Abstract
OBJECTIVE
A genetic risk score (GRS) comprised of single nucleotide polymorphisms (SNPs) and metabolite biomarkers have each been shown, separately, to predict incident type 2 diabetes. We tested whether genetic and metabolite markers provide complementary information for type 2 diabetes prediction and, together, improve the accuracy of prediction models containing clinical traits.
RESEARCH DESIGN AND METHODS
Diabetes risk was modeled with a 62-SNP GRS, nine metabolites, and clinical traits. We fit age- and sex-adjusted logistic regression models to test the association of these sources of information, separately and jointly, with incident type 2 diabetes among 1,622 initially nondiabetic participants from the Framingham Offspring Study. The predictive capacity of each model was assessed by area under the curve (AUC).
RESULTS
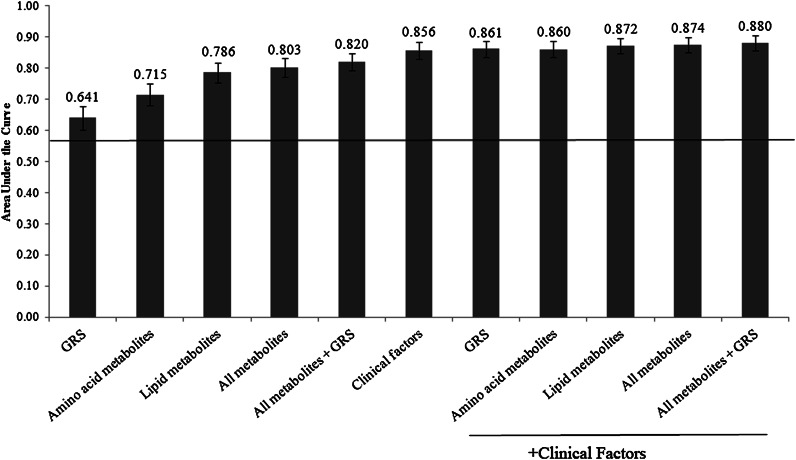
Two hundred and six new diabetes cases were observed during 13.5 years of follow-up. The AUC was greater for the model containing the GRS and metabolite measurements together versus GRS or metabolites alone (0.820 vs. 0.641, P < 0.0001, or 0.820 vs. 0.803, P = 0.01, respectively). Odds ratios for association of GRS or metabolites with type 2 diabetes were not attenuated in the combined model. The AUC was greater for the model containing the GRS, metabolites, and clinical traits versus clinical traits only (0.880 vs. 0.856, P = 0.002).
CONCLUSIONS
Metabolite and genetic traits provide complementary information to each other for the prediction of future type 2 diabetes. These novel markers of diabetes risk modestly improve the predictive accuracy of incident type 2 diabetes based only on traditional clinical risk factors.
Introduction
Type 2 diabetes is estimated to affect >550 million people worldwide by 2030 (1). Given the personal and health care system costs associated with this growing epidemic, it is critical to identify high-risk individuals as a first step in providing effective preventive interventions for diabetes (2). Although the medical history and standard laboratory testing provide clues as to an individual’s future risk of diabetes (3), many of these predictors emerge only after years of subclinical metabolic dysfunction (4). Novel markers may help to elucidate aspects of metabolic dysfunction contributing to diabetes risk and improve the early identification of individuals who might benefit from preventive therapies for diabetes.
Genetic information, specifically, single nucleotide polymorphisms (SNPs) grouped into a genetic risk score (GRS), has been shown to predict type 2 diabetes incidence alone and in the context of clinical prediction models within the Framingham Offspring Study (5,6) as well as within multiple independent cohorts (7–10). However, genetic information alone generally has low predictive performance for type 2 diabetes incidence (11) and only modestly improves accuracy of prediction by traditional clinical risk factors for type 2 diabetes. Circulating metabolite biomarkers, specifically, branched-chain and aromatic amino acids (12), lipid species of particular acyl chain length composition and saturation (13), the glutamine-to-glutamate ratio (14), and 2-aminoadipic acid (15), have also been shown to predict diabetes risk independent of clinical factors such as sex, BMI, and fasting glucose. These metabolite associations have been demonstrated in the Framingham Offspring Study (12–15) and validated in independent cohorts (12,14–17).
While the majority of diabetes-related SNPs with known physiological associations have been linked to β-cell function (18,19), amino acid and lipid metabolites have been strongly associated with measures of insulin resistance (12,13,16,20–24). We hypothesized that genetic and metabolite biomarkers of diabetes risk, by capturing different pathogenic elements for type 2 diabetes development, would provide complementary information to each other regarding future diabetes risk and possibly improve clinical models of diabetes prediction in aggregate. We therefore evaluated the ability of a type 2 diabetes GRS, containing 62 diabetes-related SNPs derived from the most recent association findings (25), and circulating metabolites involved in cardiometabolic risk pathways, separately and together, to predict diabetes and improve discrimination of incident type 2 diabetes compared with conventional clinical predictors.
Research Design and Methods
The Framingham Offspring Study was initiated in 1971 (26), and participants were examined approximately every 4 years. The fifth examination of the Framingham Offspring Study, which was conducted between 1991 and 1995, was considered the baseline assessment for the current study. Genotyping and metabolite profiling were performed on archived plasma samples collected from participants with at least one exam cycle of follow-up from the baseline examination (26). Individuals with known diabetes (defined as fasting plasma glucose ≥7.0 mmol/L, glucose at 2 h after a standard 75-g oral glucose load ≥11.1 mmol/L, or use of antidiabetes therapy) at the baseline examination or without a sample for genotyping or metabolite profiling were excluded. Diabetes incidence was defined by plasma glucose level ≥7.0 mmol/L or use of antidiabetes therapy, and time to diabetes incidence was derived from the time of the baseline examination.
Measurement methods for amino acid and intermediary metabolites (12,14,15), as well as lipid metabolites (13), have previously been reported. A previous study has documented concordance in amino acid metabolite measures between archived samples from the Framingham Offspring Study and freshly obtained samples (27). Peak areas of internal standards were monitored for quality control, and individual samples with peak areas from individual samples differing from the group mean by more than two SDs were reanalyzed. Metabolite peaks were manually reviewed for quality of integration and compared against a known standard to confirm identity. Replicates derived from a single pooled plasma sample were run after every 30 experimental samples, enabling detection of temporal drift in instrument performance.
In age- and sex-adjusted logistic regression models testing the association of amino acid metabolites or lipid metabolites with incident diabetes, backward stepwise regression was used to remove metabolite traits sequentially until each remaining trait was associated with diabetes at P value <0.05. The initial models containing all amino acid and lipid metabolite traits and those following stepwise regression are shown in Supplementary Tables 1 and 2, respectively; the metabolite traits retained following backward stepwise regression were used in subsequent prediction models.
Sixty-two autosomal SNPs, each previously associated with type 2 diabetes at genome-wide significance (25), were genotyped or imputed, including 22 new SNPs from prior diabetes prediction analyses (6). Genotypes were obtained from Affymetrix array data in the Framingham Offspring Study or from de novo genotyping on the iPLEX (Sequenom) platform. As in prior reports (5,6), minimum call rates were 97% for Affymetrix and 96.9% for iPLEX SNPs. All SNPs were in Hardy-Weinberg equilibrium. To create the 62-SNP GRS, the number of risk alleles at each SNP (0, 1, or 2) was multiplied by its published β-coefficient for diabetes risk (28), and then these weighted alleles were summed across the 62 loci (19). This 62 SNP GRS has been associated with type 2 diabetes in another study from the Framingham Offspring Study (n = 3,869) and the Coronary Artery Risk Development in Young Adults (n = 2,470) study (25). Statistical association of each SNP, independently, with type 2 diabetes in the cohort examined in the current study was not required for inclusion into the GRS. The SNP components of the GRS with published β-coefficient are displayed in Supplementary Table 3.
Association tests were performed in sex- and age-adjusted models containing the 62-SNP GRS, amino acid metabolites, lipid metabolites, and previously validated clinical predictors of incident diabetes: family history of diabetes, BMI, systolic blood pressure, HDL cholesterol, triglycerides, and fasting blood glucose (3). Logistic regression models tested the association of these variables with diabetes onset in the overall cohort and in subgroups stratified by age (<50 and ≥50 years) and BMI (<30 and ≥30 kg/m2). Within-study model validity was assessed using a jackknife procedure with 10 random samples of 90% of the cohort, as has been done previously (3). Linear regression was used to test the association of the same variables with the log-transformed homeostasis model assessment of insulin resistance (log HOMA-IR) (29) and β-cell function (log HOMA-B) (29) at the baseline assessment in age-, sex-adjusted models. Model discrimination and reclassification were evaluated using areas under the receiver operating characteristic curve (AUC) and continuous net reclassification improvement indices (NRI), respectively (30,31). An NRI of 0.2 was interpreted as weak improvement, NRI of 0.4 as intermediate moderate improvement, and NRI of 0.8 as strong improvement (31). Analyses were conducted using SAS software (version 9.2; SAS Institute, Cary, NC). A two-tailed P value <0.05 indicated statistical significance.
The institutional review boards at Boston University and the Partners Human Research Committee approved the study. All participants gave written informed consent.
Results
Baseline characteristics of the 1,622 participants with clinical, genotype, and metabolite profiling measurements and at least 4 years of follow-up information from the Framingham Offspring Study are displayed in Table 1. In total, there were 206 new diabetes cases over an average follow-up of 13.5 years. The mean (SD) time to diabetes incidence was 9.2 (4.3) years. When examined in aggregate, three amino acids (isoleucine, tyrosine, and phenylalanine) and six lipid metabolites (C18:2 lysophosphatidylcholine [LPC], C38:6 phosphatidylcholine [PC], C44:1 triacylglycerol [TAG], C48:0 TAG, C52:1 TAG, and C56:9 TAG) remained associated with incident diabetes and were included in subsequent prediction models.
Table 1.
Baseline characteristics of participants
| All | With incident type 2 diabetes | Without incident type 2 diabetes | |
|---|---|---|---|
| N | 1,622 | 206 | 1,416 |
| Age (years) | 54.7 ± 9.63 | 56.67 ± 8.94 | 54.41 ± 9.7 |
| Female | 841 (51.9) | 86 (41.8) | 755 (53.3) |
| BMI (kg/m2) | 27.32 ± 4.7 | 30.33 ± 5.03 | 26.88 ± 4.49 |
| Systolic blood pressure (mmHg) | 125.32 ± 18.33 | 134.33 ± 17.83 | 124.01 ± 18.03 |
| HDL cholesterol (mmol/L) | 1.30 ± 0.39 | 1.09 ± 0.29 | 1.33 ± 0.39 |
| Fasting triglycerides (mmol/L) | 1.63 ± 1.21 | 2.27 ± 1.44 | 1.54 ± 1.14 |
| Fasting plasma glucose (mmol/L) | 5.29 ± 0.55 | 5.83 ± 0.52 | 5.21 ± 0.51 |
| Log HOMA-IR (mmol ⋅ pmol/L2) | 1.89 ± 0.4 | 2.18 ± 0.4 | 1.8 ± 0.3 |
| Log HOMA-B (pmol/mmol) | 1.81 ± 0.3 | 1.91 ± 0.3 | 1.8 ± 0.3 |
Data are means ± SD or n (%).
The AUC for each of the prediction models is shown in Fig. 1. The AUC was greater for age- and sex-adjusted models containing amino acid or lipid measurements than for the model containing the GRS (P < 0.0001 for both comparisons). The AUC for the model containing both the GRS and metabolite measurements was greater than the AUC for models with either set of predictors alone (P < 0.0001 for GRS alone vs. GRS and metabolites; P=0.01 for metabolites alone vs. GRS and metabolites). The reclassification capacity of the metabolite prediction model was improved by the addition of the GRS (NRI = 0.421, P < 0.0001), and the reclassification capacity of the GRS prediction model was improved by the addition of the metabolites (NRI = 0.796, P < 0.0001).
Figure 1.
AUC for prediction models for incident type 2 diabetes. All models are adjusted for age and sex, and the line indicates the AUC for the model containing only age and sex as predictors. Error bars indicate 95% CI. The models include 1,622 participants with clinical, metabolite, and genetic information.
Addition of metabolomic information alone but not the GRS alone increased the AUC of the model containing clinical risk factors (P = 0.002 for addition of all metabolites and P = 0.08 for addition of GRS). Addition of metabolomic information increased the AUC of the model containing the GRS and clinical risk factors (P = 0.007). Within-study validity testing demonstrated a narrow range of AUCs for each model (≤0.22); in each case, the AUC from the model in the entire cohort was at or near the center of this range (Supplementary Table 4).
The ability of the standard clinical model in reclassifying the risk for type 2 diabetes was improved by the addition of the GRS alone (NRI = 0.247, P = 0.0009), by metabolites alone (NRI = 0.442, P < 0.0001), and by GRS and metabolites together (NRI = 0.576, P < 0.0001). The AUC of each prediction model, including that incorporating clinical factors, metabolites, and GRS, was greater for participants <50 vs. ≥50 years old (Supplementary Fig. 1) and for participants with BMI <30 vs. ≥30 kg/m2 (Supplementary Fig. 2). Within the model containing clinical, GRS, and metabolite information, participants in the lowest and highest deciles of predicted diabetes risk had actual diabetes rates of 1.2 and 56.0%, respectively (Supplementary Fig. 3).
Odds ratios (ORs) for the GRS and metabolite traits within prediction models, separately and jointly, are shown in Table 2. The OR for the association of the GRS with incident type 2 diabetes was not attenuated by the addition of metabolite data and remained significant when considered alone or in the combined GRS and metabolites model. Similarly, the OR for the association of metabolite traits was not attenuated by the addition of the GRS, and phenylalanine, C18:2 LPC, C38:6 PC, C44:1 TAG, and C52:1 TAG were significant when considered in the metabolites-only model or the combined GRS and metabolites model. The ORs for the GRS and metabolite traits with incident type 2 diabetes remained consistent when considered with clinical factors (Supplementary Table 5).
Table 2.
OR for incident type 2 diabetes associated with a GRS and serum metabolites in separate and combined models
| Parameter |
GRS | Metabolites | GRS + metabolites | |||
|---|---|---|---|---|---|---|
| OR (95% CI) |
P |
OR (95% CI) |
P |
OR (95% CI) |
P |
|
| Age | 1.02 (1.01, 1.04) | 0.002 | 1.02 (1.00, 1.04) | 0.04 | 1.02 (1.00, 1.04) | 0.023 |
| Sex (female vs. male) | 0.62 (0.46, 0.83) | 0.002 | 0.94 (0.65, 1.35) | 0.72 | 0.88 (0.61, 1.28) | 0.52 |
| 62-SNP GRS | 1.07 (1.04, 1.11) | <0.0001 | 1.08 (1.05, 1.12) | <0.0001 | ||
| Amino acid metabolites | ||||||
| Isoleucine | 0.91 (0.41, 2.00) | 0.81 | 0.85 (0.38, 1.88) | 0.68 | ||
| Tyrosine | 1.63 (0.83, 3.17) | 0.15 | 1.64 (0.83, 3.24) | 0.15 | ||
| Phenylalanine | 3.35 (1.54, 7.28) | 0.002 | 3.48 (1.58, 7.66) | 0.002 | ||
| Lipid metabolites | ||||||
| C18:2 LPC | 0.15 (0.07, 0.3) | <0.0001 | 0.14 (0.07, 0.28) | <0.0001 | ||
| C38:6 PC | 0.11 (0.04, 0.31) | <0.0001 | 0.12 (0.04, 0.33) | <0.0001 | ||
| C44:1 TAG | 0.78 (0.66, 0.93) | 0.005 | 0.79 (0.66, 0.93) | 0.006 | ||
| C48:0 TAG | 1.10 (0.86, 1.42) | 0.44 | 1.15 (0.89, 1.48) | 0.28 | ||
| C52:1 TAG | 3.01 (2.16, 4.2) | <0.0001 | 2.96 (2.11, 4.14) | <0.0001 | ||
| C56:9 TAG | 1.62 (0.94, 2.79) | 0.08 | 1.61 (0.93, 2.78) | 0.09 | ||
Lipid metabolites notation is CXX:YY, where CXX is the number of carbons in the acyl chain and YY is the number of double bonds. Significant traits within a model are in boldface. AUC (95% CI) for GRS: 0.641 (0.603, 0.679). AUC (95% CI) for metabolites: 0.803 (0.773, 0.832). AUC (95% CI) for GRS + metabolites: 0.820 (0.792, 0.848).
When considered individually, the GRS (P = 0.0005) and all metabolites, individually, were associated with log HOMA-B (P < 0.0001 for all metabolites except C38:6 PC, P = 0.03). When both sources of information were examined in the same age- and sex-adjusted model, the GRS remained associated with log HOMA-B (P < 0.0001), though fewer metabolites (P > 0.05 for phenylalanine and C44:1 TAG) remained associated with log HOMA-B (Table 3). When considered individually, all metabolites were associated with log HOMA-IR (P < 0.0001 for all metabolites except C38:6 PC, P = 0.008). In contrast, the GRS was not associated with log HOMA-IR (P = 0.43). When both sources of information were examined together in age- and sex- adjusted models, all metabolites remained associated with log HOMA-IR (P < 0.05 for all), and the GRS was not associated with log HOMA-IR (P = 0.29) (Table 3).
Table 3.
Estimates for HOMA-IR or HOMA-B associated with a GRS and serum metabolites in combination
| HOMA-B |
HOMA-IR |
|||
|---|---|---|---|---|
| Estimate (95% CI) | P | Estimate (95% CI) | P | |
| Age | −0.01 (−0.02, 0.003) | 0.16 | 0.01 (−0.001, 0.03) | 0.06 |
| Sex | 0.22 (−0.01, 0.45) | 0.06 | −0.27 (−0.56, 0.01) | 0.06 |
| 62-SNP GRS | −0.04 (−0.06, −0.02) | <0.0001 | −0.01 (−0.04, 0.01) | 0.29 |
| Isoleucine | 1.25 (0.73, 1.77) | <0.0001 | 0.82 (0.18, 1.46) | 0.01 |
| Tyrosine | 0.60 (0.16, 1.04) | 0.007 | 1.40 (0.87, 1.94) | <0.0001 |
| Phenylalanine | 0.27 (−0.30, 0.84) | 0.35 | 0.72 (0.02, 1.42) | 0.045 |
| C18:2 LPC | −1.43 (−1.82, −1.05) | <0.0001 | −2.28 (−2.75, −1.81) | <0.0001 |
| C38:6 PC | −1.01 (−1.59, −0.43) | 0.001 | −1.71 (−2.43, −0.99) | <0.0001 |
| C44:1 TAG | −0.02 (−0.11, 0.07) | 0.61 | −0.13 (−0.24, −0.02) | 0.02 |
| C48:0 TAG | 0.20 (0.02, 0.38) | 0.03 | 0.23 (0.01, 0.45) | 0.045 |
| C52:1 TAG | 0.52 (0.30, 0.74) | <0.0001 | 1.05 (0.78, 1.33) | <0.0001 |
| C56:9 TAG | 0.57 (0.21, 0.93) | 0.002 | 0.90 (0.45, 1.35) | <0.0001 |
β-Coefficient estimates provided from age- and sex-adjusted regression models. Lipid metabolites notation is CXX:YY, where CXX is the number of carbons in the acyl chain and YY is the number of double bonds. Significant traits within a model are in boldface.
Conclusions
This study integrates novel assessment of the combined effects of genetic and circulating metabolite measurements on predictive models for incident type 2 diabetes. While the association between genetic traits or metabolite levels and future type 2 diabetes has been demonstrated separately in the Framingham Offspring Study (5,6,12–15,25) and validated in several independent cohorts (7–9,12,14–17,25), we show that metabolite and genetic traits together contribute information both distinct and complementary to each other for the prediction of future type 2 diabetes among adults in the Framingham Offspring Study. Within-study replication was used to demonstrate high reliability of accuracy for incident type 2 diabetes for each of the models presented, and two different methods of assessing model performance, AUC and NRI, showed consistent results. All metabolites and clinical risk factors were measured at a common baseline examination, and the time to diabetes incidence or diabetes-free follow-up was derived from that assessment. Thus, strengths of this study's design are the measurement of genetic information and baseline metabolite concentrations in the same well-phenotyped cohort with longitudinal information (avoiding confounding from extant disease), use of the most current GRS for diabetes prediction, and use of metabolites currently validated against future risk of type 2 diabetes.
Our results extend prior studies demonstrating the capacity of the GRS (5,6,9) and metabolite measurements (12–14), separately, to predict type 2 diabetes risk. Consistent with prior studies (5,7,10) and a recent meta-analysis (11), we show that the addition of genetic information only slightly improves accuracy of type 2 diabetes prediction over traditional clinical risk factors. The increase in predictive accuracy from incorporating metabolomic data with or without genetic information to clinical risk factors is greater.
These findings suggest that both metabolomic and genetic information may have utility for type 2 diabetes prediction in specific subpopulations. In agreement with prior reports from de Miguel-Yanes et al. (6) and Vassy et al. (8,9) demonstrating that the GRS adds more information to a diabetes prediction model in younger versus older individuals, we show that both genetic and metabolomic measurements yield improved accuracy for type 2 diabetes prediction in individuals younger than 50 years versus older than 50 years and in nonobese versus obese individuals. Further work is needed to test whether genetic and metabolite markers, separately and together, are more informative with respect to type 2 diabetes risk in individuals who have not yet developed traditional clinical risk factors for the disease, such as older age and obesity, than those who have. The large difference in diabetes incidence between individuals in the lowest predicted and highest predicted risk decile suggests a model using all sources of information may have clinical applicability in the identification of individuals who are at high risk of developing type 2 diabetes.
Importantly, by comparing the effect of the GRS and metabolite measures on diabetes prediction in the same individuals, we were able to show that the discrimination and reclassification for models containing metabolites, with or without clinical predictors, were greater than for similar models containing the GRS among adults in the Framingham Offspring Study. Furthermore, we show that, of the metabolites previously associated with incident type 2 diabetes, separately, nine remain significant predictors when considered in aggregate. This subset of metabolites, along with any newly discovered metabolite biomarkers, may be sufficient to capture the currently known metabolomic information on diabetes risk in future studies. Indeed, the AUC for each predictive model is similar whether all metabolites previously associated with type 2 diabetes or only the subset of nine metabolites is used (Supplementary Fig. 4). Notably, the association between the GRS and incident diabetes was not attenuated by the addition of metabolomic information and retained significance in all prediction models tested. This may indicate that the GRS and metabolites represent different aspects of type 2 diabetes risk.
The majority of diabetes-related SNPs with known physiological associations have been linked to β-cell function (18,19). By contrast, amino acids and lipids have been associated principally with measures of insulin resistance but also with measures of insulin secretion (12,13,16,20–24). Thus, the ability of metabolites and GRS to provide information on diabetes prediction complementary to each other may be related to their capturing different aspects of type 2 diabetes development. Supporting this hypothesis, our results show that the GRS is consistently associated with an estimate of β-cell function, while all the metabolite traits are consistently associated with an estimate of insulin resistance.
In contrast to a prior report that demonstrated similar correlations between individual amino acid metabolites and either HOMA-IR or HOMA-B in a case-control experiment (12), we find that, for the subset of amino acid and lipid metabolites that were associated with both traits, the relationship of all but one metabolite (C48:0 TAG) was stronger with HOMA-IR than with HOMA-B. This difference may be explained by our examining the relationship of all metabolites with each trait in aggregate models, as opposed to separately, or by our having performed the analyses in a longitudinal cohort design as opposed to in a case-control design. Although the relationship between individual metabolites and HOMA-B could be attenuated by the additional metabolite or clinical variables, the relationship between the GRS and HOMA-B was not affected by these other sources of information. In exploratory analyses, we found that incorporating HOMA-IR into models with clinical factors and metabolites did not alter the AUC or the ORs of metabolites with type 2 diabetes risk (not shown). This finding is consistent with the prior observation that adjustment for HOMA-IR does not reduce the association of amino acid metabolites with type 2 diabetes risk (12). Together, these findings suggest that the GRS is a robust indicator of β-cell function and that circulating metabolites may be marking several aspects of diabetes risk, including the fasting insulin resistance as estimated by HOMA-IR. Still, further studies are warranted to investigate whether the genetic and metabolomic data are signaling different biological etiologies for diabetes development. Detailed physiological tools, such as insulin clamps, were not available to discern the relative contribution of the metabolite signature and GRS to either insulin resistance or β-cell function in this study, which used archived samples from an established cohort.
Other limitations of our study deserve comment. First, participants were all of European ancestry and of average risk for type 2 diabetes development. While amino acids and SNP components of the GRS have been associated with insulin resistance or diabetes incidence, respectively, in nonwhite populations (23,32–36), the relative contribution of these sources of information to type 2 diabetes prediction is not known outside of this study. Therefore, these findings should be tested in nonwhites and populations at high risk of diabetes. Second, we used within-study statistical techniques to demonstrate reliability of the prediction models and the range of AUCs that might be expected in other data sets. While the GRS and metabolite predictors have each been validated independently, formal replication in a different cohort, ideally using data collected in a prospective manner, would further support the reproducibility of these findings. Third, the SNPs and metabolites used in these analyses do not fully represent the genetic or metabolomic contribution to diabetes risk. It is anticipated that new genetic variants and circulating metabolites associated with diabetes risk will be validated, at which time they can be incorporated into the existing type 2 diabetes prediction models established here.
In summary, specific measurements from genomic and metabolomic platforms, distilled here into the most predictive determinants, provide complementary information for prediction of type 2 diabetes, and, together, modestly improve the accuracy of standard clinical models of diabetes prediction. Future studies should test the effect of a combined type 2 diabetes prediction model in high-risk and multi-ethnic populations and seek to understand how this information may be used to target clinical interventions for the prevention of type 2 diabetes in subpopulations with the highest metabolic risk.
Supplementary Material
Article Information
Funding. This work was supported by National Institutes of Health contracts NO1-HC-25195, R01-DK-HL081572, R01-DK-078616, and K24-DK-080140 and by the American Heart Association. M.D. is supported by research grants of the University of Verona. J.L.V. is supported by NIH-U01-HG006500 and L30-DK-089597. S.C. is supported in part by K99-HL-107642 and a grant from the Ellison Foundation.
The funding sources had no direct involvement in the collection, analysis, or interpretation of data; writing of the report; or decision to submit the manuscript for publication.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. G.A.W. participated in study design and data interpretation, wrote the manuscript, and finalized the draft based on comments from other authors. B.C.P. analyzed all data sources, constructed figures and tables, contributed to all discussion, and revised the manuscript. M.D., J.L.V., S.C., and E.P.R. provided substantial revisions for the manuscript and contributed to all discussion. T.J.W., J.B.M., R.E.G., and J.C.F. conceived of the study and provided overall guidance. G.A.W. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 74th Scientific Sessions of the American Diabetes Association, San Francisco, CA, 13–17 June 2014.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc14-0560/-/DC1.
G.A.W. and B.C.P. contributed equally to this study. R.E.G. and J.C.F. contributed equally to this study.
References
- 1.Whiting DR, Guariguata L, Weil C, Shaw J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract 2011;94:311–321 [DOI] [PubMed] [Google Scholar]
- 2.Knowler WC, Barrett-Connor E, Fowler SE, et al. Diabetes Prevention Program Research Group . Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB, Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med 2007;167:1068–1074 [DOI] [PubMed] [Google Scholar]
- 4.Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimäki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 2009;373:2215–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med 2008;359:2208–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Miguel-Yanes JM, Shrader P, Pencina MJ, et al. MAGIC Investigators. DIAGRAM+ Investigators . Genetic risk reclassification for type 2 diabetes by age below or above 50 years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care 2011;34:121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 2008;359:2220–2232 [DOI] [PubMed] [Google Scholar]
- 8.Vassy JL, Dasmahapatra P, Meigs JB, et al. Genotype prediction of adult type 2 diabetes from adolescence in a multiracial population. Pediatrics 2012;130:e1235–e1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vassy JL, Durant NH, Kabagambe EK, et al. A genotype risk score predicts type 2 diabetes from young adulthood: the CARDIA study. Diabetologia 2012;55:2604–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imamura M, Shigemizu D, Tsunoda T, et al. Assessing the clinical utility of a genetic risk score constructed using 49 susceptibility alleles for type 2 diabetes in a Japanese population. J Clin Endocrinol Metab 2013;98:E1667–E1673 [DOI] [PubMed] [Google Scholar]
- 11.Bao W, Hu FB, Rong S, et al. Predicting risk of type 2 diabetes mellitus with genetic risk models on the basis of established genome-wide association markers: a systematic review. Am J Epidemiol 2013;178:1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med 2011;17:448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee EP, Cheng S, Larson MG, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest 2011;121:1402–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng S, Rhee EP, Larson MG, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 2012;125:2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang TJ, Ngo D, Psychogios N, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest 2013;123:4309–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stancáková A, Civelek M, Saleem NK, et al. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes 2012;61:1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Floegel A, Stefan N, Yu Z, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013;62:639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupuis J, Langenberg C, Prokopenko I, et al. DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voight BF, Scott LJ, Steinthorsdottir V, et al. MAGIC investigators. GIANT Consortium . Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felig P, Marliss E, Cahill GF, Jr. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med 1969;281:811–816 [DOI] [PubMed] [Google Scholar]
- 21.Huffman KM, Shah SH, Stevens RD, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care 2009;32:1678–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009;9:311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai ES, Tan ML, Stevens RD, et al. Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia 2010;53:757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Würtz P, Soininen P, Kangas AJ, et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care 2013;36:648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassy JL, Hivert MF, Porneala B, et al. Polygenic type 2 diabetes prediction at the limit of common variant detection. Diabetes 2014;63:2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol 1979;110:281–290 [DOI] [PubMed] [Google Scholar]
- 27.Shaham O, Wei R, Wang TJ, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol 2008;4:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris AP, Voight BF, Teslovich TM, et al. Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- Pencina MJ, D'Agostino RB Sr, D'Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008;27:157–172 [DOI] [PubMed]
- 31.Pencina MJ, D’Agostino RB, Sr, Demler OV. Novel metrics for evaluating improvement in discrimination: net reclassification and integrated discrimination improvement for normal variables and nested models. Stat Med 2012;31:101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008;40:1098–1102 [DOI] [PubMed] [Google Scholar]
- 33.Xiang J, Li XY, Xu M, et al. Zinc transporter-8 gene (SLC30A8) is associated with type 2 diabetes in Chinese. J Clin Endocrinol Metab 2008;93:4107–4112 [DOI] [PubMed] [Google Scholar]
- 34.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008;40:1092–1097 [DOI] [PubMed] [Google Scholar]
- 35.Kooner JS, Saleheen D, Sim X, et al. DIAGRAM. MuTHER . Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet 2011;43:984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxena R, Saleheen D, Been LF, et al. DIAGRAM. MuTHER. AGEN . Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes 2013;62:1746–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.