Abstract
Activated ALK and ROS1 tyrosine kinases, through gene fusions, has been found in lung adenocarcinomas and are highly sensitive to selective kinase inhibitors. This study aimed at identifying the presence of these rearrangements in human colorectal adenocarcinoma (CRC) specimens using a 4-target, 4-color break-apart fluorescence in situ hybridization (FISH) assay to simultaneously determine the genomic status of ALK and ROS1. Among the clinical CRC specimens analyzed, rearrangement-positive cases for both ALK and ROS1 were observed. The fusion partner for ALK was identified as EML4 and the fusion partner for one of the ROS1-positive cases was SLC34A2, the partner for the other ROS1-positive case remains to be identified. A small fraction of specimens presented duplicated or clustered copies of native ALK and ROS1. In addition, rearrangements were detected in samples that also harbored KRAS and BRAF mutations in two of the three cases. Interestingly, the ALK-positive specimen displayed marked intra-tumoral heterogeneity and rearrangement was also identified in regions of high-grade dysplasia. Despite the additional oncogenic events and tumor heterogeneity observed, elucidation of the first cases of ROS1 rearrangements and confirmation of ALK rearrangements support further evaluation of these genomic fusions as potential therapeutic targets in CRC.
Implications
ROS1 and ALK fusions occur in colorectal cancer and may have substantial impact in therapy selection.
Introduction
Activation of proto-oncogenes by genomic rearrangements resulting in the fusion of two unrelated genes was identified in leukemias and lymphomas decades ago and is an extensively explored mechanism of tumorigenesis as well as a basis for classification of hematopoietic neoplasms (1, 2). More recently, similar phenomena have been identified in a variety of solid tumors. Among these, rearrangement of the anaplastic lymphoma kinase (ALK) gene, originally identified in association with anaplastic large cell lymphoma (3), has been implicated in lung adenocarcinoma. Activation of ALK through gene fusions in lung cancer has been reported in approximately 5% of unselected lung adenocarcinomas, with increasing incidence when some clinicopathologic selection criteria are applied (4-17). The importance of this molecular diagnosis is that it predicts benefit from targeted kinase inhibitors. Patients with advanced ALK+ lung cancers, when treated with ALK inhibitors (e.g., crizotinib), have shown dramatic clinical response (18). The v-ros avian UR2 sarcoma virus oncogene homolog (ROS1) encodes a tyrosine kinase which shares significant homology with ALK and is activated by fusion events in 1.2-2.6% of lung cancer. Crizotinib is also clinically effective in lung cancer patients harboring these ROS1 rearrangements (19-21). ROS1 gene fusions have also been found in many other tumor types beyond lung cancer (22).
Colorectal cancer (CRC) is a major cause of cancer deaths worldwide. However, because existing therapies can be toxic, more specific therapeutic regimens such as targeted agents have been sought to improve the outcomes and quality of life of CRC patients. Efforts to identify alterations that could predict benefit from a targeted therapy approach in CRC have proven difficult. Whilst KRAS mutation analysis is an accepted molecular approach in CRC, unlike the demonstration of EGFR mutation or ALK rearrangement in NSCLC which are used to select patients for targeted therapies, KRAS mutational status is instead used to exclude patients unlikely to benefit from monoclonal anti-EGFR therapy. Descriptions of fusion events such as ALK fusions in CRC have been rare as summarized in Table 1. In studies using reverse-transcriptase polymerase chain reaction (RT-PCR) for EML4-ALK fusions, no ALK rearrangements were found among 48 cases (8) and 96 cases (23) of CRC tested. ALK rearrangements were also not found by FISH in 12 colorectal neuroendocrine carcinoma cases (24), but ALK gene copy gain or amplification were found in 26 of 756 colorectal carcinoma cases(25). On the other hand, , EML4-ALK gene fusions were detected in 2 of 83 (2.4%) CRC specimens through exon array profiling (9), the PRKAR1A-ALK fusion was found in CRC by full exome sequencing(26), and the C2orf44-ALK fusion was found in 1 out of 40 (2.5%) CRC specimens tested by next generation sequencing (27). In this case, the in-frame fusion C2orf44-ALK resulted from a 5 megabase (MB) tandem duplication. The authors reported a ∼90-fold increase in 3′ ALK expression, suggesting that the C2orf44-ALK fusion transcript resulted in ALK kinase overexpression. Based on the presence of ALK rearrangements in CRC, and due to the extensive homology between ALK and ROS1, we hypothesized that ROS1 genes may also be activated by gene fusions in CRC. Although there has been no report of ROS1 activation in CRC to date, Lee et al (2013) recently reported 23 of 495 gastric adenocarcinoma cases (4.6%) with high level of ROS1 expression by immunohistochemistry (28). Of these 23 cases, 3 were positive for gene rearrangement by FISH break-apart, two of which were found to present the SLC34A2-ROS1 (S4:R32) fusion by RT-PCR (28). Additionally, in 2011, Gu et al reported 2 of 23 cases of cholangiocarcinoma that were positive for the GOPC (FIG)-ROS1 gene fusion using phosphotyrosine signaling profiling (mass spectrometry) followed by 5′RACE (29).
Table 1. Summary of published reports evaluating ALK and ROS1 rearrangements in tumors of the gastrointestinal system.
| Gene | Reference | Tumor Type | Technology | # Tested | # Positive | Fusion Partner* |
|---|---|---|---|---|---|---|
| ALK | Fukuyoshi et al.(23) | Colorectal Carcinoma | RT-PCR for EML4 | 96 | 0 | NA |
| Takeuchi et al.(8) | Colon Carcinoma | RT-PCR for EML4 | 48 | 0 | NA | |
| Lin et al.(9) | Colorectal Carcinoma | Exon-Array Profiling and RT-PCR for EML4 | 83 | 2 | EML4 E20 and E21 | |
| Karkouche et al.(24) | Colorectal Neuroendocrine Carcinoma | FISH | 12 | 0 | NA | |
| Lipson et al.(27) | Colorectal Carcinoma | Next Generation Sequencing | 40 | 1 | C2orf44 | |
| Bavi et al.(25) | Colorectal Carcinoma | FISH | 756 | 0 | NA | |
| Eddy et al.(26) | Full Exome Sequencing | Full Exome Sequencing | NA* | NA* | PRKAR1A, EML4 | |
| ROS1 | Lee et al.(28) | Gastric Adenocarcinoma | FISH then RT-PCR for identification of fusion partner | 495 | 3 | SLC34A2 (×2); 1 unknown |
| Gu et al.(29) | Cholangiocarcinoma | Phosphotyrosine signaling profiling (mass spectrometry) followed by 5′RACE | 23 | 2 | GOPC (FIG) E3 and E7 |
NA: Not Applicable;
NA: Data not available E: exon of the partner gene fused to ALK or ROS1
Overall, these findings suggest that an unrecognized subset of CRC may harbor genetic alterations predicting response to crizotinib and other targeted therapies. We herein analyzed the frequency of ALK and ROS1 rearrangements in specimens from patients with metastatic CRC via FISH. In addition, we sought to identify the fusions partners in rearranged specimens through RT-PCR. Moreover, patients who harbored atypical ALK FISH patterns were further analyzed by RT-PCR to determine possible rearrangements not detectable by FISH based on classically described definitions of FISH positivity (i.e. C2orf44-ALK).
Materials and Methods
Patients and Tissue Microarrays (TMA)
The tissue microarray was prepared using formalin fixed paraffin embedded (FFPE) CRC tissue specimens from 268 patients enrolled in the Australian Gastrointestinal Trials Group Randomized Phase III MAX Study(30), including all patients with adequate tissue available. These patients are representative (clinical, pathological characteristics) of the MAX phase III clinical trial population, as previously reported(31). All patients had histologically confirmed colorectal adenocarcinoma. Three tumor tissue cores per patient were distributed in 10 blocks, making up a 12×8 grid of cores on each slide. Thirty-nine of the patients were duplicated, and three were triplicated in the TMA for quality control. Institutional review board-approved informed consent was obtained by the MAX trial investigators for biomarker evaluation. Additional slides from the original pathology blocks of the positive samples were also made available for PCR and investigation of intra-tumoral heterogeneity.
FISH Assays and Analyses
The TMA slides were subjected to a FISH assay using a novel 4-color, 4-target ALK/ROS1 break-apart probe (Abbott Molecular) developed to determine genomic status of ALK and ROS1 in the same cells. ALK gene sequences were labeled in SpectrumRed (3′ALK) and SpectrumGreen (5′ALK); fused 5′/3′ ALK signals were classified as normal, whereas split 5′-3′ ALK by >2 diameters of the signal, and single 3′ALK were classified as abnormal (32). 3′ALK doublets-single green (RGR or RRG) was expected as the FISH pattern for the C2orf44-ALK fusion (27), thus this pattern was considered positive atypical. ROS1 gene sequences were labeled in SpectrumAqua (3′ROS1) and SpectrumGold (5′ROS1); fused 5′-3′ROS1 signals were classified as normal, whereas split 5′-3′ROS1 signals by >1 signal diameter, and single 5′ROS1 or single 3′ROS1 signals were classified as abnormal. For each patient, at least 50 tumor cells in two cores were scored. Variant patterns, such as the appearance of doublets or pairs, were annotated. A signal doublet was defined as the presence of two copies of the signal for a given target placed adjacently, that is, separated by ≤1 diameter of the average signal diameter; paired signals were defined as two fusion signals placed adjacently but separated by 1-2 diameters of the average signal. Doublet and paired signals were observed in hybridizations with both ALK and ROS1 probes and, when present in >10% of cells, the specimen was classified as atypical. Some specimens displayed both fusion signal doublets and pairs, in which case they were included in the doublet category.
Using the same ALK/ROS1 probe described above, additional FFPE slides of the resection blocks for the ALK+ patient were investigated for intra-tumoral heterogeneity. The FISH assays were performed using the Zymed Spot-Light Tissue Pretreatment kit (Invitrogen) in the TMAs and the Vysis Paraffin Pretreatment IV and Post-Hybridization Wash Buffer Kit (Abbott Molecular) in the sections, per manufacturers' instructions. Analysis was performed using interference filters sets for blue (DAPI), green (FITC), red (Texas Red), turquoise (Aqua) and yellow (Gold). Monochromatic images were acquired for each interference filter and merged using the CytoVision application (Leica Microsystems).
Reverse Transcriptase PCR
To identify the fusion partner for ALK and ROS1, reverse transcriptase (RT)-PCR was carried out as described using the SuperScript III First-Strand Synthesis System (Invitrogen) with previously published ALK and ROS1 primers; the ALK primer was located in exon 20 (ALK Rev20; (13)), whereas the ROS1 primer was located in exon 34 (ROS1 E34R; (33)). First-strand synthesis was carried out as above followed by a 20-minute RNaseH digestion at 37°C. Individual PCR reactions were carried out to amplify either EML4-ALK or C2orf44-ALK, using previously published primers for exon 6, exon 13 and exon 18 of EML4 and exon 20 of ALK (ALK Sanders R20 (13) and an in-house primer for C2orf44 (C2orf44fwd1)). Likewise, individual PCR reactions were carried out to amplify either SLC34A2-ROS1, CD74-ROS1, or SDC4-ROS1 using the previously published primers (SLC34A2:E4F, CD74:E5F, ROS1:E34F (20)) along with a primer to SDC4 of our design (SDC4-E2F; (33)). PCR conditions for detecting the ALK and ROS1 fusion partners included an initial denaturation at 95°C for 5 min followed by 10 cycles of touchdown PCR and 30 cycles of PCR. PCR products were resolved on a 2% agarose gel. Positive PCR products were excised from agarose gel, purified (Wizard SV Gel and PCR Clean Up Kit; Promega), and sequenced. All primer sequences are listed in Supplementary Table 1.
Microdissection and Mutation Analysis
KRAS and BRAF mutational analysis was performed initially on CRC specimens used in the TMA using high-resolution melting point (HRM) PCR as previously reported (34). Subsequently for specimens where ALK or ROS1 rearrangements were identified with KRAS or BRAF mutations, tumor areas were identified and areas for differential microdissection were mapped based on parallel Hematoxylin and Eosin stained slide. 4μm sections were deparaffinized, hematoxylin counterstained and microdissected by scalpel point under microdissecting microscope. Microdissected material was washed with 70% ethanol, air dried, and resuspended in lysis buffer and DNA extracted (Qiagen QIAamp DSP DNA FFPE Tissue Kit (#60404) using manual extraction with elution into 30 microliters of elution buffer.
For first round mutational analysis, DNA samples from selectively microdissected areas were PCR amplified with primers flanking KRAS exon 2 as previously described (35), followed by Sanger DNA sequencing. Positive samples for mutation by Sanger sequencing were not further evaluated, negative samples were evaluated by HRM curve to achieve higher analytical sensitivity. Briefly, DNA samples from selectively microdissected areas were PCR amplified with primers flanking KRAS exon 2 on the Roche LightCycler 480 using the Roche LC480 High Resolution Melting Master Kit (#04909631001). Resulting real-time PCR curves were evaluated for perturbations in the melting curve profiles with appropriate controls. The HRM assay was estimated to have an analytical sensitivity of ∼5% based on dilution studies.
Results
Demonstration of ALK and ROS1 Fusions in CRC
Of the 268 patient specimens originally included in the tissue microarray, 236 had evaluable FISH results defined as at least 50 tumor cells in two cores. The cutoff threshold for positivity was identified as ≥15% of cells displaying patterns compatible with rearrangement, based on the distribution of relevant patterns in the cohort with application of evaluation of mean + 3× standard deviation of signal and beta inverse function (data not shown). Two cases (0.8%) demonstrated FISH patterns consistent with ROS1 rearrangement, predominantly single 3′ROS1 signals (Figures 1A, B). One case (0.4%) demonstrated a pattern consistent with ALK rearrangement, and it also had predominantly single 3′ALK signals (Figure 1C). The atypical pattern 3′ ALK doublets (3′/5′/3′ALK) associated with the C2orf44-ALK fusion were identified in 7 cases (3%), subjected later to RT-PCR testing. Other signal variants were identified for both ALK and ROS1, including 25 cases with 3′/5′ fusion ALK doublets (10.6%), and 12 cases each (5.1%) with 3′ROS1 doublets (3′/5′/3′ROS1) and 3′/5′ fusion doublets for ROS1.
Figure 1.
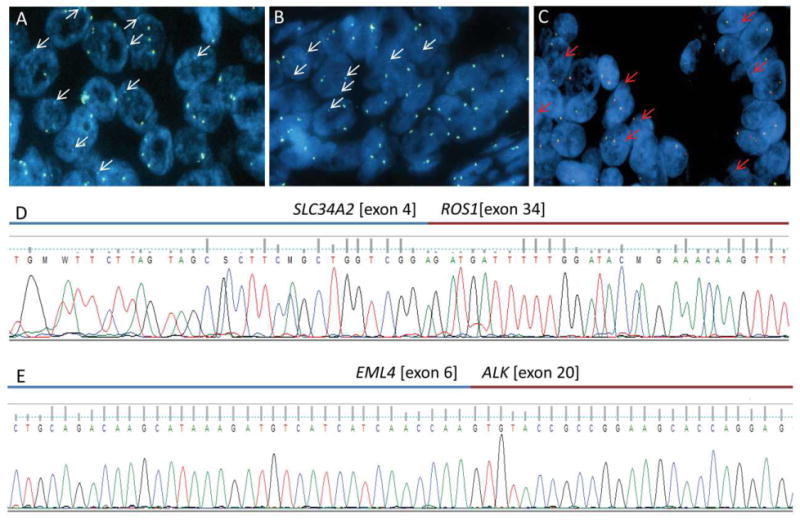
A, B) FISH images showing ROS1 rearrangement in two specimens as demonstrated by single 3′ ROS1 (aqua) signals; C) FISH image demonstrating ALK rearrangement based on single 3′ALK (red) signals; D) Sequencing of RT-PCR product from the sample depicted in panel A confirming ROS1 fusion with SLC34A2; E) Sequencing of RT-PCR product from the sample depicted in panel C confirming EML4-ALK fusion.
In patient #406 (ALK positive), the primary tumor site was the rectum with metastases of lymph nodes and lung. In patient #38 (ROS1 positive), the primary tumor site was the ascending colon with metastases in lymph nodes and liver. In patient #100 (ROS1 positive) the primary tumor site was the rectum and the sigmoid colon with metastasis in the lung. These patients were, respectively, 84 (female), 78 (male) and 69 (female) years old at the time at which their metastatic disease was diagnosed..
Original pathology blocks from the identified cases were evaluated by RT-PCR to further verify the presence of fusion events. RT-PCR spanning previously published ROS1 breakpoints paired with specific primers for known fusion partners of ROS1 was employed. Of the two cases demonstrating FISH patterns consistent with ROS1 fusion events, one was confirmed by RT-PCR to harbor a SLC34A2-ROS1 fusion (exons 4 and 34, respectively; Figure 1D) while the second case was negative for all known fusion partners of ROS1. Similarly, RT-PCR assays spanning previously published ALK breakpoints paired with specific primers for known fusion partners of ALK were employed. The case identified as consistent with ALK rearrangement demonstrated the presence of an EML4-ALK rearrangement (exons 6 and 20, respectively; Figure 1E). The seven cases identified as atypical with 3′ALK doublets (3′/5′/3′ ALK fusion) were all negative for known fusion partners of ALK, including C2orf44. Specimens with other variants were also tested, when available, by RT-PCR and no fusion was detected.
Of note, the specimen with SLC34A2-ROS1 fusion and the ALK+ case were previously classified as positive for BRAF [c.1799T>A (p.V600E)] and KRAS [c.35G>C (p.G12A)] mutations, respectively, during routine clinical testing.
Identification of Intra-Tumoral Heterogeneity
In the case identified with ALK rearrangement, three tissue cores containing tumor were subjected to analysis, however only two of the three cores demonstrated the finding of ALK rearrangement by FISH. Pathologic evaluation confirmed the presence of tumor in the core negative for ALK rearrangement, and confirmed morphology of the tumor compatible with the other two tissue cores. This finding was suggestive of intratumoral heterogeneity, which was further explored by evaluation of two tissue blocks from the source material. Multiple tissue areas from each of two tumor blocks were selected for additional FISH evaluation. Areas were marked on a parallel H&E stained section, and each region was separately evaluated for the presence of ALK rearrangement by FISH. Analysis of one block demonstrated a marked separation between areas of tumor that were positive and negative for ALK rearrangement by FISH (Figure 2A). Analysis of the second block demonstrated multiple areas of tumor with positive and negative patterns for ALK rearrangement in a more interposed distribution (Figure 2B). Histologic evaluation demonstrated that some of the tissue areas identified as positive for ALK rearrangement were pathologically best classified as high-grade dysplasia (Figures 2C, D).
Figure 2.
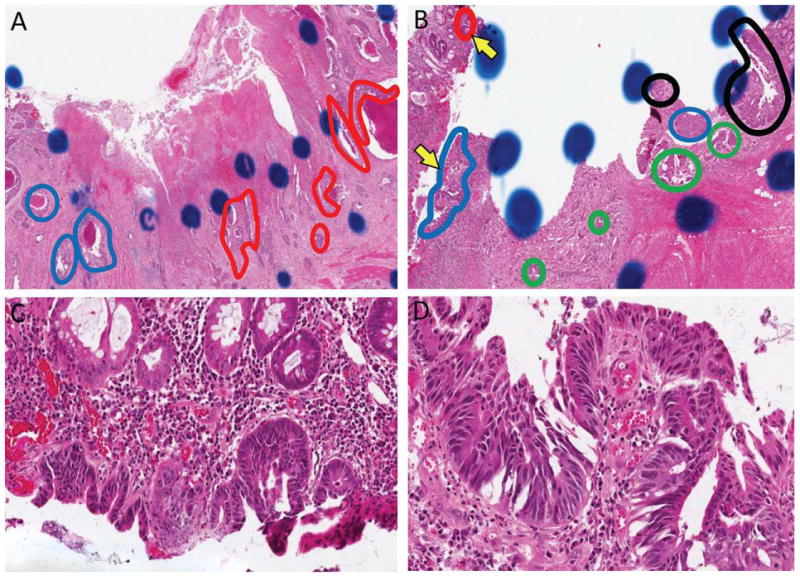
A, B) Two blocks from the original specimen utilized for TMA demonstrated areas with varying patterns of ALK rearrangement and KRAS mutation status. Red circled areas indicate ALK rearranged and mutated KRAS (ALK+/KRAS+). Blue circled areas indicate ALK wild-type and KRAS mutated (ALK-/KRAS+). Green circled areas indicated ALK rearranged and KRAS wild-type (ALK+/KRAS-). Black areas indicate wild-type for both alterations (ALK-/KRAS-). C, D) Higher magnification of the regions indicated in panel E by yellow arrows demonstrates that some regions are best classified as high-grade dysplasia. FISH analysis in region F was positive for ALK rearrangement and FISH analysis in region G was negative for ALK rearrangement (FISH images not shown).
Given that the specimen with heterogeneity for ALK rearrangement was classified as KRAS positive during routine clinical analysis, we sought to determine whether the ALK status overlapped with KRAS status within sub-regions of the tumor. Areas of tumor which were parallel to those evaluated by FISH were separately microdissected and KRAS mutation status ascertained by Sanger sequencing and, when negative, also by high-resolution melting curve analysis. These analyses demonstrated that some areas of the tumor retained positivity for KRAS mutation (Figure 3A) while other regions showed all four possible combinations of ALK/KRAS status (Figure 3B).
Figure 3.
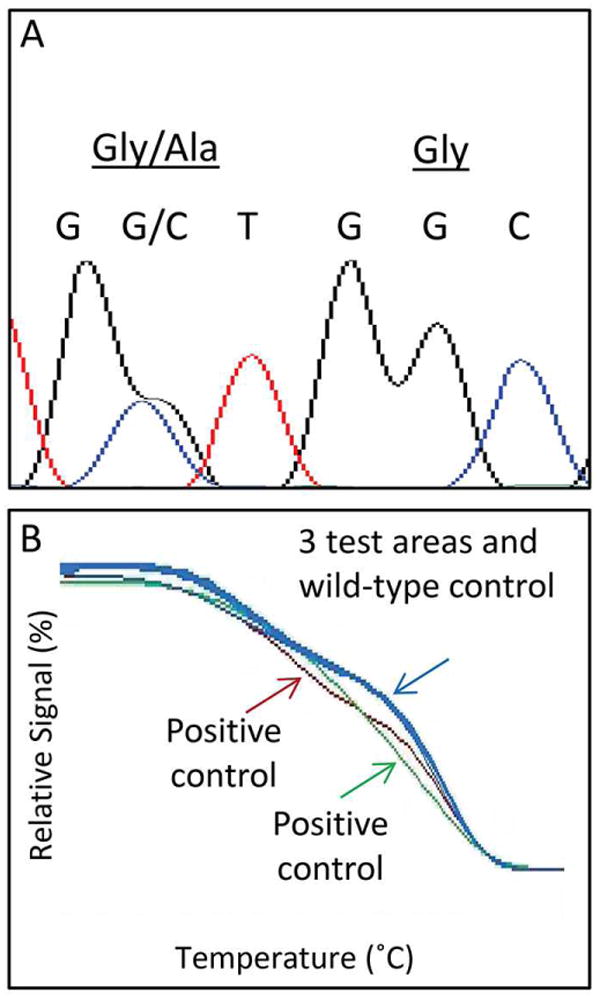
A) Representative sequencing findings of KRAS mutated area. B) Representative high-resolution melting analysis of regions negative for KRAS mutation by sequencing. Blue curves show overlap of tested regions with wild-type control, red and green show positive controls (2 distinct mutations)
The finding of intratumoral heterogeneity with respect to both ALK and KRAS alterations also led to the question of whether such heterogeneity was observed in either case with ROS1 rearrangement. However, no evidence of intratumoral heterogeneity with respect to ROS1 rearrangement was identified, therefore further analysis of intratumoral heterogeneity BRAF mutation in the ROS1+ case was not pursued.
Discussion
Previous reports of gene fusions involving ALK in CRC indicate that these events are rare, and our findings are consistent with reported studies demonstrating a low but detectable rate of ALK rearrangement in CRC. In addition, this study is the first to demonstrate a similarly low but detectable rate of ROS1 rearrangement in CRC. The demonstration of a fusion product by RT-PCR in two of the three rearrangement-positive cases confirms the FISH findings and serves to underscore the importance of further characterization of these fusion events in CRC. The absence of a detectable fusion product in the third case (ROS1 positive) is likely attributable to an unknown fusion partner. These findings have potentially significant therapeutic implications, as identification of these rearrangements may open the possibility for targeted therapy.
Of particular note, 2 of the 3 cases positive for fusion events were found in concert with oncogene point mutation events (KRAS and BRAF). This is in contrast to the predominant findings in NSCLC, which show that concurrent ‘driver’ mutations such as EGFR mutation and ALK rearrangements may occur but are uncommon (36-39). This result is of particular clinical relevance as attempts to identify CRC cases harboring these fusion events cannot benefit from an enrichment strategy where patients with KRAS or BRAF mutations are excluded from further testing.
A surprising result in this study was the demonstration of marked intratumoral heterogeneity for both KRAS mutation and ALK rearrangement status. Moreover, the identification of all four combinations of KRAS and ALK status throughout the specimen was particularly unexpected, as was the identification of a region of high-grade dysplasia harboring both molecular alterations. Multiple studies have indicated that not only is KRAS mutation an early event in colorectal carcinoma tumorigenesis, but it shows a very low discordance rate between primary tumor and corresponding metastasis (40, 41). These findings often support the notion that KRAS mutation is both homogeneously distributed and required for tumor perpetuation. However, recent studies have demonstrated that marked intratumoral heterogeneity does exist (42). Importantly, the current study was performed retrospectively, and none of the three patients identified with fusion events were treated with targeted therapy agents specific to those fusion products prior to death.
High-grade dysplasia is the precursor lesion to invasive carcinoma in the lower gastrointestinal tract, and the identification of a region of high-grade dysplasia harboring both KRAS mutation and ALK rearrangement is intriguing and creates the basis for several hypotheses explaining mechanisms by which all four combinations of KRAS and ALK status might exist through clonal evolution (Figure 3). In each of these hypotheses, the originating cell is negative for both alterations, and a gain of one alteration is the first step. One possibility is that the gain of alteration is step-wise (Figures 4A, B), in which either KRAS or ALK is sequentially gained in the neoplastic population. In order for this ‘sequential gain’ hypothesis to then yield a fourth species, the population must, by definition, undergo a ‘loss event’. Alternatively, the gains of alterations could first be in parallel, in which separate populations of cells independently gain either KRAS mutation or ALK rearrangement (Figure 4C). This ‘separate gain’ hypothesis would then require that a second event occur in order to generate a fourth species. Lastly, the possibility that the technology used to interrogate KRAS mutation status and ALK rearrangement is not sufficiently sensitive to determine whether the alterations actually occur in the same cells is a consideration, and gives rise to a ‘separate clones’ hypothesis (Figure 4D). Among these possibilities, we regard the separate gain hypothesis and the sequential gain hypothesis with ALK rearrangement occurring prior to KRAS mutation to be the least likely of these events, largely because of the substantial volume of data demonstrating KRAS mutation commonly occurring in adenomatous lesions.
Figure 4.
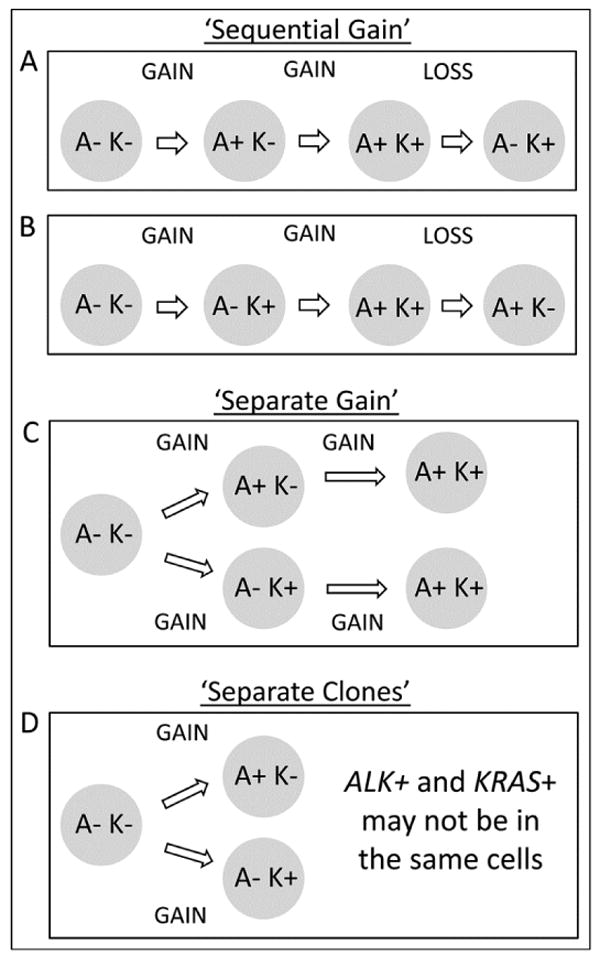
Possible mechanisms to explain findings of different combinations of ALK rearrangement and KRAS mutation status.
These potential hypotheses regarding the genesis of the observed spectrum of sub-species in this heterogeneous lesion have several putative functional considerations. One possibility is that KRAS mutation is a ‘driver’ and ALK rearrangement observed functionally acts as a ‘passenger’. This explanation does not sufficiently explain the finding of ALK+/KRAS- regions. Similarly, the ALK rearrangement may be a modulator of tumor growth, which is also difficult to reconcile with the finding of ALK+/KRAS- regions. Another possibility, best hypothesized in the ‘separate clones’ explanation is that KRAS and ALK represent dual drivers with subclonal evolution. This hypothesis is best considered in the context of underlying genomic instability, which could fuel random events expressed as subclonal heterogeneity.
The findings of this study have several specific implications with regard to future analysis of colorectal carcinoma. These data strongly support further evaluation of colorectal carcinoma for fusion events in ALK and ROS1, and further suggest the possibility that these events may serve as targets for therapy in CRC. As mentioned, the overlap of these fusions with both KRAS and BRAF mutations is a potential confounding factor, as not only does it impact approaches to screening, but these alterations may also modulate responsiveness to targeted therapy agents. Based on our findings, it may be challenging to identify a substantial number of patients with a uniform molecular profile with regard to fusion events and mutation status. The screening process itself may be impacted by these findings, which suggest that multiple regions of tumor may need to be evaluated. Furthermore, these findings highlight the technological hurdles involved in evaluation of tumor heterogeneity, and underscore the importance of methodologies to evaluate mutation status on a single-cell in situ basis.
Supplementary Material
Acknowledgments
Study was partially supported by research grants from Abbott Molecular to MVG, Boettcher Foundation to RCD, and the NCI CCSG P30CA046934 (Molecular Pathology Shared Resource). TN was supported by the Cancer Research Summer program from the University of Colorado Cancer Center; DDP was a fellow from the Brazilian Agency CAPES; AJW was supported by an NHRMC post graduate Fellowship and Roche HOTT Fellowship. Authors thank Lisa Litzenberger for graphics assistance.
Dara L. Aisner had educational speaker engagement with Abbott Molecular and Marileila Varella-Garcia received research grant and had educational speaker engagement with Abbott Molecular.
Footnotes
Conflict of Interest: No other authors have conflicts of interest to declare.
References
- 1.WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th. Lyon: International Agency for Research on Cancer; 2008. [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 3.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–4. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 4.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 5.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Choi YL, Takeuchi K, Soda M, Inamura K, Togashi Y, Hatano S, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–6. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 7.Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–83. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–24. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 9.Lin E, Li L, Guan Y, Soriano R, Rivers CS, Mohan S, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7:1466–76. doi: 10.1158/1541-7786.MCR-08-0522. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–9. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 11.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–33. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi T, Sonobe M, Kobayashi M, Yoshizawa A, Menju T, Nakayama E, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 2010;17:889–97. doi: 10.1245/s10434-009-0808-7. [DOI] [PubMed] [Google Scholar]
- 13.Sanders HR, Li HR, Bruey JM, Scheerle JA, Meloni-Ehrig AM, Kelly JC, et al. Exon scanning by reverse transcriptase-polymerase chain reaction for detection of known and novel EML4-ALK fusion variants in non-small cell lung cancer. Cancer Genet. 2011;204:45–52. doi: 10.1016/j.cancergencyto.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Wong DW, Leung EL, Wong SK, Tin VP, Sihoe AD, Cheng LC, et al. A novel KIF5B-ALK variant in nonsmall cell lung cancer. Cancer. 2011;117:2709–18. doi: 10.1002/cncr.25843. [DOI] [PubMed] [Google Scholar]
- 15.Jung Y, Kim P, Jung Y, Keum J, Kim SN, Choi YS, et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer. 2012;51:590–7. doi: 10.1002/gcc.21945. [DOI] [PubMed] [Google Scholar]
- 16.Soda M, Isobe K, Inoue A, Maemondo M, Oizumi S, Fujita Y, et al. A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin Cancer Res. 2012;18:5682–9. doi: 10.1158/1078-0432.CCR-11-2947. [DOI] [PubMed] [Google Scholar]
- 17.Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, et al. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One. 2012;7:e31323. doi: 10.1371/journal.pone.0031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camidge DR, Bang YJ, Kwak EL, Iafrate AJ, Varella-Garcia M, Fox SB, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012;13:1011–9. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–70. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies KD, Le AT, Theodoro MF, Skokan MC, Aisner DL, Berge EM, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2012;18:4570–9. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou S, Bang Y, Camidge D, Riely G, Salgia R, Shapiro G, et al. Efficacy and safety of crizotinib in patients with advanced ROS1-rearranged non-small cell lung cancer (NSCLC) ASCO Meeting Abstracts. 2013;31:8032. [Google Scholar]
- 22.Davies KD, Doebele RC. Molecular Pathways: ROS1 Fusion Proteins in Cancer. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuyoshi Y, Inoue H, Kita Y, Utsunomiya T, Ishida T, Mori M. EML4-ALK fusion transcript is not found in gastrointestinal and breast cancers. Br J Cancer. 2008;98:1536–9. doi: 10.1038/sj.bjc.6604341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karkouche R, Bachet JB, Sandrini J, Mitry E, Penna C, Cote JF, et al. Colorectal neuroendocrine carcinomas and adenocarcinomas share oncogenic pathways. A clinico-pathologic study of 12 cases. Eur J Gastroenterol Hepatol. 2012;24:1430–7. doi: 10.1097/MEG.0b013e3283583c87. [DOI] [PubMed] [Google Scholar]
- 25.Bavi P, Jehan Z, Bu R, Prabhakaran S, Al-Sanea N, Al-Dayel F, et al. ALK gene amplification is associated with poor prognosis in colorectal carcinoma. Br J Cancer. 2013 doi: 10.1038/bjc.2013.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eddy S, Tomilo M, Urick ME, Khazanov MA, Williams P, Bankhead A, et al. Expanded clinical opportunities for crixotinib from analysis of over 5,000 cancer patient exomes. AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics. 2013 abstract C256. [Google Scholar]
- 27.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–4. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Lee SE, Kang SY, Do IG, Lee S, Ha SY, et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer. 2013;119:1627–35. doi: 10.1002/cncr.27967. [DOI] [PubMed] [Google Scholar]
- 29.Gu TL, Deng X, Huang F, Tucker M, Crosby K, Rimkunas V, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One. 2011;6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tebbutt NC, Wilson K, Gebski VJ, Cummins MM, Zannino D, van Hazel GA, et al. Capecitabine, bevacizumab, and mitomycin in first-line treatment of metastatic colorectal cancer: results of the Australasian Gastrointestinal Trials Group Randomized Phase III MAX Study. J Clin Oncol. 2010;28:3191–8. doi: 10.1200/JCO.2009.27.7723. [DOI] [PubMed] [Google Scholar]
- 31.Weickhardt AJ, Williams D, Lee C, Simes J, Murone C, Wilson K, et al. Vascular endothelial growth factors (VEGF) and VEGF receptor expression as predictive biomarkers for benefit with bevacizumab in metastatic colorectal cancer (mCRC): Analysis of the phase III MAX study. ASCO Meeting Abstracts. 2011;29:3531. [Google Scholar]
- 32.Camidge DR, Kono SA, Flacco A, Tan AC, Doebele RC, Zhou Q, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res. 2010;16:5581–90. doi: 10.1158/1078-0432.CCR-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Fang R, Sun Y, Han X, Li F, Gao B, et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One. 2011;6:e28204. doi: 10.1371/journal.pone.0028204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price TJ, Hardingham JE, Lee CK, Weickhardt A, Townsend AR, Wrin JW, et al. Impact of KRAS and BRAF Gene Mutation Status on Outcomes From the Phase III AGITG MAX Trial of Capecitabine Alone or in Combination With Bevacizumab and Mitomycin in Advanced Colorectal Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:2675–82. doi: 10.1200/JCO.2010.34.5520. [DOI] [PubMed] [Google Scholar]
- 35.Franklin WA, Haney J, Sugita M, Bemis L, Jimeno A, Messersmith WA. KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn. 2010;12:43–50. doi: 10.2353/jmoldx.2010.080131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alrifai D, Popat S, Ahmed M, Gonzalez D, Nicholson AG, Parcq J, et al. A rare case of squamous cell carcinoma of the lung harbouring ALK and BRAF activating mutations. Lung Cancer. 2013;80:339–40. doi: 10.1016/j.lungcan.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 37.An SJ, Chen ZH, Su J, Zhang XC, Zhong WZ, Yang JJ, et al. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One. 2012;7:e40109. doi: 10.1371/journal.pone.0040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boland JM, Jang JS, Li J, Lee AM, Wampfler JA, Erickson-Johnson MR, et al. MET and EGFR mutations identified in ALK-rearranged pulmonary adenocarcinoma: molecular analysis of 25 ALK-positive cases. J Thorac Oncol. 2013;8:574–81. doi: 10.1097/JTO.0b013e318287c395. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Zhang S, Yang X, Yang J, Zhou Q, Yin L, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe TKT, Yamamoto Y, Matsuda K, Ishihara S, Nozawa K, Iinuma H, Shibuya H, Eshima K. Heterogeneity of KRAS status may explain the subset of discordant KRAS status between primary and metastatic colorectal cancer. Dis Colon Rectum. 2011;54:1170–8. doi: 10.1097/DCR.0b013e31821d37a3. [DOI] [PubMed] [Google Scholar]
- 41.Mariani P, Lae M, Degeorges A, Cacheux W, Lappartient E, Margogne A, et al. Concordant analysis of KRAS status in primary colon carcinoma and matched metastasis. Anticancer Res. 2010;30:4229–35. [PubMed] [Google Scholar]
- 42.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


