Background: CNTF signaling is mediated by CNTFR or IL-6R in complex with gp130 and LIFR.
Results: The CNTFR variant CV-1 is CNTFR-selective.
Conclusion: The single amino acid exchange R28E within CNTF abrogated IL-6R binding.
Significance: CV-1 allows discrimination between CNTFR- and IL-6R-mediated effects in vivo.
Keywords: Cell Surface Receptor, Cytokine Action, Interleukin, Receptor Structure-Function, Signal Transduction
Abstract
Ciliary neurotrophic factor (CNTF) is a neurotrophic factor with therapeutic potential for neurodegenerative diseases. Moreover, therapeutic application of CNTF reduced body weight in mice and humans. CNTF binds to high or low affinity receptor complexes consisting of CNTFR·gp130·LIFR or IL-6R·gp130·LIFR, respectively. Clinical studies of the CNTF derivative Axokine revealed intolerance at higher concentrations, which may rely on the low-affinity binding of CNTF to the IL-6R. Here, we aimed to generate a CNTFR-selective CNTF variant (CV). CV-1 contained the single amino acid exchange R28E. Arg28 is in close proximity to the CNTFR binding site. Using molecular modeling, we hypothesized that Arg28 might contribute to IL-6R/CNTFR plasticity of CNTF. CV-2 to CV-5 were generated by transferring parts of the CNTFR-binding site from cardiotrophin-like cytokine to CNTF. Cardiotrophin-like cytokine selectively signals via the CNTFR·gp130·LIFR complex, albeit with a much lower affinity compared with CNTF. As shown by immunoprecipitation, all CNTF variants retained the ability to bind to CNTFR. CV-1, CV-2, and CV-5, however, lost the ability to bind to IL-6R. Although all variants induced cytokine-dependent cellular proliferation and STAT3 phosphorylation via CNTFR·gp130·LIFR, only CV-3 induced STAT3 phosphorylation via IL-6R·gp130·LIFR. Quantification of CNTF-dependent proliferation of CNTFR·gp130·LIFR expressing cells indicated that only CV-1 was as biologically active as CNTF. Thus, the CNTFR-selective CV-1 will allow discriminating between CNTFR- and IL-6R-mediated effects in vivo.
Introduction
The interleukin (IL)-6 type cytokine family consists of cardiotrophin-1, cardiotrophin-like cytokine (CLC),3 CNTF, IL-6, IL-11, IL-27, IL-30, IL-31, leukemia inhibitory factor (LIF), and oncostatin M (OSM) (1). Except for IL-31, all IL-6 type cytokines bind to the β-receptor gp130, demonstrating a high degree of binding site plasticity within gp130 toward the respective cytokines. Whereas IL-6 and IL-11 bind to homodimeric gp130 receptor complexes, all other cytokines of this family bind to heterodimeric signal-transducing β-receptor complexes, e.g. CNTF and CLC to gp130·LIFR (1). IL-6 type cytokine signaling activates the intracellular signaling cascades JAK/STAT, MAPK, and PI3K. For some IL-6 type cytokines, binding to their β-receptors is dependent on the initial complex formation with an α-receptor. The following high-affinity cytokine/α-receptor pairs exist: CLC/CNTFR, CNTF/CNTFR, IL-6/IL-6R, IL-11/IL-11R, and p28/Epstein-Barr virs-induced gene 3 (1). Moreover, the IL-6R was described as alternative low affinity α-receptor for CNTF and p28 (2, 3), highlighting receptor plasticity also at the α-receptor/cytokine binding interface.
IL-6 type cytokines have a typical four-helix bundle fold that links the four α-helices (A-D) by two long loops (A-B and C-D) and one short loop (B-C) placing the four helices in an up-up-down-down orientation. Here, site I of the cytokine binds to the α-receptor, whereas site II and site III are needed for the interaction with the β-receptors. Site I is constituted by the C-terminal A-B loop (site Ia) and the C-terminal D helix (site Ib), site II by parts of the A and C helix and site III by the C-terminal A helix and the N-terminal A-B loop (site IIIa), the B-C loop with adjacent parts of the B and C helix (site IIIb), and the C-terminal C-D loop with the adjoining NH2-terminal D helix (site IIIc).
Binding of IL-6 type cytokines to their receptors is mainly mediated by ionic and hydrophobic interactions (4). Close inspection of the electrostatic surface potential of site I, II, and III of IL-6 type cytokines, however, reveals almost no similarity. Cross-reactivity is likely entropy driven by water exclusion from the interacting surfaces (5).
Interestingly, species-specific differences of cytokine/receptor interactions within the IL-6 type cytokine family were described. This is exemplified by differences for human and murine IL-6 and OSM. Human IL-6 binds to human and murine IL-6R·gp130 receptor complexes, but murine IL-6 exclusively binds to murine IL-6R·gp130 receptor complexes (2). Furthermore, human OSM bind to both human LIFR and OSM receptor, but murine OSM exclusively interacts with murine OSM receptor (6, 7). Rat CNTF is able to engage signaling via a heterodimer of gp130/LIFR in the absence of CNTFR (8), which has not been observed for murine or human CNTF.
Furthermore, transfer of the binding site III between IL-6 type cytokines led to chimeric cytokines with novel and unique receptor specificities (9). IC-7 is a chimera of IL-6 and CNTF, in which site III of IL-6 is exchanged by site III of CNTF. Following binding to the IL-6R via site I, IC-7 recruits gp130 to site II and the LIFR by site III. On cells expressing the IL-6R, gp130, and the LIFR, IC-7 displays biological activities comparable with that of LIF or IL-6, but it is inactive on cells that do not express the IL-6R (9). Viral IL-6, from the human herpesvirus 8 has ∼25% sequence homology with human IL-6. Viral IL-6 signals via a gp130 receptor homodimer but in contrast to IL-6 without the need of the IL-6Rα (10). IV-9 is a chimera of IL-6 and viral IL-6, in which an extended site III of IL-6 is exchanged by site III of viral IL-6. Consequently, IV-9 signaling via a gp130 homodimer was independent of the IL-6R (11).
The transfer of site I or II has, however, not been described as of yet. Moreover, abrogation of promiscuous cytokine/receptor usage by introduction of single point mutations within the cytokine or cytokine receptor has also not been achieved. Generation of CNTF or p28 variants specific for a single receptor would pave the way to analyze the function of CNTF/CNTFR- versus CNTF/IL-6R-induced or p28/EBI3 versus p28/IL-6R-induced signal transduction in vivo.
A CNTFR-selective CNTF might be achieved by transfer of site I from the CNTFR-selective cytokine CLC to CNTF or by introduction of specific amino acid exchanges in CNTF. Importantly, because even though the binding to the low affinity α-receptor will be abrogated, this must not result in a major reduction of binding affinity of CNTF to the high-affinity CNTFR.
Here, we describe the development the human CNTF variant (CV)-1, which binds with high affinity to the CNTFR and induces signal transduction via LIFR and gp130. Due to the single point mutation R28E, interaction of CV-1 with the IL-6R was abrogated. CV-1 might be used to discriminate between IL-6R and CNTFR-mediated effects of CNTF in vivo.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Ba/F3-gp130 cells transduced with human gp130 were obtained from Immunex (Seattle, WA) (12). Ba/F3-gp130-hIL-6R cells (13), BaF/3-gp130-LIFR cells, BaF/3-gp130-LIFR-IL-6R, and BaF/3-gp130-LIFR-CNTFR cells were generated as described previously (9). HepG2 and COS-7 cells were from ATCC (Manassas, VA). All cells were grown in DMEM high-glucose culture medium (PAA Laboratories, Cölbe, Germany) supplemented with 10% fetal bovine serum, penicillin (60 mg/liter), and streptomycin (100 mg/liter) at 37 °C with 5% CO2 in a water-saturated atmosphere. All Ba/F3 cells were cultured using 10 ng/ml recombinant Hyper IL-6, which is a fusion protein of IL-6 and soluble IL-6R (sIL-6R) acts as a growth factor for Ba/F3-gp130 cells (14, 15). Hyper IL-6 was expressed and purified as described previously (14). Human LIF was purchased from R&D Systems (Minneapolis, MN). Phospho-STAT3 mAb (Tyr-705) and STAT3 mAb (124H6) were purchased from Cell Signaling Technology (Frankfurt, Germany). The peroxidase conjugated secondary antibodies were purchased from Pierce (Thermo Scientific). All restriction enzymes were obtained from Fermentas (Thermo Scientific).
Cloning of CNTF, CV-1 to CV-5 and sCNTFR-Fc
The bacterial expression plasmid pet23a(+)-CNTF (d14d15-C17A-Q63R-CNTF) coding for a N- and C-terminal shortened version of CNTF, including the amino acid exchanges C17A and Q63R was generated using standard PCR and mutagenesis procedures. pet23a(+)-CNTF and pet23a(+)-CLC were used to generate the expression plasmids pet23a(+)-CV-1 to pet23a(+)-CV-5 using site-directed mutagenesis (pet23a(+)-CV-1) and splicing by overlapping extension PCR (pet23a(+)-CV-2 to pet23a(+)-CV-5). Initially, PCR products were cloned into pPCR-Script Amp SK(+) and subcloned into pet23a(+) via NdeI and NotI restriction sites. The sequences of all primers used are available on request.
To generate a fusion protein of human soluble CNTFR (sCNTFR) with an Fc portion, the coding sequence of CNTFR (corresponding to amino acids 1–346) was amplified by PCR. In 5′ and 3′ of sCNTFR SalI BglII restriction sites were inserted (5′ primer: AAAGCGTCGACGCGTGCTTGGTGTGTGGCC and 3′ primer AAAGTCACAAGATCTGGGCTCTACGGGTCC). CNTFR was subcloned via SalI and BglII into pDC409-sIL-6R-Fc (10).
Expression, Purification, and Renaturation of CV-1 to CV-5, CLC, IC-7, IL-6, and CNTF
Expression of CV-1 to CV-5, CLC, IC-7, IL-6, and CNTF was performed in Escherichia coli BL21(DE3) (Merck KGaA). In detail, protein expression was induced at an A600 of 0.8 with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested after 4 h at 37 °C. The pelleted bacterial cells were lysed by sonification (Sonopuls HD2200, Bandelin, Berlin, Germany). CV-1 to CV-5, CLC, IC-7, and CNTF inclusion bodies were purified by repeated washing and sonification steps (three times) in 50 mm Tris-HCl, 0.2% Tween 20, pH 8.0, followed by washing with (three times) 50 mm Tris-HCl, pH 8.0. Purified inclusion bodies were solved in 6 m guanidine hydrochloride, 50 mm Tris-HCl, pH 8.0. Denatured CV-1 to CV-5, IC-7, IL-6, and CNTF were refolded at a protein concentration of 0.1 mg/ml by two-step 24-h dialysis against 50 mm Tris-HCl, pH 8.0, 1 m GuHCl, 3 mm oxidized glutathione, 0.6 mm reduced glutathione and against 50 mm Tris-HCl, pH 8.0, 3 mm reduced glutathione, and 0.9 mm oxidized glutathione. CLC was refolded by dialysis against 50 mm CAPS, pH 11. Dialyzed CV-1 to CV-5, CLC, IC-7, IL-6, and CNTF were centrifuged for 30 min at 40,000 × g at 4 °C. CV-1 to CV-5, CLC, IC-7, IL-6, and CNTF were sterile filtered and concentrated with an Amicon stirred ultrafiltration cell equipped with an YM10 filter membrane, nominal molecular weight limit of 10,000 (Merck Millipore, Merck KGaA, Germany). CV-1 to CV-5, CLC, IC-7, IL-6, and CNTF were purified by size exclusion chromatography with an ÄKTAexplorer chromatography system and a HiLoad 16/60 Superdex 200 prep grade column (both from GE Healthcare). The fractions containing monomeric CV-1 to CV-5, CLC, IC-7, IL-6, and CNTF were concentrated by a Vivaspin 20 column, molecular weight cut-off of 10,000 (GE Healthcare). Pure CV-1 to CV-5, CLC, IC-7, IL-6, and CNTF proteins were analyzed by Coomassie Blue staining and Western blotting using a standard SDS-PAGE under reduced conditions using biotinylated mouse c-Myc (9E10) antibody (Santa Cruz Biotechnology). Folding of pure monomeric CV-1 to CV-5, CLC, IC-7, IL-6, and CNTF was verified by circular dichroism in a quartz cuvette (Helma, Mühlheim, Germany) with a Jasco J-720 CD-spectropolarimeter (Jasco Corp., Tokyo, Japan). The instrument was calibrated with an aqueous solution of 10-camphosulfonic acid at 25 °C. The spectral bandwidth was 1.5 nm.
Proliferation Assays
The different transduced Ba/F3 cell lines were washed three times with sterile PBS and suspended in DMEM containing 10% FBS at a final concentration of 5 × 103 cells per well in a 96-well microtiter plate. Cells were incubated for 72 h as indicated with the cytokines and cytokine receptors in a final volume of 100 μl. After 72 h, cell growth was measured using the CellTiter Blue cell viability assay reagent (Promega, Karlsruhe, Germany) following the manufacturer's instructions. The extinction was measured using a Lambda FLUORO 320 microplate fluorescence reader (excitation, filter 530/25; emission, filter 590/35; Software KC4, Bio-Tek Instruments). Normalization of relative light units was achieved by subtraction of negative control values. One representative example of two to three experiments was chosen for the figures. For clarity, no standard deviations were included in the diagrams; the S.D. were, as common for Ba/F3 cells, only minimally and usually below 5%.
Immunoprecipitation with sCNTFR-Fc and sIL-6R-Fc
For co-precipitation, supernatants of p409DC-IL-6R-Fc and pDC-CNTFR-Fc transiently transfected COS-7 were collected. For co-precipitation supernatants (transfected or co-transfected as indicated) were mixed with recombinant proteins (1 μg) incubated 2 h at 4 °C under gentle agitation. As control, a further tube containing the corresponding cytokine was incubated without receptor containing supernatant. 150 μl of protein A-agarose (Roche Diagnostics GmbH, Mannheim, Germany) was added, and the mixture was incubated at 4 °C for 2 h under gentle agitation. The samples were washed six times with PBS, and proteins were eluted by adding 50 μl of Laemmli buffer and incubated for 10 min at 95 °C. The resulting supernatants were subjected to Western blot analysis.
Western Blotting
For Western blotting, ∼2 × 107 cells per experiment were washed three times with sterile PBS. The cells were distributed to 2-ml tubes and starved in FBS-free medium for 4–6 h (Ba/F3 cell lines) or overnight (HepG2 cells) at 37 °C and CO2 saturation. Cells were stimulated with the indicated cytokines for 10 min followed by centrifugation at 4 °C and 2,000 rpm for 10 min. 150 μl of 5× Laemmli buffer (312 mm Tris-HCl, pH 6.8, 50% glycerol, 10% sodium dodecyl sulfate, 5% β-mercaptoethanol, and 0.13% bromphenol blue) were added to each tube, and cells were lysed by sonification (Sonopuls HD2200, Bandelin, Berlin, Germany) for 10 s and boiling at 95 °C for 10 min. Proteins were separated by SDS-PAGE and transferred to a PVDF membrane using a Trans-Blot SD semi-dry transfer cell (Bio-Rad). The membrane was blocked in 5% low fat milk in TBS-T (10 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 1% Tween 20) and probed with the primary antibody in 1% low fat milk in TBS-T (STAT3-mAb) or 5% BSA (pSTAT3-mAb) at 4 °C overnight. The membranes were washed and incubated with the secondary peroxidase-conjugated antibody for 1 h before applying the ECL-plus peroxidase substrate (GE Healthcare). The Fluor ChemQ system (Cell Biosciences, Santa Clara, CA) was used for signal detection according to the manufacturer's instructions. The membranes were stripped with stripping buffer (20 ml of 10% SDS, 12.5 ml of 0.5 m Tris-HCl, pH 6.8, 67.5 ml of ultra-pure water, 0.8 ml of β-mercaptoethanol), blocked again, and probed with another primary antibody. The STAT3 phosphorylation assays were reproduced three times with one representative experiment shown.
RESULTS
Cloning, Bacterial Expression, Refolding, and Purification of CNTF and the CNTF Variants CV-1 to -5
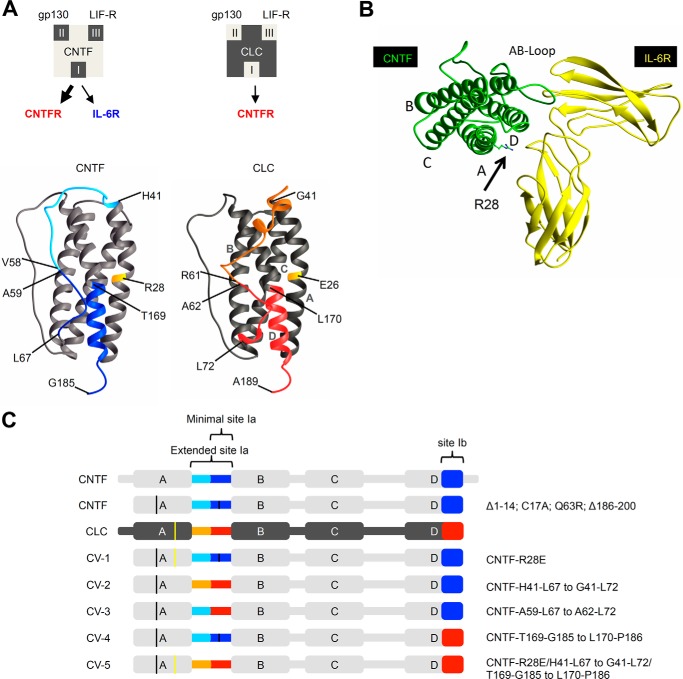
Binding of CNTF to its α-receptors CNTFR or IL-6R is mediated via site I of CNTF and the cytokine binding module of the α-receptors. CNTF has a higher affinity to the CNTFR than to the IL-6R (3). Also, CLC needs the CNTFR for signaling via gp130 and LIFR, but cannot bind to the IL-6R. CLC has, however, a lower efficiency to signal via CNTFR·gp130·LIFR complexes as compared with CNTF (16). Fig. 1A illustrates the binding site I of CLC and CNTF, respectively. In this study, we aimed to generate a CNTFR-selective CNTF variant without influencing the affinity of CNTF to the CNTFR·gp130·LIFR complex. In the past, successful transfer of the binding site III of CNTF to IL-6 led to the chimeric IL-6 variant IC-7, which signals via the artificial IL-6R·gp130·LIFR complex (9). Here, this strategy was also applied to site I of CNTF. We generated potential CNTFR-selective CNTF variants by transfer of amino acid residues of site I from CLC to CNTF (Fig. 1A). Alternatively, using molecular modeling of CLC·CNTFR and CNTF·CNTFR complexes, we predicted that the amino acid Glu-26 in CLC might be involved in CNTFR selectivity because it is in close contact to the CNTFR (Fig. 1B). The respective amino acid in CNTF was identified to be Arg28.
FIGURE 1.
CNTF and CLC receptor-binding sites and schematic illustration of CNTF variants CV-1 to CV-5. A, illustration of CLC and CNTF binding site I to CNTFR/IL-6R, site II to gp130, and site III to LIFR. The minimal binding site Ia of CLC is highlighted in red, with the extended site Ia in red and orange. For CNTF, minimal site Ia is colored light blue, extended site Ib is colored light and dark blue. B, model of the CNTF·IL-6R (D2/D3) complex; Arg28 is highlighted. C, schematic illustration of CNTF (light gray), CLC (dark gray), and CV-1 to CV-5. Site I from CLC is marked in orange and red; site I from CNTF is marked in light and dark blue. The exact mutations are shown on the right.
As a starting point, we generated a shortened CNTF variant to improve recombinant CNTF protein expression in E. coli and to support the refolding efficiency from CNTF inclusion bodies. Therefore, the cDNA coding for CNTF was modified to code for a N-terminal deletion of the amino acids 1–14 and a C-terminal deletion of the amino acids 186–200 (17). Moreover, two point mutations coding for the amino acid exchanges C17A and Q63R were introduced (Fig. 1C). With the exception of the C-terminal deletion, all other modifications were described previously for the CNTF variant Axokine. Axokine was shown to have an improved biological activity as compared with CNTF (18). A sequence coding for a C-terminal c-Myc and His tag was introduced in all cytokine-encoding cDNAs, including CNTF, CLC, IL-6, LIF, and IC-7. The cDNA coding for CNTF served as a platform for the transfer of amino acids from CLC to CNTF in CV-1 to CV5. CV-1 contained the single amino acid substitution R28E, located in close proximity to site I, CV-2 contained the extended site Ia (CNTF-His41–Leu67 to CLC-Gly41–Leu72) and CV-3 the minimal site Ia (CNTF-Ala59–Leu67 to CLC-Ala62–Leu72), CV-4 contained the site Ib (CNTF-Thr169–Gly185 to CLC-Leu170–Pro186) and CV-5 contained all exchanges, the extended site Ia and site Ib and the amino acid substitution R28E (CNTF-His41–Leu67 to CLC-Gly41–Leu72/CNTF-Thr169–Gly185 to CLC-Leu170–Pro186/R28E) (Fig. 1C).
CLC, CNTF, CV-1, CV-2, CV-3, CV-4, CV-5, IL-6, and IC-7 were expressed in E. coli as inclusion bodies, refolded, and purified. Purification was completed by nickel-nitrilotriacetic acid affinity chromatography and size-exclusion chromatography. All cytokines were produced as monomers (Fig. 2A and supplemental Fig. 1) and correctly folded as verified by circular dichroism revealing the typical spectra of α helical proteins (Fig. 2B and supplemental Fig. 1). Purity and identity of correctly folded and monomeric CLC, CNTF, CV-1 to CV-5, IL-6, and IC-7 was shown by Coomassie staining of SDS-PAGE gels and Western blotting against the C-terminal cytokine Myc tag, respectively (Fig. 2, C and D, and supplemental Fig. 1, A–H).
FIGURE 2.
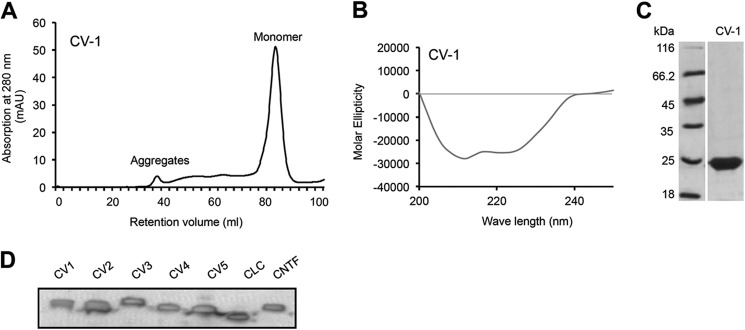
Pure CV-1 is monomeric and correctly folded. A, CV-1 was expressed in E. coli, purified via nickel-nitrilotriacetic acid agarose, and refolded. Finally, CV-1 was purified and analyzed by size-exclusion chromatography. CV-1 was eluted as a monomer. B, CD revealed the correct folding of CV-1. C, Coomassie stains of CV-1. CV-1 was separated by SDS-PAGE and visualized by Coomassie stain. D, Western blot analysis of purified CV-1 to CV5, CLC, and CNTF. Recombinant proteins were separated by SDS-PAGE and detected by Myc-mAb.
The CNTF Variants (CV-1, CV-2, CV-3, CV-4, CV-5) Bind to CNTFR, but only CV-3 and CV-4 Bind to IL-6R
First, we tested whether CNTF, CLC, IL-6, and the CNTF variants CV-1 to CV-5 were able to bind to CNTFR or IL-6R by co-precipitation. To this end, cDNAs coding for sCNTFR and sIL-6R C-terminally fused to an Fc part of an IgG1 antibody were subcloned and transiently transfected into COS-7 cells. sIL-6R-Fc or sCNTFR-Fc conditioned cell culture supernatants were used for Protein A-mediated co-precipitation of recombinant CNTF, CLC, IL-6, and CV-1 to CV-5. Precipitated cytokines were detected by Western blotting using a mAb against the C-terminal c-Myc tag. Non-precipitated input served as positive control, precipitation without sCNTFR-Fc or sIL-6R-Fc as negative control. As expected, co-precipitation of CLC and CNTF but not of IL-6 was mediated by sCNTFR-Fc (Fig. 3A). In the CNTF analog Axokine and in CNTF, Gln-63 of human CNTF was exchanged to Arg63 of rat CNTF, which increased the specific activity of human CNTF (18). Because rat CNTF does not need to bind to the human IL-6R (8), it remained open, whether this mutation also abolished the binding of human CNTF to signal via CNTFR or IL-6R. Molecular modeling, however, indicated that Arg63 is not involved in the interaction area between human CNTF and the human IL-6R or CNTFR. Here, our co-precipitation experiments showed that the Q63R did not abolish binding of CNTF to the IL-6R and the CNTFR (Fig. 3, A and B).
FIGURE 3.
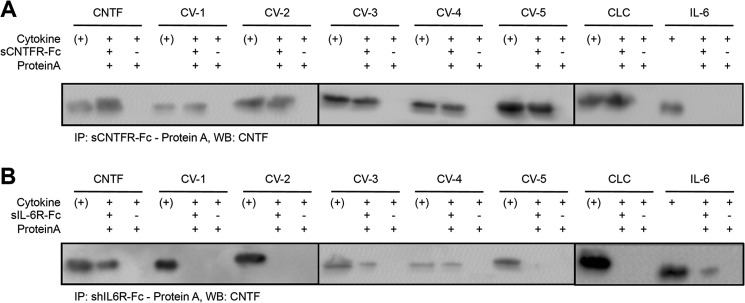
Analysis of the interaction of CV-1 to CV-5, CNTF, CLC and IL-6 with sCNTFR and sIL-6R by co-precipitation. A, co-precipitation of CV-1 to CV-5, CNTF, CLC, and IL-6 by sCNTFR-Fc conjugated to protein A-agarose beads and detection by Myc-mAb. B, co-precipitation of CV-1 to CV-5, CNTF, CLC, and IL-6 by sIL-6R-Fc conjugated to protein A-agarose beads and detection by Myc-mAb. (+), input control of the cytokine. WB, Western blot.
Importantly, all CNTF variants CV-1, CV-2, CV-3, CV-4, and CV-5 were also precipitated by sCNTFR-Fc, indicating that CV-1, CV-2, CV-3, CV-4, and CV-5 were able to form heterodimeric complexes with the CNTFR (Fig. 3A). As expected, co-precipitation of IL-6 and CNTF but not of CLC was also mediated by sIL-6R-Fc (Fig. 3B). Interestingly, only the CNTF-variant CV-3 and CV-4 were co-precipitated by sIL-6R-Fc, indicating that the amino acid exchange(s) in CV-1, CV-2 and CV-5 were sufficient to abrogate interaction with the IL-6R (Fig. 3B). We concluded that CV-1, CV-2 and CV-5 have CNTFR-selective binding properties.
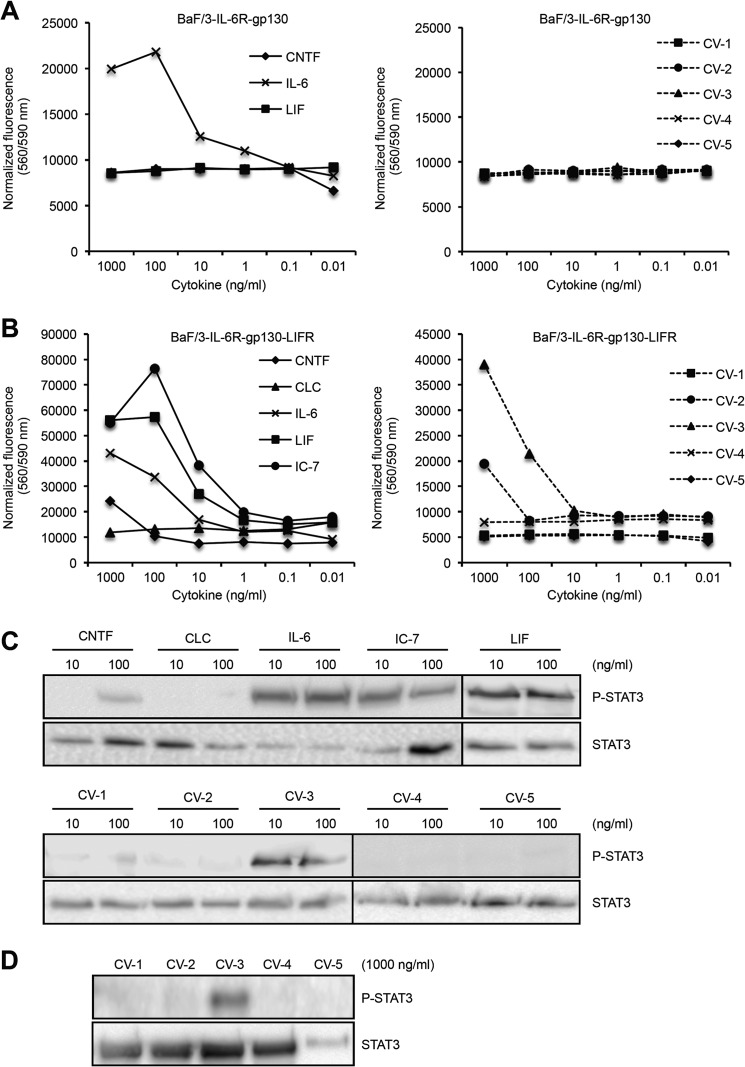
CV-1, -4, and -5 Have Lost the Ability to Induce Signaling via gp130-LIFR-IL-6R
To test CV-1 to CV-5 in a biological system, we generated Ba/F3 cells expressing IL-6R and gp130, or IL-6R, gp130, and LIFR. Proliferation of Ba/F3-IL-6R-gp130 cells is dependent on IL-6 via an IL-6R·gp130 receptor complex. As expected, only IL-6 but not LIF, CNTF and CV-1 to -5 was able to induce cellular proliferation of Ba/F3-IL-6R-gp130 cells (Fig. 4A). After stable transfection of Ba/F3 cells with human gp130, LIFR, and IL-6R (referred to as Ba/F3-IL-6R-gp130-LIFR cells), proliferation of these cells was dependent on CNTF, LIF, and IC-7 via the IL-6R·gp130·LIFR receptor complex. As a control, CLC was not able to induce cellular proliferation of Ba/F3-IL-6R-gp130-LIFR cells because CLC specifically signals via a CNTFR·gp130·LIFR complex. As described previously, CNTF-induced proliferation of Ba/F3-IL-6R-gp130-LIFR cells was ∼100-fold lower compared with IL-6-induced cellular proliferation (3). The CNTF-variants CV-1, -4, and -5 completely failed to induce cellular proliferation of Ba/F3-IL-6R-gp130-LIFR cells, whereas CV-3-induced proliferation was induced by 100–1000 ng/ml and was stronger as compared with CNTF. Surprisingly, 1000 ng of CV-2/ml induced proliferation of Ba/F3-IL-6R-gp130-LIFR cells (Fig. 4B).
FIGURE 4.
Biological activity of CV-1 to CV-5, CNTF, CLC, IC-7, IL-6, and LIF on cells expressing IL-6R/gp130 and IL-6R/gp130/LIFR. A, Ba/F3-IL-6R-gp130 cell proliferation assay comparing the bioactivities of CNTF, IL-6, LIF, and CV-1 to CV-5 (1000–0.01 ng/ml). B, Ba/F3-IL-6R-gp130-LIFR cell proliferation assay comparing the bioactivities of CNTF, CLC, IL-6, LIF, IC-7, and CV-1 to CV-5 (1000–0.01 ng/ml). C, analysis of phosphorylation of STAT3 in HepG2 cells by Western blotting comparing the bioactivities of CNTF, CLC, IL-6, LIF, IC-7, and CV-1 to CV-5 (10 and 100 ng/ml). D, analysis of phosphorylation of STAT3 in HepG2 cells comparing the bioactivities of CV-1 to CV-5 (1000 ng/ml).
HepG2 cells are responsive to CNTF via the CNTFR·gp130·LIFR complex (3, 19). Therefore, we analyzed STAT3 phosphorylation in HepG2 cells after stimulation with CNTF, CLC, LIF, IL-6, IC-7, and CV-1 to CV-5. As expected, LIF, IL-6, and IC7 but not CLC induced STAT3 phosphorylation in HepG2 cells at 10 and 100 ng/ml, whereas only 100 but not 10 ng/ml were sufficient to induce STAT3 phosphorylation by CNTF (Fig. 4C). Only CV-3 but not CV-1, CV-2, -4, and -5 were able to induce STAT3 phosphorylation in HepG2 cells (Fig. 4C). Of note, the lower concentration of CV-3 (10 ng/ml) induced STAT3 phosphorylation, supporting the notion that CV-3 is more efficient than CNTF to induce signaling via IL-6R·gp130·LIFR complexes. In addition, at higher concentrations of CV-1 to CV-5 (1000 ng/ml), only CV-3 induced detectable STAT3 phosphorylation in HepG2 cells (Fig. 4D). Our findings showed that the three CNTF variants CV-1, CV-4, and CV5 completely lost their ability to signal via the IL-6R·gp130·LIFR complex.
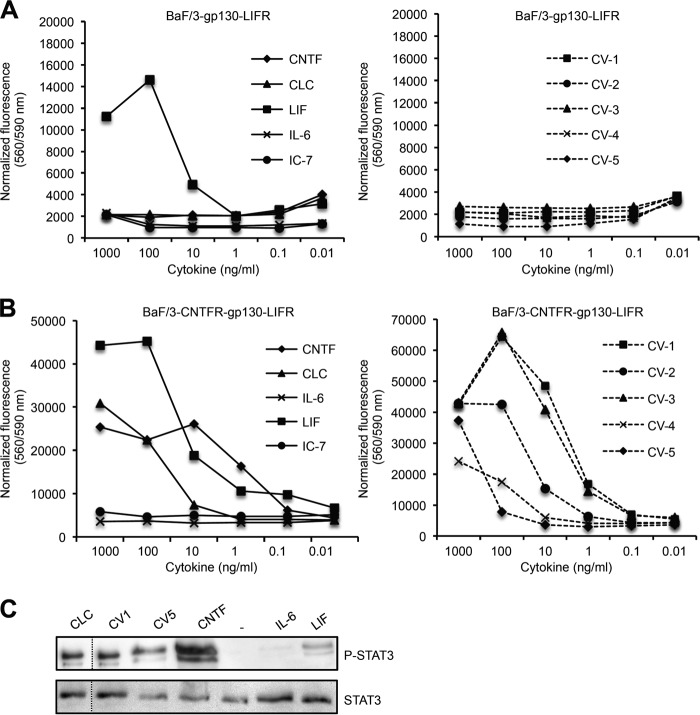
The CNTF Variant CV-1 Signals via the CNTFR·gp130·LIFR Complex with Equal Efficacy as Compared with CNTF
Next, we generated Ba/F3 cells expressing gp130·LIFR and CNTFR·gp130·LIFR. After stable transfection with cDNAs coding for human gp130 and LIFR (referred to as Ba/F3-gp130-LIFR cells), proliferation of Ba/F3-gp130-LIFR cells was dependent on IL-6 trans-signaling (Hyper-IL-6; IL-6/sIL-6R fusion protein) via a gp130 homodimer or on LIF via a gp130/LIFR heterodimer. As expected, CLC, CNTF, and CV1–5 were not able to induce cellular proliferation of Ba/F3-gp130-LIFR cells (Fig. 5A), again indicating that the Q63R in CNTF did not lead to activation of gp130 and LIFR without CNTFR. After stable transfection of Ba/F3 cells with human CNTFR/gp130/LIFR cDNAs (referred to as Ba/F3-CNTFR-gp130-LIFR cells), dose-dependent proliferation of Ba/F3-CNTFR-gp130-LIFR cells was also dependent on CNTF or CLC via a CNTFR·gp130·LIFR receptor complex (Fig. 5B). To compare the biological activity of the CNTF variants CV-1 to CV-5, dose-dependent cytokine-induced proliferation of Ba/F3-CNTFR-gp130-LIFR cells was performed. Although all CNTF variants were precipitated with sCNTFR-Fc, the CNTF variants differ in their potency to induce cellular proliferation by a factor of 100 (Fig. 5C). Whereas the biological activity of CV-1 and CV-3 was comparable with CNTF, CV-2 and especially CV-4 and CV-5 revealed ∼10- to 100-fold reduced biological activity as compared with CNTF (Fig. 5C). Furthermore, we also verified that CNTF, CV1, and CV5 induced STAT3 phosphorylation in Ba/F3-CNTFR-gp130-LIFR cells (Fig. 5D). Our data demonstrated that only CV-1 fulfilled the initial requirements, namely CNTFR selectivity and high biological activity.
FIGURE 5.
Biological activity of CV-1 to CV-5, CNTF, CLC, IC-7, IL-6, and LIF on cells expressing gp130/LIFR and CNTFR/gp130/LIFR. A, Ba/F3-gp130-LIFR cell proliferation assay comparing the bioactivities of CNTF, IL-6, LIF, and CV-1 to CV-5 (1000–0.01 ng/ml). B, Ba/F3-CNTFR-gp130-LIFR cell proliferation assay comparing the bioactivities of CNTF, CLC, IL-6, LIF, IC-7, and CV-1 to CV-5 (1000–0.01 ng/ml). C, analysis of phosphorylation of STAT3 in Ba/F3-CNTFR-gp130-LIFR cells comparing the bioactivities of CNTF, CLC, IL-6, LIF, CV-1, and CV-5 (100 ng/ml).
DISCUSSION
CNTF prevents degeneration of axotomized peripheral motor neurons and retrograde cell death of neurons in thalamic nuclei after dissection of intracerebral neuronal circuits (20, 21). Intraperitoneal implantation of a CNTF-producing cell line improved survival in a mouse model of motor neuropathy (22). In models of Huntington disease, CNTF has neuroprotective effects on striatal cells (23–25). CNTF also enhanced the survival of sensory, hippocampal, and cerebellar neurons (26–28) and increased survival of retinal photoreceptors in animal models of retinal degeneration (29–31). On denervated rat skeletal muscles, CNTF reduced denervation-induced atrophy and increased muscular strength (32, 33).
Moreover, CNTF is still unique in its ability to overcome leptin resistance and reduce food intake and body weight in obese, leptin-resistant humans and rodents. Importantly, weight loss was sustained after discontinuation of the drug (34). Because leptin resistance is still a major problem in obesity, CNTF was suggested as therapeutic principle to combat human obesity (35). It was hypothesized that CNTF has leptin-like actions in the hypothalamus. Activation of either leptin receptor or CNTFR in the hypothalamus leads to local STAT3 and mTOR activation (36–38). Both cytokines also reduce AMP-activated kinase phosphorylation (39, 40). Additionally, CNTFR and leptin receptor expression is overlapping in the hypothalamus (41), including the paraventricular nucleus and arcuate nucleus (42–44). However, a recent report indicates that despite anatomical overlap of CNTFR and leptin receptor expression, CNTF and leptin act within distinct neuronal populations to elicit anorectic effects (45).
Here, we describe the development of the CNTFR-selective CNTF variant CV-1, which is characterized by the single amino acid exchange R28E from CLC to CNTF. Biological activity of CV-1 was comparable with CNTF on Ba/F3-CNTFR-gp130-LIFR cells, indicating that the affinity of CV-1 to CNTFR·gp130·LIFR complex was unaffected.
CV-4 contained the exchange of site Ib. Although this variant appeared to bind to the IL-6R, proliferation assays revealed that CV-4 was CNTFR-selective. Biological activity of CV-4 was, however, ∼100-fold reduced as compared with CNTF and appeared to be similar to CLC. We have, however, no explanation why CV-4 bind to IL-6R but did not induce proliferation of Ba/F3-IL-6R/gp130/LIFR cells. CV-2 and CV-3 contained the extended or the minimal site Ia from CLC, respectively. Exchange of the minimal site Ia did not affect receptor selectivity and biological activity of CV-3 toward CNTFR/gp130/LIFR but, surprisingly, increased the biological activity toward IL-6R·gp130·LIFR complexes. Exchange of the extended site Ia in CV-2 abrogated interaction with IL-6R, but at the highest concentration applied, CV-2 still induced cytokine-dependent proliferation via the IL-6R·gp130·LIFR receptor complex, albeit no detectable STAT3 phosphorylation. Moreover, the biological activity toward CNTFR·gp130·LIFR complexes was reduced by a factor of 10 for CV-2. From these data, we concluded that simple combination of site Ia and site Ib would result in CNTFR-selectivity but with overall reduced biological activity toward CNTFR·gp130·LIFR complexes. Therefore, we decided to combine all site exchanges, including the amino acid substitution R28E in CV-5. Albeit CV-5 was CNTFR-selective, exchange of the complete site Ia and Ib from CLC to CNTF abolished the positive affinity effect of R28E observed in CV-1.
All CNTF variants were generated using the human CNTF cDNA. Murine CNTF has about 82% sequence homology with human CNTF. In CNTF, Arg28 and the surrounding amino acids are identical between mice and humans; therefore, we assume that either receptor selectivity will be transferrable to murine CNTF and/or CV-1 will maintain its receptor selectivity also on murine receptor complexes. This must, however, to be studied before CV-1 will be tested in in vivo studies.
The CNTFR-selective CV-1 may be of particular interest in view of the therapeutic potential of CNTF in neurodegenerative diseases and obesity and may even allow the reinvestigation of the therapeutic effects of CNTF at higher dosages.
Supplementary Material
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Bonn, Germany) DFG SCHE 907/2-1 and SFB877 Project A1, A6 and by the Cluster of Excellence “Inflammation at Interfaces.”

This article contains supplemental Fig. 1.
- CLC
- cardiotrophin-like cytokine
- CNTF
- ciliary neurotrophic factor
- LIF
- leukemia inhibitory factor
- OSM
- oncostatin M
- gp130
- glycoprotein 130
- CV
- CNTF variant
- sIL-6R
- soluble IL-6R
- sCNTFR
- soluble CNTFR
- LIFR
- LIF receptor.
REFERENCES
- 1. Garbers C., Hermanns H. M., Schaper F., Müller-Newen G., Grötzinger J., Rose-John S., Scheller J. (2012) Plasticity and cross-talk of interleukin 6-type cytokines. Cytokine Growth Factor Rev. 23, 85–97 [DOI] [PubMed] [Google Scholar]
- 2. Garbers C., Spudy B., Aparicio-Siegmund S., Waetzig G. H., Sommer J., Hölscher C., Rose-John S., Grötzinger J., Lorenzen I., Scheller J. (2013) An interleukin-6 receptor-dependent molecular switch mediates signal transduction of the IL-27 cytokine subunit p28 (IL-30) via a gp130 protein receptor homodimer. J. Biol. Chem. 288, 4346–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schuster B., Kovaleva M., Sun Y., Regenhard P., Matthews V., Grötzinger J., Rose-John S., Kallen K. J. (2003) Signaling of human ciliary neurotrophic factor (CNTF) revisited: the interleukin-6 receptor can serve as an alpha-receptor for CTNF. J. Biol. Chem. 278, 9528–9535 [DOI] [PubMed] [Google Scholar]
- 4. Grötzinger J., Kurapkat G., Wollmer A., Kalai M., Rose-John S. (1997) The family of the IL-6-type cytokines: specificity and promiscuity of the receptor complexes. Proteins 27, 96–109 [PubMed] [Google Scholar]
- 5. Wang X., Lupardus P., Laporte S. L., Garcia K. C. (2009) Structural biology of shared cytokine receptors. Annu. Rev. Immunol. 27, 29–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ichihara M., Hara T., Kim H., Murate T., Miyajima A. (1997) Oncostatin M and leukemia inhibitory factor do not use the same functional receptor in mice. Blood 90, 165–173 [PubMed] [Google Scholar]
- 7. Mosley B., De Imus C., Friend D., Boiani N., Thoma B., Park L. S., Cosman D. (1996) Dual oncostatin M (OSM) receptors: cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J. Biol. Chem. 271, 32635–32643 [DOI] [PubMed] [Google Scholar]
- 8. Gearing D. P., Ziegler S. F., Comeau M. R., Friend D., Thoma B., Cosman D., Park L., Mosley B. (1994) Proliferative responses and binding properties of hematopoietic cells transfected with low-affinity receptors for leukemia inhibitory factor, oncostatin M, and ciliary neurotrophic factor. Proc. Natl. Acad. Sci. U.S.A. 91, 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kallen K. J., Grötzinger J., Lelièvre E., Vollmer P., Aasland D., Renné C., Müllberg J., Myer zum Büschenfelde K. H., Gascan H., Rose-John S. (1999) Receptor recognition sites of cytokines are organized as exchangeable modules: transfer of the LIFR binding site from CNTF to IL-6. J. Biol. Chem. 274, 11859–11867 [DOI] [PubMed] [Google Scholar]
- 10. Müllberg J., Geib T., Jostock T., Hoischen S. H., Vollmer P., Voltz N., Heinz D., Galle P. R., Klouche M., Rose-John S. (2000) IL-6 receptor independent stimulation of human gp130 by viral IL-6. J. Immunol. 164, 4672–4677 [DOI] [PubMed] [Google Scholar]
- 11. Adam N., Rabe B., Suthaus J., Grötzinger J., Rose-John S., Scheller J. (2009) Unraveling viral interleukin 6 binding to gp130 and activation of STAT-signaling pathways independent of interleukin 6-receptor. J. Virol. 83, 5117–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gearing A. J., Beckett P., Christodoulou M., Churchill M., Clements J., Davidson A. H., Drummond A. H., Galloway W. A., Gilbert R., Gordon J. L. (1994) Processing of tumor necrosis factor-α precursor by metalloproteinases. Nature 370, 555–557 [DOI] [PubMed] [Google Scholar]
- 13. Chalaris A., Rabe B., Paliga K., Lange H., Laskay T., Fielding C. A., Jones S. A., Rose-John S., Scheller J. (2007) Apoptosis is a natural stimulus of IL6R shedding and contributes to the pro-inflammatory trans-signaling function of neutrophils. Blood 110, 1748–1755 [DOI] [PubMed] [Google Scholar]
- 14. Fischer M., Goldschmitt J., Peschel C., Brakenhoff J. P., Kallen K. J., Wollmer A., Grotzinger J., Rose-John S. (1997) A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 15, 142–145 [DOI] [PubMed] [Google Scholar]
- 15. Schroers A., Hecht O., Kallen K. J., Pachta M., Rose-John S., Grötzinger J. (2005) Dynamics of the gp130 cytokine complex: a model for assembly on the cellular membrane. Protein Sci. 14, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tormo A. J., Letellier M. C., Lissilaa R., Batraville L. A., Sharma M., Elson G., Crabé S., Gauchat J. F. (2012) The cytokines cardiotrophin-like cytokine/cytokine-like factor-1 (CLC/CLF) and ciliary neurotrophic factor (CNTF) differ in their receptor specificities. Cytokine 60, 575–582 [DOI] [PubMed] [Google Scholar]
- 17. Krüttgen A., Grötzinger J., Kurapkat G., Weis J., Simon R., Thier M., Schröder M., Heinrich P., Wollmer A., Comeau M. (1995) Human ciliary neurotrophic factor: a structure-function analysis. Biochem. J. 309, 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Preti A. (2003) Axokine (Regeneron). IDrugs 6, 696–701 [PubMed] [Google Scholar]
- 19. Schooltink H., Stoyan T., Roeb E., Heinrich P. C., Rose-John S. (1992) Ciliary neurotrophic factor induces acute-phase protein expression in hepatocytes. FEBS Lett. 314, 280–284 [DOI] [PubMed] [Google Scholar]
- 20. Sendtner M., Kreutzberg G. W., Thoenen H. (1990) Ciliary neurotrophic factor prevents the degeneration of motor neurons after axotomy. Nature 345, 440–441 [DOI] [PubMed] [Google Scholar]
- 21. Clatterbuck R. E., Price D. L., Koliatsos V. E. (1993) Ciliary neurotrophic factor prevents retrograde neuronal death in the adult central nervous system. Proc. Natl. Acad. Sci. U.S.A. 90, 2222–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sendtner M., Schmalbruch H., Stöckli K. A., Carroll P., Kreutzberg G. W., Thoenen H. (1992) Ciliary neurotrophic factor prevents degeneration of motor neurons in mouse mutant progressive motor neuronopathy. Nature 358, 502–504 [DOI] [PubMed] [Google Scholar]
- 23. Anderson K. D., Panayotatos N., Corcoran T. L., Lindsay R. M., Wiegand S. J. (1996) Ciliary neurotrophic factor protects striatal output neurons in an animal model of Huntington disease. Proc. Natl. Acad. Sci. U.S.A. 93, 7346–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emerich D. F., Winn S. R., Hantraye P. M., Peschanski M., Chen E. Y., Chu Y., McDermott P., Baetge E. E., Kordower J. H. (1997) Protective effect of encapsulated cells producing neurotrophic factor CNTF in a monkey model of Huntington's disease. Nature 386, 395–399 [DOI] [PubMed] [Google Scholar]
- 25. de Almeida L. P., Zala D., Aebischer P., Déglon N. (2001) Neuroprotective effect of a CNTF-expressing lentiviral vector in the quinolinic acid rat model of Huntington's disease. Neurobiol. Dis. 8, 433–446 [DOI] [PubMed] [Google Scholar]
- 26. Simon R., Thier M., Krüttgen A., Rose-John S., Weiergräber O., Heinrich P. C., Schröder J. M., Weis J. (1995) Human CNTF and related cytokines: effects on DRG neurone survival. Neuroreport 7, 153–157 [PubMed] [Google Scholar]
- 27. Lärkfors L., Lindsay R. M., Alderson R. F. (1996) Characterization of the responses of Purkinje cells to neurotrophin treatment. J. Neurochem. 66, 1362–1373 [DOI] [PubMed] [Google Scholar]
- 28. Ip N. Y., Li Y. P., van de Stadt I., Panayotatos N., Alderson R. F., Lindsay R. M. (1991) Ciliary neurotrophic factor enhances neuronal survival in embryonic rat hippocampal cultures. J. Neurosci. 11, 3124–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. LaVail M. M., Yasumura D., Matthes M. T., Lau-Villacorta C., Unoki K., Sung C. H., Steinberg R. H. (1998) Protection of mouse photoreceptors by survival factors in retinal degenerations. Invest. Ophthalmol. Vis. Sci. 39, 592–602 [PubMed] [Google Scholar]
- 30. Chong N. H., Alexander R. A., Waters L., Barnett K. C., Bird A. C., Luthert P. J. (1999) Repeated injections of a ciliary neurotrophic factor analogue leading to long-term photoreceptor survival in hereditary retinal degeneration. Invest. Ophthalmol. Vis. Sci. 40, 1298–1305 [PubMed] [Google Scholar]
- 31. Peterson W. M., Wang Q., Tzekova R., Wiegand S. J. (2000) Ciliary neurotrophic factor and stress stimuli activate the Jak-STAT pathway in retinal neurons and glia. J. Neurosci. 20, 4081–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Helgren M. E., Squinto S. P., Davis H. L., Parry D. J., Boulton T. G., Heck C. S., Zhu Y., Yancopoulos G. D., Lindsay R. M., DiStefano P. S. (1994) Trophic effect of ciliary neurotrophic factor on denervated skeletal muscle. Cell 76, 493–504 [DOI] [PubMed] [Google Scholar]
- 33. Guillet C., Auguste P., Mayo W., Kreher P., Gascan H. (1999) Ciliary neurotrophic factor is a regulator of muscular strength in aging. J. Neurosci. 19, 1257–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ettinger M. P., Littlejohn T. W., Schwartz S. L., Weiss S. R., McIlwain H. H., Heymsfield S. B., Bray G. A., Roberts W. G., Heyman E. R., Stambler N., Heshka S., Vicary C., Guler H. P. (2003) Recombinant variant of ciliary neurotrophic factor for weight loss in obese adults: a randomized, dose-ranging study. JAMA 289, 1826–1832 [DOI] [PubMed] [Google Scholar]
- 35. Kalra S. P. (2001) Circumventing leptin resistance for weight control. Proc. Natl. Acad. Sci. U.S.A. 98, 4279–4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lambert P. D., Anderson K. D., Sleeman M. W., Wong V., Tan J., Hijarunguru A., Corcoran T. L., Murray J. D., Thabet K. E., Yancopoulos G. D., Wiegand S. J. (2001) Ciliary neurotrophic factor activates leptin-like pathways and reduces body fat, without cachexia or rebound weight gain, even in leptin-resistant obesity. Proc. Natl. Acad. Sci. U.S.A. 98, 4652–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vaisse C., Halaas J. L., Horvath C. M., Darnell J. E., Jr., Stoffel M., Friedman J. M. (1996) Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat. Genet. 14, 95–97 [DOI] [PubMed] [Google Scholar]
- 38. Cota D., Matter E. K., Woods S. C., Seeley R. J. (2008) The role of hypothalamic mammalian target of rapamycin complex 1 signaling in diet-induced obesity. J. Neurosci. 28, 7202–7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andersson U., Filipsson K., Abbott C. R., Woods A., Smith K., Bloom S. R., Carling D., Small C. J. (2004) AMP-activated protein kinase plays a role in the control of food intake. J. Biol. Chem. 279, 12005–12008 [DOI] [PubMed] [Google Scholar]
- 40. Steinberg G. R., Watt M. J., Fam B. C., Proietto J., Andrikopoulos S., Allen A. M., Febbraio M. A., Kemp B. E. (2006) Ciliary neurotrophic factor suppresses hypothalamic AMP-kinase signaling in leptin-resistant obese mice. Endocrinology 147, 3906–3914 [DOI] [PubMed] [Google Scholar]
- 41. Gloaguen I., Costa P., Demartis A., Lazzaro D., Di Marco A., Graziani R., Paonessa G., Chen F., Rosenblum C. I., Van der Ploeg L. H., Cortese R., Ciliberto G., Laufer R. (1997) Ciliary neurotrophic factor corrects obesity and diabetes associated with leptin deficiency and resistance. Proc. Natl. Acad. Sci. U.S.A. 94, 6456–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kordower J. H., Yaping C., Maclennan A. J. (1997) Ciliary neurotrophic factor receptor α-immunoreactivity in the monkey central nervous system. J. Comp. Nerol. 377, 365–380 [DOI] [PubMed] [Google Scholar]
- 43. MacLennan A. J., Vinson E. N., Marks L., McLaurin D. L., Pfeifer M., Lee N. (1996) Immunohistochemical localization of ciliary neurotrophic factor receptor α expression in the rat nervous system. J. Neurosci. 16, 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee M. Y., Deller T., Kirsch M., Frotscher M., Hofmann H. D. (1997) Differential regulation of ciliary neurotrophic factor (CNTF) and CNTF receptor α expression in astrocytes and neurons of the fascia dentata after entorhinal cortex lesion. J. Neurosci. 17, 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stefater M. A., MacLennan A. J., Lee N., Patterson C. M., Haller A., Sorrell J., Myers M., Woods S. C., Seeley R. J. (2012) The anorectic effect of CNTF does not require action in leptin-responsive neurons. Endocrinology 153, 2647–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.