Summary
The organization of DNA within the HSV-1 capsid has been determined by cryoelectron microscopy and image reconstruction. Purified Ccapsids, which are fully packaged, were compared with A-capsids, which are empty. Unlike A-capsids, C-capsids show fine striations and punctate arrays with a spacing of ~2.6 nm. The packaged DNA forms a uniformly dense ball, extending radially as far as the inner surface of the icosahedral (T = 16) capsid shell, whose structure is essentially identical in A-capsids and Ccapsids. Thus we find no evidence for the inner T=4 shell previously reported by Schrag et al. to be present in C-capsids. Encapsidated HSV-1 DNA closely resembles that previously visualized in bacteriophages T4 and γ, thus supporting the idea of a close parallelism between the respective assembly pathways of a major family of animal viruses (the herpesviruses) and a major family of bacterial viruses.
Introduction
Packaging of viral genomes into viral particles represents a broad class of nucleic acid condensation reactions. In the packaged state, the nucleic acid is usually not metabolically active, but is securely sequestered from the external milieu. Moreover, the configuration assumed by the packaged nucleic acid must be such as to allow its release subsequent to infection, so that transcription or replication may ensue. Although the packaging process has been elucidated, at least in part, in a few viral systems (Black, 1989) in most others, including many medically important and otherwise well-studied viruses, little is known about thle structural organization of the packaged genome or the mlechanisms responsible for its packaging and release.
In packaging, a high degree of condensation must be achieved in order to make efficient use of the limited space available within the viral capsid; and in so doing, the electrostatic repulsion between strands of nucleic acid molecules must somehow be overcome. In some cases this is accomplished by complexing the genome with basic proteins, for instance, cellular histones in the case of papova-viruses. However, since these proteins occupy a significant fraction of the available space, their presence reduces the overall packing efficiency. Particularly high packing efficiencies are achieved in the icosahedral double-stranded DNA bacteriophages, where the DNA duplexes are close packed at a center-to-center spacing of about 2.6 nm (Earnshaw et al., 1976). This constraint has been incorporated into several recent models (Black et al., 1985; Serwer, 1986; Lepault et al., 1987), in all of which the rods of duplex DNA are configured in more-or-less ordered parallel bundles.
Herpesviruses, whose genomes consist of 150-225 kbp of lineardouble-stranded DNA, are among the most genetically complex and structurally elaborate of animal viruses. The herpesvirus capsid is a thick-shelled icosahedron of triangulation number T= 16 (Wildy et al., 1960; Steven et al., 1986) whose structure has been analyzed in some detail by recent studies employing cryoelectron microscopy(Booyet al., 1988) and three-dimensional reconstruction (Baker et al, 1989, 1990; Schrag et al., 1989). Based on electron microscopy of thin sections of both purified and intracellular capsids, and negatively stained isolated capsids, several proposals have been made for the organization of the packaged chromosome (Furlong et al., 1972; Nazerian, 1974; Nii and Yashuda, 1975; Friedman et al., 1975; O'Callaghan and Randall, 1976; Okada et al., 1980). Many of these envisage an arrangement first proposed by Furlong and coworkers (1972) whereby the DNA is wound in a toroidal manner around a cylindrical structure that has different staining properties and presumably a different chemical composition from the spooled DNA. The recent cryoelectron microscopy study by Schrag et al. (1989) reported the presence inside the filled capsid of an inner icosahedral protein shell with a different triangulation number (T=4), with the DNA residing inside that shell, i.e., inside a radius of 31 nm. Taking into account the size of the herpes simplex virus type 1 (HSV-1) genome (McGeoch et al., 1988), the internal dimensions of the capsids of phages T4 and h (Lepault et al., 1987) and the genome sizes of these two phages, we may calculate that the proposal of Schrag et al. (1989) requires that the packing density of DNA (in kbp per unit volume) inside the inner cavity of the HSV-1 capsid should be some 2.5 times higher than in either phage.
To test this proposition, and in the hope of obtaining further information concerning the configuration(s) in which herpesvirus chromosomes are packaged, we have prepared highly purified fractions of HSV-1 capsids of types A (empty) and C (fully packaged). This material has been analyzed by cryoelectron microscopy and image processing, including three-dimensional reconstruction. In addition, the respective protein compositions of the two species of capsid have been compared by SDS-polyacryl-amide gel electrophoresis.
Results
Fingerprint Motif on DNA-Containing HSV-1 Capsids
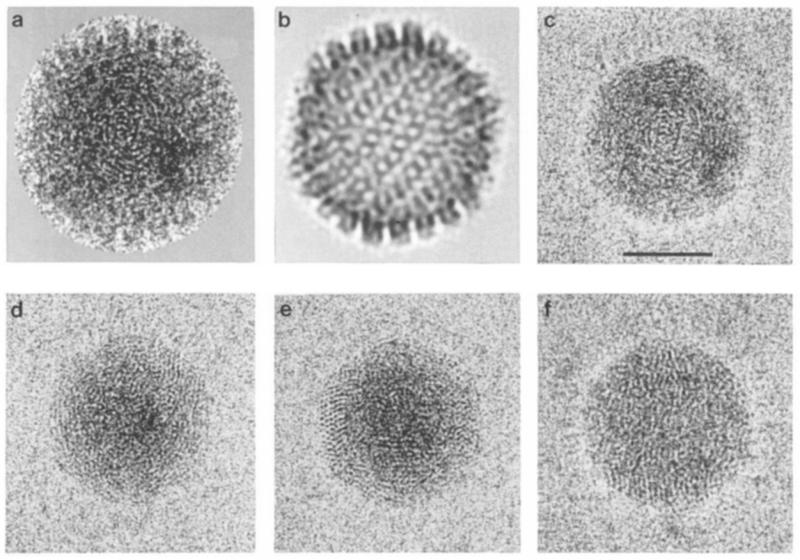
Fields of A-capsids and C-capsidsvisualized in the frozen, hydrated state are shown in Figures 1 a and 1 b, respectively. Viewed at higher magnification (Figure 1c), the C-capsid images are seen to be overlaid with “fingerprints” of finely spaced (~2.5 nm) striations. This pattern varies somewhat from particle to particle: in some cases it resembles a ball of yarn, and the striations may be erther straight or curvilinear. In others the pattern is more punctate but with the same predominant spacing, and in some places suggests a quasi-hexagonal packing. Fingerprints are not discernible on A-capsids that are present in the same field (and thus imaged under the same conditions of ice thickness, defocus, electron dose, etc.). Thus the fingerprint motif is associated only with capsids that contain fully packaged DNA.
Figure 1. Cryoelectron Micrographs of purified Preparatopms of A-Capsids and C-Capsids of HSV-1.
(a) A-capsids. (b) C-capsids. Viewed at higher magnification (c), C-capsids show a distinctive “fingerprint” motif of punctate arrays or finely spaced, curvilinear striations. Bar = 100 nm
The lcosahedral Shells of A-Capsids and C-Capsids Are Indistinguishable
Several three-dimensional reconstructions based on independent data sets were performed for both C-capsids and A-capsids. The signal-to-noise ratios were lower for the filled C-capsids, at least for the signal component that conforms to icosahedral symmetry, which made it somewhat more difficult to determine their orientational parameters (see Experimental Procedures). However, stable solutions were obtained for about 90% of the particles analyzed. The molecular topography of the outer surface of the C-capsid is shown in Figure 2. It bears a very close resemblance to the reconstruction of equine herpesvirus 1 (Baker et al., 1989, 1990) and is similar in most respects to the HSV-1 capsid structure published by Schrag et al. (1989); viz., it shows the hollow hexon and penton protrusions, and “triplexes” at the sites of local 3-fold symmetry between the capsomer protrusions. In this reconstruction the shape of the hexon protrusion is markedly hexagonal, and in this respect matches the equine herpesvirus 1 reconstruction of Baker et al. (1990) more closely than the HSV-1 recon struction of Schrag et al. (1989) which shows rather annular protrusions. On the basis of these reconstructions, we find no significant differences between the outer topography of the C-capsid and that of the A-capsid (the latter data not shown).
Figure 2. Stereo Pairs of the Outer Surface of a Three-Dimensional Reconstruction of C-Capsids of HSV-I.
The particles are viewed along a 5-fold axis of symmetry (upper panels) and a 3-fold axis of symmetry (lower panels).
Internal Structure of HSV-1 Capsids
To allow examination of their internal structures, thin (0.8 nm) central sections from the respective reconstructions are shown in Figure 3. The capsid shells are 14-15 nm thick in both cases, and their structural features are essentially indistinguishable; i.e., such differences as exist are extremely subtle and are no greater than the minor distinctions encountered when two independent reconstructions of the same kind of capsid are compared. This holds true when central sections corresponding to other orientations, e.g., the 3-fold and P-fold axial views, and noncentral sections are compared. We conclude that, to the effective resolution of thisstudy(see Experimental Procedures), the T = 16 icosahedral outer shells of A-capsids and C-capsids are identical.
Figure 3. Transverse Central Sections Taken from the Three-Dimensional Density Maps of C-Capsids and A-Capsids of HSV-1, Reconstructed from Cryoelectron Micrographs.
(a) C-capsid. (b) A-capsid. The particles are oriented as if viewed along a2-fold symmetry axis. The respective outer shell structures closely resemble each other. In the empty A-capsids, the density inside is typically at the same level as the solvent outside. In C-capsids, the density inside is much higher than background but lower than in the shell on account of averaging (see Discussion). In both cases, although the internal structure (or solvent) is not icosahedrally symmetric, the sections show, with low contrast, internal features whose symmetry was imposed by the icosahedral reconstruction procedure. Bar = 50 nm.
These data clearly demonstrate that A-capsids are, as expected, empty (Figure 3b). For C-capsids, inside a radius of about 43 nm, the distribution of positive density is approximately uniform and contains no strongly contrasted features (Figure 3a). These are the properties that one would expect if icosahedral symmetry were to be imposed-as it has been in the reconstruction–on acontinuous distribution of density that does not observe that symmetry. In particular, in the cross-section of the C-capsid (Figure 3a), we see no evidence of the 12 nm thick inner shell reported by Schrag et al. (1989). The same conclusion follows from inspection of serial sections immediately above and below the one shown in Figure 3a, so that the absence of a clearly defined inner shell in that section may not be attributed to sampling the density map in one particular plane. Similar observations also apply to other central and noncentral sections from both of our C-capsid reconstructions.
Filtered Images of Encapsidated Viral DNA
To visualize the internal contents of the C-capsid more clearly, i.e., without interference from both sides of the capsid shell which are viewed in coprojection with the internal structures in the original images, we calculated core images from which the contribution of the capsid shell has been filtered away. In this context it is noteworthy that the capsid contribution is preponderant, since it accounts for about two-thirds of the C-capsid mass (the shell mass being about 200 Md [Newcomb et al., 19891 and that of the genome, about 100 Md [Kieff et al., 1971; McGeoch et al., 19881). In the three-dimensional map, densities inside a radius of 43 nm–corresponding to the inner margin of the capsid shell, which is unambiguously icosahedral–were set to the background solvent level (zero). This modified density map was then reprojected into the correct viewing geometry for the particle of current interest, giving the corresponding two-dimensional projection of its surface shell. This projection was scaled to match the original image and then subtracted from it.
In effect, this computational procedure allows one to peel off the capsid shell and thus obtain an unimpeded view of its internal structure. Several examples are shown in Figure 4. There are no visual indications of icosahedral symmetry in these images, and this conclusion was substantiated by the high values that were obtained when common-line residuals were calculated. The resulting images all show ball-like entities that contain locally ordered bundles of parallel strands, extending to a radius of ~43 nm, i.e., right up to the inner surface of the surrounding icosahedral shell. Indistinguishable results were obtained when the A-capsid reconstruction was substituted in the shell-filtering operation. These structures, with their fingerprint motif, are highly reminiscent of the internal organization of bacteriophages T4 and γ (Richards et al., 1979; Lepault et al., 1987).
Figure 4. Computer-Filtered images of Cores of C-Capsids of HSV-I.
For a typical particle (a), panel (b) represents the contribution of the icosahedral shell to this image, calculated by reprojecting its three-dimensional reconstruction into this viewing geometry. The shell contribution is then digitally subtracted from the original to expose the core (c). All such filtered images show the characteristic “fingerprint” motif of packaged HSV-1 DNA. Panels (d)–(f) show three more examples. The vortex-like aspect of fingerprints is illustrated in (c) and (d), punctate arrays in (d) and (e), and linear striations as well as punctate formations in (f). Bar = 50 nm.
Determination of the Average Interduplex Spacing by Diffraction Analysis
To obtain an objective measurement of the interstrand spacings, we calculated averaged diffraction patterns (powder patterns) from sets of images of both A-capsids and C-capsids (Figure 5). Both patterns show systems of rings at relatively low angles that relate to the most prominent features of the shell structure. However, the pattern intensityfallsoff at high angles more rapidly in the A-capsid pattern (compare Figures 5b and 5c). When azimuthally averaged density scans from the two patterns are appropriately scaled and the resulting difference spectrum is calculated (Figure 5d), it is found to show a single, quite broad peak that covers the range of spacings from (2.4 nm)−1 to (2.9 nm)−1 and centered on (2.6 nm)−1. We conclude, therefore, that the fingerprints represent projections of locally ordered, “liquid-crystalline,” parallel packings of DNA duplexes, whose nearest-neighbor spacings lie in this range.
Figure 5. Powder Pattern Diffractograms Calculated from Cryoelectron Micrographs of HSV-1 Capsids.
(a) Composite pattern from 16 C-capsids, showing a system of concentric rings. Like the individual patterns (not shown), the composite pat-Iern is azimuthally symmetric, indicating a stigmatic micrograph. The intensity falls to zero at a spatial frequency of about (2.3 nm)−1, which is the effective resolution limit of these data, there being only a very faint secondary lobe of contrast transfer. The composite patterns were azimuthally averaged to reduce the noise, and the resulting C-capsid pattern (c) is compared with the A-capsid pattern (b). The particles used in this analysis were all present in the same untilted micrograph, so that this comparison is not biased by any differences in defocus, magnification, etc. The difference spectrum (d) represents primarily the contribution of the packaged DNA. Its peak, at (2.6 nm)−1, marks the average interduplex spacing (arrow).
A-Capsids and C-Capsids Have Indistinguishable Protein Compositions
The respective protein contents of A-capsids and C-capsids were compared by SDS–PAGE analysis of highly purified preparations (Figure 6). No significant differences were detected between them.
Figurd 6. Protein Compositions of A-Capsids and C-Capsids.
SDS-polyacrylamide gel electrophoretogram, stained with Coomassie blue, comparing the protein compositions of A-capsids and C-capsids of HSV-1. The major components are indicated. The two protein compositions are essentially identical.
Discussion
One feature common to most earlier electron microscopic studiesof DNA-containing herpesviruscapsids has been a pronounced variability in appearance among the particles observed. At least part of this variability has been attributable to adverse effects of conventional specimen prepara tion procedures, viz., fixation, dehydration, and selective staining of thin sections, and air drying-induced distortions and variable stain penetration for negatively stained specimens. It is likely that the full range of observed variation also reflects the existence of genuinely different kinds of capsids, and this factor will be discussed further below. In contrast to earlier studies, the C-capsids that were studied here were morphologically homogeneous when visualized by cryoelectron microscopy (Figure 1). Furthermore, in suitably defocused micrographs, C-capsids invariably exhibit the fingerprint motif, depicting the organizational state of the packaged DNA.
The Fingerprint Motif
The fingerprint motif does vary from particle to particle. Some particles exhibit curved azimuthal striations, giving avortex-like pattern (e.g., Figures 4c and 4d); in others the pattern is more punctate (e.g., Figures 4d and 4e) or has sets of striations that run neither azimuthally nor radially (e.g., Figure 4f. It is possible that these distinctions may reflect a toroidal, rather than a totally random, winding of the viral chromosome (Furlong et al., 1972) with the different types of fingerprint representing projections of the toroid in different directions. Alternatively, the images–particularly those that show quasi-hexagonal punctate arrays–are suggestive of arrays of parallel rods in which the linear DNA molecule may be folded back sharply, and perhaps kinked, between neighboring rods (Black et al., 1985; Serwer, 1986).
It should, however, be borne in mind that any given fingerprint represents a complex superposition pattern resulting from the coprojection of a large number of duplexes that are only locally ordered. To determine the complete folding pattern of a viral chromosome would require three-dimensional reconstructions based on tilt series of individual particles, since there is no reason to suppose that the DNA is packed in exactly the same way in each C-capsid (which would allow averaging of different particles). However, to achieve an adequate resolution, i.e. ~2.5 nm, on a core that is ~83 nm in diameter would require approximately 100 views (Klug and Crowther, 1972). In view of the acute radiation sensitivity of frozen-hydrated specimens, this is hardly a feasible proposition.
No Inner Capsid Is Present in C-Capsids of HSV-1
Several lines of evidence lead us to the firm conclusion that the inner icosahedral (T = 4) shell reported by Schrag et al. (1989) to occupy the space between radii 31 nm and 42 nm is not present in the capsids studied here. First, such a structure would require large amounts of some protein(s). However, the protein composition of the C-cap sid is no different from that of the A-capsid (Figure 6), leaving no potential constituents for the putative inner shell. Second, transverse thin sections of our C-capsid reconstructions show no inner shell (e.g., Figure 3). Third, the shell-filtered core images show no indication of icosahedral symmetry, either visually or according to common-lines residuals. Fourth, the DNA-related striations or punc-tate patterns are generally visible beyond a radius of 31 nm, as far as the outer margin of the core at 43 nm, whereas they should be excluded from this region if the putative inner shell were present. Fifth, if all the space out to 43 nm radius were to be available for packaged DNA, then the packing density (total amount of DNA divided by the available volume) would be very similar to that calculated for T4 and γ (see Introduction). This outcome is entirely in agreement with the observation that the average interduplex spacing is very similar (~2.6 nm) in all three viruses (Lepault et al., 1987; see above).
To consider possible explanations for the inner shell reported by Schrag et al. (1989) we note that their filled capsids were drawn from a mixed population of particles. Some of these may possibly have represented different types of capsid (see below), and their inclusion in the reconstruction may have contributed to the interpretation of an inner icosahedral shell. Consistent with this notion, fingerprint motifs were not observed on their filled particles, although this may just as well be a consequence of the microscopy, e.g., degree of defocus, or state of preservation of the particles. This latter consideration, coupled with the fact that they had to combine data from several different micrographs, leads us to suspect that the reconstruction of Schrag et al. (1989) was at significantly lower resolution than the present study. However, it seems equally plausible to us that the “inner shell” may have been generated artifactually by the reconstruction procedure, which tends to impose icosahedral symmetry even if the object analyzed (here, the capsid interior) does not conform to this symmetry. Finally, there is the following consideration: although protein is less dense than DNA, when an icosahedrallysymmetric protein distribution undergoes icosahedral averaging, the resulting density is higher than is obtained when an asymmetric DNA distribution is averaged in the same way, because the higher density of the DNA is diluted through averaging with lower solvent densities. This effect is clearly illustrated in Figure 3. Thus if there were indeed an icosahedral inner shell, one would expect its density in the reconstruction to be as high as that of the protein in the outer shell and substantially higher than that of the underlying, smeared-out, DNA, which is evidently not the case.
The Toroidl Cylinder Model
Furlong et al. (1972) have described the internal structure of the filled capsid of HSV-1 as a toroidal winding of DNA around a central cylindrical structure, and similar interpretations have been reported for electron micrographs of several other herpesviruses (Nazerian, 1974; Nii and Yashuda, 1975; O'Callaghan and Randall, 1976). As discussed above, our data may be consistent with a toroidal winding of the DNA, but are not conclusive on this point. However, the data are stronger to the effect that there are no inner cylinders in our capsids. In terms of this model, the vortex-like fingerprints should presumably represent projections of the toroid along its axis, in which case the DNA-related striations should be excluded from their central regions by the cylinder, but this prediction is not borne out by our data (e.g., Figures 4c and 4d). Moreover, if the cylinder were to be proteinaceous (to account for its different staining properties compared with the toroid), there are no candidate proteins present in our C-capsid preparations. It has been suggested by Friedman et al. (1975) that the toroidlcylinder morphology may represent a developmental intermediate, with a uniformly staining species of particle representing the mature capsid. This proposal could certainly reconcile the two sets of observations, although we are not aware of any compelling argument against the alternative possibility that the toroidal aspect may be an artifact generated when the liquid-crystalline ball of DNAissubjected to fixation, dehydration, etc. In this regard, recent experiments have shown that the morphology of the viral nucleoid in thin sections of HSV may be considerably affected by changes in the dehydration procedure (Puvion-Dutilleul et al., 1987).
Other Capsid Morphologies: Packaging Intermediates or Abortive Assemblies?
Earlier observations have been made of filamentous substructure in HSV capsids that have been penetrated by negative stain (Furlong et al., 1972; Hayet al., 1987). However, the width (5 nm) and appearance of these structures are difficult to reconcile with the much finer (~2.6 nm) filaments described here.
Herpesvirus assembly initiates with the polymerization of a capsid species that is devoid of DNA but has a protein aceous core (Rixon et al., 1988; Lee et al., 1988). (In this discussion, observations pertaining to several different herpesviruses are considered together.) In situ, this core appears as a hollow inner shell that is approximately spherical and may conceivably be icosahedral (discussed by Baker et al., 1990). Its major constituent is the viral protein VP22 (also known as VP22a or ~40) (Newcomb and Brown, 1989; Baker et al., 1990), which is expelled during the course of DNA packaging (Rixon et al., 1988; Sherman and Bachenheimer, 1988). It seems to us likely that there will be some intermediate stages before VP22 has been fully expelled and the DNA fully packaged, in which morphologically distinctive structures may be assumed. The toroidlcylinder model, discussed above, and the 5 nm filament coil are clearly candidates for such intermediate states. Alternatively, different morphological types may represent aberrant particles produced in diversions from the normal assembly pathway.
Capsid Assembly of HSV and of dsDNA Bacteriophages
There are already several lines of evidence that indicate a basic similarity between the respective capsid assembly pathways of herpesviruses and of the icosahedral double-stranded DNA-containing bacteriophages (the latter reviewed by Casjens and Hendrix, 1988). Like phages–but unlike such other animal viruses as the papovaviruses (Baker et al., 1988) or adenovirus (Nermut, 1987)–the major capsid protein of HSV forms hexamers (Furlong, 1978; Steven et al., 1986; Newcomb et al., 1989). As with phages, HSV capsid assembly initiates with polymerization of a DNA-free precursor particle into which DNA is subsequently packaged (Lee et al., 1988). Moreover, the major core protein of the precursor capsid is also expelled during packaging (see foregoing Discussion), just as in phage assembly. The present observations reveal that the internal organization of encapsidated DNA is essentially identical in HSV-1 and phages T4 and γ (Lepault et al., 1987; our unpublished data). Moreover, the average 2.6 nm interduplex spacing closely resembles that observed for liquid crystals of DNA in vitro by cryoelectron microscopy (Richter and Dubochet, 1990) or X-ray diffraction (Rau et al., 1984). It will be acutely interesting to find out how much further this resemblance extends: at any rate, the phage system prototype suggests certain specific avenues that may be explored to further elucidate capsid assembly in the herpesvirus system.
Experimental Procedures
Virus Growth and Capsid Purification
All experiments were carried out with HSV-1 strain 17MP, which was grown at 37°C on monolayer cultures of BHK-21 cells as previously described (Newcomb and Brown, 1989). Capsids were isolated from Infected cells by a modification of the procedure of Perdue et al. (1975). The infected cells were disrupted by sonication in 1% Triton X-100, followed by separation of A-, B-, and C-capsids by centrifugation on linear 20%–50% sucrose gradients prepared in TNE buffer (0.5 M NaCI, 1 mM EDTA, 20 mM Tris-HCI [pH 7.41). After centrifugation, capsid-containing bands were removed from the gradient with a Pasteur pitette, diluted in TNE, and pelleted by centrifugation for 1 hr at 24,000 rpm in an SW28 rotor. Typical preparations beginning with five850cm* roller bottlesof infected cells yielded approximately0.5 mg each of purified A- and C-capsids and l–2 mg of B-capsids. Electron microscopic examination revealed that preparations of each capsid type contained less than 15% contamination with heterologous capsids.
Cryoelectron Microscopy
In a typical experiment, a 2 μl droplet of capsids suspended at ~2 mgl ml in phosphate-buffered saline (PBS) was adsorbed to a thin carbon film for IO s. The grid was then briefly blotted from both sides simultaneously with filter paper to reduce the drop to a thin film, and immediately plunged into liquid ethane at – 180°C in the reservoir of a Reichert KF-80 cryostation (Reichert Scientific Instruments. Buffalo, NY). The grid was transferred into the precooled tip of a Gatan 626 cryoholder (Gatan Inc., Warrendale, PA), which was then inserted into a Philips EM400T electron microscope. After waiting an appropriate interval (usually 10–60 min) to allow for thermal stabilization of the cryoholder and restoration of the vacuum, the grid was searched to locate fields of particles in an ice layer of suitable thickness. During this procedure, the microscope was operated in diffraction mode with the beam defocused to provide a low-magnification (~2000×) image. Micrographs were recorded from suitable areas at a nominal magnification of 36,000 × (Booy et al., 1985), at defocus settings such that the first zero of the phase-contrast transfer function of the microscope occurred at about (2.0 nm)−1. Low-dose techniques, utilizing the Philips beam deflection unit, were used, so that the specimens imaged received exposures of 500–1000 e−/nm2. Magnifications were calibrated according to the 2.49 nm spacing of Olive-T crystallites, and we estimate that the dimensions reported are reliable to within about 1%.
Image Processing
Original negatives were scanned on a Perkin Elmer IOIOMG flat-bed microdensitometer using a scan step and aperture size corresponding to 0.83 nm at the specimen. From the digitized micrographs. all particles that were not visibly damaged or clumped were excised for analysis as images of 165 × 165 pixels. The analysis was performed on a VAX 8350 computer (Digital Equipment Corporation, Maynard, MA) with a Gould 8500 image processor (Vicom, Fremont, CA). The data were preprocessed by subtracting a linear density gradient calculated from the area immediately surrounding each particle, to compensate for slowly varying changes in the thickness of the embedding ice layer. Each image was then normalized to constant mean and variance (Carrascosa and Steven, 1978). Preprocessing and general image processing operations were performed with the PIC system (Trus and Steven, 1981).
An initial centering of each particle was achieved with reference to the cross-correlation function calculated between the image and its inversion (Olson and Baker, 1989). Particle orientations were determined by iterative use of the”crossed common-lines” technique (Fuller, 1987; Baker et al., 1988, 1989), and subsequent refinement of the translational and orientational settings of each particle was performed essentially as described by Baker et al. (1990). In this analysis the orientational parameters of about 90% of the particles were found to converge to stable solutions as assessed by consistently low values of their common-lines phase residuals. These solutions were also confirmed by visual comparison of appropriately oriented reprojections from the reconstruction with the original images. Stable solutions could not be found for the remaining lo%, and these particles were eliminated from the data set. Most likely, these anomalous particles are distorted or damaged in ways that are not overtly apparent.
Three-dimensional reconstructions were performed according to Fouriertechniques (Crowther, 1971; Baker et al., 1990). For A-capsids, two independent reconstructions were performed with data from 16 particles each (at 36,000 × magnification), and the resulting data were finally pooled. ForC-capsids, two reconstructionswere performed: one based on 28 particles from a micrograph at 36,000 × , and the other using 15 particles from a micrograph at 60,000 × Each reconstruction was based on particles from a single micrograph, and all micrographs chosen for analysis had the first zero of their phase-contrast transfer functions at approximately (2.2 nm)−1. The separate data sets of like particles exhibited a high degree of mutual consistency as judged by the consistency observed between independent reconstructions. Moreover, after appropriate adjustment of the scaling, it proved possible to exchange particles between like data sets without incurring a significant deterioration of the common-lines phase residuals.
Calculated according to the conservative criterion of the 45O threshold for the phase residual calculated between two independent three-dimensional data sets, the resolution of the capsid reconstructions was about 3.5 nm. Clearly, the nonicosahedral component of the images contains information that extends to higher resolution, since neighboring DNA strands 2.6 nm apart are clearly resolved. The spatial frequency limit of the data is, in effect, about (2.2 nm)−1 (see Figure 5).
To calculate the powder pattern diffractograms, each capsid image, preprocessed as described above, was floated to zero background value, and its digital diffraction pattern was calculated. Many such patterns were added together with equal weighting. The composite patterns were azimuthally averaged–a valid operation since the micrographs were highly stigmatic–to obtain the spectra shown in Figures 5b and 5c.
The filtered pictures of balls of DNA were obtained as follows: from the C-capsid reconstruction the average radial location of the inner edge of the capsid shell was determined, and all density values inside this radius were set to the solvent value (zero). This modified reconstruction, in which the average core density had thus been computationally “scooped out,” was then reprojected according to the orientational parameters of a particle of interest, to give our best estimate of the capsid's contribution to that image. This capsid projection was then computationally subtracted from the original image after the two images (original and capsid reprojection) were adjusted to the same scaling as described by Baker et al. (1990).
Acknowledgments
This work was supported in part by Public Health Service grants GM34036 (to J. C. 6.) and GM33050 (to T. S. B.) from the National Institutes of Health.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
References
- Baker TS, Drak J, Bina M. Reconstruction of the three-dimensional structure of simian virus 40 and visualization of the chromatin core. Proc. Natl. Acad. Sci. USA. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. The capsid of small papova viruses contains 72 pentameric capsomeres: direct evidence from cryo-electron microscopy of simian virus 40. Biophys. J. 1989;55:243–253. doi: 10.1016/S0006-3495(89)82799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Newcomb WW, Booy FP, Brown JC, Steven AC. Three-dimensional structures of maturable and abortive capsidsofequine herpesvirus 1 from cryoelectron microscopy. J. Virol. 1990;64:563–573. doi: 10.1128/jvi.64.2.563-573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW, Newcomb WW, Boring JW, Brown JC. Ion etching of bacteriophage T4: support for a spiral-fold model of packaged DNA. Proc. Natl. Acad. Sci. USA. 1985;82:7960–7964. doi: 10.1073/pnas.82.23.7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LW. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbial. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Booy FP, Ruigrok RWH, Van Bruggen EFJ. Electron microscopy of influenza virus. A comparison of negatively stained and ice-embedded particles. J. Mol. Biol. 1985;784:667–676. doi: 10.1016/0022-2836(85)90312-2. [DOI] [PubMed] [Google Scholar]
- Booy FP, Newcomb WW, Brown JC, Steven AC. Proc. 46th Annu. Mtg. EMSA. San Francisco Press; San Francisco: 1988. Herpesvirus nucleocapsids visualized in the frozen-hydrated state. pp. 164–165. [Google Scholar]
- Carrascosa JL, Steven AC. A procedure for evaluation of significant structural differences between related arrays of protein molecules. Micron. 1978;9:199–206. [Google Scholar]
- Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The Bacteriophages. Vol. 1. Plenum Publishing Corp.; New York: 1988. pp. 15–91. [Google Scholar]
- Crowther RA. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micro-graphs. Proc. Roy. Sot. Lond. B. 1971;267:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Casjens S, Harrison SC. Assembly of the head of bacteriophage P22: X-ray diffraction from heads, pro-heads, and related structures. J. Mol. Biol. 1976;704:387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- Friedman D, Coward JE, Rosenkranz HS, Morgan C. Electron microscopic studies on the assembly of herpes simplex virus upon removal of hydroxyurea block. J. Gen. Virol. 1975;26:171–181. doi: 10.1099/0022-1317-26-2-171. [DOI] [PubMed] [Google Scholar]
- Fuller SD. The T =4 envelope of sindbis virus is organized by interactions with a complementary T = 3 capsid. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- Furlong D. Direct evidence for 6-fold symmetry of the herpesvirus hexon capsomere. Proc. Natl. Acad. Sci. USA. 1978;75:2764–2766. doi: 10.1073/pnas.75.6.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong D, Swift H, Roizman B. Arrangement of herpesvirus DNA in the core. J. Virol. 1972;70:1071–1074. doi: 10.1128/jvi.10.5.1071-1074.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay J, Roberts CR, Ruyechan WT, Steven AC. Herpesviridae. In: Nermut MV, Steven AC, editors. Animal Virus Structure. Elsevier; Amsterdam: 1987. pp. 391–405. [Google Scholar]
- Kieff ED, Bachenheimer SL, Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus, subtypes 1 and 2. J. Virol. 1971;8:125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A, Crowther RA. Three-dimensional image reconstruction from the viewpoint of information theory. Nature. 1972;238:435–440. [Google Scholar]
- Lee JY, Irmiere A, Gibson W. Primate cytomegalovirus assembly: evidence that DNA packaging occurs subsequent to B-capsld assembly. Virology. 1988;167:87–96. doi: 10.1016/0042-6822(88)90057-8. [DOI] [PubMed] [Google Scholar]
- Lepault J, Dubochet J, Baschong W, Kellenberger E. Organization of double-stranded DNA in bacteriophages: a study by cryo-electron microscopy of vitrified samples. EMBO J. 1987;6:1507–1512. doi: 10.1002/j.1460-2075.1987.tb02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, Perry LJ, Scott JE, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Nazerian K. DNA configuration in the core of Marek's disease virus. J. Virol. 1974;73:1148–1150. doi: 10.1128/jvi.13.5.1148-1150.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nermut MV. Adenoviridae. In: Nermut MV, Steven AC, editors. Animal Virus Structure. Elsevier; Amsterdam: 1987. pp. 373–390. [Google Scholar]
- Newcomb WW, Brown JC. Use of Ar+ plasma etching to localize structural proteins in the capsid of herpes simplex virus type 1. J. Virol. 1989;63:4397–4402. doi: 10.1128/jvi.63.11.4697-4702.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC, Booy FP, Steven AC. Nucleocapsid mass and capsomer protein stoichiometry in equine herpes virus type 1: a scanning transmission electron microscopic study. J. Virol. 1989;63:3777–3783. doi: 10.1128/jvi.63.9.3777-3783.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nii S, Yashuda I. Detection of viral cores having toroid structures in eight herpesviruses. Biken J. 1975;78:41–46. [PubMed] [Google Scholar]
- O'Callaghan DJ, Randall CC. Molecular anatomy of herpesviruses: recent studies. Prog. Med. Virol. 1976;22:152–210. [PubMed] [Google Scholar]
- Okada K, Fujimoto Y, Baba N. Computerized reconstruction of the core of herpesvirus of turkey. J. Electron Microsc. 1980;29:401–402. [PubMed] [Google Scholar]
- Olson NH, Baker TS. Magnification calibration and the determination of spherical virus diameters using cryo microscopy. Ultramicroscopy. 1989;30:281–297. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue ML, Cohen JC, Kemp MC, Randall CC, O'Callaghan DJ. Characterization of three species of nucleocapsids of equine herpes virus type 1. Virology. 1975;64:187–205. doi: 10.1016/0042-6822(75)90091-4. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Pichard E, Laithier M, Leduc EH. Effect of dehydrating agents on DNA organization in herpes viruses. J. Histochem. Cytochem. 1987;35:635–645. doi: 10.1177/35.6.3033063. [DOI] [PubMed] [Google Scholar]
- Rau DC, Lee BK, Parsegian VA. Measurement of the repulsive force between polyelectrolyte molecules in solution: hydration forces between parallel DNA double helices. Proc. Natl. Acad. Sci. USA. 1984;87:2621–2625. doi: 10.1073/pnas.81.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards K, Williams R, Calendar R. Mode of DNA packing within bacteriophage heads. J. Mol. Biol. 1979;78:255–259. doi: 10.1016/0022-2836(73)90114-9. [DOI] [PubMed] [Google Scholar]
- Richter K, Dubochet J. Proc. XIIth Int. Cong. Electron Microsc. Vol. 1. San Francisco Press; San Francisco: 1990. High resolution study of DNA in vitrified sections. pp. 488–489. [Google Scholar]
- Rixon FJ, Cross AM, Addison C, Preston VG. The products of herpes simplex virus gene UL26 which are involved in DNA packaging are strongly associated with empty but not with full capsids. J. Gen. Virol. 1988;69:2879–2891. doi: 10.1099/0022-1317-69-11-2879. [DOI] [PubMed] [Google Scholar]
- Schrag JD, Venkataram Prasad BV, Rixon FJ, Chiu W. Three-dimensional structure of the HSVl nucleocapsid. Cell. 1989;56:851–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- Serwer P. Arrangement of double-stranded DNA in bacteriophage capsids. J. Mol. Biol. 1986;790:509–512. doi: 10.1016/0022-2836(86)90019-7. [DOI] [PubMed] [Google Scholar]
- Sherman G, Bachenheimer SL. Characterization of intranuclear capsids made by ts morphogenetic mutants of HSV-1. Virology. 1988;763:461–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- Steven AC, Roberts CR, Hay J, Bisher ME, Pun T, Trus BL. Hexavalent capsomers of herpes simplex virus type 2: symmetry, shape, dimensions, and oligomeric status. J. Virol. 1986;57:578–584. doi: 10.1128/jvi.57.2.578-584.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Steven AC. Digital image processing of electron micrographs-the PIC system. Ultramicroscopy. 1981;6:383–386. doi: 10.1006/jsbi.1996.0011. [DOI] [PubMed] [Google Scholar]
- Wildy P, Russell WC, Horne RW. The morphology of herpes virus. Virology. 1960;72:204–222. doi: 10.1016/0042-6822(60)90195-1. [DOI] [PubMed] [Google Scholar]