Background: Kelch proteins are required for cell morphogenesis, but their molecular functions have remained elusive.
Results: S. cerevisiae Kel1 and Kel2 form a stable complex with the formin-binding protein Bud14 to regulate actin cable assembly.
Conclusion: Kel1 and Kel2 directly regulate formins to control the actin cytoskeleton.
Significance: These findings help resolve the roles of S. cerevisiae Kel1 and Kel2 in morphogenesis.
Keywords: Actin, Cell Biology, Cell Polarity, Cytokinesis, Formin, S. cerevisiae, Kelch
Abstract
Formins perform essential roles in actin assembly and organization in vivo, but they also require tight regulation of their activities to produce properly functioning actin structures. Saccharomyces cerevisiae Bud14 is one member of an emerging class of formin regulators that target the FH2 domain to inhibit actin polymerization, but little is known about how these regulators are themselves controlled in vivo. Kelch proteins are critical for cell polarity and morphogenesis in a wide range of organisms, but their mechanistic roles in these processes are still largely undefined. Here, we report that S. cerevisiae Kelch proteins, Kel1 and Kel2, associate with Bud14 in cell extracts to form a stable 520-kDa complex with an apparent stoichiometry of 2:2:1 Bud14/Kel1/Kel2. Using pairwise combinations of GFP- and red fluorescent protein-tagged proteins, we show that Kel1, Kel2, and Bud14 interdependently co-localize at polarity sites. By analyzing single, double, and triple mutants, we show that Kel1 and Kel2 function in the same pathway as Bud14 in regulating Bnr1-mediated actin cable formation. Loss of any component of the complex results in long, bent, and hyper-stable actin cables, accompanied by defects in secretory vesicle traffic during polarized growth and septum formation during cytokinesis. These observations directly link S. cerevisiae Kelch proteins to the control of formin activity, and together with previous observations made for S. pombe homologues tea1p and tea3p, they have broad implications for understanding Kelch function in other systems.
Introduction
Formins are a widely conserved family of proteins that directly nucleate, elongate, and/or bundle actin filaments and perform a variety of biological roles ranging from the control of cell motility and adhesion to the regulation of intracellular transport and formation of cytokinetic actin rings (1, 2). It has recently become clear that most formins interact with additional cellular factors that govern their activities in distinct ways, including the release of formins from autoinhibition (e.g. Rho GTPases), the displacement of formins from growing ends of actin filaments (e.g. Bud14), and the transient inhibition of formin-mediated actin filament elongation (e.g. Smy1) (3, 4). Through these regulatory mechanisms, formins are controlled with a high level of spatial and temporal precision to produce actin structures with specific architectures and dynamics tailored to their different functions.
Mammals have 15 different formin genes, whereas budding yeast have only two, BNI1 and BNR1, that share an essential role in polarized cell growth and cytokinesis (5, 6). The relative simplicity of having only two formin genes combined with the tremendous genetic amenability of yeast has greatly accelerated our ability to define the in vivo functions and regulation of formins. In order for a yeast cell to grow a bud (daughter cell) and divide, the establishment and maintenance of a polarity axis are required. This in turn requires the assembly of actin cables, which serve as linear tracks for myosin V-based transport of secretory vesicles, organelles, and daughter-specific transcripts to the bud, as well as the guidance of astral microtubule plus ends to the cell cortex to position and orient the mitotic spindle (7, 8). Complementary sets of actin cables are nucleated by Bni1 and Bnr1, which reside at the bud tip and neck, respectively (9). These cables are dynamically assembled and turned over on a time scale of 5–15 s and continuously extend from the bud cortex and neck toward the rear of the mother cell at rates of 0.3–1.0 μm/s (10, 11). Myosins move rapidly on cables in the opposite direction, transporting vesicles toward the bud tip at a rate of ∼3 μm/s (12). Recently, we identified Bud14 as a cellular factor that localizes to the bud neck and cortex and binds to the FH2 domain of Bnr1, displacing Bnr1 from the growing ends of actin filaments to control actin cable architecture and function in vivo (13). In this study, we investigated whether Bud14 functions alone or together with other factors in vivo, and we linked its functions in formin regulation to two Kelch family proteins, Kel1 and Kel2.
Kelch family proteins are widely expressed, from yeast to mammals, and have conserved genetic roles in regulating cell polarity and morphogenesis (14). However, the molecular functions and mechanisms of Kelch proteins have in large part remained unclear. Members of the Kelch family are defined by the presence of 44–56 amino acid consensus pattern motifs, most often repeated in a 6-fold pattern (15). Structural studies on Kelch proteins indicate that this common motif folds into a four-stranded β-sheet and that each repeat forms one blade of a larger propeller structure with a central axis similar to WD repeat proteins (16–18). Many Kelch proteins bind directly to F-actin through the β-propeller domains and additionally bundle actin filaments (19–22). Genetic analyses in diverse model organisms have established roles for Kelch proteins in a variety of actin-based processes. In Drosophila, Kelch is required for proper formation of circular actin structures called ring canals, which connect nurse cells to oocytes and allow the delivery of nutrients and other materials to the oocyte (23–25). Ring canals require formins for their assembly and become highly disorganized and overgrown in Kelch mutants (20). Similarly, genetic data obtained from mammalian systems have demonstrated roles for at least five different Kelch proteins in actin-based processes, e.g. cell adhesion, membrane protrusion, and stress fiber formation (15). Collectively, these observations have indicated that Kelch proteins play an important role in maintaining proper actin architecture and/or dynamics but without a clear mechanistic understanding of their roles.
In Saccharomyces cerevisiae, two Kelch family members Kel1, and to a lesser extent Kel2, are required for normal cell polarity and morphology (26–30). kel1Δ cells exhibit hyper-elongated buds, and overexpression of either Kel1 or Kel2 causes highly aberrant cell morphologies (26). In addition, Kel1 and Kel2 physically associate with each other in cell extracts, and numerous potential binding partners of Kel1 and Kel2 have been identified in proteomic studies (28, 29, 31–33). In Schizosaccharomyces pombe, it has been shown that the Kelch proteins tea1p and tea3p are critical for polarized cell growth (34–37) and that tea1p is required for localization of the formin for3p to cell tips (38). These observations suggest that at least in fission yeast, Kelch proteins directly influence actin cytoskeleton organization via an association with formins. However, until now, this has not been extended to other systems, and it has remained unclear whether formin regulation is a Kelch protein function that is widely conserved.
Here, we demonstrate that S. cerevisiae Kel1 and Kel2 are required for proper Bnr1-dependent assembly of actin cables. We show that Kel1, Kel2, and Bud14 are integral components of a stable 520-kDa complex that directly regulates Bnr1 to control cable formation, polarized cell growth, and cytokinesis. This direct link between Kelch proteins and formin regulation, combined with observations from previous studies in S. pombe, suggests that formin regulation may be a common function of Kelch proteins by which they influence the actin cytoskeleton organization and in turn cell polarity and morphogenesis.
EXPERIMENTAL PROCEDURES
Strain and Plasmid Construction
Standard methods were used for molecular biology and growth of yeast cultures (39). Wild type w303 strains of opposite mating type (BGY12 and BGY10) were used to generate new strains carrying gene deletions and/or integrated tags by the methods described (40). All gene deletions and tags were verified by PCR analysis of genomic DNA.
Protein Fractionation by Sedimentation Velocity and Gel Filtration Analysis
For the analyses shown in Fig. 1, A–D, crude cell extracts from yeast strains expressing epitope-tagged proteins were prepared as follows. Cells were grown to late log phase (OD = 1) in YPD medium, then pelleted, washed once with water, resuspended in 0.25× (w/v) water, and frozen dropwise in liquid nitrogen. Frozen cells were then lysed using liquid nitrogen and mechanical sheering as described (41), and the powdered lysates were stored at −80 °C until use. Frozen powder was resuspended and thawed in 5× HEK buffer (1× HEK buffer = 25 mm HEPES (pH 7.5), 1 mm EDTA, 50 mm KCl) and protease inhibitor mixture (Roche Applied Science), using a 5:1 (w/v) ratio of powder to buffer. The lysate was cleared by centrifugation for 20 min at 80,000 rpm in a TLA100.3 rotor, and then protein concentration in the supernatant was quantified by the BCA assay (Thermo Scientific, Rockford, IL) and adjusted to 20 mg/ml. 8–10 mg of lysate was loaded onto a sucrose gradient (12.6 ml volume, 3–30% sucrose in HEK buffer) and centrifuged for 18 h at 4 °C in an SW40 Ti rotor at 30,000 rpm (Beckman Coulter, Brea, CA). 400-μl fractions were collected, and samples of each fraction were analyzed by gel fractionation and immunoblotting with antibodies recognizing the tags. Size standards were fractionated in parallel. To determine the Stokes radii of protein complexes, peak fractions from sucrose gradients were pooled, concentrated to 500 μl, and fractionated on a Superose 6 column (10/30; AP Biotech) in HEK buffer. Column fractions were analyzed by immunoblotting. Molecular masses of native proteins were calculated using the formula M = (6 πη0 Nas)/(1 − nρ) as described previously (42), where M is molecular mass; a is Stokes radius; s is the sedimentation coefficient; η0 (viscosity of water) = 1.002 × 102g/(cm·s); n is the partial specific volume of an average protein = 0.725 cm3/g; ρ is density of water = 0.998 g/cm3; and N is Avogadro's number. These values are tabulated in Fig. 1E. For the two “recombinant proteins” listed in Fig. 1E (untagged Bud14 or Kel2-His6), a similar analysis was performed as above, except 1–5 μg of purified protein was fractionated on sucrose gradients and gel filtration columns. Bud14 was followed on Coomassie-stained gels of fractions, whereas Kel2-His6 was followed on immunoblots probed with anti-His6 antibodies.
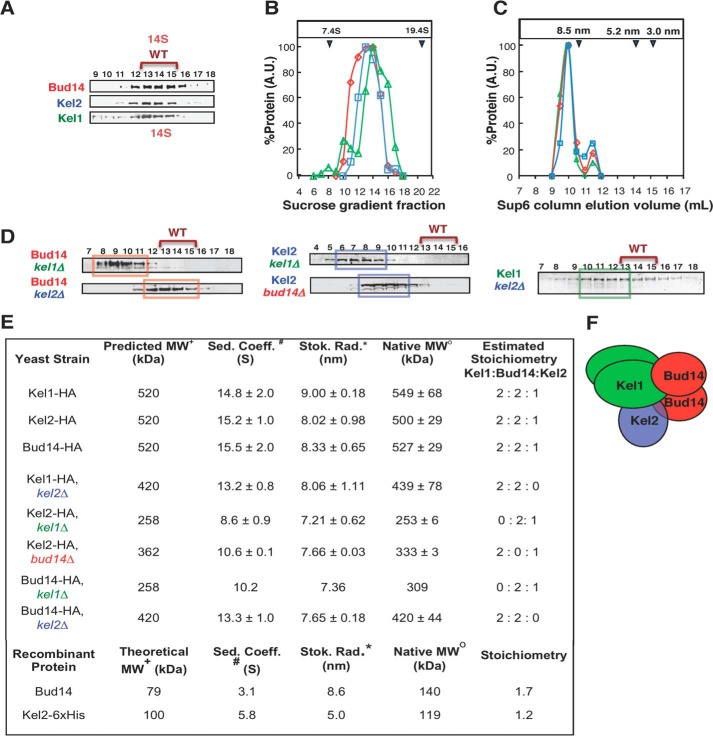
FIGURE 1.
Endogenous Bud14, Kel1, and Kel2 form a stable complex in cell extracts. A, velocity sedimentation analysis. Cell lysates from strains with integrated C-terminal 3×HA tags on Bud14, Kel1, and Kel2 were fractionated in parallel on sucrose gradients. Samples of each fraction were separated by SDS-PAGE and analyzed by immunoblotting with anti-HA antibodies. B, relative levels of each tagged protein from blots in A were determined by densitometry. The sedimentation coefficients (S values) for native Bud14-HA, Kel1-HA, and Kel2-HA were determined by comparison with protein standards run in parallel (indicated by blue arrowheads at the top of the graph). C, peak fractions from A were pooled, concentrated, and fractionated on a Superose 6 column. Fractions were immunoblotted to detect tagged proteins, and Stokes radii were determined. A small fraction of each protein migrated in a shoulder peak, consistent with some dissociation of components during the fractionation process. D, velocity sedimentation analysis of tagged proteins in cell lysates from Bud14-3×HA kel1Δ, Bud14-3×HA kel2Δ, Kel1–3×HA kel2Δ, Kel2-3×HA bud14Δ, and Kel2-3×HA kel1Δ strains, which were fractionated and analyzed as in A. E, summary of hydrodynamic properties for native proteins (top rows, isolated from the indicated strains), and purified proteins (bottom rows), including calculated molecular mass (MW), sedimentation coefficient (Sed. Coeff.), Stokes radius (Stok. Rad.), and estimated molar stoichiometry. Key to symbols: +, predicted mass is calculated from the estimated stoichiometry and accounts for the added mass of the 3×HA tag; #, determined by velocity sedimentation analysis; *, determined by analytical gel filtration analysis. o, calculated from the Stokes radius and sedimentation coefficient (see “Experimental Procedures”). F, working model for the 520-kDa Bud14-Kel1-Kel2 complex with a 2:2:1 stoichiometry.
Protein Purification
Bud14 was expressed with an N-terminal GST-TEV tag in Escherichia coli strain BL21 (DE3) in LB broth. Kel2 was expressed with an N-terminal GST-TEV tag and a C-terminal His6 tag in E. coli strain BL21 (DE3) Star in TB broth. In both cases, cell cultures were grown to log phase at 37 °C, induced for 3–4 h with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside, pelleted by centrifugation, and frozen at −80 °C. Cells were lysed by addition of zymolase and sonication and were centrifuged for 15 min at 16,000 rpm in an SA600 rotor (DuPont). Lysis buffer for Bud14 was 1× PBS plus protease inhibitors, and the buffer for Kel2 was 25 mm Tris (pH 8), 1× PBS, 100 mm NaCl, 1% Sarkosyl, and protease inhibitors. The supernatant was incubated for 2 h with 250 μl of glutathione-agarose resin (Qiagen, Valencia, CA), and the beads were washed two times with 15 ml of PBS + 0.4 m NaCl, two times with 15 ml of PBS, and two times with HEK buffer (20 mm HEPES, 1 mm EDTA, 50 mm KCl). Beads were transferred to a 2-ml tube, incubated for 2 h at room temperature with 10 μg of tobacco etch virus protease, and then pelleted. The supernatant was harvested, and tobacco etch virus-released Bud14 was concentrated in a cen-30 Microcon device (Millipore, Bedford, MA), aliquoted, snap-frozen in liquid N2, and stored at −80 °C.
Yeast Cell Imaging
For imaging actin organization, cells were grown to log phase in YPD, fixed in formaldehyde, and stained with Alexa 488 phalloidin (Invitrogen). To monitor secretory vesicle movements, cells were transformed with a plasmid expressing GFP-Sec4 under the control of its own promoter and grown in selective medium to log phase. To localize Kel1, Kel2, Bud14, Bni1, and Bnr1, cells carrying integrated GFP or red fluorescent protein tags were grown to log phase and imaged. For all live cell imaging experiments, cells were mounted on slides and observed at room temperature. Movies of single focal planes were acquired through consecutive 300-ms exposures. All images were captured on a Zeiss Axioskop-2 mot plus microscope (Carl Zeiss, Thornwood, NY) with a Hamamatsu ORCA-ER digital CCD camera (Hamamatsu Photonics, Bridgewater, NJ) running OpenLab software (Improvision, Lexington, MA). Unless noted, all quantifications score at least 200 cells. Error bars are S.D. For scoring latrunculin A (LatA)2-resistant actin cables, 5-ml cultures of wild type, kel1Δ, kel2Δ, and bud14Δ strains were grown in YPD to log phase. Cells were pelleted, resuspended in 1 ml of YPD, and allowed to recover for 30 min. 200 μl of cells was incubated with control buffer or buffer containing 20 μm LatA at 30 °C. At 60 s, cells were fixed, stained with Alexa 488 phalloidin, imaged as above, and scored for visible actin cables as described (43).
Analysis of Cytokinesis Defects by Light and Electron Microscopy
To visualize chitin distribution, yeast cells were grown to mid log phase, stained with Calcofluor white, and imaged by fluorescence light microscopy as above. To examine septum ultrastructure, cells were placed on copper EM grids (400 mesh) coated with Formvar. Grids were rapidly submersed in liquid propane and held at −180 °C with liquid nitrogen. Then grids were transferred to a pre-cooled solution of 4% osmium tetroxide in dry acetone in a 1.8-ml polypropylene vial at −82 °C for 72 h (substitution) and warmed gradually to room temperature, followed by three washes in dry acetone and infiltrated with Araldite (epoxy resin; Sigma). Ultrathin sections (80 nm) were incubated for 20 min with periodic acid (1%), washed five times with water (10 min each), and incubated for 30 min in thiocarbohydrazide (0.2% in acetic acid 20%). Then cells were washed five times for 30 min, contrasted for 30 min in silver proteinate (1%), and finally washed three times with water. Specimens were observed with a Hitachi 7650 (80 kV) transmission electron microscope at the Bordeaux Imaging Center.
RESULTS
Bud14, Kel1, and Kel2 Interact to Form a Stable 520-kDa Complex
To gain further insights into the mechanism of Bud14 regulation of Bnr1, we examined the native state of Bud14 (with an integrated C-terminal 3×HA tag) in crude cell extracts fractionated on sucrose gradients and gel filtration columns (Fig. 1A). This analysis showed that Bud14 forms a large stable complex with a sedimentation coefficient (S) of 15.5 ± 2.0 and a Stokes radius of 8.33 ± 0.65. From these values, we calculated the mass of the complex to be 527 ± 29 kDa (Fig. 1E). Because Bud14 has an apparent molecular mass of 79 kDa and purified Bud14 forms dimers (see below), the high molecular mass of the native Bud14 complex suggested that it contains additional molecular components.
We next integrated C-terminal 3×HA tags on Kel1 and Kel2 to investigate whether these proteins might be part of the Bud14 complex, because they have been identified in proteomic studies as potential Bud14-binding partners (31–33) and because S. pombe Kelch proteins tea1p and tea3p interact with tea4p, the closest homologue of Bud14 (38). Velocity sedimentation and analytical gel filtration analysis of Kel1–3×HA and Kel2–3×HA cell extracts revealed a striking overlap in their migration patterns with each other and with Bud14-3×HA (Fig. 1, A–C). The estimated native complex molecular masses for Kel1-3×HA and Kel2-3×HA were 549 ± 68 and 500 ± 20 kDa, respectively (Fig. 1E). These data suggested that Bud14, Kel1, and Kel2 might be components of a single stable complex of ∼520 kDa.
To further test this model, we deleted individual components of the proposed complex and looked for expected shifts in the molecular masses of the remaining components. Indeed, when KEL2 was deleted, we observed a loss of ∼100 kDa from the remaining complex formed by Kel1 and Bud14 (Fig. 1, D and E), which corresponds to the mass of one molecule of Kel2 (100 kDa). Furthermore, when BUD14 was deleted, we observed a loss of ∼170 kDa from the remaining complex, which is approximately the mass of two molecules of Bud14 (79 kDa), consistent with dimerization of purified Bud14 (see below). Upon deletion of KEL1, an even larger shift was observed, corresponding to a loss of ∼230 kDa from the remaining complex. This is approximately the size of two molecules of Kel1 (131 kDa), consistent with dimerization of purified Kel1 (see below).
To address the stoichiometry of the Bud14-Kel1-Kel2 complex, we investigated the oligomerization states of purified individual components (full-length Kel2 and Bud14) by velocity sedimentation and analytical gel filtration (Fig. 1E). This showed that recombinant Bud14 has a native mass of ∼140 kDa, which is approximately twice its predicted mass (79 kDa), consistent with the formation of dimers. In contrast, Kel2 (100 kDa) had a native mass of ∼100 kDa, matching its predicted mass (100 kDa), and therefore suggesting it is monomeric (Fig. 1E). Despite repeated efforts, we were unable to purify sufficient quantities of recombinant full-length Kel1 for hydrodynamic analyses. However, previous co-immunoprecipitation and two-hybrid studies support homotypic Kel1-Kel1 interactions but failed to detect Kel2 self-interactions (26), which agrees well with our model.
Based on these observations, we propose that the native ∼520-kDa complex formed by Bud14, Kel1, and Kel2 consists of one Bud14 dimer (158 kDa), one Kel1 dimer (262 kDa), and one Kel2 monomer (100 kDa) (as depicted in Fig. 1F). Furthermore, our data suggest that no single component of the complex is critical for maintaining interactions between the remaining two components (Fig. 1E).
Bud14 and the Kelch Proteins Co-localize at Polarity Sites in a Kel1-dependent Fashion
To investigate whether Bud14, Kel1, and Kel2 function together in vivo, we first carefully examined their cellular localization patterns. Previously, we reported that endogenously tagged GFP-Bud14 localizes to sites of polarized growth, i.e. the bud neck and bud tip/cortex (13). Specifically, GFP-Bud14 was found in multiple puncta enriched at the bud neck and cortex. GFP-tagged Kel1 and Kel2 have also been shown to localize as puncta at polarity sites (26). Importantly however, Bud14 and Kelch have never been localized in the same cells to determine whether their localization patterns overlap. To accomplish this, we performed live cell imaging on yeast strains expressing mCherry-Bud14 in combination with Kel1-GFP or Kel2-GFP, as well as strains expressing individually tagged proteins (Fig. 2). As we reported previously for GFP-Bud14, mCherry-Bud14 localized as puncta enriched at the bud neck and bud tip (Fig. 2, A and B). Additionally, we observed a second set of fainter and more dynamic puncta of mCherry-Bud14 in both the mother and daughter cells (Fig. 2, A and B). Kel1-GFP and Kel2-GFP displayed similar patterns of localization, with bright puncta localizing primarily to the bud tip or neck, and a second set of faint and dynamic puncta localizing throughout cells (Fig. 2A). In strains co-expressing mCherry-Bud14 with Kel1-GFP or Kel2-GFP, the signals overlapped strongly at polarity sites (arrows, Fig. 2, B and C). However, we were unable to determine the degree to which the fainter puncta of Bud14, Kel1, and Kel2 co-localized because they were highly dynamic and moved faster than the two-color acquisition rate.
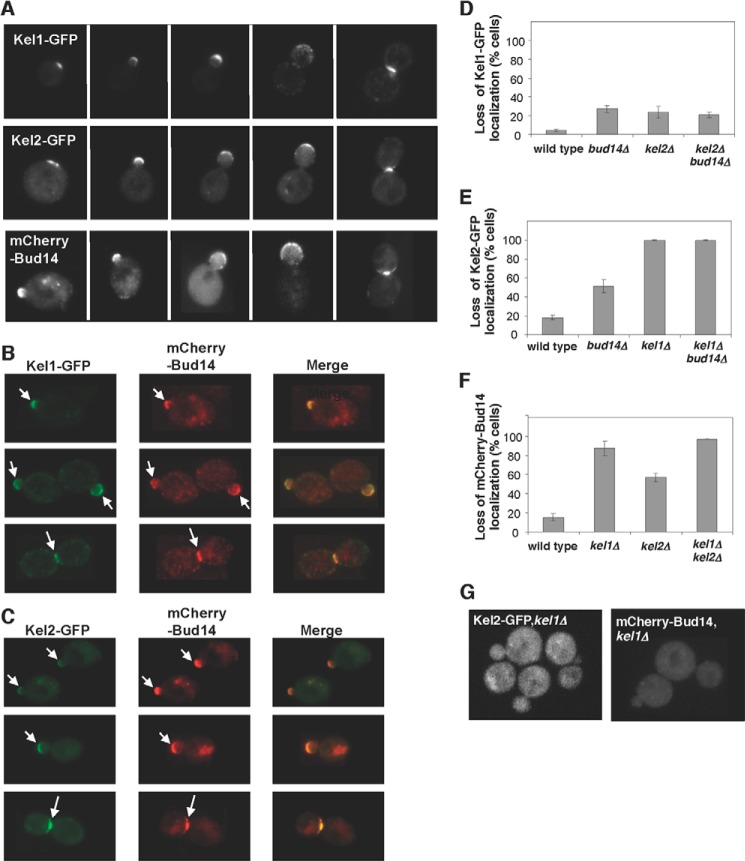
FIGURE 2.
Localization of Bud14, Kel1, and Kel2 in cells. A–C, images of cells expressing the indicated combinations of Kel1-GFP, Kel2-GFP, and mCherry-Bud14 at different stages of the cell cycle (arrows mark sites of polarized growth). D–F, comparison of Kel1-GFP, Kel2-GFP, and mCherry-Bud14 localization in wild type and mutant cells. Data were averaged from three experiments (>200 cells scored per strain) and graphed as percentage of cells with loss of localization at polarity sites. Error bars represent S.D. G, examples of mutant cells showing loss of mCherry-Bud14 or Kel2-GFP signal at polarity sites.
We next characterized the localization interdependencies among Bud14, Kel1, and Kel2 using the tagged proteins expressed from their endogenous promoters. Previous studies have reported that Bud14-GFP and Kel2-GFP localization depends on KEL1 (26, 30). However, the potential localization dependence of Kel1-GFP and Kel2-GFP on Bud14 has not been tested. Therefore, we tested all combinations in the same experiments using an isogenic strain set. We found that mCherry-Bud14 and Kel2-GFP are each severely mislocalized in kel1Δ cells, with less than 10% of cells retaining detectable localization at polarity sites (Fig. 2, E–G). In contrast, Kel1-GFP localization was only subtly perturbed in kel2Δ, bud14Δ, or kel2Δbud14Δ cells, with 75% of cells retaining normal Kel1-GFP localization (Fig. 2D). Kel2-GFP localization was modestly disrupted in bud14Δ cells, with about 50% of cells retaining normal localization (Fig. 2E). Similarly, mCherry-Bud14 localization was partially disrupted in kel2Δ cells, with about 50% of cells retaining normal localization (Fig. 2F). These results are in good agreement with the previous studies showing that Bud14 and Kel2 localization depends on KEL1 (26, 30). From these observations, we conclude that Kel1 is the primary localization determinant within the Bud14-Kel1-Kel2 complex and is chiefly responsible for directing it to polarity sites. Bud14 and Kel2 have lesser though still significant effects on each other's localization.
kel1Δ and kel2Δ Cells Display Actin Cable Defects Similar to bud14Δ Cells
Next we investigated the potential roles of KEL1 and KEL2 in actin cable regulation by comparing cellular F-actin staining in wild type, kel1Δ, kel2Δ, and bud14Δ strains. We previously showed that deletion of BUD14 leads to defects in actin cable architecture and function, with some of the cables in the mother cell appearing abnormally bent or buckled and causing defective secretory vesicle trafficking (13). Our analysis here revealed that kel1Δ and kel2Δ mutants each display bud14Δ-like bent cables (Fig. 3A, yellow arrows), with these defects visible in 48 ± 5% of bud14Δ cells, 49 ± 4% of kel2Δ cells, and 29 ± 1% of kel1Δ cells. We also observed modest depolarization of actin patches, which often accompanies cable defects; this was most evident in kel2Δ and bud14Δ strains (Fig. 3A).
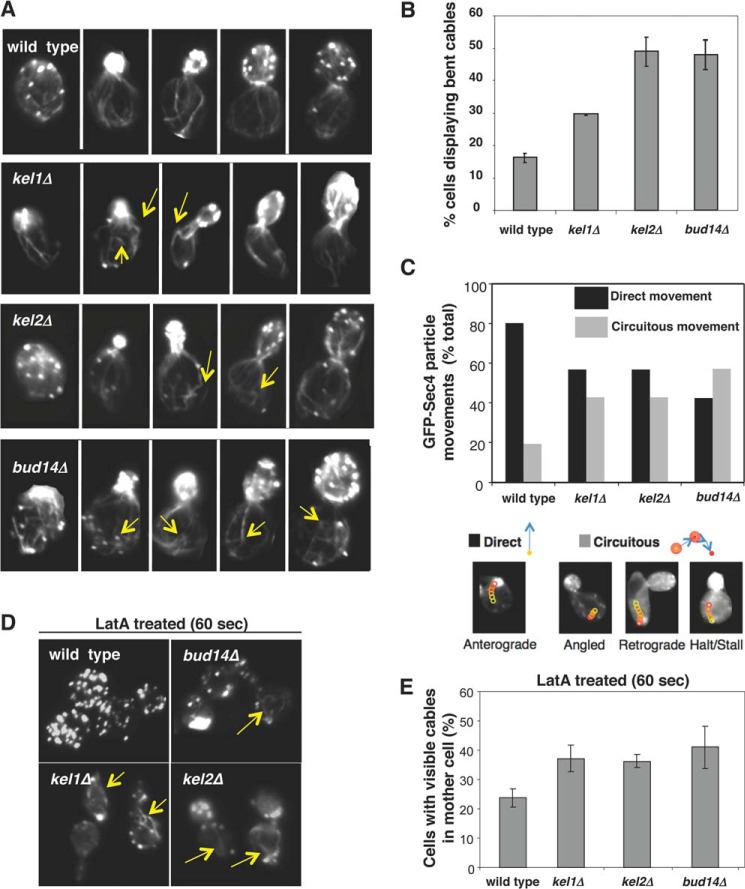
FIGURE 3.
Defects in actin cable morphology and transport of secretory vesicles in kel1Δ and kel2Δ mutants. A, cells were grown to log phase, fixed, and stained with Alexa 488 phalloidin. Yellow arrows highlight some of the “bent” cables in mutant cells. B, quantification of cable phenotype. Cables exhibiting a change in direction of at least 75 degrees at the cell cortex were scored as bent. Data averaged from two experiments (scoring >100 cells per strain in each experiment). C, comparison of GFP-Sec4 secretory vesicle movements in wild type, kel1Δ, kel2Δ, and bud14Δ cells (> 200 vesicles per strain). Vesicle movements classified as “direct” were rapid anterograde movements toward the neck. Movements classified as “circuitous” were (i) stalled, (ii) retrograde (away from the bud), or (iii) highly angled off the mother-bud axis. D, representative images of wild type, bud14Δ, kel1Δ, and kel2Δ cells treated with LatA for 60 s, fixed, and stained with Alexa 488 phalloidin. E, graphs show the average from two experiments as in D, with >100 cells scored per strain per experiment. Error bars represent S.D.
One of the essential functions of actin cables is to direct myosin-based intracellular transport of secretory vesicles (8), and defects in actin cable architecture often leads to abnormal traffic of vesicles (13). Therefore, to further examine the defects in kel1Δ, kel2Δ, and bud14Δ cells, we used live cell imaging and compared the movements of secretory vesicles, marked with GFP-Sec4, in the mother cell compartments (Fig. 3C). In each case, the movements of vesicles in cells were monitored for 5 min and categorized as either “direct,” defined as normal rapid, anterograde transport toward the bud neck without visible pauses, or as “circuitous,” defined as transverse, retrograde, and/or stalled movements (no movement for at least 90 s). Circuitous vesicle movements were observed more frequently in kel1Δ, kel2Δ, and bud14Δ cells compared with wild type cells (Fig. 3C). These results indicate that Kel1, Kel2, and Bud14 are each required for normal actin cable architecture, and in turn for proper directed transport of secretory vesicles.
As another test of actin cable defects, we measured cable sensitivity to depolymerization by the drug LatA. Cables are dynamic structures composed of short cross-linked filaments, and are rapidly and continuously assembled at the bud neck and disassembled in the cytosol. Cables in wild type cells are short lived and almost completely disappear within 60 s of treating cells with 20 μm LatA, an actin monomer-sequestering agent (11). Alterations in the lengths and/or cross-linking of filaments in cables can alter their LatA sensitivity, and previously we showed that the aberrant bent cables in bud14Δ cells are resistant to LatA (13). Therefore, we compared cable staining in wild type, bud14Δ, kel1Δ, and kel2Δ cells after treatment with 20 μm LatA for 60 s (Fig. 3D, arrows). Each mutant had approximately twice as many cells with LatA-resistant actin cables compared with wild type cells (Fig. 3E).
All of these observations taken together indicate that kel1Δ and kel2Δ cells have actin cable defects that are strikingly similar to those found in bud14Δ cells, and therefore they are consistent with Kel1, Kel2, and Bud14 functioning together to regulate actin cable formation.
Kel1 and Kel2 Function in the Same Genetic Pathway as Bud14 to Regulate Bnr1
To better understand the functional relationship between Kelch proteins and Bnr1, we considered whether the cable defects in kel1Δ and kel2Δ cells might be due to mislocalization of Bnr1, particularly because S. pombe tea1p is required for formin (for3p) localization (44). To address this, we generated a strain expressing Bnr1-RFP and Bni1–3GFP, each from their endogenous promoters, which enabled us to visualize the localization of both formins in the same cells. As reported previously (45), Bnr1-RFP localized to the bud neck through most stages of the cell cycle, whereas Bni1–3GFP localized to dynamic puncta that become enriched at the bud tip early in the cell cycle and then at the bud neck later in the cell cycle. In kel1Δ and kel2Δ cells, Bnr1 localized to the bud neck and Bni1 localized to cytosolic puncta, similar to wild type cells (representative images in Fig. 4A). From these data, we conclude that deletion of either KEL1 or KEL2 does not alter the localization of Bnr1 in polarized cells, and previously we showed that deletion of BUD14 does not alter Bnr1 localization (13). However, our data do not rule out the possibility of kel1Δ and/or kel2Δ affecting the dynamics of Bni1 punctae, which are difficult to track.
FIGURE 4.
KEL1 and KEL2 function with BUD14 to regulate BNR1. A, deletions of KEL1 and/or KEL2 do not alter Bni1 or Bnr1 localization in cells. Strains with the genotypes indicated and expressing integrated Bnr1-RFP and Bni1–3×GFP were imaged. B, kel1Δ and kel2Δ, like bud14Δ, suppress the growth defects of bnr1ΔDAD. Strains were grown to log phase, serially diluted, spotted on YPD plates, and compared for growth at 25 and 37 °C. C, representative images of the same strains as in B at different stages of the cell cycle. Cells were grown to log phase, fixed, and stained with Alexa 488 phalloidin. D, quantification of bent cable phenotype (as in Fig. 3B). Data averaged from two experiments (scoring >100 cells per strain per experiment). Error bars represent S.D. E, example images from D. Yellow arrows indicate the presence of kinked cables in these strains.
The observations above suggest that Kel1 and Kel2 function closely with Bud14 to govern Bnr1 activity in vivo and thereby influence actin cable architecture and function. To further test this model, we asked whether kel1Δ or kel2Δ mutations could suppress the defects of a hyperactive bnr1ΔDAD allele. Previously, we showed that bnr1ΔDAD leads to disorganized arrays of short cables, with accompanying defects in secretory vesicle traffic and cell growth at elevated temperatures (13). Furthermore, these defects could be suppressed by bud14Δ, because bnr1ΔDAD and bud14Δ have opposite effects on cable architecture (13). Therefore, we tested whether kel1Δ and/or kel2Δ might similarly suppress bnr1ΔDAD. Our results show that kel1Δ, kel2Δ, and kel1Δkel2Δ mutations each suppressed the temperature-sensitive growth of bnr1ΔDAD and rescued the short, disorganized actin cable arrays (Fig. 4, B and C). In addition, we compared the phenotypes of kel1Δ, kel2Δ, and bud14Δ single mutants to double and triple mutants, to determine whether or not Kelch proteins and Bud14 make independent or related contributions to cable regulation. We found that kel1Δbud14Δ, kel2Δbud14Δ, kel1Δkel2Δ, and kel1Δkel2Δbud14Δ mutants do not have compounded cable defects and instead display the same prevalence of kinked or bent cable defects as single mutants (Fig. 4, D and E). These results provide strong genetic support for the view that Kel1 and Kel2 function in the same pathway as Bud14 in governing Bnr1-mediated actin cable assembly.
Roles for Bud14, Kel1, and Kel2 in Cytokinesis
We next asked whether Kel1, Kel2, and Bud14 also regulate Bnr1 functions in cytokinesis. Bnr1 and Bni1 have genetically redundant yet mechanistically distinct roles during cytokinesis. Bni1 is critical for generating an actomyosin ring capable of contracting, and both formins contribute to the formation of actin cables, which direct vesicle traffic to the bud neck to promote septation (5, 46, 47). More specifically, the delivery of chitin synthase leads to formation of a chitin-rich structure called the “primary septum,” and next flanking “secondary septa” are laid down, which more closely resemble the normal cell wall (46, 48, 49). Finally, the primary septum is degraded by chitinase to complete cell separation. For this reason, defects in formation of the primary septum often impair cell separation.
To address the potential roles of KEL1, KEL2, and BUD14 in cytokinesis, we examined the effects of deleting each gene in a bni1Δ background, where Bnr1 becomes essential for cell division (5, 46). A hallmark of cytokinesis defects is the “chained cell phenotype,” in which cells fail to separate and therefore remain connected. Even though Bni1 is required for formation of a functional actomyosin ring, this structure is not essential for cytokinesis (50, 51). As a result, bni1Δ cells exhibit partial defects in cytokinesis, with ∼15% of cells being “chained” (Fig. 5, A and B). In bud14Δbni1Δ, kel1Δbni1Δ, and kel2Δbni1Δ double mutants, we observed a striking increase in this phenotype compared with bni1Δ single mutants (∼70% of double mutant cells in each case were connected) (Fig. 5, A and B). These results suggest that Kel1, Kel2, and Bud14 make Bni1-independent contributions to cytokinesis, consistent with a role in regulating Bnr1-mediated cable formation to facilitate cytokinesis. In addition, we compared the primary septa in single and double mutants by Calcofluor staining for chitin. This revealed that kel1Δ, kel2Δ, and bud14Δ mutations in a bni1Δ background greatly increase the percentage of cells with abnormally accumulated chitin (Fig. 5, C and D), further supporting the view that Kel1, Kel2, and Bud14 regulate Bnr1-dependent cable formation to facilitate septation.
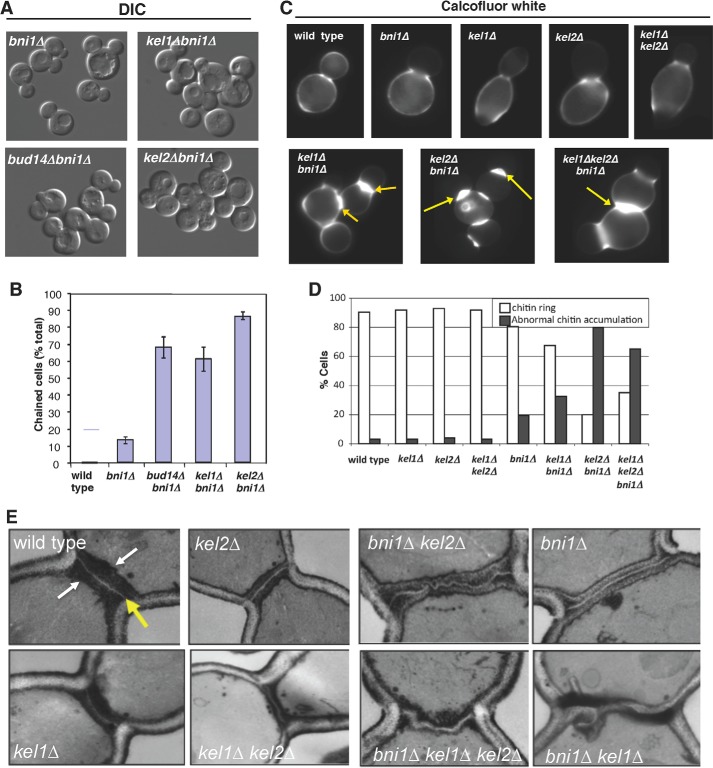
FIGURE 5.
Roles for Kel1, Kel2, and Bud14 in cytokinesis. A and B, differential interference contrast (DIC) imaging and quantification of chained phenotype in bni1Δ, kel1Δbni1Δ, kel2Δbni1Δ, and bud14Δbni1Δ cells. Data averaged from two experiments, scoring >200 cells per strain in each experiment. Error bars represent S.D. C, cell images of chitin defects. Strains were grown to log phase, fixed, and stained with Calcofluor white to visualize chitin. D, quantification of chitin defects. White bars are the percentage of cells with normal chitin rings, resembling those in wild type cells. Gray bars are the percentage of cells with abnormal chitin rings, which are much thicker than in wild type cells, e.g. yellow arrows in differential interference contrast images. E, ultrastructural analysis of septum defects. Cells were grown to log phase, fixed, thin sectioned, stained by the Thiery method, and visualized by electron microscopy. Representative images are shown for each strain.
To further analyze the septum defects caused by loss of Kel1 and Kel2, we examined the ultrastructure of the division plane in select mutant strains by electron microscopy (Fig. 5E). Wild type, kel1Δ, and kel2Δ cells each had normal primary septa (Fig. 5E, yellow arrow) and secondary septa (Fig. 5E, white arrows), consistent with the observations described above. In contrast, bni1Δ cells had primary septa that were abnormally “curly” and secondary septa that were abnormally thick. These observations are in good agreement with Bni1 being partially required for septation. Consistent with our observations by chitin staining, kel2Δbni1Δ cells showed similar septation defects to bni1Δ cells but with more cells failing to separate. Unexpectedly, kel1Δbni1Δ mutants showed very irregular and misshapen primary and secondary septa (Fig. 5E), which were not observed in kel2Δbni1Δ mutants or single mutants. The septa in kel1Δbni1Δ double mutants were unusually thick and gnarled. We speculate that the loss of Kel2 causes a more complete loss of cable function at cytokinesis, explaining the higher percentage of cells with aberrant chitin staining and chained cell phenotype, whereas loss of Kel1 causes a more partial loss of cable function that leads to more septum deposition, but with a highly altered organization (see “Discussion”). As a further test of this model, we examined the ultrastructure of kel1Δkel2Δbni1Δ mutants and found that the triple mutant defects resembled kel2Δbni1Δ mutants, supporting the view that kel2Δ causes a more complete loss of Bnr1-dependent function during cytokinesis compared with kel1Δ. These results are also consistent with the higher penetrance of chitin defects and chained cell defects in bni1Δkel2Δ versus bni1Δkel1Δ cells (Fig. 5D).
DISCUSSION
Our initial goal in this study was to determine whether Bud14 functions alone or together with other cellular factors in regulating Bnr1-mediated actin assembly. In pursuing this question, we discovered that Bud14 and the two S. cerevisiae Kelch proteins Kel1 and Kel2 form a stable, discrete (∼520 kDa) complex that localizes to sites of polarized cell growth. From a comparison of the mass of the native complex in extracts from wild type, bud14Δ, kel1Δ, and kel2Δ cells, and a determination of the oligomerization states of purified Bud14 and Kel2, we are able to assemble a working model for the complex, in which it has a 2:2:1 stoichiometry of Bud14, Kel1, and Kel2, respectively. Our data suggest that these three proteins account for the majority of the mass of the complex, while leaving open the possibility that there may be additional components of lower mass and/or transiently associated interaction partners in vivo. Indeed, along with their interactions with each other, a number of other potential interaction partners of Bud14, Kel1, and Kel2 have been identified (26, 27, 30–32). However, our data indicate that within this network of physical interactions, the triad of Bud14, Kel1, and Kel2 forms a stable hub.
Our data show that Bud14, Kel1, and Kel2 co-localize at the bud neck and the bud cortex. In addition, each protein was detected as faint, highly dynamic punctae that were difficult to track. Localization of the Bud14-Kel1-Kel2 complex to the neck is consistent with their functional role in regulating Bnr1, because Bnr1 is stably anchored at the neck by interactions with septins and/or septin-associated proteins (45, 52, 53). What function(s) might be played by the Bud14-Kel1-Kel2 complex at the bud cortex is less clear and will require more investigation. However, it is known that Bud14 regulates dynein function at the bud cortex, by directly recruiting protein phosphatase 1 (Glc7), which in turn controls dynein activity in mitotic spindle positioning (27, 30). Thus, the Bud14-Kel1-Kel2 complex appears to interact with Bnr1 at the bud neck and with Glc7 at the bud cortex. How the BKK complex potentially coordinates these two processes, actin cable formation and spindle positioning, is an important question for the future.
By analyzing the phenotypes of single, double, and triple mutants (kel1Δ, kel2Δ, and bud14Δ), we determined that these three proteins function in the same pathway regulating Bnr1-mediated actin cable formation. These results have at least three important implications. First, they demonstrate that although in vitro Bud14 is sufficient to regulate Bnr1 activity, in vivo it functions as part of a large multicomponent complex. Second, they shed new light on how Kel1 and Kel2 mechanistically contribute to directing cell morphogenesis, directly linking Kel1 and Kel2 to the control of actin assembly. Third, they point to a common mechanism by which members of the Kelch family may regulate the actin cytoskeleton in diverse species. Genetic studies in a wide range of model organisms have shown that Kelch proteins are required for proper formation of diverse actin-based structures (see Introduction); however, the mechanisms underlying these effects have remained obscure. Our results, together with key observations made for Kelch proteins in S. pombe (see Introduction and below), suggest that formin regulation may be a common function of the Kelch family in diverse systems.
What Mechanistic Roles Do Kelch Proteins Play in Regulating Formin Activities?
In vivo, Bud14 is required for proper morphology of actin cables generated by Bnr1, which in turn is required for efficient delivery of secretory vesicles to the bud neck (13). Here, we found that deletions of KEL1 and/or KEL2 caused very similar phenotypes, i.e. abnormal cable architecture and defective secretory traffic. How does each component of the Bud14-Kel1-Kel2 complex contribute to this function? Bud14 directly inhibits the FH2 domain of Bnr1 in vitro and in the presence of capping protein is sufficient to displace Bnr1 from the growing ends of actin filaments (13). What then are the roles of Kel1 and Kel2 within the Bud14-Kel1-Kel2 complex? For Kel1, our data, and that from previous studies (26, 30), indicate that it is required for normal localization of Bud14 and Kel2 to polarity sites. Therefore, subcellular targeting may be one of the key roles of Kel1 in the complex, although this certainly does not rule out additional contributions to formin regulation, including potential direct interactions with F-actin. For Kel2, we speculate that it may regulate Bud14's displacement effects on Bnr1 based on the observation that a kel2Δ complex is as nonfunctional in vivo as a bud14Δ complex, yet the kel2Δ complex (consisting of Kel1 and Bud14) partially localizes to polarity sites. Kel2 (and/or Kel1) could help control the timing of Bud14 displacement activities and/or “reset” the inhibitory effects of Bud14 on Bnr1, ensuring that not all Bnr1 molecules are bound by Bud14.
Defining the precise functions of Kel2 (and Kel1) on Bud14-Bnr1 activities will require more in depth biochemical analysis. To date this was not possible because, despite repeated attempts, we were not able to isolate sufficient quantities of Kel1 and Kel2 to perform these tests. We were able to purify only small quantities of Kel2-His6, sufficient to analyze its hydrodynamic properties by tracking the protein on immunoblots (Fig. 1E). Nonetheless, we propose the following working model. Kel1 functions to localize the complex, and Kel2 plays a key role in controlling Bud14-Bnr1 activities. This would explain why the kel1Δ phenotype is less severe than kel2Δ and bud14Δ phenotypes. If the primary role of Kel1 is to localize the complex to polarity sites, yet Kel2 and Bud14 can remain associated in kel1Δ mutants (Fig. 1D), then the remaining Bud14-Kel2 complex might weakly interact with Bnr1 at the neck and retain partial function. Indeed, this view is consistent with our data showing that Bud14 and Kel2 each partially contribute to localization of the complex to the bud neck (Fig. 2, D–F).
Actin Regulation by Kelch Proteins in Diverse Organisms
Drosophila Kelch, the founding member of the Kelch family, plays an important role in formation and stabilization of the actin-based ring canals (20, 23, 24). This observation, combined with subsequent results from studies on a variety of Kelch homologues in other species, led to the general view that Kelch proteins control the architecture of F-actin structures in vivo (15). Here, we have extended this view by uncovering new roles for the S. cerevisiae Kel1 and Kel2 in actin cable formation and directly linking them to formin regulation. How widespread is formin regulation by Kelch proteins in other systems? In S. pombe, the Kelch homologue tea1p is part of a large complex that contains the formin for3p, as well as the Bud14 homologue tea4p (38, 44). Interestingly, tea1p and tea4p are required for proper localization of for3p to the cell tip during polarized growth (38). However, in S. cerevisiae, Bnr1 localizes to the bud neck independent of Kel1 and Kel2 (this study) or Bud14 (13). This key difference may stem from the fact that S. pombe formin for3p is dynamically recruited to polarity sites by microtubules (54), and therefore, tea1p and tea4p could regulate for3p activity by controlling its localization (note: potential direct effects of tea1p and tea4p on for3p activity have not been investigated). In contrast, S. cerevisiae Bnr1 is stably anchored at the bud neck through interactions with septin-associated proteins (45, 52, 53); however, Kel1, Kel2, and Bud14 are recruited to the neck by an independent mechanism and appear to control formin activity rather than localization. The common denominator in these two systems is that they use a set of homologous proteins to generate properly functioning formin-dependent actin cables used to direct secretion and polarized cell growth. Thus, in each case the specific roles of the Kelch proteins (and their interaction partner, tea4p or Bud14) in regulating formin function appear to have been tailored to the individual system, suggesting a degree of plasticity in how this machinery can be harnessed to the control of formins.
Importantly, because Kelch proteins in both budding yeast and fission yeast have now been directly linked to formin regulation, this raises the intriguing possibility that animal Kelch proteins perform related functions. Indeed, several observations already point to such a possibility. Two Kelch proteins (KLEIP and human Keap1) localize to and are required for the formation of actin-rich cell-cell junctions, which depend on formins for their assembly (55–57). Knockdown of Muskelin, a mouse Kelch protein, leads to excessive cell protrusions, membrane ruffles, and stress fibers (58), phenotypes that also arises from formin hyperactivity (59). The mouse Kelch protein Nd1 functions in Rho-dependent rearrangements of the actin cytoskeleton, and formins are prominent effectors of Rho (60, 61). Finally, the rat Kelch protein Krp1 co-localizes with F-actin rich ruffle-like structures at cell tips and is critical for pseudopod elongation, a formin-dependent structure required for cell motility and invasion (62).
In summary, our results combined with observations made in other systems, described above, suggest that Kelch family proteins are conserved multifunctional regulators of the actin cytoskeleton that govern the dynamics and organization of actin-based structures not only through their associations with F-actin but also their emerging effects on formins.
Acknowledgments
We thank Brian Graziano and Casey Ydenberg for critical reading of the manuscript. We thank the Bordeaux Imaging center facility for assistance with the electron microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant GM083137 (to B. G.).
- LatA
- latrunculin A.
REFERENCES
- 1. Schönichen A., Geyer M. (2010) Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim. Biophys. Acta 1803, 152–163 [DOI] [PubMed] [Google Scholar]
- 2. Breitsprecher D., Goode B. L. (2013) Formins at a glance. J. Cell Sci. 126, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chesarone-Cataldo M., Guérin C., Yu J. H., Wedlich-Soldner R., Blanchoin L., Goode B. L. (2011) The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev. Cell 21, 217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chesarone M. A., DuPage A. G., Goode B. L. (2010) Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11, 62–74 [DOI] [PubMed] [Google Scholar]
- 5. Vallen E. A., Caviston J., Bi E. (2000) Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. (2002) Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 260–269 [DOI] [PubMed] [Google Scholar]
- 7. Bretscher A. (2003) Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J. Cell Biol. 160, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. (2004) Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20, 559–591 [DOI] [PubMed] [Google Scholar]
- 9. Pruyne D., Gao L., Bi E., Bretscher A. (2004) Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15, 4971–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu J. H., Crevenna A. H., Bettenbühl M., Freisinger T., Wedlich-Söldner R. (2011) Cortical actin dynamics driven by formins and myosin V. J. Cell Sci. 124, 1533–1541 [DOI] [PubMed] [Google Scholar]
- 11. Yang H. C., Pon L. A. (2002) Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 99, 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schott D. H., Collins R. N., Bretscher A. (2002) Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J. Cell Biol. 156, 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chesarone M., Gould C. J., Moseley J. B., Goode B. L. (2009) Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev. Cell 16, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hudson A. M., Cooley L. (2008) Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. Subcell. Biochem. 48, 6–19 [DOI] [PubMed] [Google Scholar]
- 15. Adams J., Kelso R., Cooley L. (2000) The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10, 17–24 [DOI] [PubMed] [Google Scholar]
- 16. Ito N., Phillips S. E., Yadav K. D., Knowles P. F. (1994) Crystal structure of a free radical enzyme, galactose oxidase. J. Mol. Biol. 238, 794–814 [DOI] [PubMed] [Google Scholar]
- 17. Li X., Zhang D., Hannink M., Beamer L. J. (2004) Crystal structure of the Kelch domain of human Keap1. J. Biol. Chem. 279, 54750–54758 [DOI] [PubMed] [Google Scholar]
- 18. Li X., Zhang D., Hannink M., Beamer L. J. (2004) Crystallization and initial crystallographic analysis of the Kelch domain from human Keap1. Acta Crystallogr. D Biol. Crystallogr. 60, 2346–2348 [DOI] [PubMed] [Google Scholar]
- 19. Schmid M. F., Agris J. M., Jakana J., Matsudaira P., Chiu W. (1994) Three-dimensional structure of a single filament in the Limulus acrosomal bundle: scruin binds to homologous helix-loop-beta motifs in actin. J. Cell Biol. 124, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robinson D. N., Cooley L. (1997) Drosophila kelch is an oligomeric ring canal actin organizer. J. Cell Biol. 138, 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim I. F., Mohammadi E., Huang R. C. (1999) Isolation and characterization of IPP, a novel human gene encoding an actin-binding, kelch-like protein. Gene 228, 73–83 [DOI] [PubMed] [Google Scholar]
- 22. Soltysik-Espanola M., Rogers R. A., Jiang S., Kim T. A., Gaedigk R., White R. A., Avraham H., Avraham S. (1999) Characterization of Mayven, a novel actin-binding protein predominantly expressed in brain. Mol. Biol. Cell 10, 2361–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xue F., Cooley L. (1993) kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72, 681–693 [DOI] [PubMed] [Google Scholar]
- 24. Tilney L. G., Tilney M. S., Guild G. M. (1996) Formation of actin filament bundles in the ring canals of developing Drosophila follicles. J. Cell Biol. 133, 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson D. N., Cooley L. (1997) Genetic analysis of the actin cytoskeleton in the Drosophila ovary. Annu. Rev. Cell Dev. Biol. 13, 147–170 [DOI] [PubMed] [Google Scholar]
- 26. Philips J., Herskowitz I. (1998) Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. J. Cell Biol. 143, 375–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cullen P. J., Sprague G. F., Jr. (2002) The Glc7p-interacting protein Bud14p attenuates polarized growth, pheromone response, and filamentous growth in Saccharomyces cerevisiae. Eukaryot. Cell 1, 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Höfken T., Schiebel E. (2002) A role for cell polarity proteins in mitotic exit. EMBO J. 21, 4851–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seshan A., Bardin A. J., Amon A. (2002) Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 12, 2098–2110 [DOI] [PubMed] [Google Scholar]
- 30. Knaus M., Cameroni E., Pedruzzi I., Tatchell K., De Virgilio C., Peter M. (2005) The Bud14p-Glc7p complex functions as a cortical regulator of dynein in budding yeast. EMBO J. 24, 3000–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 32. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 33. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 34. Snaith H. A., Sawin K. E. (2005) Tea for three: control of fission yeast polarity. Nat. Cell Biol. 7, 450–451 [DOI] [PubMed] [Google Scholar]
- 35. Behrens R., Nurse P. (2002) Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J. Cell Biol. 157, 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Glynn J. M., Lustig R. J., Berlin A., Chang F. (2001) Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11, 836–845 [DOI] [PubMed] [Google Scholar]
- 37. Mata J., Nurse P. (1997) tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89, 939–949 [DOI] [PubMed] [Google Scholar]
- 38. Martin S. G., McDonald W. H., Yates J. R., 3rd, Chang F. (2005) Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell 8, 479–491 [DOI] [PubMed] [Google Scholar]
- 39. Guthrie C., Fink G. R. (1991) Guide to Yeast Genetics and Molecular Biology, Academic Press, New York [Google Scholar]
- 40. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 41. Moseley J. B., Maiti S., Goode B. L. (2006) Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 406, 215–234 [DOI] [PubMed] [Google Scholar]
- 42. Schuyler S. C., Pellman D. (2002) Analysis of the size and shape of protein complexes from yeast. Methods Enzymol. 351, 150–168 [DOI] [PubMed] [Google Scholar]
- 43. Okada K., Ravi H., Smith E. M., Goode B. L. (2006) Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol. Biol. Cell 17, 2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feierbach B., Verde F., Chang F. (2004) Regulation of a formin complex by the microtubule plus end protein tea1p. J. Cell Biol. 165, 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buttery S. M., Yoshida S., Pellman D. (2007) Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol. Biol. Cell 18, 1826–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. VerPlank L., Li R. (2005) Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell 16, 2529–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tolliday N., VerPlank L., Li R. (2002) Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr. Biol. 12, 1864–1870 [DOI] [PubMed] [Google Scholar]
- 48. Lesage G., Bussey H. (2006) Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cabib E., Roh D. H., Schmidt M., Crotti L. B., Varma A. (2001) The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276, 19679–19682 [DOI] [PubMed] [Google Scholar]
- 50. Lippincott J., Li R. (1998) Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rodriguez J. R., Paterson B. M. (1990) Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil. Cytoskeleton 17, 301–308 [DOI] [PubMed] [Google Scholar]
- 52. Gao L., Liu W., Bretscher A. (2010) The yeast formin Bnr1p has two localization regions that show spatially and temporally distinct association with septin structures. Mol. Biol. Cell 21, 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Buttery S. M., Kono K., Stokasimov E., Pellman D. (2012) Regulation of the formin Bnr1 by septins and a MARK/Par1-family septin-associated kinase. Mol. Biol. Cell 23, 4041–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martin S. G., Chang F. (2006) Dynamics of the formin for3p in actin cable assembly. Curr. Biol. 16, 1161–1170 [DOI] [PubMed] [Google Scholar]
- 55. Young K. G., Copeland J. W. (2010) Formins in cell signaling. Biochim. Biophys. Acta 1803, 183–190 [DOI] [PubMed] [Google Scholar]
- 56. Hara T., Ishida H., Raziuddin R., Dorkhom S., Kamijo K., Miki T. (2004) Novel kelch-like protein, KLEIP, is involved in actin assembly at cell-cell contact sites of Madin-Darby canine kidney cells. Mol. Biol. Cell 15, 1172–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Velichkova M., Hasson T. (2003) Keap1 in adhesion complexes. Cell Motil. Cytoskeleton 56, 109–119 [DOI] [PubMed] [Google Scholar]
- 58. Adams J. C., Seed B., Lawler J. (1998) Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J. 17, 4964–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang C., Czech L., Gerboth S., Kojima S., Scita G., Svitkina T. (2007) Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5, e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ohta Y., Fujimura L., Nishio S., Arima M., Sakamoto A., Shimada H., Ochiai T., Tokuhisa T., Hatano M. (2010) A kelch family protein Nd1-L functions as a metastasis suppressor in cancer cells via Rho family proteins mediated mechanism. Int. J. Oncol. 36, 427–434 [PubMed] [Google Scholar]
- 61. Sasagawa K., Matsudo Y., Kang M., Fujimura L., Iitsuka Y., Okada S., Ochiai T., Tokuhisa T., Hatano M. (2002) Identification of Nd1, a novel murine kelch family protein, involved in stabilization of actin filaments. J. Biol. Chem. 277, 44140–44146 [DOI] [PubMed] [Google Scholar]
- 62. Spence H. J., Johnston I., Ewart K., Buchanan S. J., Fitzgerald U., Ozanne B. W. (2000) Krp1, a novel kelch related protein that is involved in pseudopod elongation in transformed cells. Oncogene 19, 1266–1276 [DOI] [PubMed] [Google Scholar]