Abstract
Background
Studies of simple RNA animal viruses show that cell attachment, particle destabilization and cell entry are complex processes requiring a level of capsid sophistication that is difficult to achieve with a shell containing only a single gene product. Nodaviruses [such as Flock House virus (FHV)] are an exception. We have previously determined the structure of FHV at 3 A resolution, and now combine this information with data from cryo-electron microscopy in an attempt to clarify the process by which nodaviruses infect animal cells.
Results
A difference map was computed in which electron density at 22Å resolution, derived from the 3.0Å resolution X-ray model of the FHV capsid protein, was subtracted from the electron density derived from the cryo-electron microscopy reconstruction of FHV at 22 Å resolution. Comparisons of this density with the X-ray model showed that quasi-equivalent regions of identical polypeptide sequences have markedly different interactions with the bulk RNA density. Previously reported biphasic kinetics of particle maturation and the requirement of subunit cleavage for particle infectivity are consistent with these results.
Conclusions
On the basis of this study we propose a model for nodavirus infection that is conceptually similar to that proposed for poliovirus but differs from it in detail. The constraints of a single protein type in the capsid lead to a noteworthy use of quasi-symmetry not only to control the binding of a 'pocket factor' but also to modulate maturation cleavage and to release a pentameric helical bundle (with genomic RNA attached) that may further interact with the cell membrane.
Keywords: cryo-electron microscopy, nodaviruses, RNA–protein interactions, quasi-equivalence, viral infection
Introduction
Quasi-equivalence
Genetic economy in viruses is evident from the prevalence of icosahedral particles used to transport many different viral genomes. A shell formed by 60 identical gene products arranged with the symmetry of the icosahedron provides the largest container in which all subunits are in identical environments. The principles by which icosahedral virus shells are constructed with more than 60 subunits was first explained by the concept of quasi-equivalence [1,2]. Quasi-equivalent shells are denoted by the triangulation number, T, which, for most simple viruses, specifies the number of chemically identical subunits (gene products) in each of the 60 icosahedral asymmetric units. The total number of subunits in the capsid is 60T Each subunit within the icosahedral asymmetric unit must occupy a slightly different environment and in those T= 3 shells, where structures are known to atomic or near atomic resolution, the quasi-equivalent states of the subunits are determined by a switching mechanism that is directed by the protein subunit itself or the packaged RNA or both [2,3].
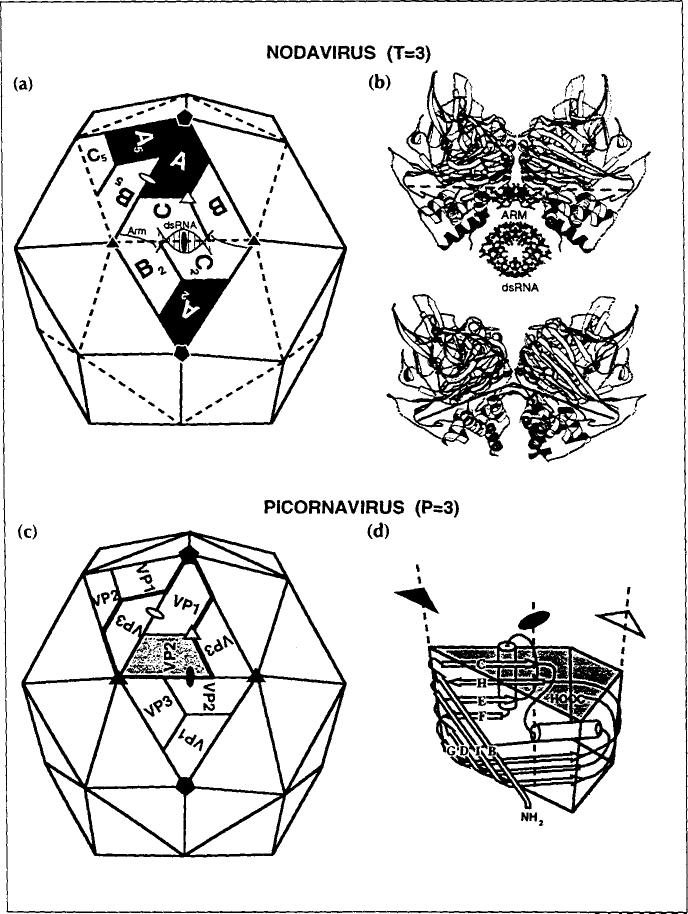
Capsids with T= 3 symmetry are common among RNA plant viruses, but there are few examples of quasi- equivalence among the simple RNA viruses that infect animals. The dominant capsid form in the latter category of viruses is the P= 3 (pseudo T= 3) picomavirus shell. The T= 3 and P= 3 shells are clearly related (Fig. 1), but the latter contains three different gene products while the former contains one. The trapezoids labeled A, B and C in Fig. 1 correspond to the same gene product located in slightly different environments within one icosahedral asymmetric unit (the central triangle). The tertiary folds of A, B and C are virtually identical, but the quasi-equivalent C-C 2 contact and A-B5 contact are flat and bent respectively (side views shown in Fig. 1b), although the same protein surfaces are juxtaposed. A protein polypeptide ('arm' in Fig. 1b) and a portion of duplex RNA (dsRNA) are ordered only at the C–C 2 contact and serve as a wedge to prevent bending. These structures are disordered at the A–B5 interface and the contact is bent about an axial hinge that is conserved in both flat and bent contacts. Viral protein 1 (VP1), VP2 and VP3 are different gene products in the picornavirus capsid and therefore the pseudo-equivalent VP2–VP2 interface and VP1-VP3 interface have different protein surfaces juxtaposed and there is no need for a molecular switch as observed in the T= 3 shell. In both capsid types, the tertiary structure of the subunit is almost always an eight-stranded, antiparallel β-barrel with a jelly roll topology (Fig. 1d) [3].
Fig. 1.
A comparison of a T=3 nodavirus capsid with a P=3 picornavirus capsid, showing that the two capsids are similar in overall shape, size and organization. (a) Nodavirus capsid. (b) Side views of the C–C 2 contact (top) and A–Bs contact (bottom). (c) Picornavirus capsid. (d) Schematic representation of the eight-stranded β-barrel structure of the capsid subunits. Closed symbols identify icosahedral axes of symmetry whereas open symbols identify quasi (lo-cal) symmetry axes.
Fundamental differences which accompany the infection of plant and animal cells may skew the distribution of capsid types for viruses infecting the different hosts. First, higher animals are protected by a circulating immune system which may exert selective pressure for a P= 3 capsid. The P= 3 capsid favors the formation of escape mutations because, with three gene products in the icosahedral asymmetric unit instead of one, the combinatorial possibility for altering epitopes is much greater. Furthermore, a single mutation in the T=3 gene generates three changes in the icosahedral asymmetric unit, thereby increasing the probability that a function of the capsid will be affected by the mutation [4]. Second, viruses infecting an animal host enter the cell through a receptor-mediated path. Considerable progress has been made recently in understanding the process by which rhinovirus [5] and poliovirus [6] bind to receptors and enter cells. In these viruses it is clear that there are distinct roles for VP1 compared with VP2 and VP3 (Fig. 1), suggesting that the multiple gene products in the capsid are important to achieve the level of sophistication needed for cell entry.
In this report we identify some structural features in the T=3 nodaviruses [as exemplified by the Flock House virus (FHV), an insect nodavirus] that are different from any other T= 3 structures reported and which probably relate to early events in cell entry. We suggest that FHV has a mechanism of entry related to that observed in the polio picomavirus system, and that quasi-equivalent interactions facilitate this mechanism with just a single gene product.
Nodavirus structure
The nodaviruses are an excellent model system for investigating animal virus assembly, maturation, and factors that affect viral stability and infectivity. They are single-stranded RNA viruses with 180 identical gene products (Mr = 44kDa) forming their capsids. Nodaviruses infect insects [7], mammals [8] and fish [9], and they are among the simplest biological replicating systems. The nodavirus genome contains only three genes located on two RNA molecules which are both packaged in the same particle. The capsid proteins undergo a post assembly cleavage:
This cleavage is required for infectivity [10,11].
We have determined the structures of black beetle virus (BBV) at 2.8Å [12] and FHV at 3.2Å [13] resolution by X-ray crystallography. The structure of FHV has been subsequently refined to 3.0Å resolution (V Reddy, A Fisher andJ Johnson, unpublished data). The FHV and BBV atomic structures showed that duplex RNA contributes to the formation of the T= 3 capsid (Fig. 1a), with 10 well-ordered base pairs packed against the protein shell near each icosahedral two-fold axis. This highly ordered RNA accounts for ~ 20 % of the genome. These studies also showed that the subunit cleavage site (Asn363/Ala364) is on the interior of the protein shell and inaccessible to cellular enzymes. Residues 364–407 (the y-peptide) are on the capsid interior with amino acids 364–379 in an amphipathic ta-helical configuration. The 28 carboxy-terminal residues (380–407) are not visible in the X-ray electron density map.
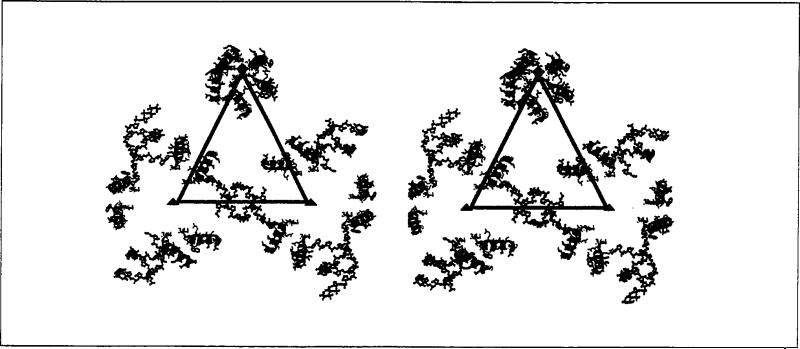
The three subunits in the icosahedral asymmetric unit have nearly identical tertiary structures with their γ-helices bound to the interior surface of the β-barrel by hydrophobic interactions. However, in the assem bled particle, the γ-helices are located in dramatically different environments because of the quasi-equivalent contacts in the shell. The γ-helices in the A subunits (γA) form a pentameric helical bundle that is stabilized by interactions among the hydrophilic surfaces of the helices. In contrast, the γc-helices interact with the ordered duplex RNA and with the γB-helices in pairs about the icosahedral three-fold axes. The γB- and γc-helices are the innermost protein features in the X-ray electron density map and hydrophilic residues of these helices interact with each other, point towards the particle interior, or (in γc) interact with the ordered RNA. Although the primary structure of the γ-peptides is the same in all subunits, the presence or absence of RNA dramatically changes the relative positions of the γ-peptides near the symmetry axes. The relative disposition of the γ-helices and ordered duplex RNA are displayed in Fig. 2.
Fig. 2.
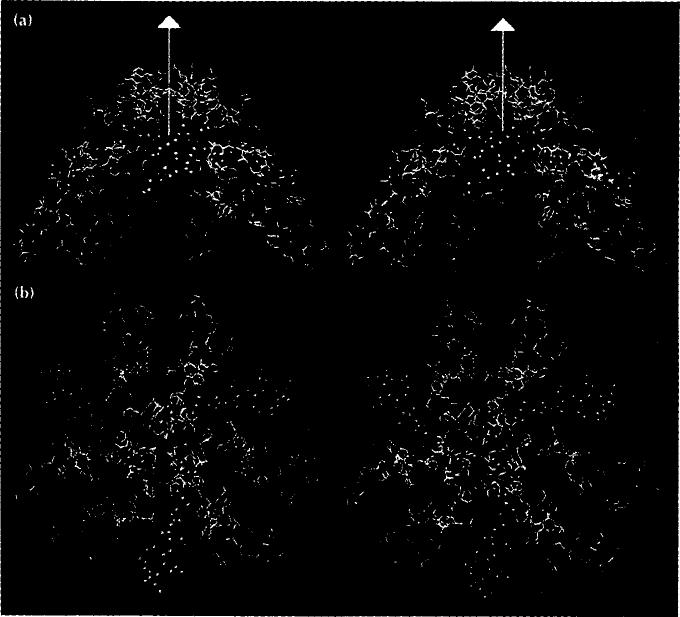
Stereoview of the γ-peptides and ordered RNA segments of the FHV particle structure. The triangle represents the border of the icosahedral asymmetric unit containing A, B and C in the T= 3 particle in Fig. 1. Blue helices are associated with the A subunits (γA), red with the B subunits (γB), and green with the C subunits (γc). Quasi six-fold axes are shown as red triangles, the five-fold axes as a red pentagon. Duplex RNA is represented as ball-and-stick models.
The differences in the surroundings of the γA, γB, and γc-peptides with respect to each other, the ordered RNA, and the bulk RNA [which is invisible in the X-ray map because it is not icosahedrally ordered to a high enough resolution (15-3.0Å)] suggests that the roles of these peptides depend on the differences in the quasi-equivalent environments. The data from the X-ray analysis alone did not allow us to confirm that these helices had different interactions with the bulk RNA. Such interactions were significant at two levels. First, kinetic studies suggest that 120 subunits cleave at a faster rate than the remaining 60 subunits [10]. Based on the subunit tertiary structures, the environments at the cleavage points of the A, B and C proteins appeared to be identical. We wanted to see if the bulk RNA affected the cleavage site in a way that could explain the kinetic result. Second, a hypothesis that we developed for the role of the γA-helices in the early interaction of the virus with the cell surface predicts that these helices will have a different interaction with the bulk RNA than the γB- and γc-helices. Based on an analogy with the picornaviruses [14,15], the γA pentameric helical bundle may be released upon interaction of the virion with the cellular receptor, or with the membrane, or both. This hypothesis predicts minimal interaction of the γA-helices with the RNA in order to facilitate release of the cleaved y-peptides along the pentamer axis.
We investigated features of the protein-RNA interface in FHV by combining data from the X-ray and the cryo-electron microscopy (cryoEM) experiments in a manner similar to that used to study complexes between viruses and Fab fragments [16,17], whole antibodies [18], and receptor molecules [5]. The fidelity of the procedure used to visualize the RNA density was tested with two control experiments that demonstrated the extraordinary level of detail available from the electron microscopy data when density for the protein and RNA in the virus can be assigned on the basis of the X-ray model.
Results and discussion
In addition to the biological relevance of the results derived from the combined use of cryoEM and an X-ray model to analyze the bulk RNA-protein interface in FHV, we also provide a brief description of the procedures used for this analysis. Because this is the first application of this method to study detailed protein-nucleic acid interactions, we emphasize the use of controls used to establish the significance of the difference electron density map.
CryoEM -X-ray model difference map
Two electron density maps (from cryoEM and from the X-ray model) were used to compute a difference map which showed only features that were present in the cryoEM reconstruction and missing in the high resolution model derived from the 3.0Å X-ray electron density map of FHV [13]. Images of FHV particles embedded in vitreous ice (Fig. 3) were used to compute a three-dimensional reconstruction (Figs 4a and 4b). A second density map was computed with amplitudes and phases for all data to 22Å resolution calculated with the atomic model of the protein that was derived from the 3.OÅ resolution X-ray map (Figs 4c and 4d). This map exhibits excellent fidelity with the outer shell of the cryoEM reconstruction. Comparison of the data in cross-section shows that a 16Å diameter cavity, which lies on the quasi three-fold symmetry axes, was visible in both maps (Figs 4b and 4d). A difference electron density map, which was computed by subtracting the calculated map based on the X-ray model from the cryoEM reconstruction, revealed density for only those regions not accounted for by the X-ray model (Figs 4e, 5 and 6). To test the validity of the method, the ordered RNA in the X-ray model was not included in the structure factor calculation. The prominent high radius region of the difference density (90-110Å) corresponds exactly with the ordered RNA observed in the X-ray structure (Fig. 5b). The excellent fit of the RNA duplex model in the difference map (Figs 5b and 6b) adds confidence to the interpretation of difference density appearing elsewhere in the map and at lower contour levels. Indeed, the helical portion of the γ-peptide associated with the B subunits (γB), which was not apparent at the contour level used for building the FHV X-ray model [13], was clearly visible in the difference map, and re-examination of the X-ray map at lower contour confirmed its presence in a semi-ordered configuration. It was included in the final model used for computing the difference map.
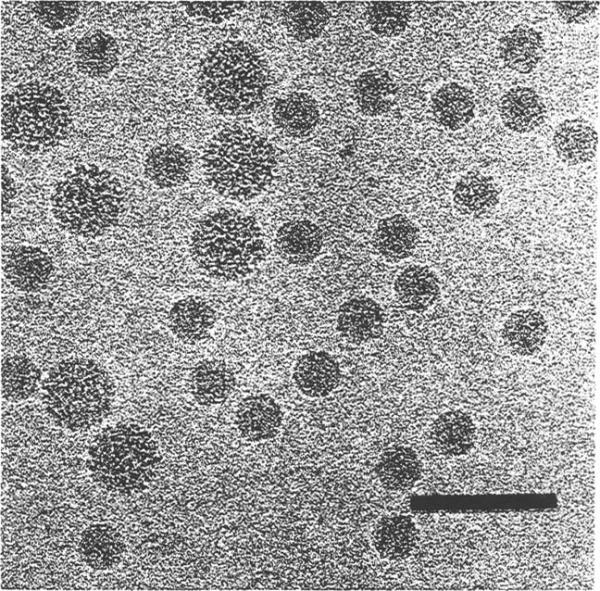
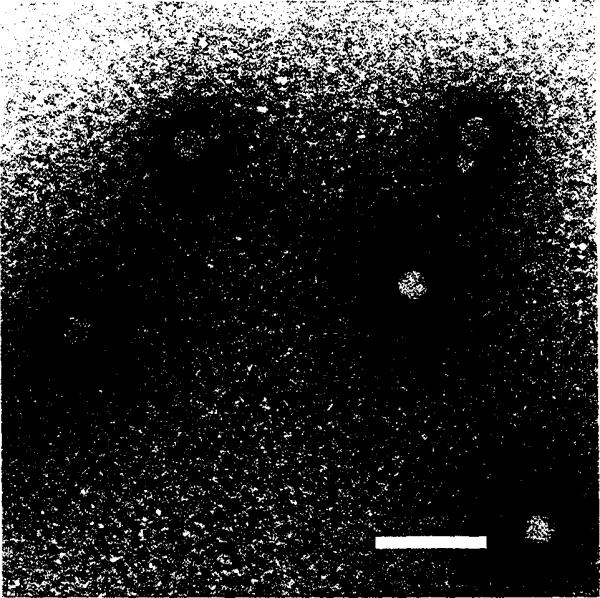
Fig. 3.
Frozen-hydrated Flock House virus. Polyoma virus (larger particles, 495 Å diameter) was mixed with the FHV sample prior to the vitrification for size comparison and as a familiar specimen which aided assessment of image quality. Bar = 1000Å.
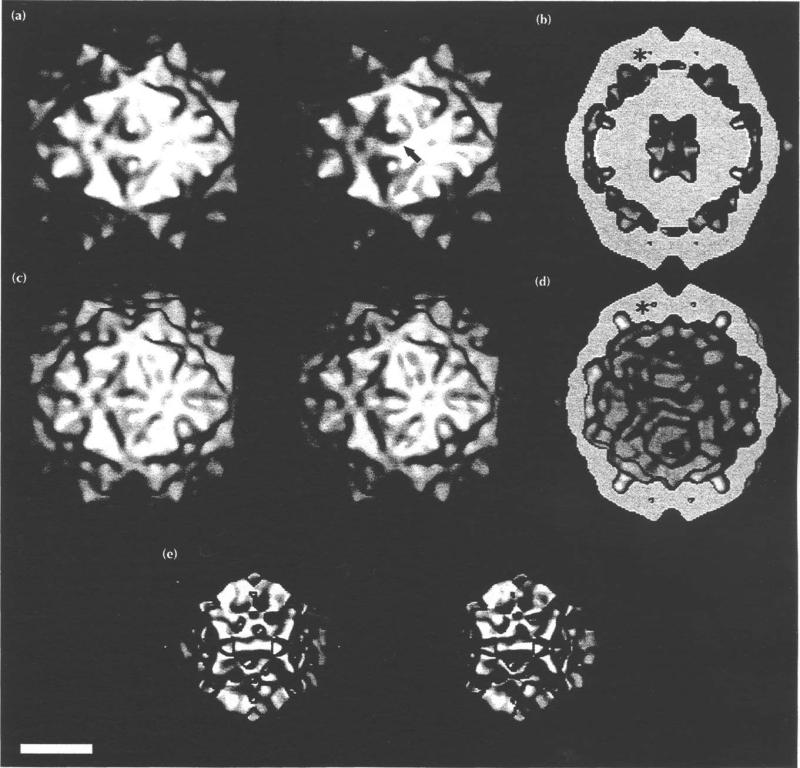
Fig. 4.
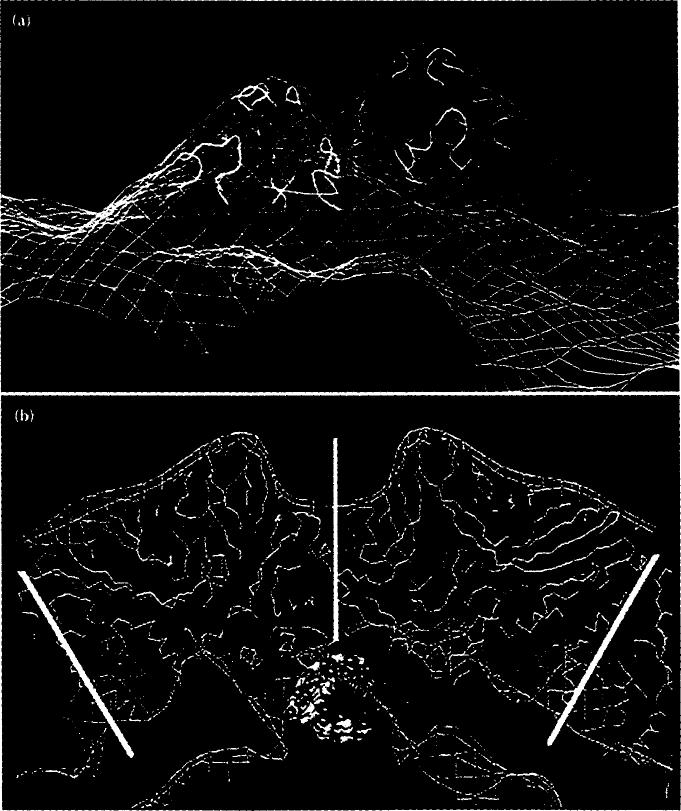
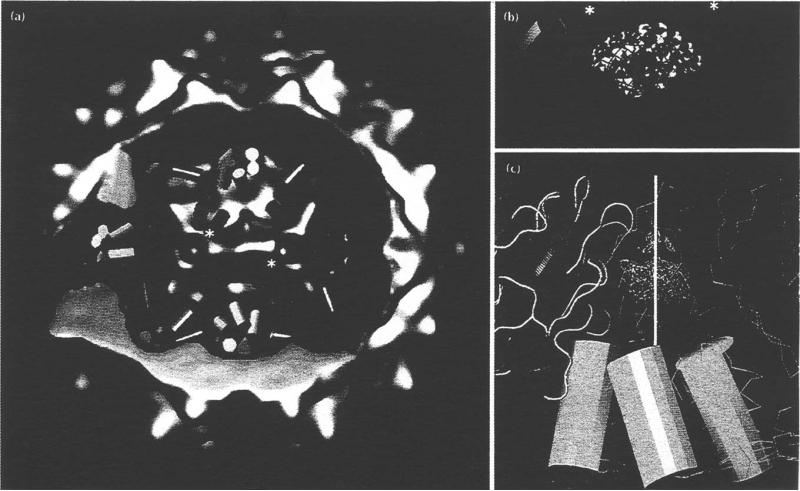
Electron density distributions displayed as surface shaded images of FHV. Bar = 100 Å (a) Stereoview of FHV from the cryoEM reconstruction. The arrow indicates the direction of view in Fig. 5. (b) Equatorial section of (a), showing the interior RNA density. The large center cavity may be an artificial feature because the noise in image reconstruction of icosahedral particles is greatest in this region. One of the 16 A cavities is labeled with an asterisk. Four of the 60 cavities are visible. (c) Stereoview of the 22 Å electron density map, computed with structure factors based on the 3.0 Å resolution model [13]. (d) Equatorial section of (c). Note the 16A cavity (asterisk) and the absence of interior RNA density. (e) Stereoview derived from the cryoEM -X-ray model difference map. This electron density not accounted for by the X-ray model is largely RNA.
Fig. 5.
Close up of the two-fold related protrusions at the quasi threefold axes of the surface (arrowed in Fig. 4a) showing the consistency of the cryoEM and X-ray data. (a) CryoEM electron density (blue contours) superimposed on the FHV X-ray model (yellow and green ribbon diagram). (b) Cross-section view of the full virion (blue). Superimposed on this are; FHV protein Cα atomic coordinates (yellow), the contoured difference electron density map (red lines), and the locally ordered RNA (yellow, red and blue space-filling representation). Vertical white line represents icosahedral two-fold axis; other white lines capped with pentagon symbols identify two five-fold axes.
Fig. 6.
Positions of the γ-helices. (a) Cut away view of the whole FHV virion. Electron density derived from the cryoEM reconstruction is blue, density derived from the difference map is red. γy8-helices are red cylinders, γc-helices green and γA-helices light blue. Asterisks indicate the helices shown in close up in (b). (b) Close up of γB and γc and two-fold related mates showing the close association with bulk RNA (red contours) and ordered RNA (red, blue and yellow space-filling model). (c) Side view of a bundle of γA-helices. The five-fold axis (vertical white line with red pentagon) passes through the ellipsoidal density (red contours indicate the density as plotted in the difference map, white contours the density from the X-ray map and blue contours the cryoEM envelope). One A subunit is shown in ribbon representation (green, yellow and red), others as thin yellow lines (Coc backbone trace).
A second feature found in the difference map served as an unexpected control for these experiments. The X-ray electron density maps of both FHV and BBV revealed an ellipsoid-shaped density on the five-fold symmetry axes that was not interpretable as protein. It lies in a hydrophobic environment and the density is not seen at the analogous location on the quasi six-fold axes. Because this feature was not included in the X-ray model structure factor calculation, its appearance in the cryoEM - X-ray difference map (Fig. 6c) confirmed the fidelity of both types of data. The only significant density to appear within the protein shell is coincident with the ellipsoidal density observed in the X-ray maps. The density appears larger in the difference map suggesting that a portion of this chemical entity is more highly ordered than the rest and therefore is all that is visible in the X-ray map. As discussed below, this unmodeled density may play an important role in uncoating FHV.
Protein-RNA and protein-protein interactions
The γ-helices of the three quasi-equivalent subunits and their relation to the bulk RNA are shown in Figs 5 and 6. The cleavage site in all the helices (Asn363/Ala364) is at the end that lies at higher radius and closest to the symmetry axes of the structure. Both γB and γc directly contact the bulk RNA over their lengths and the region in each of these helices that lies close to the cleavage site is also adjacent to the RNA. However, only the carboxyl terminus of each γA-helix contacts the bulk RNA, and the cleavage site for the A subunit is 35Å from the bulk RNA. This dramatic difference in the environment of the cleavage sites of YB and YC on the one hand and γÅ on the other is consistent with Gallagher and Rueckert's interpretation [10] that 120 subunits are cleaved at a relatively fast rate (t½ = 2.5 h) and the remaining 60 subunits are cleaved at a slower rate (t½ = 13 h). We have proposed an acid-catalyzed, proteolytic cleavage mechanism for this reaction which requires a nucleophilic attack of the main chain carbonyl carbon at residue 363 in order for cleavage to occur [19]. The phosphates of the RNA that lie close to the γB, γC cleavage sites may increase the rate of the reaction by polarizing water molecules for this purpose. At γA the nucleophile may be the occasional hydroxide ion present at pH 5.8 (the pH optimum for the reaction) which would explain the reduced rate of the reaction at this site.
The γA-helices form pentamer bundles about each of the five-fold symmetry axes with interactions between the amphipathic helices occurring through the hydrophilic sides (Fig. 6). The hydrophobic side of the γA-helix packs against the body of the β-barrel. The amino terminus of γA is adjacent to the ellipsoid-shaped density that appeared in the X-ray and difference maps. All of the residues that are close to the five-fold symmetry axes are hydrophobic, forming a potential channel for the escape of the cleaved y-peptide bundle which has a hydrophobic surface and a hydrophilic core (Fig. 6).
Viral uncoating
The structure reported here, as well as other biochemical studies of nodaviruses, suggest that the mechanism of FHV infection has similarities to that proposed for poliovirus. The details, however, are different because FHV is a T= 3 virus and must employ subtleties of quasi-equivalence to achieve what is more straightforward in picomaviruses where three times the genetic information is used to achieve similar results.
The disassembly of rhinovirus 14 and poliovirus is initiated when each virus binds to its respective cellular receptor [5,6]. For poliovirus it has been shown that the amino terminus of VP1 is exposed at this stage, probably as an amphipathic helix, which, together with the myristic acid associated with VP4, interacts with the lipid portion of the cellular membrane [14]. Poliovirus and rhinovirus both have hydrophobic pockets within the VP1 subunits that form the capsid pentamers. These pockets may be occupied by fatty acids in the wild-type virus (e.g. poliovirus, rhinovirus 16) or the so-called ‘pocket factor’ may be absent in the purified virus (e.g. rhinovirus 14). A variety of studies have shown that when this pocket is filled with a tightly binding drug, the particle is stabilized and acquires resistance to uncoating and therefore becomes non-infectious [3]. Thus, the endogenous pocket factor, which stabilizes the isolated virus particle, is probably lost early in the normal infection process and this permits the necessary flexibility required for the protein shell to disassemble. Although the picornavirus VP2 and VP3 subunits also contain β-barrel domains like VP1, they do not have an endogenous pocket factor and do not bind drugs.
The picornavirus studies suggest that one of the roles for the multiple subunit types in P= 3 viruses is to differentiate pentamers and hexamers. Such a distinction can not be made in a T= 3 virus because the subunits at the hexamer and pentamer are chemically identical. We hypothesize that the ellipsoidal density seen in FHV and BBV is another form of pocket factor. It occurs only at the five-fold symmetry axes because of a significant difference in quasi-equivalent contacts at the five-fold and quasi six-fold axes. The ellipsoidal density is bordered at high radius by five leucine residues (Leu81) and by the aliphatic portion of five lysine residues (Lys237) (Fig. 7). The red 'belt' (Fig. 7a) is a ring of five salt-linked pairs of Lys237 and Gly233 residues. It is ex posed to the interior of the shell at lower radius and is directly adjacent to the γA-helices. Thus, the entire lining of the pentamer cavity is hydrophobic. Methionine residues are at the top of the pentamer with side chains extended into the solvent. At the quasi six-fold axes, which are true three-fold axes, the corresponding region is occupied not by the pocket factor but by the side chains of the three-fold-related Thr20 residues that are part of the molecular switch that alters quasi-equivalent contacts between subunits (Fig. 8). Although formally quasi-equivalent to the five-fold axis and expected to be structurally similar, the organization of the protein is significantly different from that seen in Fig. 7. The helices do not form a bundle, and the region occupied by the pocket factor on the five-fold axis is occupied here by Thr20 of the peptide arm. Also, the methionines, which are exposed to the solvent at the pentamer axis, are folded inside at the hexamer axis and Glu183 is exposed. Thus, the protein portion of the molecular switch (Thr20-Va130), which is ordered only at the icosahedral three-fold axes, contributes not only to the particle architecture but also determines its chemical composition by selectively occupying a region on the quasi six-fold axes that is occupied by the pocket factor on the five-fold symmetry axes.
Fig. 7.
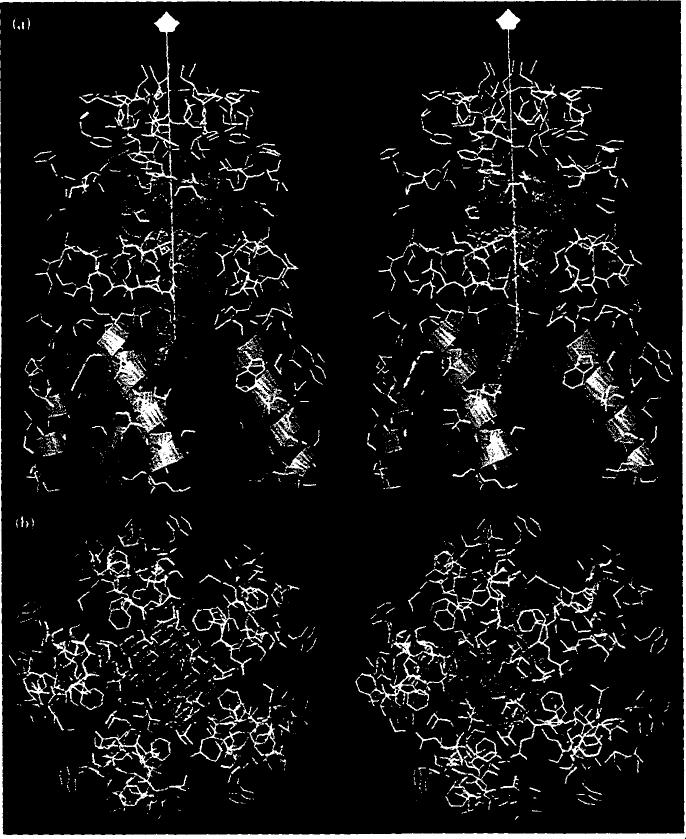
Stereoviews of the helical bundle of γ-peptides at the pentamer contact and residues from the subunits adjacent to the five-fold axis (labeled A in Fig. 1a). (a) Side view (five-fold axis is shown as a white line with a pentagon symbol at the end furthest from the virus center). (b) View directly down the five-fold axis. Residues in yellow are hydrophobic and those in red are charged. Blue helices are the γA-peptides. Blue contours are the ellipsoidal density seen in the X-ray map, red contours correspond to this density from the difference map.
Fig. 8.
Stereoviews of the γB- and γc-peptides (red and green helices, respectively) and the peptide arms (green, space-filling atoms) at the quasi six-fold contact and residues from the subunits adjacent to the quasi six-fold axis (labeled B and C in Fig. 1a). (a) Side view with the quasi six-fold axis shown as a white line with a triangle at the end. (b) A view directly down the quasi sixfold axis.
We propose a mechanism for the initial interaction of the virus with the cell surface which includes the release of the pocket factor either upon binding the cellular receptor, or interacting with the cell membrane, or both. The associated destabilization of the intersub-unit contacts along the pentamer axis would then allow the γA-peptides to emerge along the axis, dragging the RNA with them, via their carboxy-terminal ends. Upon reaching the capsid surface, the hydrophobic portion of the γA-helix might interact with the membrane and participate in the invagination process. The release of the γA-peptides, as proposed in this model, explains the requirement for the cleavage reaction. The cleavage of all three subunit types may be necessary for stabilizing the mature virion [10], but the cleavage of the γA-helix would be all that is required for the interaction with the cell surface in the proposed mechanism.
Another difference in the hexamer and pentamer axes that supports this mechanism was noticed after the initial hypothesis was formulated. The side chains of Met182 in the A subunits are totally exposed to the particle exterior. This creates a substantial hydropho bic patch at the five-fold symmetry axes and thus a favorable chemical environment for interaction with the membrane. The contacts between A subunits near Met182 are primarily hydrophobic, thus the proposed entry of Met182 into the membrane might initiate the destabilization process described above by 'dissolving' quaternary structure interactions at the pentamer axes in the membrane. Consistent with this hypothesis, Metl82 in the B and C subunits (at the quasi six-fold symmetry axes) are not as exposed as they are in the A subunits. The difference in quaternary structure at the hexamer axes exposes the side chain of Glu183 to the viral surface and folds the Met182 side chains inside the capsid surface where they interact with each other near the symmetry axis.
The addition of a helical domain to the interior of the FHV subunit, the mechanism for placing the pocket factor and exposed methionines only on the five-fold axes, and the post-assembly cleavage all are critical to the proposed model. These variations from the more common T= 3 plant viruses provide the nodaviruses with the level of sophistication required to meet the more complex demands of animal virus infectivity. We note that, in spite of the ability to infect animal cells, the nodaviruses will infect only animals with either no circulating immune system or a compromised immune system, suggesting that the disadvantages of the T= 3 capsid type for avoiding the immune system (as discussed in the Introduction) are of considerable significance.
The possible release of the γA-peptides was examined further by heating the virus. Studies of poliovirus showed that heating the virus generated the same antigenic site that emerged by the binding and release of the virus from the cellular receptor [20]. That antigenic site is now known to be the amino terminus of VP1 [14]. We postulated that a similar process in FHV might be initiated by heating. The virus was heated at 65°C for 10 minutes (under these conditions 2 logs of infectivity are lost on the basis of a plaque assay; B McK-inney and J Johnson, unpublished data) and electron micrographs of samples stained with 1%, unbuffered uranyl acetate were recorded. Virus particles remain essentially intact after this treatment, but a distinct single 'puff of density appears associated with most of the particles (Fig. 9). Each puff may be one bundle of γA-peptides associated with RNA, consistent with the hypothesis presented above. The apparent single site release of the RNA was an unexpected result from the heating experiments, although such specificity might be expected in the interaction of the virus with the cell. These phenomena have at least two explanations.
Fig. 9.
Micrograph of negatively-stained FHV particles that were heated to 65°C. Most of the particles exhibit a 'puff of density that appears to protrude from a single site on the capsid. The novel structure seen at each of the FHV pentamer axes makes it a strong candidate for the location of release of a y-helical bundle and associated RNA. Bar = 1000 Å.
Release of one 'puff may lead to a configurational relaxation of the virus structure such that further 'puffs' cannot be released. Alternatively, a specific sequence of RNA may prime one pentamer site for release upon perturbation of the particle by destabilizing the pen-tamer with which it is associated. This possibility can be tested by sequencing RNA from heat-treated particles that have been exposed to ribonuclease. We also aim to analyze the heat-treated particles by cryoEM in order to identify the exact location of the 'puff.
The combined cryoEM and X-ray approach described here provides a powerful tool for relating the low resolution order of the bulk RNA to the high resolution order of RNA and protein visible by the X-ray method. Beyond these qualitative comparisons we have shown that the X-ray model can be effectively refined as a rigid body in the EM density with the program X-PLOR [21]. After intentionally shifting the model of the icosahedral asymmetric unit by 6Å, it was refined to within 0.8A root mean square difference from the original model when correlation of the normalized observed (from cryoEM) and calculated (from the 3.0Å X-ray model) structure factors was used as the target function for the rigid-body refinement. The availability of an infectious clone of FHV [22] and the spontaneous assembly of virus-like particles when the FHV capsid protein is expressed in a baculovirus system [23], will allow our hypotheses to be tested with molecular biology experiments.
Biological implications
Nodaviruses, such as Flock House Virus, are among the simplest of agents that infect animals. Their single-stranded RNA genome is encapsulated in a shell assembled from 180 copies of a single type of protein. Geometrical considerations require that not every protein molecule in the shell has the same pattern of contacts with its neighbors. Capsid proteins are therefore in three different environments, and this may enable them to play different roles in the life-cycle of the virus.
By combining cryo-electron microscopy and X-ray crystallography data, we have modeled the interactions of capsid proteins with the viral genome in greater detail than was possible with our high-resolution X-ray study alone. We have confirmed that different packing environments lead capsid proteins to interact with the viral RNA in distinct ways. Our model leads us to suggest that the mechanism of infection used by nodaviruses is similar to that of the picornaviruses (which have coats composed of 60 copies each of three similar but different proteins), in which membrane contact destabilizes the virus coat and exposes an internal portion of the capsid protein.
A preliminary study of the thermal stability of virions is consistent with this idea, which can be further tested using molecular biological experiments.
Most viruses with a single coat protein infect plants rather than animals, perhaps because of constraints imposed by the simplicity of the coat structure. While nodaviruses can overcome some of these limitations by packing their simple coat components together in a sophisticated way, they only infect animals with no circulating immune system (e.g. insects) or with a compromised immune system (e.g. suckling mice). This may be because viruses formed with only one type of coat protein are limited in their ability to mutate to evade sophisticated immune responses.
Material and methods
Cryo-electron microscopy and image analysis
The cryoEM and image analysis procedures that were used to obtain the three-dimensional reconstructions of FHV were essentially similar to those previously reported [24–26]. The sample was maintained at near liquid nitrogen temperature in a Gatan 626 cryotransfer stage (Gatan Inc., Warrendale, PA) in a Philips EM420 transmission electron microscope (Philips Electronics Instruments, Mahwah, NJ). Micrographs were recorded under minimal dose conditions ( ~ 20 e– Å– 2), at an objective lens defocus of about 1.2-1.4 μm, and at an instrument magnification setting of X49 000. Micrograph selection and digitization, and particle image selection were performed as previously described [24-26].
Initial estimates of the centers of the particle images (phase origins) were made by means of a cross-correlation method [27], with a circular-average of the projected X-ray model used as the reference. A total of 374 projected images of the X-ray model, representing all unique views of the structure in 1° increments within the icosahedral asymmetric unit, were used as a reference data base for determining the orientation (θ, Φ ω), of each of the selected particles (TS Baker and RH Cheng, unpublished data). All model and particle images were converted by interpolation from Cartesian (x, y) to polar (r, φ) coordinate representations. The data at each radius were separately Fourier-transformed for all images, and the resulting data (Fourier amplitudes) for each particle were cross-correlated with the corresponding data for each of the 374 model images to obtain a best estimate (to within 1°) of the particle view direction (0, ± Φ)). The relative rotation (ω) of the particle about the view direction and the relative hand ( + Φ, – Φ ambiguity) were determined by means of real-space rotational correlations of the raw and reference polar images. The orientation search procedure was optimized by correlating data that had been filtered in both real space (only data between radii of 120–170A were used) and reciprocal space [band pass between (1/22 Å) and (1/90 Å)]. A preliminary three-dimensional reconstruction was computed [28]. This cryoEM reconstruction was subsequently used as the reference model in the same way as the X-ray model to further refine the orientation and origin parameters of the particle images. These methods, along with cross-common lines procedures, were used to select out a set of 17 particle images from which the final three-dimensional reconstruction, with full 532 icosahedral symmetry was computed [29]. An R-factor calculation [30] indicated that the data were reliable out to ~ 22 Å
Computation of X-ray model and cryoEM–X-ray difference map
The FHV electron density model was computed with structure factors calculated from the partially refined ~ 442 000 non-hydrogen, protein atomic coordinates that were derived from the X-ray map computed with data between 15Å and 3.0Å resolution. The Fourier synthesis was truncated at 22 Å resolution (~ 12400 structure factors). An artificial temperature factor of 2 000 Å2 was applied to reduce the effect of Fourier termination ripples.
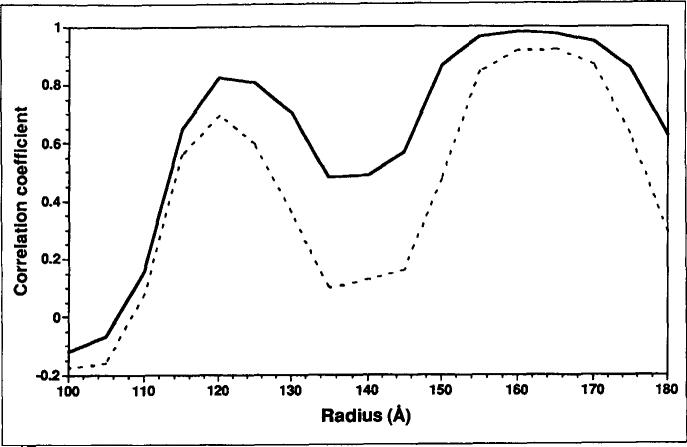
The initial difference map was computed after scaling the size and the contrast of the cryoEM map to the X-ray model on the basis of densities between radii 120Å and 170Å. Because the X-ray model only included contributions from protein atoms that could be assigned in the 3.0Å map, the initial difference map mainly contained density features attributed to the bulk viral RNA [31]. This difference map was subtracted from the rescaled cryoEM reconstruction to produce a 'cryoEM capsid map'. Corrections to reduce the effects of the microscope contrast transfer function were applied to the cryoEM capsid map by means of a least-square fit between the Fourier amplitudes. This two-step process (scaling the cryoEM map to the X-ray model in real space followed by scaling the cryoEM capsid map to the X-ray model in Fourier space) was carried out iteratively until no further improvement of Pearson's correlation coefficients in real space could be obtained (Fig. 10). A final difference map and cryoEM capsid map were then computed based on the best scaling parameters.
Fig. 10.
Plot of correlation coefficients as a function of virus radius, comparing the X-ray model with cryoEM reconstructions before (dashed line) and after (solid line) corrections were made to reduce the effects of the microscope contrast transfer function. The plots clearly demonstrate marked improvement in the data within the capsid shell after compensation for the defocus effects were included.
Accuracy of fit between X-ray model and cryoEM density
The structure factors corresponding to the cryoEM capsid map were calculated by inverse Fourier transformation. These calculated structure factor amplitudes and phases were used to measure the fit of the X-ray model to the cryoEM capsid map. The R-factor and the correlation coefficient of the target function ElE1 [21] for the data between 950Å and 22 Å resolution were 40% and 0.76, respectively. To further validate the fit of the model to the cryoEM capsid map, the model in an icosahedral asymmetric unit was radially translated up to 6Å, and this model was then subjected to rigid-body minimization with X-PLOR. The displaced model had an R-factor of 55 %, a correlation coeffi cient of 0.55, and a root mean square deviation (rmsd) of 6A with respect to the original model. After 20 cycles of rigid-body minimization, the model returned to a position, which was close to the original, with a resulting R-factor of 39 % and correlation coefficient of 0.78. The final rmsd of the Cc atoms to the original model was 0.8 Å. Additional cycles of refinement led to no further improvement in the fit.
Acknowledgments
We thank Bonnie McKinney for preparation of the FHV samples, W Murakami for polyoma virus samples, C Music for photographic assistance, and Roland Rueckert, Annette Schneemann, and Adam Zlotnick for stimulating and helpful discussions. The work was supported by NSF grant MCB-9206305 (TSB), NIH grants GM-34220 (JEJ) and GM-33050 (TSB), and a grant from the Lucille P Markev Charitable Trust for the development of structural studies at Purdue.
References
- 1.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JE, Fisher AJ. Principles of virus structure. In: Webster RG, Granoff A, editors. Encyclopedia of Virology. Academic Press; London: 1994. in press. [Google Scholar]
- 3.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu. Retr Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, et al. Johnson JE. Protein-RNA interactions in an icosahedral virus at 3.OÅ resolution. Science. 1989;245:154–159. doi: 10.1126/science.2749253. [DOI] [PubMed] [Google Scholar]
- 5.Olson NH, et al. Rossmann MG. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad Sci. USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yafal AG, Kaplan G, Racaniello VR, Hogle JM. Characterization of poliovirus conformational alteration mediated by soluble cell receptors. Virology. 1993;197:501–505. doi: 10.1006/viro.1993.1621. [DOI] [PubMed] [Google Scholar]
- 7.Hendry D. Nodaviridae of invertebrates. In: Kurstak E, editor. Viruses of Invertebrates. Marcel Dekker, Inc.; New York: 1991. pp. 227–276. [Google Scholar]
- 8.Newman JFE, Brown FJ. Evidence for a divided genome in nodamura virus, an arthropod-bome picornavirus. J. Gen. Virol. 1973;21:371–384. [Google Scholar]
- 9.Mori K-I, Nakai T, Muroga K, Misao A, Mushiake K, Furusawa I. Properties of a new virus belonging to nodaviridae found in larval stripped jack (Pseudocaranx dentex) with nervous necrosis. Virology. 1992;187:368–371. doi: 10.1016/0042-6822(92)90329-n. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher TM, Rueckert RR. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 1988;62:3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneemann A, Zhong W, Gallagher TM, Rueckert RR. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosur MV, et al. Rueckert RR. Structure of an insect virus at 3.0Å resolution. Proteins. 1987;2:167–176. doi: 10.1002/prot.340020302. [DOI] [PubMed] [Google Scholar]
- 13.Fisher AJ, Johnson JE. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 14.Fricks CE, Hogle JM. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roivainen M, Pürainen L, Rysa T, Närvänen A, Hovi T. An immunodominant N-terminal region of VP1 protein of poliovirion that is buried in crystal structure can be exposed in solution. Virology. 1993;195:762–765. doi: 10.1006/viro.1993.1427. [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Porta C, Chen Z, Baker TS, Johnson JE. Identification of a Fab interaction site (footprint) on a spherical virus by cryo-electron microscopy and X-ray crystallography. Nature. 1992;355:275–278. doi: 10.1038/355275a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith TJ, et al. Baker TS. Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody. J Viro. 1993;67:1148–1158. doi: 10.1128/jvi.67.3.1148-1158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith TJ, Olson NH, Cheng RH, Chase ES, Baker TS. Structure of a human rhinovirus-bivalent antibody complex: implications for virus neutralization and antibody flexibil-ity. Proc Natl Acad Sci USA. 1993;90:7015–7018. doi: 10.1073/pnas.90.15.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zlotnick A, et al. Johnson JE. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J Biol Chem. 1994 in press. [PubMed] [Google Scholar]
- 20.Breindl M. The structure of heated poliovirus particles. J Gem Virol. 1971;11:147–156. doi: 10.1099/0022-1317-11-3-147. [DOI] [PubMed] [Google Scholar]
- 21.Brünger AT. Extension of molecular replacement: a new search strategy based on Patterson correlation refinement. Acta Crystallogr. A. 1990;46:46–57. [Google Scholar]
- 22.Dasmahaptra B, Dasgupta R, Saunders K, Selling B, Gallagher T, Kaesberg P. Infectious RNA derived by transcription from cloned cDNA copies of the genome RNA of an insect virus. Proc Natl Acad Sci USA. 1986;83:63–66. doi: 10.1073/pnas.83.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneemann A, Dasgupta R, Johnson J, Rueckert R. Use of recombinant baculovirus in synthesis of morphologically distinct virus-like particles of Flock House virus, a nodavirus. J. Virol. 1993;67:2756–2763. doi: 10.1128/jvi.67.5.2756-2763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional reconstruction. Biopbys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng RH, Olson NH, Baker TS. Cauliflower mosaic virus: a 420 subunit (T=7), multilayer structure. Virology. 1992;186:655–668. doi: 10.1016/0042-6822(92)90032-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dryden KA, et al. Baker TS. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson NH, Baker TS. Magnification calibration and the determination of spherical virus diameters using cryo-electron microscopy. Ultramicroscopy. 1989;30:281–298. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crowther RA. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Pbilos Trans R Soc. Lond. [Biol] 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 29.Fuller SD. The T4 envelope of sindbis virus is organized by interactions with a complementary T= 3 capsid. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 30.Winkelmann DA, Baker TS, Rayment I. Three-dimensional structure of myosin subfragment-1 from electron microscopy of sectioned crystals. J Cell Biol. 1991;114:701–713. doi: 10.1083/jcb.114.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker TS, Cheng RH, Johnson JE, Olson NH, Wang GJ, Schmidt TJ. Organized packing of RNA inside viruses as revealed by cryo-electron microscopy and X-ray diffraction analysis. Proc. Elec Microsc. Soc. Am. 1992;50:454–455. [Google Scholar]