Summary
There are 80 spikes on the surface of Sindbis virus arranged as an icosahedral surface lattice. Each spike consists of three copies of each of the glycoproteins E1 and E2. There are two glycosylation sites on E1 and two on E2. These four sites have been located by removal of the glycosylation recognition motifs using site-specific mutagenesis, followed by cryoelectron microscopy. The positions of these sites have demon strated that E2 forms the protruding spikes and that E1 must be long and narrow, lying flat on the viral surface, forming an icosahedral scaffold analogous to the arrangement of the E glycoprotein in flaviviruses. This arrangement of E1 leads to both dimeric and tri meric intermolecular contacts, consistent with the ob served structural changes that occur on fusion with host cell membranes, suggesting a similar fusion mechanism for alpha- and flaviviruses.
Introduction
Sindbis virus (SINV), Ross River virus (RRV), and Semliki Forest virus (SFV) are enveloped ssRNA viruses belong ing to the Alphavirus genus of the Togaviridae family. These viruses, usually transmitted by mosquitoes, cause fevers, rashes, arthritis, and other diseases (Strauss and Strauss, 1994). The structural and func tional simplicity of alphaviruses compared to other enve loped viruses permits an excellent opportunity for struc tural studies of a lipid-containing virus. As no well diffracting crystals of any alphavirus have yet been formed (Wiley and von Bonsdorff, 1978; Harrison, 1992), previous three-dimensional studies have been based on cryoelectron microscopy (cryoEM) image reconstruc tions of the whole virus (Paredes et al., 1993; Cheng et al., 1995; Fuller et al., 1995; Mancini et al., 2000) and X-ray crystallography studies of component proteins (Choi et al., 1991, 1997).
Alphaviruses encapsidate an ~11,700 base genome within an icosahedral particle. A 26S subgenomic messenger RNA encodes a structural polyprotein that is posttranslationally cleaved into five individual proteins: capsid, E3, E2, 6K, and E1 (Cancedda et al., 1975; Garoff et al., 1980; Schlesinger, 1980; Rice and Strauss, 1981). The capsid protein accumulates in the cytoplasm where it associates with the genomic RNA into spherical core particles. In the mature virion, the nucleocapsid is orga nized into a T = 4 (Caspar and Klug, 1962) symmetric shell, which contains 240 protein subunits and the RNA genome, surrounded by a lipid bilayer that is penetrated by 80 glycoprotein spikes. Each spike is a trimer of E1- E2 heterodimers of the E1 (439 amino acids in SINV) and E2 (423 amino acids in SINV) glycoproteins. N-linked glycosylation sites, each identified by an Asn-X-Ser/Thr motif, occur at residues Asn139 and Asn245 in E1 and at residues Asn196 and Asn318 in E2 (Rice and Strauss, 1981). Virus mutants will be identified by the glycopro tein (E1 or E2), followed by a hyphen (-), and then the amino acid number corresponding to the site of substitu tion (e.g., E1-139). For mutants with multiple substitu tions, each mutation will be separated by a slash (e.g., E1-139/E2-196).
The E1 and E2 glycoproteins undergo N-linked glyco sylation with mannose-rich oligosaccharides (Sefton, 1977) in the lumen of the rough endoplasmic reticulum (Garoff et al., 1978; Bonatti et al., 1979). The glycopro teins are then processed through the Golgi, where their carbohydrate chains are modified, and transported to the plasma membrane (Robbins et al., 1977; Schachter and Roseman, 1980; Schlesinger and Schlesinger, 1986). The budding process involves the interaction of the cytoplasmic domain of E2 with the nucleocapsid core surface, resulting in the release of mature virus from the cell. The E2 glycoprotein has a cell recognition function, whereas E1 facilitates fusion of the viral and cellular membranes (Garoff et al., 1980; Dubuisson and Rice, 1993), which requires an acidic environment (Kielian, 1995) and a cholesterol membrane component (Lu et al., 1999). Cleavage of the p62-E1 precursor into E1, E2, and E3 is the maturation step that makes the virus fusion competent. The E3 protein is absent in mature SINV and RRV particles, but remains noncovalently associated with the spikes in SFV. The initial fusion event results in exposure of new epitopes on E1 and the formation of stable E1 trimers (Schmaljohn et al., 1983; Anthony and Brown, 1991; Ekstrom et al., 1994).
Earlier cryoEM results had shown that the glycopro tein spikes of alphaviruses are arranged with T = 4 symmetry. There are 60 spikes in general positions that have quasi-3-fold (q3) symmetry and 20 spikes situated on icosahedral 3-fold (i3) symmetry axes (Figure 1). Thus, the icosahedral asymmetric unit contains four chemically identical, although structurally slightly differ ent, E1-E2 heterodimers, each of which interacts with a different capsid protein. Three heterodimers comprise one q3 spike and one contributes to the i3 spike. A consequence of the T = 4 symmetry arrangement within the icosahedral shell is that the two different spikes have similar, but not identical, environments and chemical properties (Helenius and Kartenbeck, 1980). Although the choice of delineating any one of the 60 icosahedral asymmetric units is arbitrary, the boundaries shown in Figure 1 are used here.
Figure 1. CryoEM Images of the Wild-type Reconstructions Sindbis Virus.
(a) Depth queued representation overlaid with a T-4 lattice, (b) surface shaded exterior, and (c) cross section. Shown is the icosahedral surface lattice and the chosen icosahedral asymmetric unit. The coordinate system used to define the position of the carbohydrate sites (Table 3) is also indicated. The membrane is seen clearly in (c) and is situated between 205 and 245Å radii.
We have used site-specific mutagenesis to alter the four potential glycosylation sites, either individually or in groups of two or more. We report here preliminary characterizations of some of these mutants and the three-dimensional localization of the carbohydrate moi eties on the virus using cryoEM image difference maps. Knowledge of their positions now shows that the E1 glycoprotein of alphaviruses lies approximately parallel to the viral surface, whereas the E2 glycoprotein forms the protruding spikes. Thus, E1 forms an icosahedral scaffold that organizes the T = 4 architecture of the mature particle. Comparison of alphaviruses with the distantly related flaviviruses suggests that the E1 of alphaviruses might be functionally similar to the E sur face glycoprotein of flaviviruses and, therefore, E1 should undergo a structural transition similar to the E glycoprotein of flaviviruses during the fusion process (Kenney et al., 1994; Ferlenghi et al., 1998).
Results and Discussion
Characterization of the Deglycosylated Mutant Viruses
All of the 15 possible mutational combinations were created among the four potential glycosylation sites on the SINV glycoproteins (Table 1). All substitutions were verified by sequencing the cDNA in the vicinity of the mutated sites. Plaque size and growth rates were mea sured for the wild-type virus and for the majority of the mutant viruses. The single site mutants were found to have either large or medium size plaques; double mu tants had medium size plaques; and triple or quadruple mutants had small or no plaques. The growth properties of the single mutants were slightly reduced relative to wild-type virus. Mutant viruses having two glycosylation sites removed had growth rates reduced by about two orders of magnitude, whereas triple and quadruple sub stituted viruses produced even lower titers.
Table 1.
Properties of Rescued Mutants
| Virus | Plaque Size* | % Relative Growth† | CryoEM |
|---|---|---|---|
| Wild-type | l | 100 | Y |
| Single mutants | |||
| E1-139 | m | 42.8 | Y |
| E1-245 | l | 44.3 | Y |
| E2-196 | l | 47.4 | Y |
| E2-318 | m | 45.5 | Y |
| Double mutants | |||
| E1-139/E1-245 | m | 0.5 | N |
| E1-139/E2-196 | m | 0.5 | N |
| E1-139/E2-318 | m | 3.4 | Y |
| E1-245/E2-196 | m | 0.7 | N |
| E1-245/E2-318 | m | 0.9 | N |
| E2-196/E2-318 | m | 0.8 | N |
| Triple mutants | |||
| E1-139/E1-245/E2-196 | s | ND | N |
| E1-139/E1-245/E2-318 | s | ND | N |
| E1-139/E2-196/E2-318 | np | – | – |
| E1-245/E2-196/E2-318 | s | ND | N |
| Quadruple mutants | |||
| E1-139/E1-245/E2-196/E2-318 | np |
Plaque size: l, large, 4–6 mm; m, medium, 2–4 mm; s, small, 1–2 mm; np, no plaques, after 48 hr in BHK cells.
The growth properties, measured as virus released from BHK cells after 18 hr at an MOI of 1, relative to native virus taken as 100%.
ND, not determined.
Location of the Glycosylation Sites
Although the SINV cryoEM reconstructions shown here contain structural information about all parts of the virion, our analysis and discussion focus on the structure of the glycoprotein spikes. Difference maps (Figures 2 and 3) between the native SINV minus any one of the four single SINV mutant structures showed, in each case, four (A, B, C, and D) well-defined peaks consistent with T = 4 quasi-symmetry within the icosahedral asymmetric unit. These peaks are at least 2.3 times higher than any other peak in the difference map (Table 2), and they were also well resolved with a roughly spherical profile of about 14Å radius. A difference map between the wild-type SINV and the (E1-139/E2-318) double mutant structures showed eight peaks in each asymmetric unit, consistent with the peaks found in the corresponding single mutant difference maps. The peak-to-noise ratio (Table 2) for the carbohydrate difference peaks in the double mutant difference map was similar to that in the single mutant difference maps. The volume of the peaks and their peak heights suggest the presence of about twelve sugar residues per glycosylation site, consistent with the expected molecular masses of about 2 kDa per site (Mayne et al., 1985).
Figure 2. Radial Sections Showing the Native Virus Densities (Top) and Difference Map Densities (Bottom) at the Radii Corresponding to the Different Carbohydrate Sites.
As T = 4 related peaks will be at roughly the same radius, these maps demonstrate the quasi-symmetry for each carbohydrate site. The maps are color coded to correspond to the colors in Figure 3. Shown also is the icosahedral lattice and the outline of the chosen icosahedral asymmetric unit. A 200Å scale bar is shown at the bottom right.
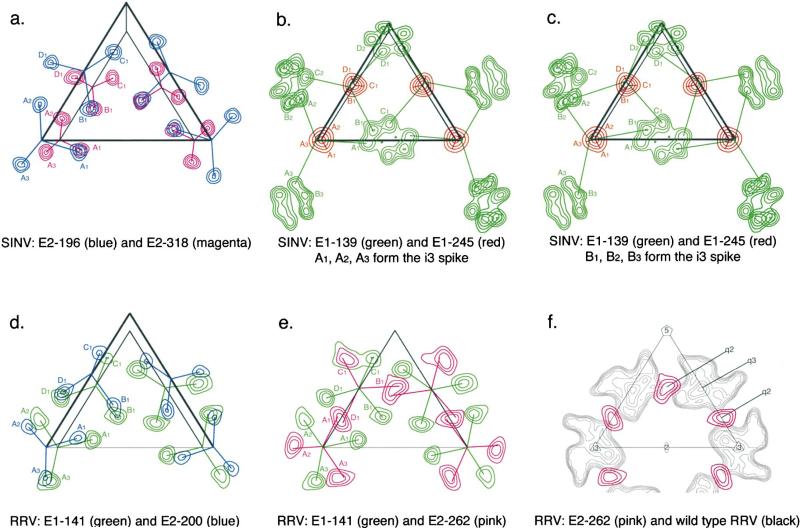
Figure 3. Composite of Difference Map Sections Normal to the Central Icosahedral Fold Axis for SINV (a, b, and c) and RRV (d, e, and f).
(a) E2-196 (blue contours) and E2-318 (magenta contours) mutation sites.
(b and c) E1-139 (green contours) and E1-245 (red contours) mutation sites.
(d) E1-141(green contours) and E2-200 (blue contours).
(e) E1-141 (green contours) and E2-262 (pink contours).
(f) E2-262 (pink contours) and wild-type density (black contours). Appropriately colored lines indicate with which spike each difference peak is associated in (a) through (e). The triangle delineates the icosahedral asymmetric unit (see Figure 1) projected down an icosahedral 2-fold axis on planes that are between 260 and 335Å from the center of the virus. Contours are at 2s intervals. The four quasi-equivalent peaks are marked A, B, C, and D for the T = 4 related mutation sites. Symmetry axes surrounding the icosahedral asymmetric unit and the identification of the i3 and q3 spikes are marked in (f).
Table 2.
EM Data Collection and Processing of Sindbis Virus
| Sample | Defocus levels in microns* | No. of micrographs/no. of particles boxed (used) | Mean correlation coefficient (std.)† | Resolution of the map (Å) | Diff. Map with Respect to Native SINv |
||
|---|---|---|---|---|---|---|---|
| (Carbo. den.)/σ‡ | (Highest bkgd.)/σ‡ | S/N§ | |||||
| Native | 1.42–2.40 | 38/5637 (2926) | 0.407 (0.091) | 22.0 | – | – | – |
| E1-139 | 1.68–2.09 | 15/1366 (933) | 0.426 (0.082) | 23.2 | 15.3 | 3.8 | 4.0 |
| E1-245 | 1.26–2.22 | 7/1462 (928) | 0.407 (0.127) | 22.0 | 27.2 | 3.0 | 9.1# |
| E2-196 | 1.04–2.47 | 11/1024 (754) | 0.410 (0.104) | 21.6 | 7.9 | 3.5 | 2.3 |
| E2-318 | 1.27–2.11 | 17/5986 (3845) | 0.414 (0.125) | 15.0 | 16.4 | 5.4 | 3.0 |
| E1-139/E2-318 | 1.72–2.39 | 10/980 (575) | 0.417 (0.095) | 24.5 | 10.5 | 3.5 | 3.0 |
| Native RRV | 1.20–2.32 | 15/1684 (1111) | 0.394 (0.135) | 22.0 | 5.2 | 1.8 | 2.9 |
Determined from the contrast transfer function of the microscope.
Average real space correlation coefficient for all particles computed between 165 and 350 Å radii of the final model. CC = Σ(ρ1ρ2 - < ρ1 > < ρ2 >) / [Σ(ρ12 - < ρ1 >2) Σ(ρ22 - < ρ2 >2)]0.5
σ is the rms deviation from the mean taken over all grid points for each map.
S/N is signal-to-noise ratio.
SINV E1-245 peaks are higher because three peaks overlap near the spike threefold axes.
The structures of the i3 and q3 spikes were compared by minimizing the sum of the squared distances between equivalent sites (Supplemental Table S1, see below). This required the prior identification, for each mutant virus, as to which of the four quasi-equivalent carbohy drate sites belonged to which spike. The identification was straightforward for the E1-245, E2-196, and E2-318 sites as each of these were associated with the cryoEM density of a specific spike. However, the E1-139 sites were more difficult to assign because they occurred at a relatively large distance (~70Å) from the center of any spike. Calculations showed two reasonable assignments (Table S1) that each gave approximately equal distances of the carbohydrate sites from their associated spike and approximately 3-fold rotational symme try about the axis of the q3 spike.
The best solution (Figure 3c) has site B1 assigned to an i3 spike and sites C1, A2, and D2 to the nearest q3 spike. A slightly poorer solution (Figure 3b) has site A1 assigned to an i3 spike and sites B1, C2, and D1 to the q3 spike. The carbohydrate moieties had the expected T = 4 symmetry to within 1.6Å for the E1-245 and E2-318 sites, 3.4Å for the E2-196 sites, and 7.4Å for the E1-139 sites (Table S1). The larger variation of the E1-139 sites from T = 4 symmetry can be attributed to the nature of quasi-symmetry. One of these four sites (site D associated with a q3 spike) is close to an icosahedral 5-fold axis, whereas the other two sites around the q3 spike are near an icosahedral 2-fold axis, which is also a quasi-6-fold axis (Figures 3b and 3c). Thus, the four sites (A, B, C, and D) have rather different environments in spite of their quasi-equivalence, causing distortions in the spike structures. The E2-196 sites deviate slightly from T = 4 symmetry because they are situated on the extremity of the somewhat flexible spikes, where changes in conformation can be readily accommodated. In contrast, the accurate fit of the E1-245 sites is a result of being near the base and within 4Å from the 3-fold axis of the spike (Figures 3b and 3c). The deviations should be compared with the experimental error of locating the carbohydrate sites in the difference maps, which had an rms error of 1.7Å determined by superposing the independently located i3 sites onto themselves after rotation of 1208 (Table S1). The q3 axes were found to be almost radial in contrast to the orientation of quasi- symmetry axes in other viruses (Rossmann et al., 1983).
RRV and SINV glycoproteins have about 47% amino acid identity to each other. However, RRV has only three potential glycosylation sites per E1-E2 heterodimer, which do not align with the glycosylation sites of SINV. Significant carbohydrate difference peaks were ob tained by computing a SINV minus RRV difference map (Figures 3d–3f), thus providing further positional mark ers on E1 and E2. This map had positive peaks at each of the sets of four, T = 4 related, SINV glycosylation sites and sets of four negative peaks at the three (E1-141, E2-200, and E2-262) RRV glycosylation sites. The RRV peaks could be assigned to the known carbohydrate sites because the E1-141 sites of RRV were close, but not identical, to the E1-139 sites of SINV and, similarly, because the E2-200 sites of RRV were close to the E2-196 sites of SINV. Thus, the remaining, unidentified negative peaks must correspond to the set of T = 4 equivalent RRV E2-262 glycosylation sites. The E2-262 carbohydrate peaks were situated in pairs, close to the quasi-2-fold axes relating the i3 to the q3 spikes and relating the 5-fold-rotated neighboring q3 spikes (Figure 3f). The carbohydrate moieties at E2-262 possibly provide stabilizing interactions between neighboring spikes in RRV, a function absent in SINV. As in SINV, the carbohydrate sites more distant from the center of the spike and, hence, closer to other icosahedral axes, have greater variation from T = 4 symmetry.
The RRV glycosylation site at E1-141 is two residues away from the corresponding site at E1-139 in SINV (sequence alignment of SINV and RRV E1 shows no insertion or deletions), but has much smaller deviations from T = 4 symmetry. RRV carbohydrate associated with residue E2-200, four residues from the SINV E2- 196 site (there are no insertions or deletions between SINV and RRV E2 prior to residue 200), is located on the extremity of the spike like E2-196, deviating about 8.6Å from T = 4 symmetry (Table S1). The distal loca tions of these residues from the viral center might be the cause of disorder in alphavirus crystals (Harrison, 1992).
The distance between the carbohydrate binding sites at E1-139 in SINV and E1-141 in RRV should be the same as for each of the four quasi-equivalent E1 monomers. The mean distance between these sites is 33.6Å, with an rms deviation of 7.6Å for the assignment of E1-139 sites as shown in Figure 3b and 31.5Å and 3.9Å, respectively, for the assignment as shown in Figure 3c. This, therefore, establishes that the assignment of E1-139 carbohydrate sites as shown in Figure 3c is most likely the correct choice.
Superposition of the carbohydrate difference density onto the corresponding wild-type virus density showed that, in every case, the carbohydrate falls well outside the density of the protein (Figure 3f). Indeed, the center of the carbohydrate difference density is about 20Å on average from the nearest significant protein density. The carbohydrate sites associated with E1-139 in SINV and E1-141 in RRV are about 30Å apart, which appears large for two residues that are separated by only two amino acids in the sequence. However, these sites are on op posite sides of an intervening positive protein electron density in the wild-type cryoEM map of SINV. Considering the observed 20Å gap between carbohydrate and protein, it must be concluded that the observed center of gravity of the carbohydrate sites are at least 10Å removed from the Nd atom of the glycosylated Asn.
The absence of any significant difference peaks in any of the difference maps, other than those associated with the carbohydrate sites, strongly suggests that re moval of carbohydrate moieties, or changes in sequence between SINV and RRV, do not induce detectable con formational changes in the structure of the viruses. As the accuracy of placing sites had an error margin of about 2Å for a mass of about 2 kDa (see Experimental Procedures), any structural changes in the viruses that occurred must have been on a considerably smaller scale than that represented by the carbohydrate substi tution. Nevertheless, as described above, there is an approximate two log reduction of virus production con current with removal of two glycosylation sites. Possibly, the absence of carbohydrate hinders the correct folding of the viral proteins and only the correctly folded proteins can be incorporated into infectious virions. Further analysis is required to determine whether there was a decrease in the total number of particles released or an increase in the particle-to-pfu ratio, which would indicate that less infectious virus was being released.
The E1 and E2 Glycoproteins Form an External Scaffold
Given the position of the four SINV and three RRV glycosylation sites (Table 3; Figure 4), it is clear that E1 adopts an extended conformation and lies horizontally over the virus surface between the E1-245 and E1-139 sites, spanning a distance of about 70Å, and that E2 is oriented in a radial direction between E2-318 and E2-196. Thus, E1 can be ascribed to the density identified as the viral “skirt” around the base of each spike (Fuller et al., 1995), whereas E2 forms much of the 80Å long spike protrusions. Residue E2-196 was found on the external side of the petal that forms a receptor recognition motif of alphaviruses (Smith et al., 1995), whereas the site of E2-200 in RRV maps to the lower side of the petal. The carbohydrate moieties associated with RRV residue E2- 200 and SINV residue E2-318 (the next carbohydrate marker along the polypeptide chain) are separated by 77Å, a distance that can be covered comfortably by the 118 amino acids that comprise the stem of the pro truding spike. Residues 413 to 436 of E1 and 365 to 390 of E2 (Rice and Strauss, 1981) form helical transmem brane regions of each glycoprotein (Mancini et al., 2000), which explains the presence of the E2-318 carbohydrate site being only about 24Å above and 47 amino acids away from the lipid bilayer. Indeed, the cryoEM map of SINV shows a strong density feature running from E2-318 to the surface of the membrane. Therefore, this region and the transmembrane domain may be the only sites of association between the E1 and E2 polypeptides that determine the assembly of the heterodimer.
Table 3.
Location of Carbohydrates Sites
| A1 |
B1 |
C1 |
D1 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | x | y | z | x | y | z | x | y | z | x | y | z |
| Sindbis | ||||||||||||
| E1-139 | –5.8 | 33.3 | 296.2 | 15.4 | 23.5 | 294.4 | 26.0 | 0.0 | 298.1 | 135.8 | 8.7 | 266.8 |
| E1-245 | –10.6 | 97.1 | 265.0 | 74.6 | 57.7 | 266.8 | 83.7 | 40.8 | 266.8 | 92.3 | 57.7 | 261.3 |
| E2-196 | –17.3 | 75.0 | 338.6 | 50.6 | 56.3 | 338.6 | 133.5 | 18.8 | 320.7 | 117.3 | 112.9 | 305.4 |
| E2-318 | –15.8 | 71.2 | 268.6 | 49.2 | 50.4 | 268.6 | 97.1 | 21.7 | 259.4 | 96.9 | 78.8 | 250.2 |
| Ross River | ||||||||||||
| E1-141 | 12.5 | 56.2 | 298.1 | 49.0 | 19.2 | 305.4 | 144.6 | 31.7 | 276.0 | 77.9 | 105.8 | 277.3 |
| E2-200 | 15.0 | 80.8 | 338.6 | 64.4 | 25.0 | 334.9 | 149.6 | 48.1 | 310.5 | 86.5 | 103.8 | 312.8 |
| E2-262 | 51.9 | 87.9 | 296.2 | 96.2 | –6.7 | 298.1 | 140.5 | 83.7 | 268.6 | 36.5 | 80.8 | 298.1 |
Figure 4. Stereo View of Carbohydrate Sites (Shown as Difference Density) Associated with One Spike.
The color code is as in Figure 3. Black lines between E1-139 and E1-245 and between E2-196 and E2-318 correspond to E1 and E2 monomers, respectively. The cryoEM density of the wild-type map is shown in light blue for the spike and in yellow for the phospholipid membrane.
If it is assumed that most of the ectodomain of E1 is roughly cylindrical, at least 80Å long and of average density, then the radius of the cylinder would be about 14Å. These cylinders will lie roughly tangential to the exterior of the viral membrane, touching the carbohydrate moieties at E1-245 on one side of the cylinder to avoid crowding three E1 molecules on top of each other at the center of the trimeric spikes. The cylinders can be placed on the viral surface (Figure 5) according to the best assignment of carbohydrate sites in which the E1-139 difference density peak B is associated with an i3 spike (Figure 3c) to give a reasonable fit to the cryoEM density of wild-type SINV. Part of the cylinder representing E1 superimposes onto the E2-318 carbohydrate dif ference density. Not only is it impossible to have E1 and E2 intersect in this way, but the wild-type SINV density at the site of the E2-318 carbohydrate moiety is low (see above). Thus, a cylinder is an oversimplification of the E1 structure, which must have a more curved structure to avoid the E2 polypeptide.
Figure 5. The E1 Icosahedral Scaffold Consistent with the Carbohydrate Sites Corresponding to Figure 3.
Shown also are the sites of the carbohydrate moieties at E1-139, E1-245, and E2-318 colored as described in Figure 3. The orthogonal projection for the whole top hemisphere of the virus (top left) is viewed down an icosahedral 2-fold axis. Details about one spike are shown in the right-hand and bottom panels, viewed down a quasi-3-fold axis and fitted into the slabbed electron density (grey) of the wild-type cryoEM reconstruction. The top panels show the E1 molecules as cylinders, 80Å long and 28Å diameter. The bottom panel shows a stereographic view of the E1 molecule Ca backbone tracing, assuming its homology with the TBEV E glycoprotein. The putative site of the fusion peptide (E1 79 to 97) is shown in red. Note that the latter assumption (as opposed to the cylindrical simplification) avoids steric conflict with the carbohydrate site associated with E2-318.
The arrangement of the E1 glycoproteins shows a pattern of interactions within the surface lattice con sisting of dimers and trimers (Figure 5). The organization can be considered either as being composed of two types of dimers or two types of trimers. One type of dimer forms the interface between the i3 and q3 spikes, whereas the other dimer forms the interface between two neighboring q3 spikes (Figure 5). Each monomer in the two types of dimers represents one of the quasi-equivalent units in the T = 4 lattice. Alternatively, the assembly can be considered as an association of three identical E1 monomers around the i3 spikes and three nonidentical monomers about the q3 spikes. Dimers consist of E1 molecules contacting one another between spikes, promoting lateral spike interactions, whereas trimers consist of E1 molecules within a spike, possibly promoting E2 interactions. The presence of dimers is supported by the observation that lateral interactions between spikes are important for particle formation (von Bonsdorff and Harrison, 1975, 1978; Ekstrom et al., 1994; Strauss and Strauss, 1994). In addition, the role of E1 in forming an icosahedral lattice was suggested on the basis of chemical cross-linking studies that showed the stability of the E1 scaffold to be dependent upon intra- molecular disulfide bridges (Anthony and Brown, 1991).
Many attempts have been made at determining the structure of SINV and RRV nucleocapsid cores by cryoEM (unpublished data) and X-ray crystallography (Harrison et al., 1992). Although cores exhibit a consistent diameter of 410 ± 10Å, no cryoEM reconstructions or crystallization attempts have yielded any success to date. Despite these failures, the T = 4 nucleocapsid core is well ordered and clearly delineated in cryoEM reconstructions of mature virions (Cheng et al., 1995). Thus, isolated cores may lack accurate icosahedral symmetry and, presumably, only become better ordered following their association with the glycoproteins during budding and maturation. Similar conclusions were reached in the study of an SFV capsid mutant that was unable to form cores except in association with the viral glycoproteins during budding (Forsell et al., 2000). The surface of the SINV capsid protein has a hydrophobic pocket (Lee et al., 1996; Skoging et al., 1996). There is good evidence that residues 400 and 402, in the short cytoplasmic end of E2, bind into this pocket. Presumably, it is this association with the T = 4 scaffold structure described above that catalyzes the proper alignment of the nucleocapsid subunits to form the well- ordered, T = 4 cores in the mature virions generated during the budding process.
Comparison of E1 with the TBEV E Surface Glycoprotein
The long, thin, E1 glycoprotein, lying on the surface of the virus, is analogous to the structure of the tick-borne encephalitis virus (TBEV) surface glycoprotein E (Rey et al., 1995). TBEV is a member of the family of flaviviruses previously thought to be structurally similar to alphavi ruses (Murphy, 1980). Flaviviruses possess only one gly coprotein (E), but, instead of a second glycoprotein, they have a small, 8 kDa membrane protein (M). Attempts to align the amino acid sequences of SINV E1 with TBEV E were not very convincing, but did suggest that 32 additional amino acids in TBEV E were an insertion be tween the end of the extracellular region and the transmembrane domain. Thus, if there were a structural similarity between the alphavirus E1 and TBEV E glyco- proteins, then the analogous portions would correspond to the part of TBEV E whose crystal structure has been determined.
Rey et al. (1995) suggested that one of two possible dimers of TBEV E found in their crystal structure was biologically relevant. However, the above amino acid sequence alignment would place residue E1-245 in the very center of the dimer. As the E1-245 site is located very close to the i3 and q3 axes (Figures 3b and 3c), an impossible steric overlap of three different dimers would be created. However, if the other possible crystallo- graphic TBEV E dimer was chosen, then E1-245 would lie at the periphery of the dimer in a position suitable to form E1 trimers around the i3 and q3 positions. Further more, whereas the simple assumption of cylinders rep resenting the E1 structure leaves the carbohydrate sites associated with E2-318 overlapping the E1 cylinders, the structure of the homologous TBEV E1 glycoprotein avoids the steric clash (Figure 5, bottom panel). This possible relationship of SINV E1 and TBEV E is interest ing, but in no way establishes structural homology. Nevertheless, the location of monomers in the proposed structure of TBEV (Ferlenghi et al., 2001) would corre spond to the structure suggested by Figures 3c and 5, which gives the most accurate fit to T = 4 symmetry for the carbohydrate sites. The similarity of the organization of E1 in alphaviruses and E in flaviviruses provides some evidence that at least part of the genome of these related virus families originated from a common primordial an cestor.
A critical event in the life cycle of enveloped viruses is fusion of the viral membrane with that of the host's plasma membrane, allowing the internalization of the viral pathogen into the host cell. The most detailed studies to date of membrane fusion have been reported for influenza virus. Upon lowering of pH, the hemagglutinin glycoprotein of influenza virus undergoes very large conformational changes that result in fusion of the viral and host cell membranes (Carr et al., 1997; Rosenthal et al., 1998; Baker et al., 1999). Fusion of both flaviviruses and alphaviruses is similarily initiated by lowering the pH and results in the reorganization of E or E1 glycoproteins into trimers (Schmaljohn et al., 1983; Kenney et al., 1994; Vénien-Bryan and Fuller, 1994; Allison et al., 1995; Fuller et al., 1995; Kielian, 1995; Ferlenghi et al., 1998; Phinney et al., 2000). The E1 amino acid sequence contains a putative, highly conserved, hydrophobic fusion peptide (residues E1 71 to 97) that has been hypothesized to insert itself into the host cell membrane and thereby trigger fusion (Garoff et al., 1980; Levy-Mintz and Kielian, 1991). Mutational changes within this peptide diminish the interaction between E1 and E2 and inhibit fusion (Kielian et al., 1996).
The structure presented here gives some insight into the fusion process. Assuming the rough sequence align ment of SINV E1 to TBEV E suggested above, the fusion polypeptide would be immediately adjacent to the E2- 318 carbohydrate site. The E2 amino acid sequence immediately upstream to E2-318 contains the Trp-Ile- Val region conserved in alphavirus, suggesting a hy drophobic interaction of the fusion peptide with E2. Low ering the pH will alter the ionic interaction of E1 with its environment and may, therefore, enhance the possibility of conformational changes that had been stabilized by the hydrophobic interaction of the fusion peptide with E2. The presence of both dimeric and trimeric interfaces between E1 monomers in SINV virions, as demonstrated in this study (Figure 5), would make the transition from strong dimer to strong trimer interaction feasible. Fur thermore, diminished interaction of E2 with E1 on low ering pH would possibly allow E2 monomers within a spike to move away from the i3 and q3 axes to permit the trimerization of E1 and the presentation of the fusion peptide to the opposing cellular membrane. Because the organization of E1 and E2 in the SINV surface lattice as determined here has a remarkable similarity to the organization of E and M in a flavivirus, respectively (Fer lenghi et al., 2001), E and E1 may also have functional equivalence for both the fusion process and the forma tion of an icosahedral scaffold. In addition, the mem brane proximal end of the E2 ectodomain may have a function similar to the M protein in flaviviruses.
Experimental Procedures
Mutant Construction and Virus Production
SINV TE12 was used as the wild-type strain of virus because of its high replication efficiency in BHK cells (Lustig et al., 1988). TE12 cDNA was digested with EcoRI and HindIII endonucleases to remove a DNA fragment containing the structural genes of the virus. This fragment was inserted into a similarly digested pGEM-3Zf(+) vector (Promega, Madison, WI) and mutations were introduced into this plasmid by site-directed oligonucleotide mutagenesis using the QuickChange System (Stratagene, La Jolla, CA). The four potential glycosylation sites were independently modified at their respective asparagine residues (Asn-X-Thr/Ser) and converted to glutamine. Due to the possibility of creating a new glycosylation site when E1 Asn245 was substituted with glutamine, a second asparagine at 246 was also mutated to glutamine. In all cases, two nucleotides were substituted at the site of interest to decrease the possibility of rever sion during virus propagation. Positive clones containing the mutations were identified by DNA sequencing and were then digested with restriction endonucleases EcoRI and HindIII. The modified DNA fragment was isolated and used to replace the corresponding wild- type fragment of TE12. Full-length cDNAs, containing two, three, or four mutations, were constructed using an appropriate series of restriction enzyme digestions followed by ligation. Following large-scale preparation of plasmid DNAs, in vitro transcription was carried out as previously described (Rice et al., 1987). RNA transcripts were then used to transfect BHK-15 cells and virus mutants were rescued (Rice et al., 1987; Kuhn et al., 1991). Each mutant virus was plaque purified at least twice (Table 1). Growth of the virus was compared to wild type (TE12) by plaque assay in BHK cells. Virus production for cryoEM was carried out essentially as described by Cheng et al. (1995).
Electron Microscopy
Small (~2.8 ml) aliquots of purified virus samples containing single or double SINV deglycosylated mutants or wild-type SINV or RRV at a concentration of 5–7 mg/ml equilibrated in TNE buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, and 1 mM EDTA) were adhered to carbon-coated electron microscope grids and vitrified in liquid eth ane (Baker et al., 1999). Images were recorded on Kodak SO-163 film in a Philips CM200 field emission gun transmission electron microscope (Philips, Eindhoven, The Netherlands) under low dose conditions (~15 e−/Å) at a nominal magnification of 38,000, except for the E1-196 mutant which was examined using a dose of about 24 e−2/Å and magnification of 50,000 (Table 2). Micrographs were digitized on a Zeiss SCAI scanner using 7 mm intervals, correspond ing to 1.84Å pixels, or 1.40Å pixels for the E1-196 mutant data. The digitized data were used after bin averaging to give 3.68Å (or 2.80Å) size pixels, respectively.
The defocus level was determined for each micrograph and was used to compute the phase-contrast transfer function corrections (Baker et al., 1999). The common lines method (Fuller et al. 1996) was used to initiate reconstruction models of the native and mutant viruses. The orientation and origin of each particle image was determined by the model-based, polar-Fourier transform method (Baker and Cheng, 1996). Orientation refinement was monitored with real- and reciprocal-space correlation coefficients. Eigenvalue spectra were used to assess the conditioning of the linear, least-squares equations (Fuller et al., 1996). The resolution of each map (Table 2) was estimated by splitting the image data into two roughly equal sets and comparing the structure factors, calculated independently for each data set. Reasonable phase agreement was attained to ~22Å resolution.
Computational Analysis
Difference maps (Figures 2 and 3) between pairs of cryoEM recon structions were calculated in real space by subtracting the map of the mutant virus from the map of the wild-type virus after appropriate scaling. Scaling involved three separate linear factors: k, the magnifi cation, which determines the relative pixel size in each map; a, a “float” value, which scales the average height of the density in both maps; and b, which scales the range of density values in each map. Determination of these scale factors required minimization of Σ[ρ1 – (a + b × ρ2)]2, where ρ1 and ρ2 are the densities at equivalent positions in the first and second map, respectively, and the sum is taken over all map grid points within an annulus bounded by radii at 250 and 346Å, representing the ectodomains of the glycoproteins. The magnification factor was determined by adjusting k to give the best correlation between densities within the defined shell (Ross-mann, 2000).
The significance of the carbohydrate difference peaks was established not only by the height of the peaks above the background noise, but also by the accuracy with which the four independent peaks within the icosahedral asymmetric unit obeyed the quasi T = 4 symmetry. This was accomplished with a least-squares minimization between the positions of the T = 4 equivalenced sites using the program HOMOlogy (Rossmann and Argos, 1975). Various types of comparisons were made. The first, i3-i3, was one of fitting the sites A1, A2, A3, B1, B2, B3, ... around the i3 axes onto the sites A2, A3, A1, B2, B3, B1, ..., thus essentially rotating the sites by 1208 (Table S1). As these sites are related by the icosahedral 3-fold axes imposed in the production of the EM maps, the error in superposition was only the result of the measurement of the positions of the carbohydrate difference peaks viewed in slightly different orienta tions on the sections perpendicular to a 2-fold axis (Figure 3). These superpositions, therefore, determined the accuracy of measuring the peak positions. The second comparison, q3-q3, was similar, but related sites around the q3 spikes (Table S1). The differences between equivalenced sites in these comparisons are larger because of a slight deviation from 3-fold symmetry around the q3 spikes. This analysis was repeated in a calculation that restrained the orientation of the quasi-3-fold axis to pass through the center of the virus. The third set of comparisons, i3-q3, relates the peaks around an i3 spike with the peaks around a q3 spike (Table S1). This established the degree of similarity of the two types of spikes and determined the quasi-2-fold relationship between them.
Supplementary Material
Acknowledgments
We are indebted to Felix Rey for information on the SFV E1 crystal structure before publication, and also to Steve Fuller and Erika Man cini for helpful and stimulating discussions on their cryoEM studies of the SFV and TBEV structures and fusion mechanisms. We thank Norm Olson and Rob Ashmore for technical advice regarding the cryoEM data gathering and analysis, respectively. We are grateful to Cheryl Towell and Sharon Wilder for help in the preparation of the manuscript. The work was supported by an NIH Program Project Grant AI45976 that includes T. S. B., R. J. K., and M. G. R., an NIH grant GM56279 to R. J. K., and an NSF shared instrumentation grant to T. S. B. and M. G. R. We also thank Purdue University for an instrumentation reinvestment grant to the Purdue Structural Biology faculty.
Footnotes
Supplemental Table
The Supplemental Table cited above (Table S1) is available online (http://www.cell.com/cgi/content/full/105/1/127/DC1).
References
- Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 1995;69:695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RP, Brown DT. Protein-protein interactions in an alphavirus membrane. J. Virol. 1991;65:1187–1194. doi: 10.1128/jvi.65.3.1187-1194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonatti S, Cancedda R, Blobel G. Membrane biogenesis. In vitro cleavage, core glycosylation, and integration into microsomal membranes of Sindbis virus glycoprotein. J. Cell Biol. 1979;80:219–224. doi: 10.1083/jcb.80.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancedda R, Villa-Komaroff L, Lodish HF, Schlesinger M. Initiation sites for translation of Sindbis virus 42S and 26S messenger RNA. Cell. 1975;6:215–222. doi: 10.1016/0092-8674(75)90012-4. [DOI] [PubMed] [Google Scholar]
- Carr CM, Chaudhry C, Kim PS. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the con struction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Cheng RH, Kuhn RJ, Olson NH, Rossmann MG, Choi HK, Smith TJ, Baker TS. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Lu G, Lee S, Wengler G, Rossmann MG. The structure of Semliki Forest virus core protein. Proteins. 1997;27:345–359. doi: 10.1002/(sici)1097-0134(199703)27:3<345::aid-prot3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Choi HK, Tong L, Minor W, Dumas P, Boege U, Rossmann MG, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- Dubuisson J, Rice CM. Sindbis virus attachment: isola tion and characterization of mutants with impaired binding to vertebrate cell. J. Virol. 1993;67:3363–3374. doi: 10.1128/jvi.67.6.3363-3374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom M, Liljestrom P, Garoff H. Membrane protein lateral interactions control Semliki Forest virus budding. EMBO J. 1994;13:1058–1064. doi: 10.1002/j.1460-2075.1994.tb06354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlenghi I, Gowen B, de Haas F, Mancini EJ, Garoff H, Sjöberg M, Fuller SD. The first step: activation of the Semliki Forest virus spike protein precursor causes a localized con formational change in the trimeric spike. J. Mol. Biol. 1998;283:71–81. doi: 10.1006/jmbi.1998.2066. [DOI] [PubMed] [Google Scholar]
- Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, Harrison SC, Rey FA, Fuller SD. Molecular organization of recombinant subviral particle from tick-borne en cephalitis virus. Mol. Cell. 2001;7:593–602. doi: 10.1016/s1097-2765(01)00206-4. [DOI] [PubMed] [Google Scholar]
- Forsell K, Xing L, Kozlovska T, Cheng RH, Garoff H. Membrane proteins organize a symmetrical virus. EMBO J. 2000;19:5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller SD, Berriman JA, Butcher SJ, Gowen BE. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles—the uncom mon line. J. Struct. Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- Garoff H, Frischauf AM, Simons K, Lehrach H, Delius H. Nucleotide sequences of cDNA coding for Semliki Forest virus membrane glycoprotein. Nature. 1980;288:236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- Garoff H, Simons K, Dobberstein B. Assembly of the Semliki Forest virus membrane glycoproteins in the membrane of the endoplasmic reticulum in vitro. J. Mol. Biol. 1978;124:587–600. doi: 10.1016/0022-2836(78)90173-0. [DOI] [PubMed] [Google Scholar]
- Harrison SC. Viruses. Curr. Opin. Struct. Biol. 1992;2:293–299. [Google Scholar]
- Harrison SC, Strong RK, Schlesinger S, Schlesinger MJ. Crystallization of Sindbis virus and its nucleocapsid. J. Mol. Biol. 1992;226:277–280. doi: 10.1016/0022-2836(92)90141-6. [DOI] [PubMed] [Google Scholar]
- Helenius A, Kartenbeck J. The effects of octylglucoside on the Semliki Forest virus membrane: evidence for a spike-protein-nucleocapsid interaction. Eur. J. Biochem. 1980;106:613–618. doi: 10.1111/j.1432-1033.1980.tb04609.x. [DOI] [PubMed] [Google Scholar]
- Kenney JM, Sjöberg M, Garoff H, Fuller SD. Visualization of fusion activation in the Semliki Forest virus spike. Structure. 1994;2:823–832. doi: 10.1016/s0969-2126(94)00083-2. [DOI] [PubMed] [Google Scholar]
- Kielian M. Membrane fusion and the alphavirus life cycle. Adv. Virus Res. 1995;45:113–151. doi: 10.1016/s0065-3527(08)60059-7. [DOI] [PubMed] [Google Scholar]
- Kielian M, Klimjack MR, Ghosh S, Duffus WA. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 1996;134:863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Niesters HGM, Hong Z, Strauss JH. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182:430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- Lee S, Owen KE, Choi HK, Lee H, Lu G, Wengler G, Brown DT, Rossmann MG, Kuhn RJ. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- Levy-Mintz MP, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J. Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YE, Cassese T, Kielian M. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J. Virol. 1999;73:4272–4278. doi: 10.1128/jvi.73.5.4272-4278.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig S, Jackson AC, Hahn CS, Griffin DE, Strauss EG, Strauss JH. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 1988;62:2329–2336. doi: 10.1128/jvi.62.7.2329-2336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini EJ, Clarke M, Gowen B, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- Mayne JT, Bell JR, Strauss EG, Strauss JH. Pattern of glycosylation of Sindbis virus envelope proteins synthesized in hamster and chicken cells. Virology. 1985;142:121–133. doi: 10.1016/0042-6822(85)90427-1. [DOI] [PubMed] [Google Scholar]
- Murphy FA. Togavirus morphology and morphogenesis. In: Schlesinger RW, editor. The Togaviruses. Academic Press; New York: 1980. pp. 241–316. [Google Scholar]
- Paredes AM, Brown DT, Rothnagel R, Chiu W, Schoepp RJ, Johnston RE, Prasad BVV. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney BS, Blackburn K, Brown DT. The surface conformation of Sindbis virus glycoproteins E1 and E2 at neutral and low pH, as determined by mass spectrometry-based mapping. J. Virol. 2000;74:5667–5678. doi: 10.1128/jvi.74.12.5667-5678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rice CM, Strauss JH. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc. Natl. Acad. Sci. USA. 1981;78:2062–2066. doi: 10.1073/pnas.78.4.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CM, Levis R, Strauss JH, Huang HV. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PW, Hubbard SC, Turco SJ, Wirth DF. Proposal for a common oligosaccharide intermediate in the synthesis of membrane glycoproteins. Cell. 1977;12:893–900. doi: 10.1016/0092-8674(77)90153-2. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Zhang X, Formanowski F, Fitz W, Wong C-H, Meier-Ewert H, Skehel JJ, Wiley DC. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG. Fitting of atomic models into electron microscopy maps. Acta Crystallogr. 2000;D56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Argos P. A comparison of the heme binding pocket in globins and cytochrome b5. J. Biol. Chem. 1975;250:7525–7532. [PubMed] [Google Scholar]
- Rossmann MG, Abad-Zapatero C, Hermodson MA, Erickson JW. Subunit interactions in southern bean mosaic virus. J. Mol. Biol. 1983;166:37–83. doi: 10.1016/s0022-2836(83)80049-7. [DOI] [PubMed] [Google Scholar]
- Schachter H, Roseman S. Mammalian glycosyltransferases: their role in the synthesis and function of complex carbohydrates and glycolipids. In: Lennarz WJ, editor. The Biochemistry of Glycoproteins and Proteoglycans. Plenum Press; New York: 1980. pp. 85–160. [Google Scholar]
- Schlesinger RW. Biology, Structure, Replication. Academic Press; New York: 1980. The Togaviruses. [Google Scholar]
- Schlesinger MJ, Schlesinger S. Formation and assembly of alphavirus glycoproteins. In: Schlesinger S, Schlesinger MJ, editors. The Togaviridae and Flaviviridae. Plenum; New York: 1986. pp. 121–148. [Google Scholar]
- Schmaljohn AL, Kokubun KM, Cole GA. Protective monoclonal antibodies define maturation and pH-dependent antigenic changes in Sindbis virus E1 glycoprotein. Virology. 1983;130:144–154. doi: 10.1016/0042-6822(83)90124-1. [DOI] [PubMed] [Google Scholar]
- Sefton BM. Immediate glycosylation of Sindbis virus membrane protein. Cell. 1977;10:659–668. doi: 10.1016/0092-8674(77)90099-x. [DOI] [PubMed] [Google Scholar]
- Skoging U, Vihinen M, Nilsson L, Liljeström P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Cheng RH, Olson NH, Peterson P, Chase E, Kuhn RJ, Baker TS. Putative receptor binding sites on alpha-viruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA. 1995;92:10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vénien-Bryan C, Fuller SD. The organization of the spike complex of Semliki Forest virus. J. Mol. Biol. 1994;236:572–583. doi: 10.1006/jmbi.1994.1166. [DOI] [PubMed] [Google Scholar]
- von Bonsdorff CH, Harrison SC. Sindbis virus glycoproteins form a regular icosahedral surface lattice. J. Virol. 1975;16:141–145. doi: 10.1128/jvi.16.1.141-145.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bonsdorff CH, Harrison SC. Hexagonal glycoprotein arrays from Sindbis virus membranes. J. Virol. 1978;28:578–583. doi: 10.1128/jvi.28.2.578-583.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley DC, von Bonsdorff CH. Three-dimensional crystals of the lipid-enveloped Semliki Forest virus. J. Mol. Biol. 1978;120:375–379. doi: 10.1016/0022-2836(78)90425-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.