Background: Single immunoglobulin interleukin-1 receptor-related molecule (SIGIRR) is a negative inflammatory regulator whose regulatory mechanism is mainly described in epithelial tissues.
Results: Higher monocytic and neutrophilic SIGIRR expression is maintained by Sp1.
Conclusion: Lipopolysaccharide decreases SIGIRR expression by suppressing Sp1 via the TLR4-p38 pathway.
Significance: The LPS-dependent SIGIRR down-regulation may be critical for optimal inflammatory responses in immune cells.
Keywords: Gene Expression, Lipopolysaccharide (LPS), Monocyte, Neurotrophin, p38, Promoter, Specificity Protein 1 (Sp1), Toll-like Receptor (TLR), Single Immunoglobulin IL-1R-related Molecule (SIGIRR)
Abstract
Single immunoglobulin interleukin-1 receptor-related molecule (SIGIRR) is one of the immunoglobulin-like membrane proteins that is crucial for negative regulation of toll-like receptor 4 (TLR4) and interleukin-1 receptor. Despite the importance of understanding its expression and function, knowledge is limited on the regulatory mechanism in the epithelial tissues, such as the liver, lung, and gut, where its predominant expression is originally described. Here, we found expression of SIGIRR in non-epithelial innate immune cells, including primary peripheral blood monocytes, polymorphonuclear neutrophils, monocytic RAW264 cells, and neutrophilic-differentiated HL-60 cells. Consistent with previous findings in epithelial tissues, SIGIRR gene and protein expression were also down-regulated by LPS treatment in a time-dependent manner in primary blood monocytes and polymorphonuclear neutrophils. A reduction was also observed in RAW264 and differentiated HL-60 cells. Notably, exogenous introduction of the dominant negative form of TLR4 and siRNA of p38 resulted in inhibition of LPS-induced SIGIRR down-regulation, whereas treatment with p38 activator anisomycin showed a dose-dependent decrease in SIGIRR expression, suggesting TLR4-p38 signal as a critical pathway for LPS-induced SIGIRR down-regulation. Finally, reporter gene and chromatin immunoprecipitation assays demonstrated that Sp1 is a key factor that directly binds to the proximal promoter of SIGIRR gene and consequently regulates basal SIGIRR expression, which is negatively regulated by the LPS-dependent TLR4-p38 pathway. In summary, the data precisely demonstrate how LPS down-regulates SIGIRR expression and provide a role of LPS signal that counteracts Sp1-dependent basal promoter activation of SIGIRR gene via TLR4-p38 pathway in non-epithelial innate immune cells.
Introduction
Toll-interleukin 1 receptor (TIR)2 superfamily is a group of receptors that participates in innate immune and inflammatory responses (1). The superfamily is defined by the presence of an intracellular TIR domain, which generally activates several common signaling pathways such as NF-κB and MAPKs. TIR superfamily can be divided into two main subgroups including the leucine-rich repeat motif-containing toll-like receptors (TLRs) and the immunoglobulin (Ig) domain-containing receptors such as interleukin-1 (IL-1) receptor and IL-18 receptors. TLRs directly sense exogenous pathogen-associated molecular patterns on a variety of pathogens (2, 3), whereas Ig domain-containing receptors accept endogenous cytokines (4), and these activate similar signaling pathways that can result in elimination of the invading pathogens. Consistently, the mice lacking TIR superfamily proteins are more susceptible to infection with various types of pathogen (1). These reports support the idea that optimal expression and signaling of TLRs and Ig domain-containing receptors is required to activate the innate immune system during infection with invading pathogens. On the other hand, the pathways that negatively regulate TIR signaling, including T1/ST2 (5), single immunoglobulin IL-1R-related molecule (SIGIRR) (6), splicing variant of myeloid differentiation factor 88 (MyD88s) (7), suppressor of cytokine signaling 1 (SOCS1) (8), and Triad3A (9), are also crucial for appropriate immune responses because the mice lacking these negative regulators typically exhibit enhanced inflammatory responses leading to tissue damage. These suggest the importance of understanding the expression and function of the negative regulators of TIR signaling.
In theory most of the negative regulators are expressed at low level in steady state, and their expression is up-regulated during TIR signaling for the successful resolution of inflammation (3). However, the regulatory profiles for the SIGIRR expression are likely different. Thomassen et al. (10) demonstrated that expression levels of SIGIRR is generally kept high in organs such as the liver, lung, and gut, which may contribute to maintain an activation threshold of TIR signaling, whereas SIGIRR expression is down-regulated upon treatment with pathogen-associated molecular patterns to reach maximum induction of immune responses in various organs (6). Based on the previous reports, SIGIRR seems to be dominantly expressed in epithelial tissues, but recent reports focusing on the expression and function of SIGIRR in non-epithelial immune cells such as Th2-lymphocytes (11), macrophages (12), Langerhans cells (13), and Payer's patch dendritic cells (14) suggest a fundamental role of SIGIRR in these cells. Despite the finding showing that SIGIRR proximal promoter has a binding site for Sp1, which enhances its transcription in basal conditions in epithelial tissues (15), little is known regarding the regulatory mechanism of SIGIRR expression in non-epithelial immune cells such as monocytes/macrophages and neutrophils during inflammatory responses.
In the present study, we confirm the higher expression of SIGIRR in several non-epithelial innate immune cells including cell lines and primary cells, and identify the LPS-dependent TLR4-p38 signal as a critical pathway for LPS-induced SIGIRR down-regulation in both monocytic and neutrophilic primary cells and cell lines. Our study further uncovers a role of LPS signal that counteracts Sp1-dependent basal promoter activation of SIGIRR gene via TLR4-p38 pathway.
EXPERIMENTAL PROCEDURES
Cell Culture, Isolation of Primary cells
Primary peripheral blood monocytes (MC) and polymorphonuclear neutrophils (PMN) were isolated from heparinized venous blood of healthy individuals by following the suggested protocols using Ficoll-Paque PLUS (Amersham Biosciences) as indicated before (16). Briefly, the whole blood was mixed with 0.9% sodium chloride containing 3% dextran 500 (Sigma) and incubated at room temperature for 30 min to sediment erythrocytes. After dextran sedimentation, the supernatant was centrifuged at 1800 rpm for 10 min, and cells were then resuspended in 0.9% sodium chloride, underlaid with Ficoll-Paque PLUS, and centrifuged at 2800 rpm for 30 min. The MC recovered from the buffy coat and the PMN from the pellet were washed twice in 0.9% sodium chloride and resuspended in Roswell Park Memorial Institute (RPMI)-1640 medium. The HL-60 human promyelocytic leukemia cell line and RAW264 mouse monocytic cell line (RCB0535) were obtained from University of California, San Francisco cell culture facility and RIKEN Bio Resource Center, respectively, and maintained at 37 °C in humidified 5% CO2 atmosphere in RPMI1640 medium supplemented with 10% heat-inactivated FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. Neutrophilic differentiation was induced by exposing HL-60 cells to 1.3% dimethyl sulfoxide (DMSO) for 3 days as previously described (17). For the analysis of Sp1 inhibitor mithramycin A (mitA), we confirmed that there is no obvious undesired cellular toxicity by the treatment of differentiated HL-60 (dHL60) cells, primary MC, and PMN with mitA.
Reagents and Antibodies
Escherichia coli lipopolysaccharide (LPS) (O111;B4), anisomycin, mitA, SB203580, SB239063, PD98059, SP600125, caffeic acid phenethyl ester, and wortmannin were purchased from Sigma. LPS was re-extracted as previously described (18). The following antibodies were used in this study: anti-SIGIRR antibody (C-12) and anti-γ-tubulin antibody (Santa Cruz Biotechnology, Santa Cruz, CA); anti-p38, anti-p38α, and anti-phospho-p38 antibodies (Cell Signaling, Beverly, MA); anti-β-actin (Sigma); anti-Sp1 antibody (Active Motif, Carlsbad, CA); anti-SIGIRR ectodomain antibody (AF990) (R&D systems, Minneapolis, MN).
RNA Isolation, cDNA Synthesis, and Real-time PCR
Quantitative real-time RT-PCR for human SIGIRR, SIGIRR variants, IL-8, and 18 S ribosomal RNA (18 S rRNA), and mouse SIGIRR, TNFα, and 18 S rRNA was carried out using the primers in Table 1 as described in Mizunoe et al. (19). Briefly, total RNA from the cells was isolated using RNAisoPlus (TaKaRa, Japan), and synthesis of cDNA was performed using PrimeScript RT regent kit (TaKaRa, Japan). Real-time quantitative RT-PCR analysis was performed using SYBR Premix Ex Taq (TaKaRa, Japan) in iQ5 real-time PCR detection systems (Bio-Rad). The relative quantity of target gene mRNA was normalized using human or mouse 18 S rRNA as the internal controls and expressed as the relative quantity of target gene mRNA (-fold induction). PCR amplification was performed in triplicate, and the reaction protocol included preincubation at 95 °C to activate Ex TaqHS for 30 s, amplification of 40 cycles that was set for 15 s at 95 °C, and the annealing for 60 s at 60 °C.
TABLE 1.
Primers used for quantitative RT-PCR
| Primer | Orientation | Sequence |
|---|---|---|
| Human SIGIRR | Forward | 5′-TTCAGTCCAGTGGCTGAAAGACGG-3′ |
| Reverse | 5′-ACCTCTGACAGGTTGGCCTTGAC-3′ | |
| Human IL-8 | Forward | 5′-CTGGCCGTGGCTCTCTTG-3′ |
| Reverse | 5′-CCTTGGCAAAACTGCACCTT-3′ | |
| Human 18 S rRNA | Forward | 5′-CGGCTACCACATCCAAGGAA-3′ |
| Reverse | 5′-GCTGGAATTACCGCGGCT-3′ | |
| Mouse SIGIRR | Forward | 5′-GTGGCTGAAAGATGGTCTGGCATTG-3′ |
| Reverse | 5′-CAGGTGAAGGTTCCATAGTCCTCTGC-3′ | |
| Mouse TNFα | Forward | 5′-CATCTTCTCAAAATTCGAGTGACAA-3′ |
| Reverse | 5′-TGGGAGTAGACAAGGTACAACCC-3′ | |
| Mouse 18 S rRNA | Forward | 5′-CCATCCAATCGGTAGTAGCG-3′ |
| Reverse | 5′-GTAACCCGTTGAACCCCATT-3′ | |
| Human SIGIRRv1 | Forward | 5′-CCCAAAGGCGCACCAATGACC-3′ |
| Human SIGIRRv2 | Forward | 5′-GTGGCGGTCGCCATCAGACC-3′ |
| Human SIGIRRv2v3 | Forward | 5′-CGCACCTGGCTCAGGTGAGC-3′ |
| Human SIGIRRv1,2,3 | Reverse | 5′-CTCGGCCAGCAGACTGATCCA-3′ |
Plasmid Constructs
A 1.1-kb fragment of the human SIGIRR promoter (−1030/+131) was amplified by PCR using human genomic DNA extracted from HL60 cell line and subsequently cloned into the Zero Blunt TOPO vector (Invitrogen). Full-length (−1000/+14) and a set of 5′-deletion fragments were amplified from the −1030/+131 promoter clone by PCR using an upstream primer containing a KpnI restriction site and a downstream primer containing a XhoI site. The PCR products were subsequently subcloned into the luciferase reporter vector pGL3 basic (Promega, Madison, WI). The site-directed mutations in putative Sp1 binding sites were introduced into the human SIGIRR promoter full-length construct with QuikChange II XL site-directed mutagenesis kit following the manufacturer's protocol (Stratagene, La Jolla, CA). All DNA sequencing was performed by Sigma (at the University of Akita, Japan). The expression plasmid containing mouse C3H/HeJ TLR4 inserted into pcDNA3.1 was kindly provided by Dr. Jian-Dong Li (Georgia State University). The plasmid used in transient transfection assays was purified by PureYield plasmid miniprep system (Promega).
Luciferase Reporter Gene Assay
The pGL3 basic vectors containing 1000 bp, the deleted sequences, or the mutated sequences of the proximal 5′-flanking region of human SIGIRR upstream of exon1 upstream of the firefly luciferase coding sequence were used for analysis of SIGIRR promoter activity. The pRL vector (Promega) containing the Renilla luciferase coding sequence downstream of a constitutively active cytomegalovirus promoter was co-transfected to measure transfection efficiency in each well. HeLa cells (obtained from American Type Culture Collection and maintained in minimum essential medium) were plated in 24-well tissue culture plates (1.9 × 104 cells/well) and incubated for 24 h. The cells were co-transfected with both plasmids using TransIT-LT1 (TaKaRa, Japan), and luciferase activity was determined using the Dual Luciferase assay system (Promega) according to the manufacturer's instructions. The produced light was measured using a Lumat LB 9507 luminometer (EG&G Berhold). The ratio of firefly/Renilla luciferase activity was calculated to normalize results with respect to the efficiency of transfection. In the case of mitA treatment, protein concentrations were introduced to normalize firefly luciferase activity.
Transfection of C3H/HeJ TLR4 Plasmid DNA
RAW264 cells were seeded in a 35-mm dish at a density of 2.0 × 105 cells/dish. After growth for 24 h, 1 μg/dish mouse C3H/HeJ TLR4 expression plasmid DNA into RAW264 cells was performed by using 3 μl/dish FuGENE HD transfection reagent (Roche Applied Science) following the manufacturer's protocol. Twenty-four hours after transfection, the cells were used for further experiments.
Transfection of p38α Small Interfering RNA (siRNA)
siRNA sequence targeting to mouse p38α (5′-GAACGUUGUUUCCUGGUACTT-3′) and GL2 luciferase (negative control; 5′-CGUACGCGGAAUACUUCGATT-3′) were used in this study. siRNA oligo was synthesized and obtained from Sigma Genosys Aldrich. For siRNA transfection studies, RAW264 cells were seeded in a 35-mm dish at a density of 2.0 × 105 cells/dish shortly before transfection. HiPerFect (Qiagen, Valencia, CA) was used to transfect RAW264 cells with a final siRNA concentration of 50 nm according to the manufacturer's instructions. Fourteen hours after transfection, recovery medium RPMI containing 10% heat-inactivated FBS was added. The stimulations of the cells were carried out 84 h post-transfection to ensure protein turnover. The efficiency of target gene knockdown was confirmed by Western blotting.
SDS-PAGE and Western Blot Analysis
Cell membranes were lysed at 4 °C in the appropriate volumes of radioimmunoprecipitation assay buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mg/ml sodium deoxycholate, and 1% Nonidet P-40) containing 1% protease inhibitor mixture (Sigma) and centrifuged at 15,000 × g for 10 min at 4 °C. The supernatant was prepared as cell lysates. The whole cell lysates were subjected to SDS-PAGE on 10 or 12% polyacrylamide gels. Protein was electroblotted from the gels to polyvinylidene difluoride membrane (Immun-BlotTM; Bio-Rad). The membranes were blocked with TBS containing 5% skim milk, 5% BSA, or 5% donkey serum (Sigma), 50 mm Tris-HCl (pH 7.6), 150 mm NaCl, and 0.1% Tween 20 for 1 h at room temperature. The antigen-antibody complexes were incubated with ECL detection reagents (Amersham Biosciences) and analyzed by luminescent image analyzer (VersaDocTM; Bio-Rad) to visualize HRP. In some experiments for reprobing, the membrane was incubated with 2 m glycine-HCl (pH 2.8) at room temperature for 1 h followed by 3 washes with 0.1% TBS-Tween. In each Western blot data, protein expression levels of β-actin and γ-tubulin were measured to show an equal loading in each well. For the detection of SIGIRR protein in various cell lines, Ponceau S staining was used to confirm equal loading of protein in each well. For the band quantification, the density of the bands was quantified using Image Gauge software (Version 4.23; Fujifilm).
Chromatin Immunoprecipitation (ChIP) Assay
To measure the binding activity of transcriptional factor Sp1 in dHL-60 cells, ChIP assay was conducted with ChIP-IT enzymatic kit (Active Motif) according to the manufacturer's protocol. In brief, DNA and the protein complex were cross-linked by paraformaldehyde. The DNA was sheared into small and uniform fragments by enzymatic digestion. Anti-Sp1 antibody was added to precipitate Sp1 protein-DNA complex. A negative control IgG provided by Santa Cruz Biotechnology was added to serve as a background control antibody. Input DNA, cross-linked chromatin without immunoprecipitation of Sp1 antibody was used as a positive control. After immunoprecipitation, cross-linking was reversed, the protein was digested by proteinase K, and the DNA was isolated for PCR analysis. Input DNA, Sp1 antibody-precipitated DNA, and the negative control antibody-precipitated DNA were assessed for measuring the binding activity to human SIGIRR promoter. The primer set for measuring Sp1 binding activity was as follows: forward (5′-ACAGAGGAAAGTGCCGT-3′) and reverse (5′-CACATCCTCTCCTAACCCT-3′) (179 bp product).
Statistical Analysis
For quantitative analysis, most of the result represents the mean ± S.E. performed in triplicate, and the data were analyzed by Student's t test or by one-way analysis of variance (ANOVA) with Tukey-Kramer multiple comparison test or Dunnett's test (JMP software, SAS Institute, NC) as indicated in each figure legend. A p value of <0.05 is considered statistically significant.
RESULTS
SIGIRR Expression and Function in Monocytic and Neutrophilic cells
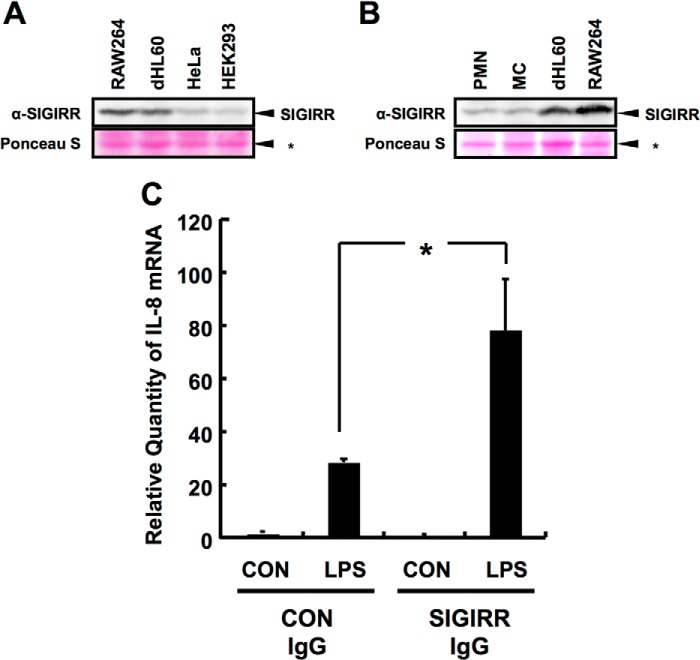
To confirm the expression levels of SIGIRR in non-epithelial cells, whole cell lysates from non-epithelial primary cells such as primary peripheral blood MC and PMN and cell lines including neutrophilic dHL60 (17), and monocytic RAW264 were subjected to Western blotting analysis. Cell lysates from epithelial cells including HeLa and HEK293 cells were also subjected to the analysis. As shown in Fig. 1A, SIGIRR protein expression was detectable in both dHL60 and RAW264 cells, and its expression was significantly higher than that in HeLa and HEK293 cells. SIGIRR protein expression was also detectable in both primary MC and PMN (Fig. 1B). Pretreatment with the antibody against SIGIRR ectodomain, which is considered to interfere with its dimerization that is crucial for its inhibitory function of LPS-TLR4 signal, resulted in the dramatic enhancement of LPS-induced IL-8 production compared with control IgG-pretreated cells (Fig. 1C). These data confirm the expression of SIGIRR in non-epithelial innate immune cells and the negative regulatory role of SIGIRR in LPS-mediated signaling as is often the case with previous observation.
FIGURE 1.
SIGIRR expression in monocytic and neutrophilic cells. A and B, Western blot analysis of lysates of RAW264, neutrophilic-dLH60, HeLa, and HEK293 cells probed for endogenous SIGIRR expression (A). SIGIRR expression in human primary MC and PMN were also loaded (B). C, dHL60 cells were treated with control IgG or Ecto-domain recognizing anti-SIGIRR antibody (2 μg/ml) for 1 h followed by treatment with 100 ng/ml LPS. Quantitative RT-PCR was carried out to detect IL-8 at the mRNA level. Data were normalized to 18 S rRNA mRNA levels as an internal control. *, p < 0.05 versus control (Con) IgG- and LPS-treated cells; Student's t test (n = 3).
LPS Down-regulates SIGIRR Expression in Monocytic and Neutrophilic Primary Cells and Cell Lines
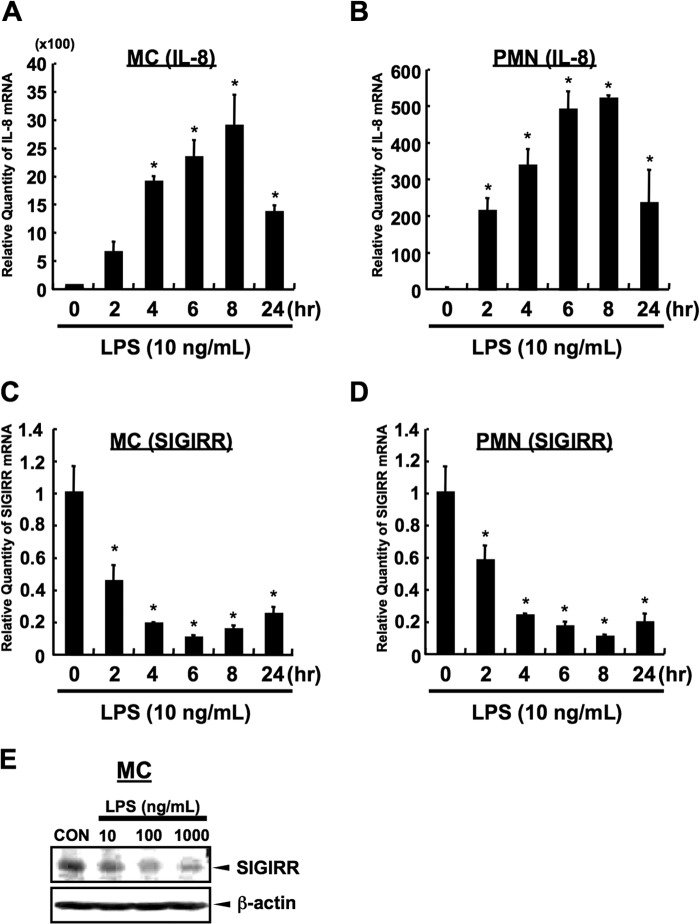
To characterize how SIGIRR expression is regulated in non-epithelial innate immune cells, we took advantage of the previous observations that SIGIRR expression is down-regulated upon treatment with bacterial components such as LPS, an inflammatory microbial ligand derived from Gram-negative bacterium. As shown in Fig. 2, A and B, LPS significantly induced cytokines expression in both primary MC and PMN. Maximum induction was observed at 6–8 h after LPS stimulation in both cells. Under the same condition, LPS decreased the SIGIRR mRNA level with a similar kinetics to that of the IL-8 expression in both cells (Fig. 2, C and D). Consistently, LPS-dependent down-regulation of SIGIRR expression at the protein level was also observed in primary MC cells (Fig. 2E).
FIGURE 2.
LPS-dependent down-regulation in primary monocytes and neutrophils. A and B, LPS-dependent signal in human primary MC and PMN. Quantitative RT-PCR was carried out to detect the up-regulation of IL-8 at mRNA level in human primary MC (A) and PMN (B), respectively. IL-8 mRNA levels were normalized to the 18 S rRNA mRNA levels as an internal control. *, p < 0.05 versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3). C and D, quantitative RT-PCR was carried out to detect the down-regulation of SIGIRR at mRNA level in human primary MC (C) and PMN (D) after LPS exposure (10 ng/ml) in the indicated time conditions. SIGIRR mRNA levels were normalized to the 18 S rRNA mRNA levels as an internal control. *, p < 0.05, versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3). E, MC were stimulated with the indicated concentrations of LPS for 12 h followed by Western blot analysis.
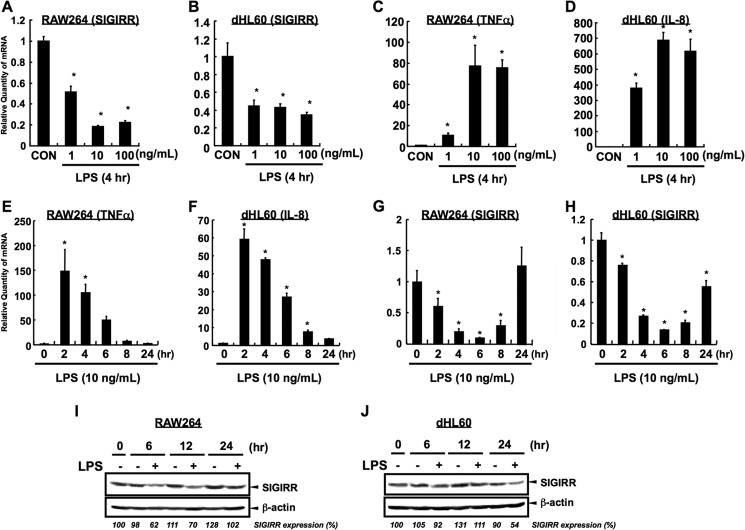
To further characterize that LPS-induced down-regulation is also observed in cell lines, the effect of LPS on the SIGIRR gene expression was evaluated in RAW264 and dHL60 cells. The analysis of different doses of LPS suggested that LPS significantly decreased SIGIRR expression with all the concentrations tested in both RAW264 and dHL60 cells (Fig. 3, A and B). In RAW264, dose-dependent reduction of SIGIRR and induction of TNFα were observed in the lower concentration range of LPS (1–10 ng/ml) (Fig. 3, A and C). On the other hand, 1 ng/ml LPS profoundly reduced SIGIRR expression and induced IL-8 expression, and it reached a plateau at higher LPS doses (10–100 ng/ml) in dHL60 cells (Fig. 3, B and D). Moreover, time course analysis revealed that maximum induction of cytokines was observed at 2 h after LPS stimulation in both cells (Fig. 3, E and F). Under the same conditions, LPS decreased the SIGIRR mRNA level, although its kinetics was different than that of the cytokine expression in both cells (Fig. 3, G and H). Maximum reduction was observed at 6 h after LPS stimulation in these cell lines. Notably, complete recovery of SIGIRR gene expression was observed at 24 h after LPS stimulation in RAW264 cells, whereas weaker recovery was observed in dHL60 cells. But overall, the kinetics of SIGIRR expression after LPS exposure was similar to that of primary cells (Figs. 2, C and D, and 3, G and H). Consistently, LPS-dependent down-regulation of SIGIRR expression at the protein level was also observed in these cells, and maximum reduction was at 6–12 and 24 h after LPS stimulation in RAW264 and dHL60 cells, respectively (Fig. 3, I and J). Together, these data demonstrate that SIGIRR gene and protein expression are also down-regulated during LPS exposure in monocytic and neutrophilic cells.
FIGURE 3.
LPS-dependent down-regulation in RAW264 and dHL60 cells. A–D, quantitative RT-PCR was carried out to detect the down-regulation of SIGIRR at mRNA level in RAW264 (A) and dHL60 (B) after LPS exposure in the indicated dose conditions. Quantitative RT-PCR was also carried out to detect the up-regulation of TNFα and IL-8 at mRNA level in RAW264 (C) and dHL60 (D), respectively. SIGIRR and cytokines mRNA levels were normalized to the 18 S rRNA mRNA levels as an internal control. *, p < 0.05, versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3). E–H, time course analysis of cytokines (E and F) and SIGIRR (G and H) at mRNA level in RAW264 (E and G) and dHL60 (F and H) after LPS exposure in indicated time condition. SIGIRR and cytokines mRNA levels were normalized to the 18 S rRNA mRNA levels as an internal control. *, p < 0.05 versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3). I and J, RAW264 (I) and dHL60 (J) cells were stimulated with 1 μg/ml LPS for indicated time periods, and the cell lysates were analyzed by Western blotting. Relative quantity of each band of SIGIRR protein was quantified and is shown as the % of band intensity of LPS non-treated group at 0 h.
TLR4-dependent p38 MAPK Pathway Plays a Critical Role in LPS-dependent Reduction of SIGIRR Expression
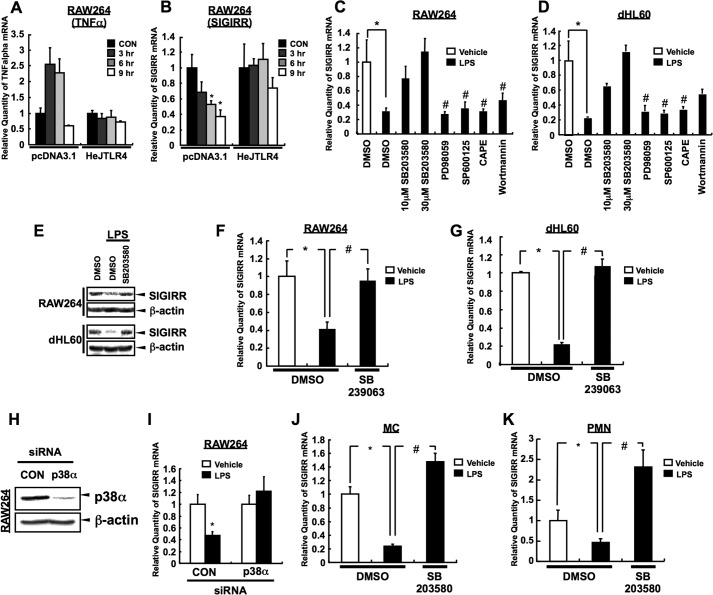
To ensure whether SIGIRR down-regulation is due to an activation of TLR4 signaling, RAW264 cells were transfected with empty vector or C3H/HeJ TLR4 (HeJ TLR4) expression plasmid, which produces the murine TLR4 protein with a point mutation (P712H) that serves as a dominant-negative inhibitor of TLR4 pathway (20). As shown in Fig. 4A, LPS-dependent TNFα induction was totally suppressed under C3H/HeJ TLR4-transfected conditions. Notably, LPS-dependent SIGIRR down-regulation was also suppressed under the same condition (Fig. 4B), suggesting that LPS-dependent SIGIRR down-regulation occurs via TLR4 signaling.
FIGURE 4.
LPS-dependent TLR4-p38 pathway is crucial for the reduction of SIGIRR expression. A and B, RAW264 cells were transiently transfected with mouse C3H/HeJ TLR4 (HeJ TLR4) or pcDNA3.1 empty vector. Twenty-four hours after transfection, the cells were stimulated with 100 ng/ml LPS for indicated time periods, mouse TNFα (A) and mouse SIGIRR (B) mRNA levels were quantified by quantitative RT-PCR, and the data were normalized to 18 S rRNA mRNA levels as an internal control. *, p < 0.05 versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3). C and D, RAW264 (C) and dHL60 (D) cells were pretreated with a variety of inhibitors for TLR4 signaling (10 and 30 μm SB203580; p38 inhibitor, 20 μm PD98059; ERK inhibitor, 10 μm SP600125; JNK inhibitor, 5 μg/ml caffeic acid phenethyl ester (CAPE); NF-κB inhibitor, 100 nm wortmannin; PI3K inhibitor). One hour after incubation the cells were stimulated with 100 ng/ml LPS (RAW264) or 10 ng/ml LPS (dHL60) for 4 h. SIGIRR mRNA levels were normalized to the 18 S rRNA mRNA levels as an internal control. *, p < 0.05, versus DMSO-treated cells without LPS conditions; Student's t test (n = 3); #, p < 0.05, versus LPS-treated cells without the inhibitor treatment condition; ANOVA with Dunnett's test (n = 3). E, RAW264 and dHL60 cells were pretreated with 30 μm SB203580 for 1 h. Then the cells were stimulated with 1 μg/ml LPS for 12 h followed by Western blot analysis. F and G, RAW264 and dHL60 cells were pretreated with 10 μm SB239063, another p38 inhibitor. One hour after incubation the cells were stimulated with the 100 ng/ml (RAW264) (F) or 10 ng/ml (dHL60) (G) of LPS for 6 h. The SIGIRR mRNA levels were measured by quantitative RT-PCR, and the data were normalized to the mRNA level of 18 S rRNA, as an internal control. *, p < 0.05 versus vehicle-treated cells; Student's t test (n = 3). #, p < 0.05 versus LPS-treated cells; Student's t test (n = 3). H and I, RAW264 cells were transfected with 50 nm of siRNA against GL2 (control) or p38α. After 84 h the cells were stimulated with 100 ng/ml LPS for 8 h followed by Western blot analysis of p38α (H) and quantitative RT-PCR that determines the expression levels of SIGIRR gene after LPS exposure (I). Data of quantitative RT-PCR were normalized to 18 S rRNA mRNA levels as an internal control. *, p < 0.05 versus vehicle-treated cells; Student's t test (n = 3). J and K, MC and PMN cells were pretreated with 30 μm SB203580. One hour after incubation the cells were stimulated with 100 ng/ml LPS for 6 h. The SIGIRR mRNA levels were measured by quantitative RT-PCR. *, p < 0.05 versus vehicle-treated cells; #, p < 0.05, versus LPS-treated cells without SB203580 treatment condition; ANOVA with Tukey-Kramer multiple comparison test (n = 3).
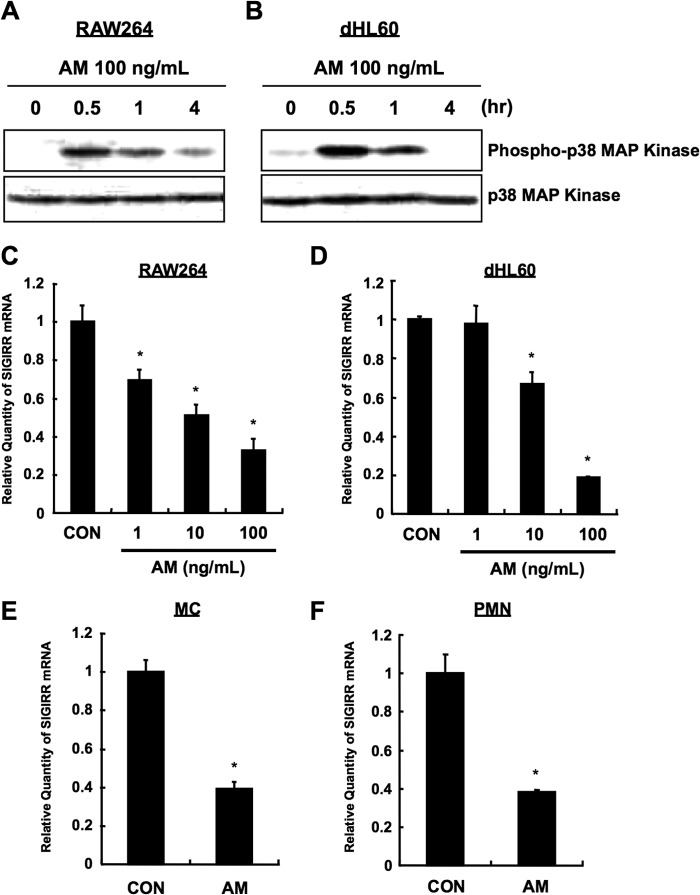
Because three MAPK (p38, ERK1/2, JNK), NF-κB, and PI3K are considered as main pathways that are activated by TLR4 (2, 3), we next sought to determine the pathway responsible for LPS-dependent SIGIRR down-regulation. RAW264 and dHL60 cells were untreated or pretreated with SB203580 (p38 inhibitor), PD98059 (ERK1/2 inhibitor), SP600125 (JNK inhibitor), caffeic acid phenethyl ester (NF-κB inhibitor), and wortmannin (PI3K inhibitor) before LPS stimulation. Interestingly, LPS-induced SIGIRR down-regulation was abolished in SB203580-treated cells but not in other inhibitor-treated cells (Fig. 4, C and D). Moreover, LPS-dependent SIGIRR down-regulation at the protein level was also dampened by SB203580 (Fig. 4E). Furthermore, pretreatment with another p38 inhibitor, SB239063 (Fig. 4, F and G), and specific knockdown (∼80%) of p38α, a p38 isoform mainly involved in the immune responses in the cells (21), also attenuated LPS-induced SIGIRR down-regulation (Fig. 4, H and I). Importantly, SB203580 pretreatment also ablated LPS-induced SIGIRR down-regulation in human primary MC and PMN (Fig. 4, J and K), strongly indicating that LPS down-regulates SIGIRR expression via TLR4-p38 pathway in monocytic and neutrophilic cells. Consistently, anisomycin (AM), a potent activator of p38 (22) (Fig. 5, A and B), decreased SIGIRR expression in RAW264 and dHL60 cells (Fig. 5, C and D) as well as in primary MC and PMN (Fig. 5, E and F), confirming a negative role of p38 signal in the regulation of basal SIGIRR gene expression.
FIGURE 5.
Anisomycin-dependent SIGIRR down-regulation in monocytes and neutrophils. A and B, AM-induced p38 activation. RAW264 (A) and dHL60 (B) cells were stimulated with 100 ng/ml AM for indicated time periods, and the cell lysates were analyzed by Western blotting. C and D, RAW264 (C) and dHL60 (D) cells were treated with indicated concentrations of AM. The SIGIRR mRNA levels were measured by quantitative RT-PCR, and the data were normalized to 18 S rRNA mRNA levels, as an internal control. *, p < 0.05 versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3). E and F, MC (E) and PMN (F) cells were treated with 100 ng/ml AM for 12 h. The SIGIRR mRNA levels were measured by quantitative RT-PCR, and the data were normalized to the mRNA level of 18 S rRNA, as an internal control. *, p < 0.05 versus Vehicle-treated cells; Student's t test (n = 3).
Identification of the Minimal Promoter Region Required for Basal SIGIRR Promoter Activity
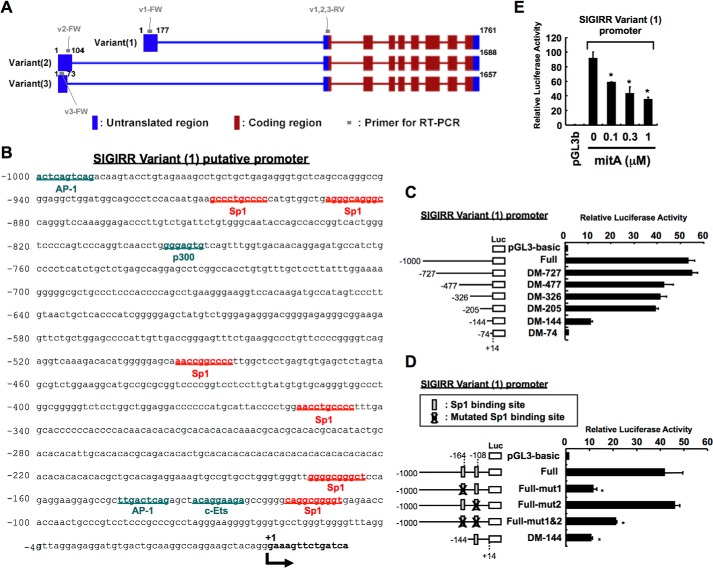
To further explore the precise mechanisms responsible for LPS-dependent SIGIRR down-regulation, we next sought to determine the critical promoter region required for basal SIGIRR promoter activity. We first performed in silico analysis to obtain the promoter sequences using National Center for Biotechnology Information (NCBI) human genome resources, a public database for genomic sequences. Based on annotated information (www.ncbi.nlm.nih.gov), three transcripts seemed to be expressed (NCBI accession numbers NM_001135054, NM_001135053, NM_021805). These three transcripts are designated as variants 1, 2, and 3, respectively (Fig. 6A). Although transcriptional start site and/or transcript lengths are different among three variants, all three variants seem to encode the same protein. To identify which variant is dominantly expressed in innate immune cells, we generated various primer sets that discriminate among three variants (Fig. 6A) and performed variants-specific quantitative RT-PCR analysis in dHL60 cells. Notably, as shown in Table 2, the primer set that specifically recognizes the sequence of variant 1 transcript amplifies much more efficiently than primer sets that detect variants 2 and 3, suggesting predominant expression of variant 1 in dHL60 cells. We then cloned the 5′-flanking 1000-bp fragment of the human SIGIRR variant 1. The fragment contains a number of potential consensus binding sites for transcription factors, including Sp1, AP1, c-Ets, and p300, but lacks a TATA box (Fig. 6B). To clarify the critical region required for basal SIGIRR promoter activity, a series of reporter plasmids containing various lengths of the promoter region was constructed, and their activities were assessed (Fig. 6C). A deletion extending to nucleotide −205 (−205/+14) had little effect on promoter activity when compared with full-length construct, whereas DM-144-luciferase activity was decreased by >70% compared with cells transfected with DM-205-luciferase plasmid (Fig. 6C), suggesting the existence of primary promoter regulatory elements between −205 and −144 bp upstream of the SIGIRR gene coding sequence. By considering the report of Kadota et al. (15) on the possible involvement of Sp1 in the regulation of SIGIRR gene in intestinal epithelial cells during inflammation and the existence of two Sp1 binding sites between −205 and −74 bp upstream of the transcriptional start site, a point mutation was introduced into each putative Sp1 binding site in SIGIRR variant 1 full-length promoter-luciferase reporter construct, and luciferase activity was analyzed. Single mutation of Sp1 binding site located between nucleotides −108 and −117 rarely affected promoter activity, whereas a single mutation located between nucleotides −164 and −173 and double mutations greatly decreased promoter activity by >50% (Fig. 6D), suggesting that the Sp1 binding site between nucleotides −164 and −173 is indispensable in SIGIRR basal promoter activity. We next examined whether mitA, a well known Sp1 inhibitor, affects the basal SIGIRR promoter activity. As shown in Fig. 6E, mitA decreases SIGIRR promoter activity in a dose-dependent manner, supporting the idea that Sp1 is a critical factor that controls SIGIRR basal promoter activity.
FIGURE 6.
Identification of the minimal promoter region required for basal SIGIRR promoter activity. A, SIGIRR gene was mapped to 11q15.5, a localization distinct from other IL-1R family members. There seems to be three different transcript variants that have been shown in NCBI Reference Sequences. Variant 1 represents the longest transcript. Variants 2 and 3 differ in the 5′-UTR compared with variant 1. All three variants encode 9 exons and the same protein. B, we cloned a 5′-flanking 1000-bp fragment of the human SIGIRR gene. This fragment contains a number of potential consensus binding sites for transcription factors, including Sp1, AP1, c-Ets, and p300, but lacks a TATA box. C and D, HeLa cells were transiently transfected with the indicated human SIGIRR variant 1 promoter constructs for 24 h. Then Luciferase activities were assessed. *, p < 0.05 versus V1 full-transfected cells (D); ANOVA with Dunnett's test (n = 3). E, HeLa cells were transiently transfected with 5′-flanking 1,000-bp fragment of human SIGIRR V1-luc construct. Twenty-four hours after transfection the cells were treated with the indicated concentrations of mitA for 20 h. Then luciferase activities were assessed. *, p < 0.05 versus vehicle-treated V1 full-transfected cells; ANOVA with Dunnett's test (n = 3).
TABLE 2.
Expression level of human SIGIRR variants in dHL60 cells analyzed by quantitative RT-PCR
FW, forward; RV, reverse.
Analysis of Basal Promoter Activity of SIGIRR Variant 2 and 3
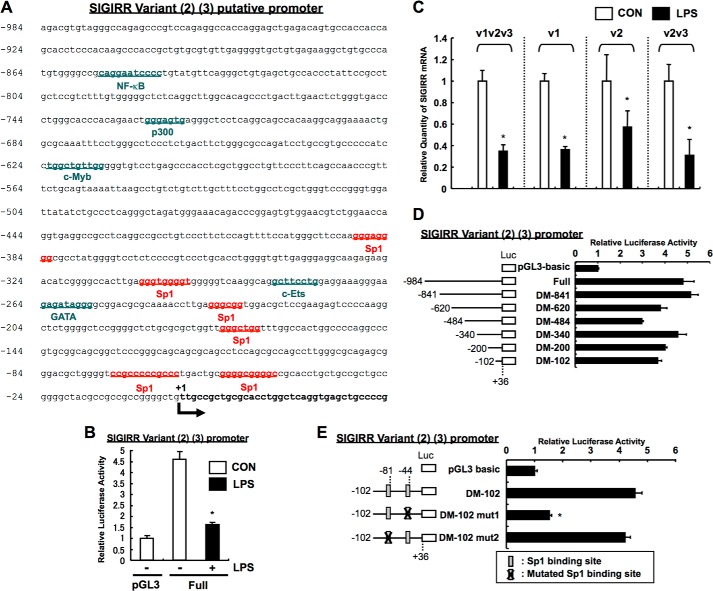
Although the SIGIRR variant 1 transcript seemed to be predominantly expressed in dHL60 cells, there may still be possible contributory roles of variants 2 and 3 transcripts for SIGIRR expression. We also cloned the 5′-flanking 984-bp fragment of the human SIGIRR variant 2/3, which contains a number of potential consensus binding sites for Sp1, c-Ets, and p300 but lacks a TATA box, as is similar to the case in variant 1 (Fig. 7A). Consistently, the basal promoter activity and gene expression of variants 2 and 3 were down-regulated after LPS treatment (Fig. 7, B and C), implying that the regulation of variant 2 and 3 transcripts may be similar to variant 1. To clarify the critical region required for basal SIGIRR variant 2/3 promoter activity, a series of reporter plasmids containing various lengths of the promoter region were constructed. Having shown that a deletion extending to nucleotide −102 (−102/+36) had little effect on promoter activity when compared with full-length construct (Fig. 7D), the existence of primary promoter regulatory elements between −102 and +36 bp upstream of the SIGIRR gene coding sequence was speculated. We further identified that a single mutation located at Sp1 site between nucleotides −44 and −53 completely suppressed promoter activity (Fig. 7E), suggesting the importance of Sp1 in the regulation of SIGIRR variant 2 and 3 transcript expressions. However, based on the fact that lower expression of SIGIRR variant 2 and 3 transcripts in dHL60 cells was observed (Table 2) and that the basal promoter activity of the variant 2/3 promoter showed only a 5-fold increase over pGL3-basic control (50-fold in variant 1) (Fig. 7D), contribution of variant 2/3 may be minimal.
FIGURE 7.
Analysis of SIGIRR variant 2 and 3 genes. A, 5′flanking region of SIGIRR variant 2 and 3 genes. We cloned a 5′-flanking 984-bp fragment of the human SIGIRR variant 2 and 3 genes. This fragment contains a number of potential consensus binding sites for transcription factors, including Sp1, c-Ets, and p300, but lacks a TATA box. B, LPS-dependent down-regulation of SIGIRR variant 2/3 promoter activity. HeLa cells were co-transfected with human SIGIRR V2V3-luc, TLR4, and MD-2 plasmids. Twenty-four hours after transfection, the cells were stimulated with 1 μg/ml LPS for 12 h. The luciferase activity was assessed. *, p < 0.05, versus control (without LPS); Student's t test (n = 3). C, variant-specific Q-RT-PCR analysis in LPS-treated cells. dHL60 cells were stimulated with 10 ng/ml LPS for 8 h. The SIGIRR variant 1 (v1), variant 2 (v2), and variants 2 and 3 (v2v3) mRNA levels were measured by quantitative RT-PCR using specific primers for each or both variants. The data were normalized to the mRNA level of 18 S rRNA as an internal control (Con). *, p < 0.05, versus vehicle-treated cells; Student's t test (n = 3). D and E, Sp1 is critical for basal promoter activity of SIGIRR variant 2/3 genes. HeLa cells were transiently transfected with the indicated human SIGIRR V2V3 promoter constructs. Luciferase activities were then assessed. *, p < 0.05 versus vehicle-treated V2V3 DM102-transfected cells) (E); ANOVA with Dunnett's multiple comparison (n = 3).
Sp1-dependent SIGIRR Gene expression and Its Inhibition by the LPS-p38 Pathway
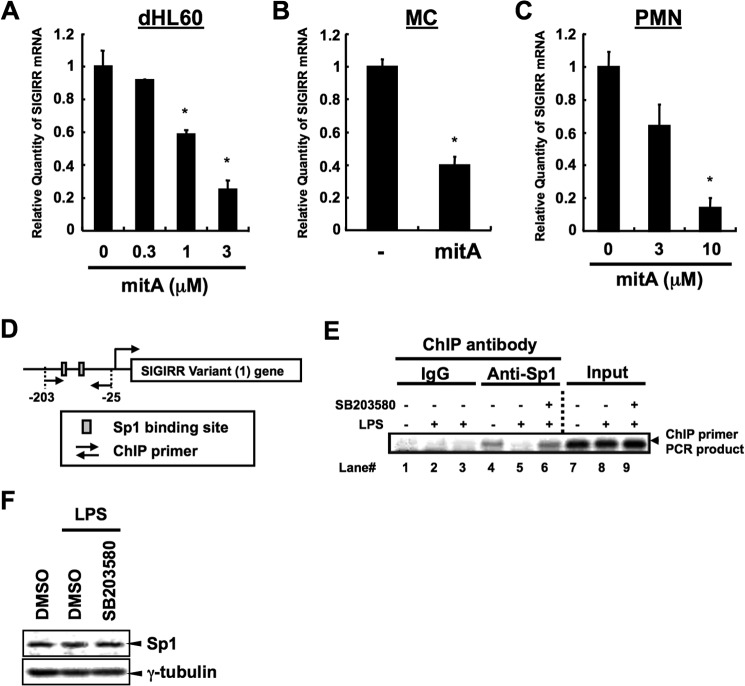
To further address whether Sp1 is positively involved in the regulation of human SIGIRR mRNA expression, dHL60, human primary MC and PMN were treated with Sp1 inhibitor mitA, and SIGIRR mRNA expression was quantified by real-time PCR. Consistent with the result of luciferase assay, mitA significantly suppressed basal human SIGIRR mRNA expressions in all cell types (Fig. 8, A–C). We next sought to determine whether LPS, which down-regulates human SIGIRR expression at the transcriptional level, influences Sp1-dependent SIGIRR basal expression via p38 activation. We performed ChIP using anti-Sp1 antibody and the primers that amplify the sequences between nucleotides −203 and −25 on human SIGIRR variant 1 promoter (Fig. 8D). We detected Sp1 binding in untreated cells (Fig. 8E, lane 4), whereas the binding was clearly decreased in LPS-treated cells (Fig. 8E, lane 5). Notably, treatment with SB203580, a p38 inhibitor, before LPS stimulation completely recovered Sp1 binding (Fig. 8E, lane 6) without affecting total Sp1 protein expression itself (Fig. 8F), suggesting that LPS inhibits Sp1 binding on human SIGIRR variant 1 promoter through p38 MAPK activation.
FIGURE 8.
Sp1-dependent SIGIRR gene expression and its inhibition by the LPS-p38 pathway. A–C, dHL60, MC, and PMN were treated with the indicated concentrations of mitA, a specific Sp1 inhibitor (dHL60, 8 h; MC, 12 h, PMN 24 h). The SIGIRR mRNA levels were measured by quantitative RT-PCR. *, p < 0.05 versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3) (A and C) or Student's t test (n = 3) (B). D and E, dHL60 cells were treated with 10 ng/ml LPS for 4 h after 30 μm SB203580 pretreatment for 1 h. Then the cells were fixed and lysed. Released chromatin was digested with enzyme, and the resulting disrupted chromatin was ChIP with Sp1 antibody or control IgG. After reversal of cross-linking, target DNA was amplified using primers with 35 cycles of PCR. The amplicon around the −203/−25 region (D), which contains two putative Sp1 binding sites on the SIGIRR promoter, were detected (E). F, dHL60 cells were pretreated with 30 μm SB203580. One hour after incubation the cells were stimulated with 10 ng/ml LPS for 4 h. Then, the nuclear extracts were subjected to Western blot analysis.
DISCUSSION
Although the importance of TIR superfamily member SIGIRR has been increasingly recognized over the decades, our understanding of the regulatory mechanism of its expression at basal levels and during inflammatory responses remains partial. Despite many reports that basal SIGIRR expression is high in epithelial cell lines and tissues (6, 10), recent studies have demonstrated that SIGIRR expression in non-epithelial lineages is also higher than originally thought (11–14). Our findings on the prevalence of functional expression of SIGIRR and its down-regulation by LPS in monocytic and neutrophilic cells alongside these studies showing a contributory role of non-epithelial systems under physiological and pathophysiological conditions further highlighted the need for a better understanding of how SIGIRR expression is regulated in this lesser-studied non-epithelial innate immune systems.
Among the three SIGIRR transcripts based on annotated information available from NCBI, we identified transcript variant 1 as a dominant transcript whose 5′-regulatory element possesses a key regulatory Sp1 binding site that maintains SIGIRR basal expression (Table 2). We further demonstrated that LPS-dependent TLR4 activation followed by inhibition of Sp1-dependent transcription is indispensable for LPS-dependent SIGIRR down-regulation. This is consistent with the finding of Kadota et al. (15) showing the LPS-dependent negative regulation of Sp1 in intestinal epithelial cells although its molecular mechanisms have not been elucidated. In the present study, we also determined that LPS-dependent SIGIRR down-regulation seems due to inhibition of Sp1-dependent SIGIRR basal expression in monocytic and neutrophilic cells by using primary cells and cell lines. Moreover, our findings emphasized that LPS-activated p38 contributes to suppress Sp1-dependent SIGIRR basal expression, although generally p38 have been assumed to be a positive regulator of Sp1 (23, 24). Thus, our findings are the first to show a negative regulatory role of p38 on Sp1-dependent transcription. On the other hand, LPS-dependent Sp1 inhibition has been implicated in the previous paper showing the Sp1 protein dephosphorylation and degradation induced by LPS, which results in reduced Sp1 binding to its target sequence (25). Although we also observed a reduction in Sp1 binding to SIGIRR promoter after LPS exposure (Fig. 8E), Sp1 protein expression in the nucleus was not affected by LPS in dHL60 cells (Fig. 8F), implying another innate immune cell-specific mechanism responsible for LPS-dependent Sp1 inhibition via TLR4-p38 pathway.
Despite numerous studies showing an alteration in genes expression by LPS, most studies analyzed only genes that are up-regulated by LPS; thereby, little is known about the molecular mechanism of LPS-dependent repression of gene expression. Apart from the involvement of p38 and Sp1, LPS-induced NF-κB activation has been considered as a strong candidate that negatively regulates transcription of LPS-repressive genes (26, 27), although this was not the case in this research (Fig. 4, C and D, caffeic acid phenethyl ester (CAPE)). There is another conceptually novel report identifying some microRNAs as negative regulators for LPS-dependent target genes (28, 29). Among these microRNAs, miR1224 is likely a key inhibitor of Sp1 mRNA expression (29). Although, as we mentioned above, LPS did not affect Sp1 expression in dHL60 cells (Fig. 8F), there may still be room to consider the involvement of LPS-induced microRNA gene regulatory network in p38-dependent negative regulation of SIGIRR in non-epithelial immune cells.
Physiological relevance of higher SIGIRR expression at basal levels in many types of cells has been gradually established by recent studies. For example, Sigirr-deficient mice exhibited a dramatic inflammation in dextran sodium sulfate colitis and endotoxin-challenged septic shock models (6). Furthermore, Sigirr-deficient mice were more susceptible to experimental autoimmune encephalomyelitis resulting from hyperactivation of Th17 cells (30). Moreover, Sigirr deficiency in the Apcmin/+ mouse, a spontaneous intestinal cancer model mimicking the familial adenomatous polyposis syndrome, led to spontaneous colonic polyposis possibly via increased IL-1- and TLRs-induced Akt-mTOR signaling (31). Based on these in vivo data, it is obvious that SIGIRR expression levels in the tissue or cells determine the activation threshold of TIR signaling, which in turn restricts the incidence of inflammation, tissue damage, autoimmunity, and cancer. On the other hand, when considering the above in vivo findings, decreased SIGIRR expression upon infectious stimuli supported the idea of a contributory role of SIGIRR down-regulation to achieve maximum induction of TIR signaling, although prolonged SIGIRR down-regulation may be detrimental to the host (6, 30). Because SIGIRR expression tended to be recovered after LPS long exposure (Figs. 2, C and D, and 3, G and H), SIGIRR must also be a critical factor for the efficient resolution of inflammation. Further study that focuses on late-phase of LPS-dependent alteration of SIGIRR expression and function is needed.
In summary, we confirmed the higher SIGIRR expression in monocytic and neutrophilic cells. Under resting conditions, basal SIGIRR gene expression is maintained by Sp1, whereas LPS-dependent TLR4-p38 signaling inhibits the Sp1 binding to SIGIRR promoter, which results in a decrease in SIGIRR expression. This study provides the first clear mechanism of LPS-dependent SIGIRR down-regulation in non-epithelial innate immune cells and identifies the LPS-activated p38 as a negative regulator of Sp1-dependent SIGIRR expression. Although how p38 regulates Sp1 and which LPS-repressive genes other than SIGIRR are regulated in this pathway in these innate immune cells remains an open question, our findings may open a new gate into SIGIRR-mediated control of TIR-mediated innate immune responses.
This work was supported in part by grants from the Ministry of Education, Science, Sport, and Culture (MEXT) of Japan.
- TIR
- Toll-interleukin 1 receptor
- TLR
- toll-like receptor
- SIGIRR
- single immunoglobulin IL-1 receptor-related molecule
- MC
- monocytes
- PMN
- polymorphonuclear neutrophils
- mitA
- mithramycin A
- dHL60
- differentiated HL-60
- AM
- anisomycin
- ANOVA
- analysis of variance.
REFERENCES
- 1. O'Neill L. A. (2008) The interleukin-1 receptor/Toll-like receptor superfamily: 10 years of progress. Immunol. Rev. 226, 10–18 [DOI] [PubMed] [Google Scholar]
- 2. Kawai T., Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 3. Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 4. Weber A., Wasiliew P., Kracht M. (2010) Interleukin-1 (IL-1) pathway. Sci. Signal. 3, cm1. [DOI] [PubMed] [Google Scholar]
- 5. Brint E. K., Xu D., Liu H., Dunne A., McKenzie A. N., O'Neill L. A., Liew F. Y. (2004) ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance. Nat. Immunol. 5, 373–379 [DOI] [PubMed] [Google Scholar]
- 6. Wald D., Qin J., Zhao Z., Qian Y., Naramura M., Tian L., Towne J., Sims J. E., Stark G. R., Li X. (2003) SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat. Immunol. 4, 920–927 [DOI] [PubMed] [Google Scholar]
- 7. Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., Tschopp J. (2003) Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J. Exp. Med. 197, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kinjyo I., Hanada T., Inagaki-Ohara K., Mori H., Aki D., Ohishi M., Yoshida H., Kubo M., Yoshimura A. (2002) SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 17, 583–591 [DOI] [PubMed] [Google Scholar]
- 9. Chuang T. H., Ulevitch R. J. (2004) Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat. Immunol. 5, 495–502 [DOI] [PubMed] [Google Scholar]
- 10. Thomassen E., Renshaw B. R., Sims J. E. (1999) Identification and characterization of SIGIRR, a molecule representing a novel subtype of the IL-1R superfamily. Cytokine. 11, 389–399 [DOI] [PubMed] [Google Scholar]
- 11. Bulek K., Swaidani S., Qin J., Lu Y., Gulen M. F., Herjan T., Min B., Kastelein R. A., Aronica M., Kosz-Vnenchak M., Li X. (2009) The essential role of single Ig IL-1 receptor-related molecule/Toll IL-1R8 in regulation of Th2 immune response. J. Immunol. 182, 2601–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lech M., Avila-Ferrufino A., Allam R., Segerer S., Khandoga A., Krombach F., Garlanda C., Mantovani A., Anders H. J. (2009) Resident dendritic cells prevent postischemic acute renal failure by help of single Ig IL-1 receptor-related protein. J. Immunol. 183, 4109–4118 [DOI] [PubMed] [Google Scholar]
- 13. Jiang X., McClellan S. A., Barrett R. P., Zhang Y., Hazlett L. D. (2012) Vasoactive intestinal peptide downregulates proinflammatory TLRs while upregulating anti-inflammatory TLRs in the infected cornea. J. Immunol. 189, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davies J. M., MacSharry J., Shanahan F. (2010) Differential regulation of Toll-like receptor signalling in spleen and Peyer's patch dendritic cells. Immunology 131, 438–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kadota C., Ishihara S., Aziz M. M., Rumi M. A., Oshima N., Mishima Y., Moriyama I., Yuki T., Amano Y., Kinoshita Y. (2010) Down-regulation of single immunoglobulin interleukin-1R-related molecule (SIGIRR)/TIR8 expression in intestinal epithelial cells during inflammation. Clin. Exp. Immunol. 162, 348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shuto T., Ono T., Ohira Y., Shimasaki S., Mizunoe S., Watanabe K., Suico M. A., Koga T., Sato T., Morino S., Sato K., Kai H. (2010) Curcumin decreases toll-like receptor-2 gene expression and function in human monocytes and neutrophils. Biochem. Biophys. Res. Commun. 398, 647–652 [DOI] [PubMed] [Google Scholar]
- 17. Shuto T., Furuta T., Cheung J., Gruenert D. C., Ohira Y., Shimasaki S., Suico M. A., Sato K., Kai H. (2007) Increased responsiveness to TLR2 and TLR4 ligands during dimethylsulfoxide-induced neutrophil-like differentiation of HL-60 myeloid leukemia cells. Leuk. Res. 31, 1721–1728 [DOI] [PubMed] [Google Scholar]
- 18. Ueno K., Koga T., Kato K., Golenbock D. T., Gendler S. J., Kai H., Kim K. C. (2008) MUC1 mucin is a negative regulator of toll-like receptor signaling. Am. J. Respir. Cell Mol. Biol. 38, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizunoe S., Shuto T., Suzuki S., Matsumoto C., Watanabe K., Ueno-Shuto K., Suico M. A., Onuki K., Gruenert D. C., Kai H. (2012) Synergism between interleukin (IL)-17 and Toll-like receptor 2 and 4 signals to induce IL-8 expression in cystic fibrosis airway epithelial cells. J. Pharmacol. Sci. 118, 512–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 21. Cuadrado A., Nebreda A. R. (2010) Mechanisms and functions of p38 MAPK signalling. Biochem. J. 429, 403–417 [DOI] [PubMed] [Google Scholar]
- 22. Hazzalin C. A., Le Panse R., Cano E., Mahadevan L. C. (1998) Anisomycin selectively desensitizes signalling components involved in stress kinase activation and fos and jun induction. Mol. Cell Biol. 18, 1844–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma W., Lim W., Gee K., Aucoin S., Nandan D., Kozlowski M., Diaz-Mitoma F., Kumar A. (2001) The p38 mitogen-activated kinase pathway regulates the human interleukin-10 promoter via the activation of Sp1 transcription factor in lipopolysaccharide-stimulated human macrophages. J. Biol. Chem. 276, 13664–13674 [DOI] [PubMed] [Google Scholar]
- 24. Lin H. H., Lai S. C., Chau L. Y. (2011) Heme oxygenase-1/carbon monoxide induces vascular endothelial growth factor expression via p38 kinase-dependent activation of Sp1. J. Biol. Chem. 286, 3829–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye X., Liu S. F. (2002) Lipopolysaccharide down-regulates Sp1 binding activity by promoting Sp1 protein dephosphorylation and degradation. J. Biol. Chem. 277, 31863–31870 [DOI] [PubMed] [Google Scholar]
- 26. Serio K. J., Johns S. C., Luo L., Hodulik C. R., Bigby T. D. (2003) Lipopolysaccharide down-regulates the leukotriene C4 synthase gene in the monocyte-like cell line, THP-1. J. Immunol. 170, 2121–2128 [DOI] [PubMed] [Google Scholar]
- 27. Baranova I., Vishnyakova T., Bocharov A., Chen Z., Remaley A. T., Stonik J., Eggerman T. L., Patterson A. P. (2002) Lipopolysaccharide down regulates both scavenger receptor B1 and ATP binding cassette transporter A1 in RAW cells. Infect. Immun. 70, 2995–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aalaei-andabili S. H., Rezaei N. (2013) Toll like receptor (TLR)-induced differential expression of microRNAs (MiRs) promotes proper immune response against infections: a systematic review. J. Infect. 67, 251–264 [DOI] [PubMed] [Google Scholar]
- 29. Niu Y., Mo D., Qin L., Wang C., Li A., Zhao X., Wang X., Xiao S., Wang Q., Xie Y., He Z., Cong P., Chen Y. (2011) Lipopolysaccharide-induced miR-1224 negatively regulates tumour necrosis factor-α gene expression by modulating Sp1. Immunology 133, 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gulen M. F., Kang Z., Bulek K., Youzhong W., Kim T. W., Chen Y., Altuntas C. Z., Sass Bak-Jensen K., McGeachy M. J., Do J. S., Xiao H., Delgoffe G. M., Min B., Powell J. D., Tuohy V. K., Cua D. J., Li X. (2010) The receptor SIGIRR suppresses Th17 cell proliferation via inhibition of the interleukin-1 receptor pathway and mTOR kinase activation. Immunity 32, 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiao H., Yin W., Khan M. A., Gulen M. F., Zhou H., Sham H. P., Jacobson K., Vallance B. A., Li X. (2010) Loss of single immunoglobulin interlukin-1 receptor-related molecule leads to enhanced colonic polyposis in Apc(min) mice. Gastroenterology 139, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]