]
- Hamatake RK, Aoyama A, Hayashi M. J. Virol. 1985;54:345–350. doi: 10.1128/jvi.54.2.345-350.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Aoyama A, Richardson DL, Jr., Hayashi MN. In: The Bacteriophages. Calendar R, editor. Vol. 2. Plenum Press; New York: 1988. pp. 1–71. [Google Scholar]
- Incardona NL. In: The Single-Stranded DNA Phages. Denhardt DT, Dressier D, Ray DS, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY.: 1978. pp. 549–555. [Google Scholar]
- Incardona NL, Blonski R, Feeny W. J. Virol. 1972;9:96–101. doi: 10.1128/jvi.9.1.96-101.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona NL, Selvidge L. J. Virol. 1973;11:775–782. doi: 10.1128/jvi.11.5.775-782.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM, Lindberg AA, Romberg A. Virology. 1975a;66:283–293. doi: 10.1016/0042-6822(75)90198-1. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM, Marco R, Kornberg A. Virology. 1975b;66:294–305. doi: 10.1016/0042-6822(75)90199-3. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Carlemalm E, Sechaud J, Ryter A, De Haller G. In: Bacterial Chromatin. Gualerzi CO, Pon CL, editors. Springer-Verlag; New York: 1986. pp. 11–25. [Google Scholar]
- Lepault J, Dubochet J, Baschong W, Kellenberger E. EMBO J. 1987;6:1507–1512. doi: 10.1002/j.1460-2075.1987.tb02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepault J, Erk I, Nicolas G, Ranck J-L. J. Microsc. 1991;161:47–57. doi: 10.1111/j.1365-2818.1991.tb03072.x. [DOI] [PubMed] [Google Scholar]
- Lepault J, Leonard K. J. Mol. Biol. 1985;182:431–441. doi: 10.1016/0022-2836(85)90202-5. [DOI] [PubMed] [Google Scholar]
- Maclean EC, Hall CE. J. Mol. Biol. 1962;4:173–178. doi: 10.1016/s0022-2836(62)80049-7. [DOI] [PubMed] [Google Scholar]
- Mandelkow E-M, Milligan R. Proc. XIIth Int. Cong. Elect. Microsc. 1990;1:502–503. [Google Scholar]
- McKenna R, Xia D, Willingmann P, Ilag LL, Krishnaswamy S, Rossmann MG, Olson NH, Baker TS, Incardona NL. Nature (London) 1992;355:137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller UW, Wells RD. J. Mol. Biol. 1980;141:25–41. doi: 10.1016/s0022-2836(80)80027-1. [DOI] [PubMed] [Google Scholar]
- Olson NH, Baker TS. Ultramicroscopy. 1989;30:281–298. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson NH, Baker TS, Johnson JE, Hendry DA. J. Struct. Biol. 1990;105:111–122. doi: 10.1016/1047-8477(90)90105-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BVV, Burns JW, Marietta E, Estes MK, Chiu W. Nature (London) 1990;343:476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- Russell PW, Miiller UW. J. Virol. 1984;52:822–827. doi: 10.1128/jvi.52.3.822-827.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes JC, Hutchison CA,, III, Slocombe PM, Smith M. Nature (London) 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sanger F, Coulson AR, Friedmann T, Air GM, Barrell BG, Brown NL, Fiddes JC, Hutchison CA,, III, Slocombe PM, Smith M. J. Mol. Biol. 1978;125:225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Shank PR, Hutchison CA, III, Edgell MA. Biochemistry. 1977;16:4545–4549. doi: 10.1021/bi00640a001. [DOI] [PubMed] [Google Scholar]
- Sinsheimer RL. J. Mol. Biol. 1959a;1:37–42. [Google Scholar]
- Sinsheimer RL. J. Mol. Biol. 1959b;1:43–53. [Google Scholar]
- Talmon Y, Burns JL, Chestnut MH, Siegel DP. J. Elect. Microc. Tech. 1990;14:6–12. doi: 10.1002/jemt.1060140103. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Unwin W. Ultramicroscopy. 1988;25:279–291. doi: 10.1016/0304-3991(88)90003-4. [DOI] [PubMed] [Google Scholar]
- Tromans WJ, Home RW. Virology. 1961;15:1–7. doi: 10.1016/0042-6822(61)90069-1. [DOI] [PubMed] [Google Scholar]
- Wang G, Porta C, Chen Z, Baker TS, Johnson JE. Nature (London) 1992;355:275–278. doi: 10.1038/355275a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingmann P, Krishnaswamy S, McKenna R, Smith TJ, Oison NH, Rossmann MG, Stow PL, Incardona NL. J. Mol. Biol. 1990;212:345–350. doi: 10.1016/0022-2836(90)90129-a. [DOI] [PubMed] [Google Scholar]
- Winkelmann DA, Baker TS, Rayment I. J. Cell. Biol. 1991;114:701–713. doi: 10.1083/jcb.114.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. J. Virol. Methods. 1981;2:159–167. doi: 10.1016/0166-0934(81)90034-3. [DOI] [PubMed] [Google Scholar]
- Yeager M, Dryden KA, Olson NH, Greenberg HB, Baker TS. J. Cell Biol. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]