Summary
Cryoelectron microscopy has been used to determine the first structure of a virus when complexed with its glycoprotein cellular receptor. Human rhinovirus 16 (HRV16) complexed with the two amino-terminal, immunoglobulin-like domains of the intercellular adhesion molecule-1 (ICAM-1) shows that ICAM-1 binds into the 12Å deep “canyon” on the surface of the virus. This is consistent with the prediction that the viral receptor attachment site lies in a cavity inaccessible to the host's antibodies. The atomic structures of HRV14 and CD4, homologous to HRV16 and ICAM-1, showed excellent correspondence with observed density, thus establishing the virus-receptor interactions.
Introduction
Human rhinoviruses are one of the major causes of the common cold. They, like other picornaviruses, are icosahedral assemblies of 60 protomers that envelope a single, positive-sense strand of RNA. Each protomer consists of four polypeptides, VP1–VP4. The three external viral proteins (VP1–VP3) each have an approximate molecular weight of 30 000 and a similar folding topology [12, 29]. The external viral radius is ~150 Å and the total molecular weight is roughly 8.5 × 106. A surface depression, or canyon, that is about 12 Å deep and 12–15 Å wide, encircles each pentagonal vertex (Fig. 1c). Residues lining the canyon are more conserved than other surface residues among rhinovirus serotypes [28]. The most variable surface residues are at the sites of attachment of neutralizing antibodies [29, 31]. It has been proposed that the cellular receptor molecule recognized by the virus binds to conserved residues in the canyon, thus escaping neutralization by host antibodies that are too big to penetrate into that region. This hypothesis [27, 29] is supported by site-directed mutagenesis of residues lining the canyon which alters the ability of the virus to attach to HeLa cell membranes [5]. Also, conformational changes in the floor of the canyon, produced by certain antiviral agents that bind into a pocket beneath the canyon floor, inhibit viral attachment to cellular membranes [26]. Conservation of the viral attachment site inside a surface depression has been observed for Mengo [14] and influenza virus [38]. On the other hand, Yeates et al. [40] suggest that for a mouse adapted poliovirus, which has ~36% amino acid identity in VP1, the canyon hypothesis may not be applicable.
Fig. 1.
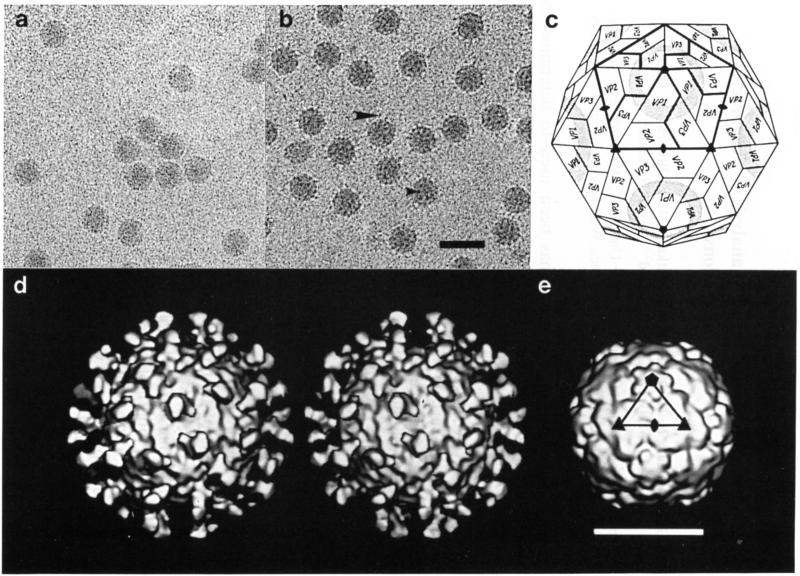
Cryoelectron microscopy of HRV16 particles and their complex with D1D2. a Native HRV16. b HRV16:D1D2 complex. D1D2 molecules (the two amino terminal domains of ICAM-1) are seen edge-on at the periphery of the virions (large arrow), or end-on in projection (small arrow). Cryoelectron microscopy was performed essentially as described by Cheng et al. [4] with images recorded at a nominal magnification of 49000X and with an electron dose of ~20e–/Å2. c Schematic diagram of HRV showing the icosahedral symmetry, subunit organization and canyon (shaded). Thick lines encircle five protomers of VP1, VP2 and VP3. The fourth viral protein, VP4, is inside the capsid. d Stereoview of the reconstruction of the HRV16:D1D2 complex, viewed along an icosahedral twofold axis in approximately the same orientation as in c. Sixty D1D2 molecules are bound to symmetry-equivalent positions at the twelve canyon regions on the virion. The reconstruction was modified to correct for defocus and amplitude contrast effects present in the original micrographs (Cheng, submitted). e Shaded-surface view of HRV14, computed from the known atomic structure [1], truncated to 20 Å resolution. The triangular outline of one icosahedral asymmetric unit corresponding to that in c is indicated. Bar = 500 Å (a,b); 200 Å (d,e)
There are well over 100 human rhinovirus serotypes, which can be divided into roughly two groups according to the cellular receptor they recognize [1]. The structures of human rhinovirus 14 (HRV14) [29] and HRV16 [25a], which belong to the major group of serotypes, and of HRV1A [13], which belongs to the minor group of serotypes, have been determined. There are at least 78 serotypes [36] that bind to intercellular adhesion molecule-1 (ICAM-1), the major group rhinovirus receptor [10, 34]. The ICAM-1 molecule has five immunoglobulin-like domains (D1 to D5 numbered sequentially from the amino end), a transmembrane portion, and a small cytoplasmic domain [33]. Domains D2, D3 and D4 are glycosylated. Unlike immunoglobulins, ICAM-1 appears to be monomeric [34]. Mutational analysis of ICAM-1 has shown that domain D1 contains the primary binding site for rhinoviruses as well as the binding site for its natural ligand, lymphocyte function-associated antigen-1 (LFA-1) [20, 23, 35]. Other surface antigens within the immunoglobulin superfamily that are utilized by viruses as receptors include CD4 for human immunodeficiency virus-1 [6, 15], the poliovirus receptor [25], and the mouse coronavirus receptor [39]. In ICAM-1, in the poliovirus receptor [8, 16], and in CD4 [2] the primary receptor-virus binding site is domain D1. The structures of the two amino-terminal domains of CD4 have been determined to atomic resolution [30. 37]. Truncated proteins corresponding to the two amino-terminal domains of ICAM-1 (tICAM-l(185)) as well as the intact extracellular portion of ICAM-1 (tICAM-l(453) or domains D1 to D5) have been expressed in CHO cells [11]. The desialated form of tICAM-l(185), which will be referred to hereafter as molecule D1D2, has recently been crystallized [17].
The attachment of rhinovirus to the receptor molecule at the cell surface is only the first step of virus uncoating. Subsequent to binding receptor, virus is apparently internalized by receptor-mediated endo-cytosis and enters the endosomal compartment. Productive rhinovirus uncoating and infection requires an intracellular low pH step [21]. In vitro, low pH treatment will convert rhinovirus to both 135S (missing VP4) and 80S (missing VP4 and RNA) subviral particles [18]. A number of studies have shown that poliovirus can be conformationally altered to a 135S form upon interaction with its receptor, and rhinovirus can be converted to an 80S empty capsid by incubation in the presence of soluble ICAM-1 [11]. Thus, both virus-receptor binding and low pH (presumably in the endosomal compartment) appear to play active roles in the controlled disassembly of virus during uncoating, although the relative contributions of these two factors and their temporal relationship in vivo are unclear.
A model of the amino-terminal domain D1 of ICAM-1, based on its homology to known structures of the constant domains of immunoglobulins, was reported by Giranda et al. [9]. Guided by mutational studies of HRV14 and ICAM-1, they were able to fit this model into the known canyon structure of HRV14. We have utilized cryoelectron microscopy and image analysis techniques to calculate a three-dimensional reconstruction of the complex of HRV16 and D1D2 to ~28 Å resolution. The reconstruction clearly shows that the receptor binds into the canyon of rhinovirus as predicted [27, 29]. In addition, we use the known structures of HRV14 and CD4 and the predicted structure of D1 of ICAM-1 to identify atomic interactions.
Structure of the virus: receptor model
Complexes between HRV16 and ICAM-1 D1D2 were prepared by incubating a 3.3 mg/ml solution of HRV16 with a 6.6 mg/ml solution of D1D2 for ~16 hours at 34°C. Under these conditions, saturated complexes of HRV16 with D1D2 can be generated, with approximately 60 moles of D1D2 per mole of virus. HRV14 could also form complexes, although these rapidly broke down to empty capsids (Hoover-Litty and Greve, in prep.) and electron micrographs of such specimens revealed severely disrupted particles in a background of protein. HRV16 complexes with D1D2 or with the complete D1 to D5 extracellular domain fragment were both used in the investigation. We present here only the results obtained on the HRV16:D1D2 complex.
Forty-four electron micrograph images for unstained HRV16:D1D2 complex were combined to compute a three-dimensional reconstruction (Fig. 1d) with an effective resolution of ~28 Å [3]. The density value of the D1D2 feature in the reconstruction was roughly the same as the density of the virion capsid, thus indicating that the D1D2 molecules nearly saturated the 60 available sites on the virion. The position of the ICAM relative to the icosahedral symmetry axes of the virus is unambiguous. Each D1D2 molecule has an approximate dumbbell shape, consistent with the presence of a two-domain structure.
A difference map between the EM model and the 20 Å HRV14 model was computed (Fig. 2). The difference map showed that the D1D2 molecule binds to the central portion of the canyon in a manner roughly as predicted by Giranda et al. [9], confirming the predictions inherent in the canyon hypothesis [27]. The D1D2 molecule is closely associated with the “southern” (see Fig. 3) wall and rim of the canyon, extending 10 Å to 12 Å into the canyon. The binding site is near the center of the triangle formed between a fivefold and two adjacent threefold axes (Fig. 3). The ICAM fragment is oriented roughly perpendicular to the viral surface and extends to a radius of ~205 Å. Its total length is ~75 Å as measured in the difference map.
Fig. 2.
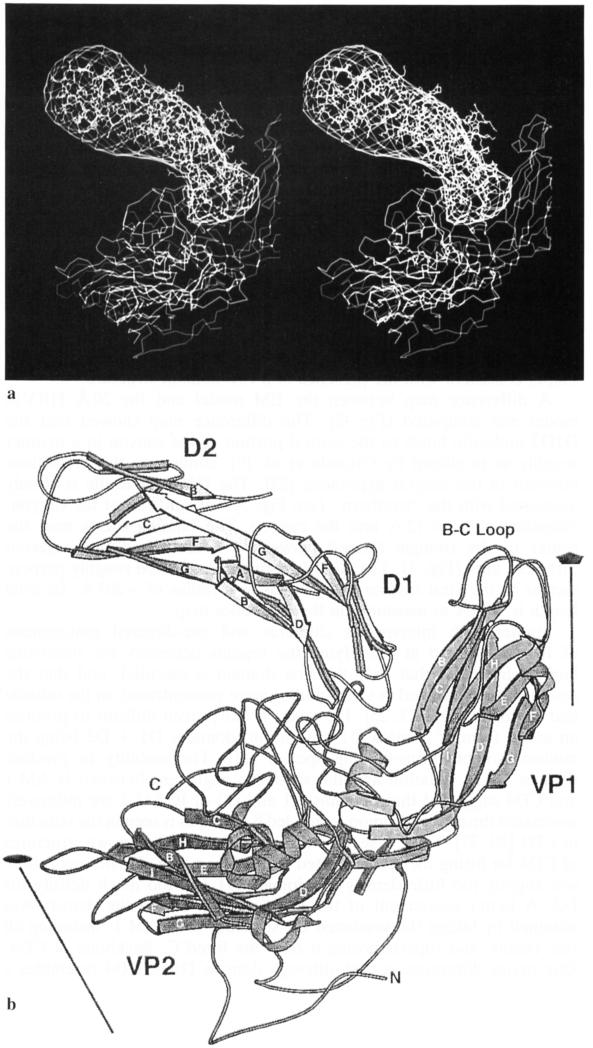
a Stereo diagram showing the fit of the CD4 structure into the difference density between the HRV16:D1D2 reconstruction (Fig. 1d) and the X-ray map of HRV14 (Fig. 1e). A radial scale factor was first determined to slightly adjust the EM model to the accurate dimensions of the X-ray model. The difference electron density has been fitted with the CD4 amino-terminal two-domain structure. A cross-sectional view of the HRV14 structure is also shown outside the ICAM-1 difference density. The additional strands βC′ and βC″ in D1 of CD4 compared to ICAM-1 lie outside the difference density, b A diagrammatic drawing [19] of the structure shown in a. Secondary structural elements of the CD4 fragment and of HRV14 (homologous structures used to represent ICAM-1 and HRV16, respectively) are identified by the standard nomenclature. The N and C termini of VP1 are also marked
Fig. 3.
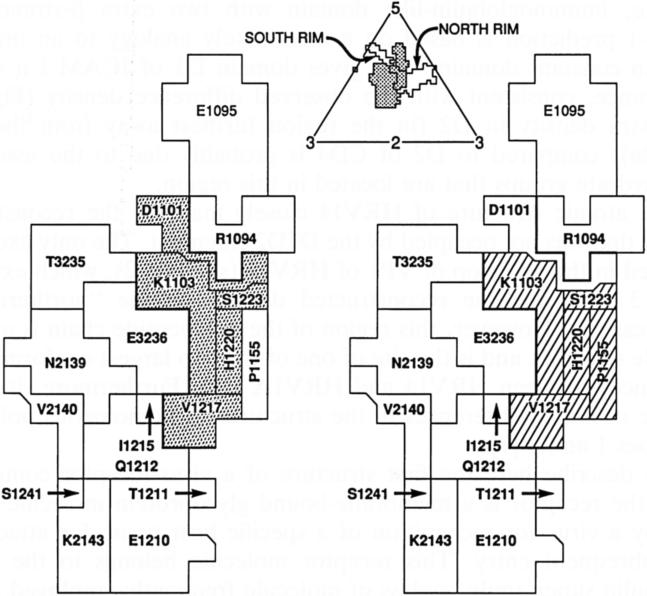
Top: View of the icosahedral asymmetric unit bounded by adjacent five- and threefold axes, outlining residues on the viral surface. Shown are the limits of the canyon, arbitrarily demarcated by a 138 Å radial distance from the viral center [28], and the ICAM-1 footprint (stippled). Improved resolution of the electron density could only marginally alter the HRV residues at the virus:receptor interface. Left and right: Enlarged view of the residues in the ICAM-1 footprint showing (right) the residues which, when mutated, affect viral attachment [5], and (left) the residues altered in structure by the binding of antiviral compounds that inhibit attachment and uncoating [32]
Studies with interspecies chimeras and site-directed mutagenesis of ICAM-1 aimed at identifying the regions necessary for rhinovirus binding indicated that only the first domain is essential, and that the residues most involved in virus binding were concentrated on the outside end of domain D1 [23, 25]. However, it has proven difficult to produce an active form of domain D1 in solution, domains D1 + D2 being the minimal soluble virus-binding species [11]. The inability to produce domain D1 in isolation and the sequence alignment between ICAM-1 and CD4 suggested that domains D1 and D2 of ICAM-1 are intimately associated through a common, extended β-strand as is seen in the structure of CD4 [30, 37]. Thus, it seemed reasonable to use the known structures of CD4 for fitting the reconstructed density map (Fig. 2), although there was slightly too little density for domain D1 and too much density for D2. A better assessment of the fit of domain D1 to the density was obtained by taking the predicted D1 structure of ICAM-1, including all side chains, and superimposing it onto the fitted Cα backbone of CD4. One major difference is that although domain D1 of CD4 resembles a variable, immunoglobulin-like domain with two extra β-strands, the ICAM-1 prediction is based on a more likely analogy to an immunoglobulin constant domain. This gives domain D1 of ICAM-1 a sleeker appearance, consistent with the observed difference density (Fig. 2a). The extra density in D2 (in the region furthest away from the virus (Fig. 2a)) compared to D2 of CD4 is probably due to the associated carbohydrate groups that are located in this region.
The atomic structure of HRV14 closely matched the reconstructed density that was not occupied by the D1D2 fragment. The only exception occurred in the BC loop of VP1 of HRV14 (see Fig. 2), which extended about 3 Å outside the reconstructed density on the “northern” rim of the canyon. However, this region of the polypeptide chain is a highly variable structure and is the site of one of the two largest conformational differences between HRV14 and HRV1A [10]. Furthermore, it is also the site of major differences in the structures of homologous poliovirus serotypes 1 and 3 [7].
We describe here the first structure of a virus-receptor complex in which the receptor is a membrane-bound glycoprotein molecule that is used by a virus for recognition of a specific host tissue for attachment and subsequent entry. This receptor molecule belongs to the immunoglobulin superfamily, a class of molecule frequently employed on cell surfaces for the recognition of other molecules (or recognized by viruses) that are subsequently transferred across the membrane.
Since the general nature of the complex described here had been predicted on the basis of the strategy used by HRV to hide its receptor attachment site, perhaps many other viruses use a similar strategy. Poliovirus is clearly homologous to HRV, and both poliovirus [25] and the major rhinovirus group use an immunoglobulin-like molecule as receptor. Thus, it would be expected that the poliovirus receptor binds into the poliovirus canyon in a manner similar to that of the complex formed for rhinoviruses [8]. The structure of a mouse-adapted chimera of human poliovirus 2 has been determined [40]. The major structural change occurs in the chimera in the BC loop, not in the canyon floor. In this instance, therefore, the BC loop might modulate the virus-receptor interaction. Knowledge of the virus-receptor interaction will illuminate various strategies currently being developed to interfere with early stages of rhinoviral and other viral infections [11, 22, 24].
Acknowledgements
We are grateful for many helpful discussions with R. Rueckert (University of Wisconsin) and M. McKinlay, F. Dutko, G. Diana and D. Pevear (Sterling Winthrop Pharmaceuticals Research Division). We are grateful to M. Kremer, C. Forte, and C. Music for technical assistance. We thank W. Hendrickson and S. Harrison for sharing coordinate information. We also thank H. Prongay and S. Wilder for help in preparation of this manuscript. The work was supported by National Institutes of Health grants to M.G.R. and to T.S.B., a National Science Foundation grant to T.S.B. and a Lucille P. Markey Foundation Award.
References
- 1.Abraham G, Colonno RJ. Many rhinovirus serotypes share the same cellular receptor. J Virol. 1984;51:340–345. doi: 10.1128/jvi.51.2.340-345.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthos J, Deen KC, Chaikin MA, Fornwald JA, Sathe G, Sattentau QJ, Clapham PR, Weiss RA, McDougal JS, Pietropaolo C, Axel R, Truneh A, Maddon PJ, Sweet RW. Identification of the residues in human CD4 critical for the binding of HIV. Cell. 1989;57:469–481. doi: 10.1016/0092-8674(89)90922-7. [DOI] [PubMed] [Google Scholar]
- 3.Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng RH, Olson NH, Baker TS. Cauliflower mosaic virus: a 420 subunit (T = 7), multilayer structure. Virology. 1992;186:655–668. doi: 10.1016/0042-6822(92)90032-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colonno RJ, Condra JH, Mizutani S, Callahan PL, Davies ME, Murcko MA. Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc Natl Acad Sci USA. 1988;85:5449–5453. doi: 10.1073/pnas.85.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalgleish AG, Beverley PCL, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 7.Filman DJ, Syed R, Chow M, Macadam AJ, Minor PD, Hogle JM. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8:1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freistadt MS, Racaniello VR. Mutational analysis of the cellular receptor for poliovirus. J Virol. 1991;65:3873–3876. doi: 10.1128/jvi.65.7.3873-3876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giranda VL, Chapman MS, Rossmann MG. Modeling of the human intercellular adhesion molecule-1, the human rhinovirus major group receptor. Proteins. 1990;7:227–233. doi: 10.1002/prot.340070304. [DOI] [PubMed] [Google Scholar]
- 10.Greve JM, Davis G, Meyer AM, Forte CP, Yost SC, Marlor CW, Kamarck ME, McClelland A. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 11.Greve JM, Forte CP, Marlor CW, Meyer AM, Hoover-Litty H, Wunderlich D, McClelland A. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J Virol. 1991;65:6015–6023. doi: 10.1128/jvi.65.11.6015-6023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 13.Kim S, Smith TJ, Chapman MS, Rossmann MG, Pevear DC, Dutko FJ, Felock PJ, Diana GD, McKinlay MA. The crystal structure of human rhinovirus serotype 1A (HRV1A). J Mol Biol. 1989;210:91–111. doi: 10.1016/0022-2836(89)90293-3. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Boege U, Krishnaswamy S, Minor I, Smith TJ, Luo M, Scraba DG, Rossmann MG. Conformational variability of a picornavirus capsid: pH-dependent structural changes of Mengo virus related to its host receptor attachment site and disassembly. Virology. 1990;175:176–190. doi: 10.1016/0042-6822(90)90198-z. [DOI] [PubMed] [Google Scholar]
- 15.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 16.Koike S, Ise I, Nomoto A. Functional domains of the poliovirus receptor. Proc Natl Acad Sei USA. 1991;88:4104–4108. doi: 10.1073/pnas.88.10.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolatkar PR, Oliveira MA, Rossmann MG, Robbins AH, Katti SK, Hoover-Litty H, Forte C, Greve JM, McClelland A, Olson NH. Preliminary X-ray crystallographic analysis of intercellular adhesion molecule-1. J Mol Biol. 1992;225:1127–1130. doi: 10.1016/0022-2836(92)90110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korant BD, Lonberg-Holm K, Yin FH, Noble-Harvey J. Fractionation of biologically active and inactive populations of human rhinovirus type 2. Virology. 1975;63:384–394. doi: 10.1016/0042-6822(75)90311-6. [DOI] [PubMed] [Google Scholar]
- 19.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J Appl Crystallogr. 1992;24:946–950. [Google Scholar]
- 20.Lineberger DW, Graham DJ, Tomassini JE, Colonno RJ. Antibodies that block rhinovirus attachment map to domain 1 of the major group receptor. J Virol. 1990;64:2582–2587. doi: 10.1128/jvi.64.6.2582-2587.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madshus IH, Olsnes S, Sandvig K. Different pH requirements for entry of the two picornaviruses, human rhinovirus 2 and murine encephalomyocarditis virus. Virology. 1984;139:346–357. doi: 10.1016/0042-6822(84)90380-5. [DOI] [PubMed] [Google Scholar]
- 22.Marlin SD, Staunton DE, Springer TA, Stratowa C, Sommergruber W, Merluzzi VJ. A soluble form of intercellular adhesion molecule-1 inhibits rhinovirus infection. Nature. 1990;344:70–72. doi: 10.1038/344070a0. [DOI] [PubMed] [Google Scholar]
- 23.McClelland A, deBear J, Yost SC, Meyer AM, Marlor CW, Greve JM. Identification of monoclonal antibody epitopes and critical residues for rhinovirus binding in domain 1 of ICAM-1. Proc Natl Acad Sei USA. 1991;88:7993–7997. doi: 10.1073/pnas.88.18.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinlay MA, Pevear DC, Rossmann MG. Treatment of the Picornavirus common cold by inhibitors of viral uncoating and attachment. Annu Rev Microbiol. 1992;46:635–654. doi: 10.1146/annurev.mi.46.100192.003223. [DOI] [PubMed] [Google Scholar]
- 25.Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptors for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin family. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 25a.Oliveira MA, Zhao R, Lee WM, Kremer MJ, Minor I, Rueckert RR, Diana GD, Pevear DC, Dutko FJ, McKinlay MA, Rossmann MG. The structure of human rhinovirus 16. Structure. 1993;1:51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 26.Pevear DC, Fancher MJ, Felock PJ, Rossmann MG, Miller MS, Diana G, Treasurywala AM, McKinlay MA, Dutko FJ. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J Virol. 1989;63:2002–2007. doi: 10.1128/jvi.63.5.2002-2007.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossmann MG. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem. 1989;263:14587–14590. [PubMed] [Google Scholar]
- 28.Rossmann MG, Palmenberg AC. Conservation of the putative receptor attachment site in picornaviruses. Virology. 1988;164:373–382. doi: 10.1016/0042-6822(88)90550-8. [DOI] [PubMed] [Google Scholar]
- 29.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, Rueckert RR, Sherry B, Vriend G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 30.Ryu SE, Kwong PD, Truneh A, Porter TG, Arthos J, Rosenberg M, Dai X, Xuong N, Axel R, Sweet RW, Hendrickson WA. Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature. 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherry B, Rueckert R. Evidence for at least two dominant neutralization antigens on human rhinovirus 14. J Virol. 1985;53:137–143. doi: 10.1128/jvi.53.1.137-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith TJ, Kremer MJ, Luo M, Vriend G, Arnold E, Kamer G, Rossmann MG, McKinlay MA, Diana GD, Otto MJ. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science. 1986;233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 33.Staunton DE, Marlin SD, Stratowa C, Dustin ML, Springer TA. Primary structure of ICAM-1 demonstrates interaction between members of the immunoglobulin and integrin supergene families. Cell. 1988;52:925–933. doi: 10.1016/0092-8674(88)90434-5. [DOI] [PubMed] [Google Scholar]
- 34.Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 35.Staunton DE, Dustin ML, Erickson HP, Springer TA. The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 36.Tomassini JE, Maxson TR, Colonno RJ. Biochemical characterization of a glycoprotein required for rhinovirus attachment. J Biol Chem. 1989;264:1656–1662. [PubMed] [Google Scholar]
- 37.Wang J, Yan Y, Garrett TPJ, Liu J, Rodgers DW, Garlick RL, Tarr GE, Husain Y, Reinherz EL, Harrison SC. Atomic structure of a fragment of human CD4 containing two immunoglobulin-like domains. Nature. 1990;348:411–418. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- 38.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 39.Williams RK, Jiang G-S, Holmes KV. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc Natl Acad Sei USA. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeates TO, Jacobson DH, Margin A, Wychowski C, Girard M, Filman DJ, Hogle JM. Three-dimensional structure of a mouse-adapted type2/type 1 poliovirus chimera. EMBO J. 1991;10:2331–2341. doi: 10.1002/j.1460-2075.1991.tb07772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]