Background: The guanine exchange factor (GEF) FGD6 is a Src substrate in osteoclasts.
Results: FGD6 is found in several protein complexes, which regulate the formation of podosomes/sealing zones and retromer-dependent membrane recycling.
Conclusion: A single GEF controls several actin-based processes to coordinate cell polarity and membrane recycling.
Significance: FGD6 is essential for osteoclast function in bone degradation.
Keywords: Actin, CDC42, Membrane Recycling, Osteoclast, Src, FGD6, Podosome
Abstract
The initial step of bone digestion is the adhesion of osteoclasts onto bone surfaces and the assembly of podosomal belts that segregate the bone-facing ruffled membrane from other membrane domains. During bone digestion, membrane components of the ruffled border also need to be recycled after macropinocytosis of digested bone materials. How osteoclast polarity and membrane recycling are coordinated remains unknown. Here, we show that the Cdc42-guanine nucleotide exchange factor FGD6 coordinates these events through its Src-dependent interaction with different actin-based protein networks. At the plasma membrane, FGD6 couples cell adhesion and actin dynamics by regulating podosome formation through the assembly of complexes comprising the Cdc42-interactor IQGAP1, the Rho GTPase-activating protein ARHGAP10, and the integrin interactors Talin-1/2 or Filamin A. On endosomes and transcytotic vesicles, FGD6 regulates retromer-dependent membrane recycling through its interaction with the actin nucleation-promoting factor WASH. These results provide a mechanism by which a single Cdc42-exchange factor controlling different actin-based processes coordinates cell adhesion, cell polarity, and membrane recycling during bone degradation.
Introduction
Osteoclasts are multinucleated cells that digest the organic and inorganic bone matrix (1, 2). To do so, osteoclasts attach onto bone surfaces. This requires the assembly of podosomes, actin-rich structures linking cell adhesion molecules with actin meshworks. Podosomes condense into podosomal belts to form a sealing zone, which segregates a bone-facing membrane domain, the ruffled border, from other membrane domains, thus determining osteoclast polarity. Bone digestion also requires the secretion of lysosomal hydrolases into these resorption lacunae and the targeting of typical lysosomal membrane proteins to the ruffled membrane, in particular to acidify the resorption lacunae. After digestion, the digested bone material is endocytosed into transcytotic vesicles that fuse with the secretory domain of osteoclasts to release their content into the extracellular space (3). This transcytotic process using large surface areas of the ruffled border suggests the possibility that internalized membrane components are recycled back to the ruffled membrane.
These cellular processes are orchestrated by the Src tyrosine kinase. The evidence comes from the targeted disruption of the corresponding gene in mice leading to osteopetrosis due to osteoclasts unable to organize sealing zones (4). In our previous work aimed at identifying genes up-regulated during osteoclastogenesis (5) and identifying Src substrates using quantitative proteomics (6), we identified FGD6 as a potential important player of osteoclast function. This protein with pleckstrin homology (PH)4 and FYVE (domain present in Fab1, YotB, Vac1p, and EEA1) domains belongs to the FGD family consisting of six Rho guanine nucleotide exchange factors (GEFs) activating Rho GTPases. FGD6 is highly homologous to FGD1, a Rho GEF regulating post-Golgi transport in bone rebuilding osteoblasts (7, 8). Mutations in the FGD1 gene are linked with the inherited disease faciogenital dysplasia or Aarskog-Scott syndrome (8, 9).
Here, we show that FGD6 plays an essential role in osteoclasts by regulating the assembly of different actin-based protein networks and activating Cdc42 at different locations; first at the plasma membrane during podosome/sealing zone formation and second at endosomes/transcytotic vesicles during membrane recycling, two highly coordinated cellular processes ensuring an efficient bone degradation.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
The following antibodies were used: mouse anti-phosphotyrosine (clone 4G10), rabbit anti-Vps35 (Millipore), mouse anti-GFP (clones 7.1 and 13.1) (Roche Applied Science), rabbit anti-GAPDH (Bioss Antibodies), mouse anti-Tubulin (clone TUB2.1), mouse anti-GST (clone GST-2), rabbit anti-Talin-1 (Sigma), mouse anti-IQGAP1 (clone 24/IQGAP1), hamster anti-CD61 (Integrin β3, clone 2C9.G2), rat anti-Lamp-1 (clone 1D4B) (BD Biosciences), mouse anti-Cdc42-GTP, rabbit anti-Cdc42 (New East Biosciences), rabbit anti-ARHGAP10 (Proteintech Group), rabbit anti-GFP (ImmunoKontact), mouse anti-Myc (clone 9E10), rabbit anti-Filamin A (clone EP2405Y) (GeneTex), and rabbit anti-FGD6 and rabbit anti-Talin-1/2 (Santa Cruz Biotechnology). Rabbit anti-WASH1 antibodies were kindly provided by Dr. Alexis Gautreau, Laboratoire d'Enzymologie et de Biochimie Structurales, Gif sur Yvette, France, and rabbit anti-EEA1 antibodies were kindly provided by Prof. Marino Zerial, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany.
HRP- or fluorescently labeled secondary antibodies were from Jackson ImmunoResearch or Molecular Probes. The following inhibitors were used at the indicated concentrations: wortmannin (500 nm, Sigma) and PP2 (10 μm, Merck). The PP2 phenotype was confirmed using another Src inhibitor (SU6656, 10 μm, Merck) or with Src knockdown using siRNA (6).
Constructs
RFP-tagged Ezrin and GFP-tagged Ezrin, Rab38, and FGD6 constructs (RhoGEF, RhoGEF+PH1, RhoGEF+PH1+FYVE, and RhoGEF+PH1+FYVE+PH2) were amplified by PCR and subcloned into the pShuttle vector (Qbiogene). GST-tagged truncated-FGD6 (aa 1–1039) was subcloned into the pGEX-4T-1 vector (GE Healthcare), and His-Myc-tagged truncated-FGD6 (aa 843–1398) was subcloned into the pET28a(+) expression vector (Merck). The pEGFP-N2 vector was from Clontech.
Cell Culture and Osteoclastogenesis
Raw264.7 cells (ATCC number TIB-71) were cultivated at 37 °C under a humidified 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (DMEM)-GlutaMAX-I (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Biochrom), 2 mm l-glutamine, 0.1 mg/ml of streptomycin, and 100 units/ml of penicillin-G (Invitrogen).
In vitro osteoclastogenesis was induced by addition of RANKL as described previously (5). At day 4 of differentiation, cells were transferred either to glass coverslips or to BD Biosciences BioCoat osteologic discs (ODs) (BD Biosciences) (now manufactured as Osteo Assay Surface by Corning).
Transient Transfection of Osteoclasts
After 4 days of differentiation osteoclasts were transiently transfected with 1 μg of pEGFP-N2 per well in a 24-well plate using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. 30 h after transfection cells were processed for subsequent analysis.
Adenovirus Production and Gene Transduction into Osteoclasts
Adenoviral vectors and recombinant adenoviruses were generated using the AdEasyTM system (Qbiogene) developed by He et al. (10, 11) using the pShuttle vector and the packaging cell line HEK293A (Qbiogene). After 4 days of differentiation osteoclasts were transduced with titered adenovirus and grown for an additional 48 h either plated on ODs or glass coverslips or plastic. Then cells were processed for subsequent analysis.
RNA Interference in Osteoclasts
Stealth siRNAs were obtained from Invitrogen. Differentiated osteoclasts were resuspended in Electroporation Isoosmolar Buffer (Eppendorf) and 1 μm predesigned stealth siRNA or Negative Control Medium GC stealth siRNA (Invitrogen) was electroporated into the cells with a single square wave pulse of 2,500 V/cm field strength and 300-μs pulse length using an ECM830 ElectroSquarePorator (BTX). Electroporated osteoclasts were resuspended in DMEM, 50 ng/ml of RANKL and allowed to recover for 46 h. Osteoclasts were processed for subsequent analysis. Stealth siRNAs used in this study (sense sequence) were: FGD6#1, CAGUGUGGAAGUUACCUCAUCCUAU; FGD6#2, AACACAUCCACAAACACUUUCUCGG; 3′ UTR FGD6, UAACCAACCACUGUAUGGAAAUUUG; Cdc42, CAGUUAUGAUUGGUGGAGAGCCAUA; IQGAP1, CCUAUGAUUGUGGUCCGAAAGUUUG; RhoGAP10, CAGCUGGAUAAGAUGGGAUUCACAA; Talin-1, CAGCUCAUUGCUGGCUACAUAGAUA; Talin-2, CAGAGAGCAUCAAUCAGCUCAUCAU; Filamin A, CAAGGCCCAUGAACCUACCUACUUU; WASH1, CAGAACAUCUCUGGAGACAUCUUCA; and Vps35, CAGCUGGCUGCUAUCACCUUGAUCA.
Quantitative RT-PCR
Total RNA was isolated from osteoclasts using the Invisorb RNA Kit I (Stratec), and reverse transcription was performed using the Moloney MLV Reverse Transcription system (Promega). Quantitative RT-PCR was performed with a Stratagene Mx3005 system and the Brilliant II SYBR Green quantitative PCR kit according to the manufacturer's instructions (Agilent Technologies). Primers used were: GAPDH forward, AGGTCGGTGTGAACGGATTTG; GAPDH reverse, TGTAGACCATGTAGTTGAGGTCA; FGD6 forward, GGGCAACTGGCAAGAGTGT; FGD6 reverse, GGGGATTTTCTTCTGCTT; Cdc42 forward, TGATGATTGCCATCTCCTTTC; Cdc42 reverse, AAAGCTTCAGCCAGTTGTTCA; IQGAP1 forward, GCTGATCTCTATCAGAAGGA; IQGAP1 reverse, CGAGATACCTTTGACAGTTT; RhoGAP10 forward, AAGAGCAGCTTGGAGCTGTA; RhoGAP10 reverse, TAGGCTCCACAAACTCGAAC; Talin-1 forward, AAGCAGAGACCCCTGAAGAT; Talin-1 reverse, TCTCGCAGTAGGGTCTTGTC; Talin-2 forward, CTACACCAAGAGGGAGCTGA; Talin-2 reverse, CGTTCTCTGCATTCAGTGTG; Filamin A forward, TTCTTATGTGCCCAGAAAGC; Filamin A reverse, CTGGCCAGCTTCAGTACAAT; WASH1 forward, AACATCAGCTCCATCAGCTC; WASH1 reverse, CAGGTCTGGCACGTAGAAGT; Vps35 forward, ACAGCTGGCTGCTATCACCT; Vps35 reverse, GGCACTCCATGACCCTTTTA.
Quantitative PCR analyses were done in triplicates, and Ct values were normalized against the internal control gene GAPDH. Fold-differences in expression levels were calculated according to the comparative Ct method (12). Quantitative PCR was confirmed by Western blotting (Figs. 4E and 5E).
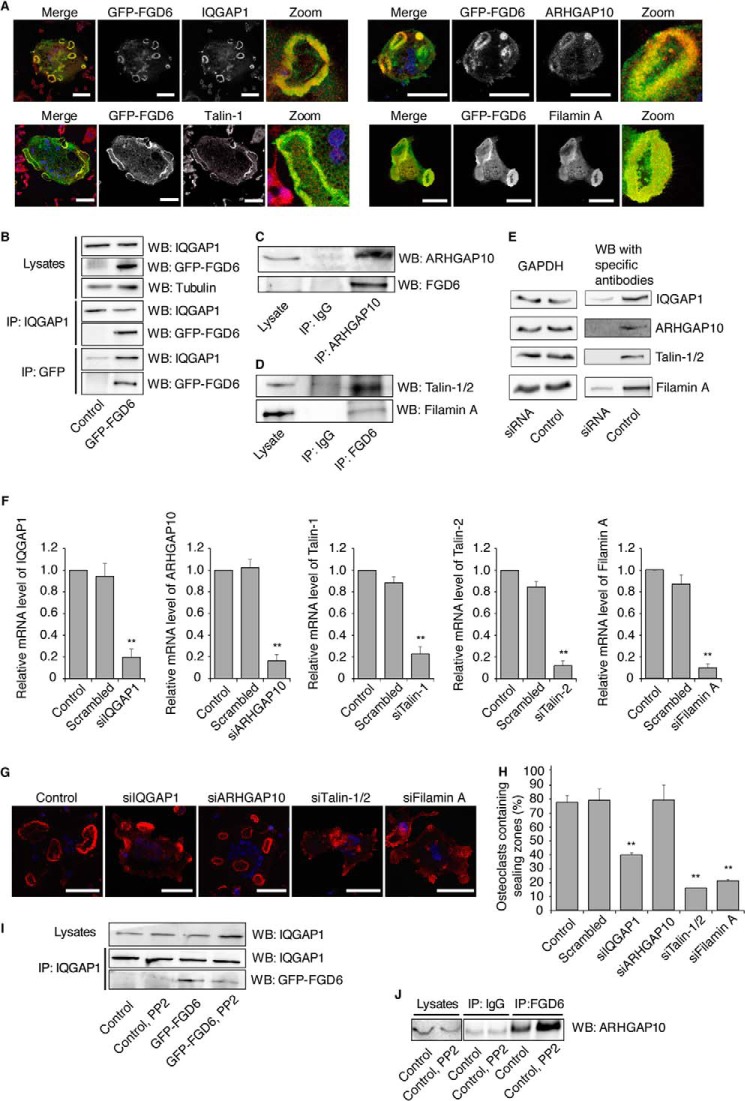
FIGURE 4.
FGD6 interacts with IQGAP1, ARHGAP10, Talin-1, and Filamin A at the plasma membrane. A, osteoclasts expressing GFP-FGD6 (green) were grown on ODs and stained with DAPI (blue) and anti-IQGAP1, anti-ARHGAP10, anti-Talin-1, or anti-Filamin A (red). Bars, 50 μm. B and C, cell lysates from control osteoclasts or osteoclasts expressing GFP-FGD6 were used for immunoprecipitation of (B) IQGAP1 or GFP-FGD6 or (C) ARHGAP10 with anti-IQGAP1, anti-GFP, or anti-ARHGAP10. After Western blotting (WB) immunoprecipitates and cell lysates were probed for IQGAP1, GFP-FGD6, or ARHGAP10. D, endogenous FGD6 was immunoprecipitated from osteoclasts lysates with anti-FGD6 and after Western blotting the immunoprecipitate (IP) was probed for Filamin A or Talin-1/2. E–H, osteoclasts were either mock-transfected, or transfected with scrambled siRNAs or with siRNAs targeting IQGAP1, ARHGAP10, Talin-1, Talin-2, or Filamin A. E, osteoclasts were collected and lysed. Lysates were analyzed by SDS-PAGE followed by Western blotting using antibodies directed against the indicated gene products. F, the expression levels were quantified by RT-PCR. Values are mean ± S.D. from 3 experiments. G, cells grown on ODs were stained with phalloidin (red) and DAPI (blue), and analyzed by confocal microscopy. Bars, 50 μm. H, the number of osteoclasts with sealing zones was quantified. Values are mean ± S.D. from 3 experiments (n = 200 osteoclasts/experiment). All groups were compared with control by applying a Dunnett one-way ANOVA test. *, p < 0.05. **, p < 0.01. See also supplemental Videos S5–S8. I and J, control and GFP-FGD6-expressing osteoclasts were left untreated or treated with 10 μm PP2 for 1 h. I, IQGAP1, or J, FGD6 was immunoprecipitated from cell lysates using the corresponding antibodies, and after Western blotting the immunoprecipitates and lysates were probed for IQGAP1, GFP-FGD6, or ARHGAP10. Each experiment was repeated at least 3 times. Representative images are shown.
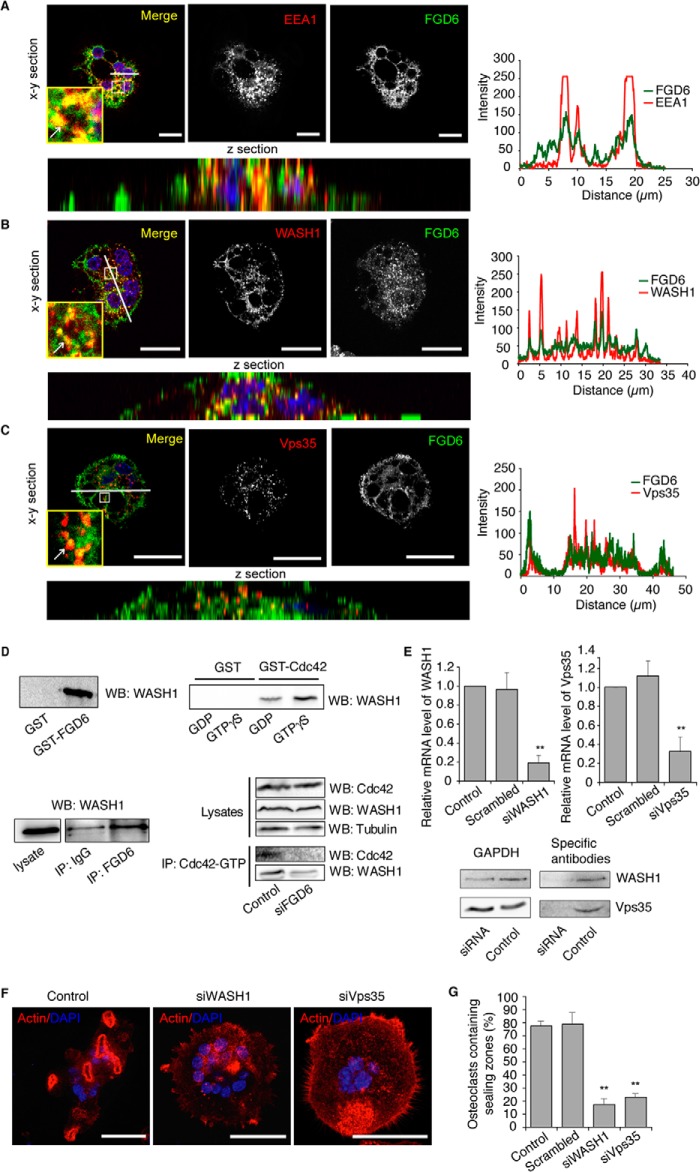
FIGURE 5.
FGD6 interacts with WASH1 on endosomes. Osteoclasts expressing GFP-FGD6 (green) were grown on ODs and stained with DAPI (blue) and anti-EEA1 (A), anti-WASH1 (B), or anti-Vps35 (C) (red), and analyzed by confocal microscopy. Bars, 20 μm. Arrows show early endosomes around transcytotic vesicles. The corresponding fluorescence intensity profiles along the white line are shown. D, osteoclast lysates were incubated with GST, GST-FGD6(1–1039), or GST-Cdc42 loaded with GDP or GTPγS, or with anti-FGD6 antibodies. After pull down, the interacting material was analyzed by Western blotting (WB) using anti-WASH1. GTP-Cdc42 was immunoprecipitated from lysates of osteoclasts depleted or not from FGD6, and after Western blotting the immunoprecipitates (IP) were probed for Cdc42 and WASH1. E–G, osteoclasts grown on ODs were mock-transfected, or transfected with scrambled siRNAs or with siRNAs targeting WASH1 or Vps35. E, the expression levels were determined by quantitative RT-PCR. Osteoclasts were also collected and lysed. Lysates were analyzed by SDS-PAGE followed by Western blotting using antibodies directed against the indicated gene products. F, osteoclasts were also stained with phalloidin (red) and DAPI (blue) and analyzed by confocal microscopy. Bars, 50 μm. G, the number of osteoclasts with sealing zones was quantified. Values are mean ± S.D. from 3 experiments (n = 200 osteoclasts per experiment). All groups were compared with control by applying a Dunnett one-way ANOVA test. *, p < 0.05. **, p < 0.01. Representative images are shown. See also supplemental Videos S9 and S10.
Calcium Uptake
Osteoclasts were infected with GFP-Rab38 adenovirus and calcium uptake was visualized as described previously using the fluorescent calcium-indicator Fura Red (13). Briefly, infected osteoclasts were grown on ODs in medium containing 5 μm cell-impermeable Fura Red (Invitrogen) for 72 h as indicated by the manufacturer. Afterward osteoclasts were fixed with 3% (w/v) paraformaldehyde in PBS for 15 min at 37 °C, washed with PBS, and mounted on glass slides using Mowiol. Imaging of calcium taken up by the cells was performed at a wavelength of 660 nm.
Immunocytochemistry
Osteoclasts grown on glass or ODs were fixed with 3% (w/v) paraformaldehyde in PBS for 15 min at 37 °C, quenched with 50 mm NH4Cl, PBS for 10 min, and permeabilized with 0.1% (w/v) Triton X-100 in PBS for 6 min. Samples were blocked with 3% (w/v) BSA in PBS for 30 min at room temperature followed by incubation with primary antibodies diluted in 3% (w/v) BSA in PBS for 1 h at room temperature. After washing staining with the appropriate secondary antibodies was performed. Staining of the actin cytoskeleton was performed with Phalloidin-Alexa 546 or Phalloidin-Alexa 633 (Invitrogen). Glass coverslips and ODs were washed and mounted on glass slides with Mowiol containing 10 μg/ml of DAPI (Invitrogen).
Confocal Laser Scanning Microscopy, Fluorescence Microscopy, and Time Lapse Microscopy
Confocal imaging was performed with a Zeiss LSM 780 inverted confocal laser scanning microscope and ZEN 2010 software (Zeiss). Fluorescence microscopy was performed with a Zeiss Axiovert 200M fluorescence microscope and Axiovision software (Zeiss). Image processing and analysis were carried out using FIJI or Volocity software. Image analysis of osteoclasts with respect to their ability to form actin rings was carried out manually. More than 200 osteoclasts per condition and per experiment were counted, the mean ± S.D. were calculated.
For time lapse microscopy Raw 264.7 cell-derived osteoclasts were transferred to uncoated glass coverslips or to glass coverslips coated with hydroxyapatite (BD Biosciences) and grown in minimum essential Eagle's medium (Sigma) supplemented with 10% FCS and 50 ng/ml of soluble recombinant RANKL. For long term imaging the cells were observed with a Zeiss Axiovert 200M inverted microscope equipped with an automated stage, an Incubator XL3 for temperature maintenance and CO2 buffering (PeCon). Images were acquired automatically and processed with MetaMorph version 6.1 imaging software (Molecular Devices).
Resorption Assay
Bone resorption was measured using bone mimicking surfaces as described previously (14). Briefly, osteoclasts were grown on ODs and 48 h after infection, cells were detached by adding 1 ml of detaching solution (∼6% NaOCl and ∼5.2% NaCl) for 5 min. ODs were rinsed twice with water and dried. ODs without osteoclasts were imaged with a Zeiss SteREO Discovery V20 microscope (Fig. 2H) and the resorbed area determined using Volocity software (PerkinElmer Life Sciences).
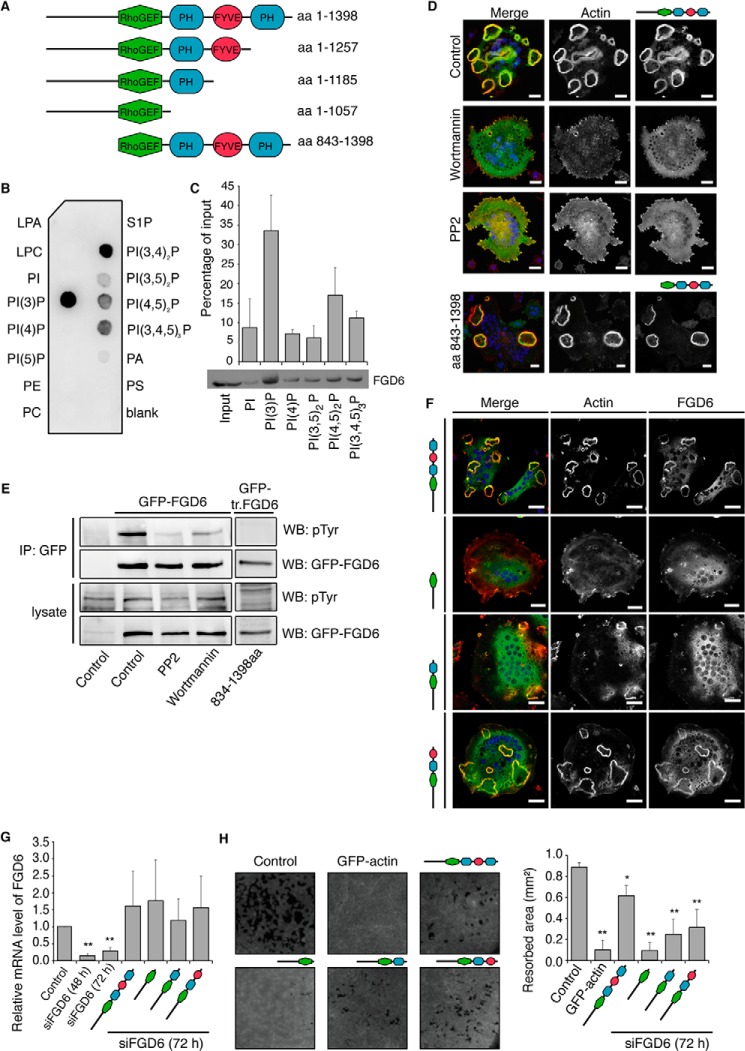
FIGURE 2.
The PH and FYVE domains of FGD6 are required for sealing zone formation and surface resorption. A, the FGD6 mutants used in this study. B and C, binding of His-Myc-tagged FGD6 lacking its N-terminal domain (aa 843–1398) to the indicated lipids present on nitrocellulose strips (B) or liposomes (C). D, osteoclasts grown on ODs expressing GFP-FGD6 full-length or truncated from its N-terminal domain (green) were left untreated or treated with either 500 nm wortmannin or 10 μm PP2 for 1 h. Cells were stained with phalloidin (red) and DAPI (blue) and analyzed by confocal microscopy. Bars, 20 μm. E, cell lysates from similarly treated osteoclasts were immunoprecipitated with anti-GFP. After Western blotting immunoprecipitates (IP) and cell lysates were probed for GFP-FGD6 or phosphotyrosine. F-H, in osteoclasts depleted from endogenous FGD6 different GFP-tagged FGD6 proteins (wild type and mutants) were expressed as indicated (green). F, cells were stained with phalloidin (red) and DAPI (blue), and analyzed by confocal microscopy. Bars, 20 μm. G, expression levels of the FGD6 full-length and mutants were quantified by RT-PCR. Values are mean ± S.D. from 3 experiments. H, the corresponding digested areas on ODs (appearing as black spots on a gray background) were quantified. Values are mean ± S.D. from 3 experiments. All groups were compared with control by applying a Dunnett one-way ANOVA test. *, p < 0.05. **, p < 0.01. Representative images are shown. The abbreviations are: PI, phosphatidylinositol; PE, phosphatidylethanolamine; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; S1P, sphingosin-1-phosphate; WB, Western blot.
Immunoprecipitation and Western Blotting
Osteoclasts were lysed using ice-cold lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1.0% Triton X-100, 0.1% sodium deoxycholate, 1 mm EDTA plus protease and phosphatase inhibitors). For immunoprecipitations of proteins, lysates (1 mg of proteins in 0.5 ml for each sample) were precleared on Protein G magnetic beads (Carl Roth) for 1 h at 4 °C. Protein G beads were incubated with antibodies for 1 h at 4 °C. Then, precleared supernatants were added for 1 h at 4 °C. Precipitates were washed two times with lysis buffer; proteins were eluted, and resolved by SDS-PAGE. For immunoblotting, gel-separated proteins were transferred to nitrocellulose membranes and incubated with the corresponding primary antibodies. After incubation with secondary antibodies conjugated with HRP (Jackson ImmunoResearch), bands were detected with enhanced chemiluminescence Western blotting Detection Reagents (GE Healthcare). Quantification of protein bands was performed using FIJI software.
GST Pull-down
Expression of GST-tagged FGD6 (aa 1–1039) or GST-tagged Cdc42 in Escherichia coli was induced with 0.1 mm isopropyl d-thiogalactoside for 20 h at 20 °C. GST-tagged Cdc42 was loaded with either GDP or GTPγS as described previously (15). Protein was pulled down from bacterial lysate by glutathione-Sepharose beads. Briefly, osteoclast lysates (2 mg of proteins for each sample) were precleared on glutathione-Sepharose beads for 1 h at 4 °C before they were added to the GST-tagged FGD6 or Cdc42 on glutathione-Sepharose beads for an additional 2 h at 4 °C. Precipitates were washed 2 times with lysis buffer; proteins were eluted and resolved by SDS-PAGE.
Mass Spectrometry Analysis
Protein bands were excised, washed, in-gel reduced, S-alkylated, and digested with trypsin (Promega) as described previously (16). NanoLC-MS/MS experiments were performed using an UltiMate3000 nanoHPLC system (Dionex) equipped with a PepMap C18 analytical column (3 μm, 100 Å, 15 cm × 75 μm inner diameter) directly coupled to the nanoelectrospray source (Proxeon) of an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Peptides were eluted with a 80-min linear gradient of 5–45% acetonitrile in 0.1% formic acid at 200 nl/min. Mass spectra were acquired in a data-dependent mode with one MS survey scan (resolution of 60,000) in an Orbitrap and MS/MS scans of the 8 most intense precursor ions in an LTQ. Data analysis was done using MaxQuant version 1.2.0.11 (17). Peak lists were searched against a database containing 16,339 entries from the UniProt-KB/Swiss-Prot mouse database (release 2011_02) and 255 frequently observed contaminants as well as reversed sequences of all entries and the following search criteria: (i) enzyme specificity, trypsin; (ii) mass accuracy, 6 ppm and 0.5 Da for precursor ion and fragment ion mass tolerance, respectively; (iii) fixed and variable modifications, cysteine carbamidomethylation and methionine oxidation, respectively; and (iv) maximum of two missed cleavage sites. Peptide identifications were accepted based on their posterior error probability until less than 1% reverse hits were retained, whereas protein false discovery rates were <1%. Proteins were considered if at least two peptides were identified.
In Vitro Protein-lipid Binding Assay
Expression of His-Myc-tagged FGD6 (aa 843–1398) in E. coli was induced with 0.5 mm isopropyl d-thiogalactoside for 20 h at 18 °C. The recombinant FGD6 was purified from bacterial lysate by immobilized metal (Ni2+) affinity chromatography (GE Healthcare). Nitrocellulose membranes containing spotted phosphoinositides and other lipids (phosphatidylinositol phosphate (PIP) strips, Echelon Biosciences) were blocked with 3% fatty acid-free BSA (Sigma) in TBS-T (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% Tween 20) for 1 h at room temperature. Blocked membranes were incubated overnight at 4 °C with 1 μg/ml of His-Myc-tagged fusion proteins in 3% BSA, TBS-T. The membranes were then washed four times for 10 min in TBS-T. After washing, membranes were incubated with anti-Myc antibody for 1 h at room temperature followed by additional washing and incubation with HRP-conjugated secondary antibody. After the final wash, lipid-bound fusion proteins were detected by chemiluminescence.
Liposomes were prepared by mixing chloroformic dissolutions of 574 nmol of 1,2-dioleoyl-sn-glycero-3-phosphocholine, 146 nmol of l-α-phosphatidylserine, 446 nmol of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 149 nmol of cholesterol, and 10 nmol of phosphatidylinositol diC16 (PI), 1,2-dioleoyl-sn-glycero-3-phosphoinositol-3-phosphate (PI(3)P), l-α-phosphatidylinositol 4-phosphate (PI(4)P), phosphatidylinositol 3,5-bisphosphate diC16 (PI(3,5)P2), l-α-phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) or 1,2-dioleoyl-sn-glycero-3-phosphoinositol-3,4,5-trisphosphate (PI(3,4,5)P3) in siliconized Eppendorf tubes. PI and PI(3,5)P2 were from Echelon Biosciences; otherwise all lipids were from Avanti Polar Lipids. The lipids were dried under a stream of gaseous nitrogen, resuspended in 450 μl of recruitment buffer (20 mm Hepes/KOH at pH 7.2, 125 mm potassium acetate, 2.5 mm magnesium acetate), and subjected to six rounds of freezing, thawing, and rigorous vortexing. 100 μl of liposomes were mixed with 400 μl of construct mixture, containing 7 μg of His-Myc-tagged FGD6 (aa 843–1398), 3.75% (w/v) ovalbumin (Sigma), and 1.25 mm tris(2-carboxyethyl)phosphine (Pierce). Binding of the peptides to the liposomes was performed for 1 h at 37 °C. The liposomes were pelleted for 5 min at 20,000 × g and washed once with recruitment buffer. The final pellets were dissolved in Laemmli sample buffer and used for Western blot analysis.
Statistical Analysis
All data were presented as mean ± S.D. Each experiment was repeated at least three times. The evaluation of the different assays was performed using the statistical software program GraphPad InStat version 3.10. The significance level was set at p < 0.05, and the Dunnett one-way ANOVA test was applied.
RESULTS
FGD6 Is Essential for Osteoclast Function in Bone Degradation
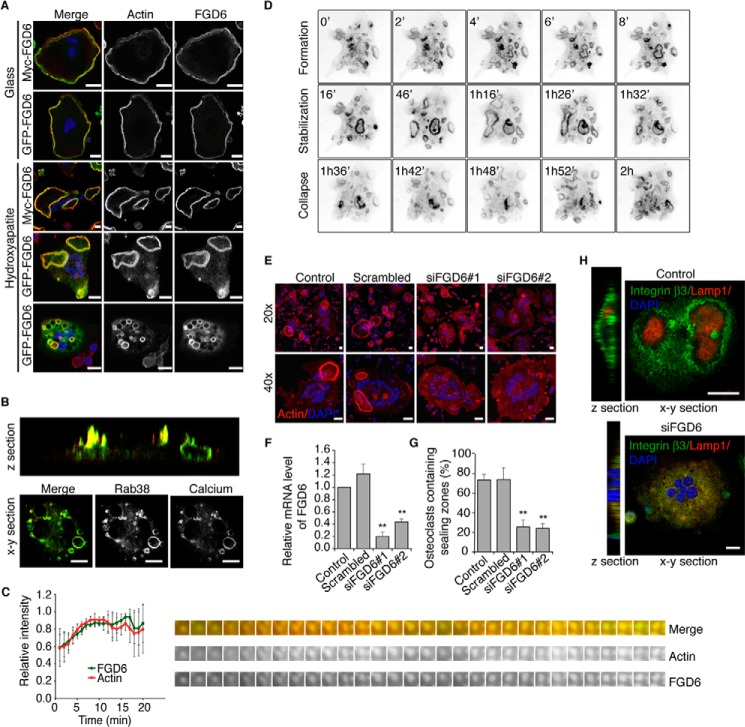
We first set out to localize FGD6 in mature osteoclasts obtained after the treatment of the murine macrophage-like Raw264.7 cells with the osteoclastogenic cytokine receptor activator of NF-κB ligand (RANKL) (18). These osteoclasts polarize onto osteological discs (ODs) (bone-mimicking surfaces made of glass coverslips coated with hydroxyapatite) and behave similarly to primary osteoclasts during cycles of bone surface degradation (supplemental Videos S1 and S2). First, we expressed GFP- or Myc-tagged FGD6 in osteoclasts using recombinant adenovirus. Expressed FGD6 localized to actin-rich podosomes/sealing zones of osteoclasts grown on ODs (Fig. 1A). FGD6 was also detected around 5–10-μm large vesicles surrounded by smaller vesicles, later shown as early endosomes (Fig. 1A). These large vesicles exhibit a high content of digested calcium indicating their trancytotic nature (Fig. 1B). In addition, the cell/tissue-specific Rab38, one of the few Rab GTPases up-regulated during osteoclastogenesis4 localized to these structures, thereby providing a new marker of transcytotic vesicles in osteoclasts. Such large vesicles are much more abundant in osteoclasts grown on ODs than in osteoclasts grown on glass indicating that their presence is linked with resorption activity (supplemental Videos S1 and S2). Time lapse microscopy showed that the dynamics of GFP-FGD6 follows that of actin at podosomes (Fig. 1C and supplemental Video S3) and remains associated with sealing zones during resorption (Fig. 1D and supplemental Video S4). Interestingly, FGD6 overexpression doubled the live time of podosomes (10 min instead of 5 min (19)). The siRNA-mediated depletion of FGD6 (up to 80% reduction, Fig. 1F) abolished the formation of podosomes and sealing zones (Fig. 1, E and G). This depletion resulted in endosomal relocalization of both integrin β3 normally enriched at sealing zones and Lamp1 normally found at the ruffled border (Fig. 1H). Altogether, these results demonstrate the essential function of FGD6 in podosome/sealing zone formation and maintenance of osteoclast polarity.
FIGURE 1.
FGD6 localizes to sealing zones and transcytotic vesicles of osteoclasts and regulates their polarity. A, intracellular distribution of Myc- or GFP-tagged FGD6 (green) expressed in osteoclasts grown on glass or ODs. Cells were stained with phalloidin (red) and DAPI (blue) and analyzed by confocal microscopy. Bars, 20 μm. B, intracellular distribution of GFP-Rab38 (green) and digested calcium (red) in osteoclasts grown on ODs. See also supplemental Videos S1 and S2. C, dynamics of GFP-FGD6 and RFP-Ezrin actin-binding domain at podosomes of osteoclasts grown on glass analyzed by time lapse video microscopy (one frame per min). Fluorescence intensities associated with GFP or RFP on single podosomes were quantified. See also supplemental Video S3. D, dynamics of GFP-FGD6 in osteoclasts grown on ODs analyzed by time lapse microscopy (one frame per min). See also supplemental Video S4. E, osteoclasts grown on ODs were either mock-transfected, or transfected with scrambled siRNAs or with siRNAs targeting FGD6. Cells were stained with phalloidin (red) and DAPI (blue) and analyzed by confocal microscopy. Bars, 20 μm. F, FGD6 expression levels were quantified by RT-PCR. G, the number of osteoclasts with sealing zones was quantified. Values are mean ± S.D. from 3 experiments (n = 300 osteoclasts per experiment). All groups were compared with control by applying a Dunnett one-way ANOVA test. *, p < 0.05. **, p < 0.01. H, distribution of integrin β3 and Lamp1 in control and FGD6-depleted osteoclasts grown on ODs. Bars, 20 μm. Representative images are shown.
Protein Domains and FGD6 Function
Beside a large N-terminal domain and a GEF domain, FGD6 contains two PH and a FYVE domain (Fig. 2A) indicating the importance of PIPs for its recruitment onto membranes. Binding assays using lipid strips and liposomes confirmed that a recombinant FGD6 truncated from its N-terminal domain mainly binds to PI(3,4)P2, PI(4,5)P2, and PI(3,4,5)P3, as expected from its two PH domains, and to PI(3)P, as expected from its FYVE domain (Fig. 2, B and C). FGD6 did not bind to other lipids. Thus, these results are in agreement with the localization of FGD6 to the PI(4,5)P2 and PI(3,4,5)P3-rich plasma membrane and to PI(3)P-rich early endosomes. Accordingly, osteoclasts treated with the pharmacological agent wortmannin, a PI 3-kinase inhibitor, previously shown to affect podosome belts (20) failed to form sealing zones as FGD6-depleted osteoclasts (Fig. 2D).
FGD6 contains several potential Src phosphorylation sites. When Src function was inhibited using the PP2 Src inhibitor, sealing zones in osteoclasts completely disappeared (Fig. 2D), and the immunoprecipitation of overexpressed GFP-FGD6 with anti-GFP showed a decrease in tyrosine phosphorylation of FGD6 (Fig. 2E). FGD6 phosphorylation on tyrosines also decreased after treatment with wortmannin (Fig. 2E). Remarkably, FGD6 truncated from its N-terminal domain was not phosphorylated on tyrosines thus suggesting that this N-terminal domain contains the Src-dependent phosphorylation sites potentially used to regulate its binding activity (Fig. 2E).
To establish the relative importance of the different FGD6 domains we expressed FGD6 mutants in osteoclasts depleted from the endogenous FGD6 by targeting its 3′ UTR mRNA with specific siRNAs (≈80% reduction, Fig. 2G). Using recombinant adenoviruses, different GFP-tagged FGD6 mutants (one lacking only the C-terminal PH domain, one lacking the C-terminal PH domain, and the FYVE domain, and one lacking all of its PH and FYVE domains (Fig. 2A)) were expressed in these FGD6-depleted osteoclasts at levels close to that of the endogenous FGD6 (Fig. 2G). Fig. 2F shows that FGD6 lacking its PH and FYVE domains was not able to induce sealing zone formation as seen in osteoclasts expressing the full-length FGD6, usually exhibiting 4–5 closed sealing zones per cell. Similarly, FGD6 without the C-terminal PH and FYVE domain could not fully rescue the wild type phenotype. These osteoclasts often exhibited a single, small sealing zone, frequently not totally closed. Apparently, the FGD6 mutant lacking its C-terminal PH domain could restore sealing zone formation. To measure the functional effectiveness of sealing zones, we measured the ability of osteoclasts expressing these FGD6 mutants to digest bone-mimicking surfaces. Fig. 2H shows that none of these mutants were able to restore a complete function of osteoclasts in degradation as seen for those expressing the full-length FGD6. Although excluding off-target effects of siRNAs, these results demonstrate that the PH and FYVE domains of FGD6 are all essential for the formation of sealing zones able to digest bone.
Identification of FGD6 Interactors
We then set out to identify FGD6 interactors using a recombinant GST-tagged FGD6 truncated from its PH and FYVE domains (aa 1–1039) as bait and an osteoclast lysate as source. In addition, we performed co-immunoprecipitation experiments using anti-GFP antibodies against the GFP-tagged FGD6 expressed in osteoclasts. FGD6 interactors were then identified by nano-LC MS/MS. The subsequent, semi-quantitative MS/MS analysis based on MS2 spectral counts identified several FGD6-interacting proteins (Table 1 and supplemental Table S1). They included the Cdc42 and Rac2 GTPases, the GTPase-activating protein (GAP) ARHGAP10 (or RhoGAP10), the Cdc42-binding actin nucleator IQGAP1, the integrin-binding and actin cross-linking proteins Talin-1 and Filamin A, several subunits of the actin nucleation promoting factor Wiskott-Aldrich Syndrome Protein and SCAR Homolog (WASH) complex, the Vps35 subunit of the retromer complex, and the actin nucleator Arp2/3.
TABLE 1.
FGD6 interactome
Recombinant FGD6 (aa 1–1039) fused to GST was expressed in E. coli. GST-FGD6 bound to glutathione beads was mixed with an osteoclast lysate, and after washing, the bound proteins were identified by MS/MS. In this table the most abundant proteins identified in this scan are seen. GFP-FGD6 was also expressed in osteoclasts and GFP-FGD6 and its bound interactors were immunoprecipitated from osteoclast lysates using anti-GFP antibodies. The eluted proteins were identified by MS/MS. The results from this scan are seen in the right column. See also supplemental Table S1.
| Protein | SwissProt accession no. | Molecular mass | Unique peptides | Sequence coverage | GFP-FGD6 co-IP |
|---|---|---|---|---|---|
| kDa | % | ||||
| GTPases | |||||
| Rac2 | Q05144 | 21.4 | 3 | 24.5 | |
| Cdc42 | P60766 | 21.3 | 12 | 85.9 | |
| GAPs | |||||
| IQGAP1 | Q9JKF1 | 188.8 | 71 | 57.6 | X |
| ARHGAP10 | Q6Y5D8 | 89.4 | 9 | 14.8 | |
| Integrin-binding proteins | |||||
| Talin-1 | P26039 | 269.8 | 57 | 39.6 | |
| Filamin A | Q8BTM8 | 281.2 | 33 | 21.8 | X |
| Actin nucleation factors (WASH complex) | |||||
| F-actin-capping protein subunit α-1 | P47753 | 32.9 | 2 | 16.8 | X |
| F-actin-capping protein subunit α-2 | P47754 | 33.0 | 1 | 10.5 | X |
| F-actin-capping protein subunit β | P47757 | 31.3 | 2 | 12.6 | |
| Actin nucleators (ARP2/3 complex) | |||||
| Actin-related protein 2 | P61161 | 44.8 | 2 | 12.4 | X |
| Actin-related protein 3 | Q99JY9 | 47.4 | 8 | 34.7 | X |
| Actin-related protein 2/3 complex subunit 1B | Q9WV32 | 41.1 | 3 | 11.0 | X |
| Actin-related protein 2/3 complex subunit 3 | Q9JM76 | 20.5 | 2 | 10.1 | X |
| Actin-related protein 2/3 complex subunit 4 | P59999 | 19.7 | 5 | 32.7 | X |
| Coat proteins | |||||
| Vps35 | Q9EQH3 | 91.7 | 4 | 9.8 | X |
FGD6 Activates Cdc42
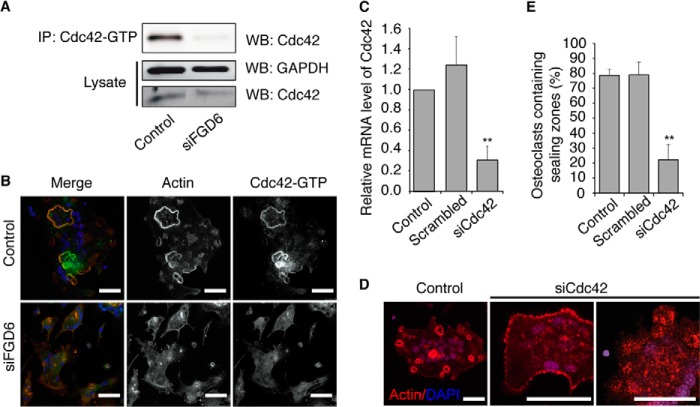
The MS analysis suggested that Cdc42 is a better FGD6 interactor than Rac2 as the amount of peptides identified and thereby also the sequence coverage was much higher for Cdc42 than for Rac2 (∼80 compared with ∼20%). This also led to a much lower posterior error probability for Cdc42 than for Rac2 (supplemental Table S1). Fig. 3A shows that indeed the amount of active, GTP-bound Cdc42 immunoprecipitated from osteoclast lysates with specific antibodies was drastically decreased upon the siRNA-mediated depletion of FGD6. Accordingly, GTP-Cdc42 was detected at the sealing zones of osteoclasts (Fig. 3B), and its siRNA-mediated depletion (≈80% knockdown efficiency, Fig. 3C) resulted in a loss of sealing zones (Fig. 3, D and E), thus mimicking FGD6 depletion. These results demonstrate that Cdc42 activation is essential for sealing zone formation and strongly argue that FGD6 is a major GEF activating Cdc42 at this location.
FIGURE 3.
FGD6 is a Cdc42 GEF. A, osteoclasts were mock-transfected or transfected with siRNAs targeting FGD6. Cell lysates were prepared and GTP-Cdc42 was immunoprecipitated with anti-GTP-Cdc42. After Western blotting (WB) immunoprecipitates (IP) and cell lysates were probed for Cdc42 and GAPDH. B, similarly treated osteoclasts were grown on ODs and stained with anti-GTP-Cdc42 (green), phalloidin (red), and DAPI (blue) and analyzed by confocal microscopy. C–E, osteoclasts were either mock-transfected, or transfected with scrambled siRNAs or with siRNAs targeting Cdc42. C, Cdc42 expression levels were quantified by RT-PCR. Values are mean ± S.D. from 3 experiments. D, similarly treated osteoclasts were grown on ODs and stained with phalloidin (red) and DAPI (blue) and analyzed by confocal microscopy. Bars, 20 μm. E, the number of osteoclasts with sealing zones was quantified. Values are mean ± S.D. from 3 experiments (n = 300 osteoclasts/experiment). All groups were compared with control by applying a Dunnett one-way ANOVA test. *, p < 0.05. **, p < 0.01. Representative images are shown.
FGD6 Regulates Sealing Zone Formation by Forming Src-dependent Protein Complexes Containing IQGAP1, ARHGAP10, Talin-1/2, or Filamin A
Our MS analysis identified a number of FGD6 interactors. To validate these interactions, we first performed co-localization studies showing that IQGAP1, ARHGAP10, Talin-1, and Filamin A localized together with FGD6 at the sealing zones of osteoclasts (Fig. 4A). Second, we performed co-immunoprecipitation experiments. The binding of FGD6 to IQGAP1 was confirmed by overexpressing GFP-FGD6 and then co-immunoprecipitating interacting components from osteoclast lysates using either anti-GFP or anti-IQGAP1 antibodies followed by Western blotting (Fig. 4B). The interaction between FGD6 and ARHGAP10 was confirmed by immunoprecipitating ARHGAP10 from osteoclast lysates with anti-ARHGAP10 antibodies followed by Western blotting to detect FGD6 (Fig. 4C). The binding of FGD6 to Talin-1 and Filamin A was confirmed by immunoprecipitating endogenous FGD6 from osteoclast lysates with anti-FGD6 antibodies followed by the detection of Talin-1 and Filamin A with the corresponding antibodies after Western blotting (Fig. 4D). Accordingly, the siRNA-mediated depletion of IQGAP1 (≈80% knockdown efficiency), Talin-1 and Talin-2 (two isoforms, ≈80% knockdown efficiency), or that of Filamin A (≈90% knockdown efficiency) (Fig. 4, E and F) resulted in the inability of osteoclasts to form sealing zones (Fig. 4, G and H), thus recapitulating FGD6 depletion. If the apparent number of sealing zones was not changed after the siRNA-mediated depletion of ARHGAP10 (≈80% knockdown efficiency, Fig. 4, E and F), they were much smaller (Fig. 4, G and H), and time lapse video microscopy showed that these sealing zones were less dynamic and had a shorter lifetime when compared with control osteoclasts (supplemental Videos S5–S8). These results demonstrate that FGD6 forms complexes with IQGAP1, AHRGAP10, Talin-1/2, or Filamin A and strongly argue that FGD6 regulates interactions between cell adhesion molecules and F-actin meshworks.
The formation of complexes containing FGD6, IQGAP1, and ARHGAP10 is regulated by Src-dependent phosphorylation. Binding of FGD6 to IQGAP1 was decreased by 60% after treatment of osteoclasts with the Src inhibitor PP2 (Fig. 4I), whereas FGD6 binding to ARHGAP10 was enhanced (a 2.3-fold increase) under those conditions (Fig. 4J). This suggests that Src regulates the activity of Cdc42 GEF and GAP in an opposite manner. These experiments demonstrate that the Src-dependent interaction of FGD6 with IQGAP1 is essential for podosome/sealing zone assembly. In contrast, ARHGAP10 appears to be a modulator of sealing zone dynamics.
FGD6 Regulates Endosomal Membrane Recycling through Interactions with WASH1
Our MS analysis of FGD6 interactors also identified several subunits of the WASH complex, which supports the retromer-dependent recycling of membrane components from early endosomes to the cell surface (21, 22). Consistent with its FYVE domain, GFP-FGD6 co-localized with EEA1, WASH1, and the Vps35 subunit of the retromer complex at early endosomes surrounding transcytotic vesicles were also labeled with GFP-FGD6 (Fig. 5, A–C), whereas GFP alone did not (data not shown). Time lapse video microscopy indicated that Rab5-positive early endosomes interact and fuse with Rab38-positive transcytotic vesicles, thus suggesting exchange of membrane components between these structures (supplemental Videos S9 and S10). Accordingly, recombinant GST-FGD6 was able to retrieve WASH1 from an osteoclast lysate (Fig. 5D) and the precipitation of endogenous FGD6 with anti-FGD6 antibodies also co-immunoprecipitated WASH1 (Fig. 5D). Interestingly, a recombinant GST-Cdc42 loaded with GTPγS was able to retrieve more efficiently WASH1 from osteoclast lysates than a GST-Cdc42 loaded with GDP (Fig. 5D). In addition, anti-Cdc42-GTP antibodies could efficiently immunoprecipitate both Cdc42 and WASH1 from lysates of control osteoclasts. This co-immunoprecipitation experiment became much less efficient (60% reduction) when FGD6 was depleted in osteoclasts (80% knockdown efficiency) (Fig. 5D). These results suggest that Cdc42-GTP, WASH1, and FGD6 belong to the same protein complex at endosomes. Accordingly, the siRNA-mediated depletion of WASH1 (≈80% knockdown efficiency) or that of Vps35 (≈70% knockdown efficiency) (Fig. 5E) resulted in the inability of osteoclasts to form sealing zones (Fig. 5, F and G). These results show that FGD6 also functions as a Cdc42-GEF at the early endosomes, where WASH triggers a Cdc42-dependent actin polymerization required for an efficient retromer-dependent recycling from early endosomes. Furthermore, they demonstrate that endosomal membrane recycling is essential for maintaining osteoclast polarity.
DISCUSSION
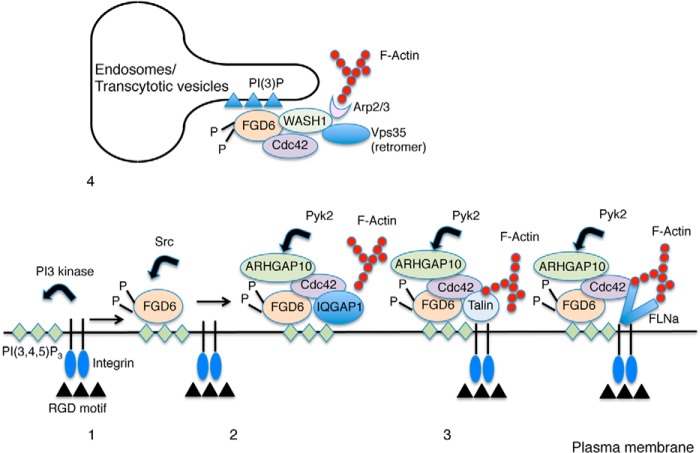
Our study demonstrates the Src-dependent function of FGD6 in coordinating cell adhesion, cell polarity, and endosomal membrane recycling in bone-digesting osteoclasts. FGD6 binds to the plasma membrane via its PH domains and coordinates podosome/sealing zone formation by forming complexes with IQGAP1, ARHGAP10, and integrin-binding proteins Talin-1/2 or Filamin A. FGD6 binds to endosomes/transcytotic vesicles via its FYVE domain and interacts with the actin nucleating promoting factor WASH, which sustains the retromer-dependent recycling of internalized membrane components. Thus, FGD6 coordinates several actin-based processes essential for osteoclast function. According to our findings, a model for FGD6 function at the plasma membrane and on endosomes/trancytotic vesicles of osteoclasts is depicted in Fig. 6.
FIGURE 6.
Model of FGD6 function in podosome dynamics and retromer-dependent recycling (1). At the plasma membrane, integrins bind to RGD motifs of extracellular bone matrix proteins. This triggers the activation of the PI 3-kinase, which produces PI(3,4,5)P3 in the vicinity of the engaged integrins (2). Upon Src-dependent phosphorylation, FGD6 binds to PI(3,4,5)P3 and potentially PI(4,5)P2, and exchanges GDP for GTP on Cdc42 (3). FGD6 interacts with IQGAP1, which binds Cdc42 and PI(3,4,5)P3. IQGAP1 can then trigger F-actin polymerization. FGD6 also interacts with Talin-1/2 or Filamin A (FLNa) and can coordinate Cdc42-dependent cell adhesion and F-actin polymerization required for podosome formation. These complexes may contain ARHGAP10 whose GAP activity is inhibited by Pyk2-dependent phosphorylation (4). On early endosomes and transcytotic vesicles, phosphorylated FGD6 binds to PI(3)P and to the WASH complex. FGD6 also exchanges GDP for GTP on Cdc42, which activates WASH. WASH triggers F-actin polymerization to sustain retromer (Vps35)-dependent recycling of membrane components.
Several GTPases including Rac1 and its exchange factor DOCK5 (23) or ARF6 and its GAP GIT2 (6) have been shown to regulate podosome/sealing zone dynamics in osteoclasts. We show that Cdc42 also regulates podosome/sealing zone dynamics, thus supporting earlier studies reporting that osteoclasts of transgenic mice overexpressing Cdc42 exhibit a higher capacity of organizing sealing zones and digesting bone (24). An interesting finding is the identification of the Cdc42-GAP ARHGAP10 in the FGD6 interactome. ARHGAP10 contains a PH domain allowing its co-localization with FGD6 at sealing zones but does not exhibit a FYVE domain consistent with its absence on endosomes. Its depletion affecting sealing zone dynamics clearly indicates its important regulatory function. A non-receptor tyrosine kinase Pyk2 inhibits its activity, thus stabilizing active Cdc42 (25). Pyk2, like Src, Syk, and the ubiquitin ligase c-Cbl, is recruited at the plasma membrane upon integrin binding to the bone matrix and regulates podosome/sealing zone dynamics (26). Integrin engagement would thus inhibit ARHGAP10 activity. On the other hand, FGD6 contains four potential Src phosphorylation sites, which regulate FGD6 activity (Fig. 6, 2). Interestingly, Src inhibition enhances FGD6 and ARHGAP10 interactions and decreases FGD6 and IQGAP1 interactions. The formation of complexes containing both GEF FGD6 and GAP ARHGAP10, whose activities and interactions are regulated by two different tyrosine kinases would provide an elegant mechanism for regulating a Cdc42-dependent cell adhesion and actin remodelling during podosome/sealing zone dynamics in osteoclasts.
At the plasma membrane, FGD6 forms complexes with the integrin-binding proteins Talin- and/or Filamin. Osteoclast adhesion is primarily mediated by the αvβ3 integrin (26, 27). Integrin binding to extracellular matrix proteins leads to an increase of PI 3-kinase activity producing PI(3,4,5)P3 (Fig. 6, 1) (28, 29). This PIP can be used as docking platforms for recruiting various proteins with phosphoinositide binding domains, in particular proteins regulating GTPase activities (30). FGD6 binds PI(3,4,5)P3, PI(3,4)P2, and PI(4,5)P2 through its PH domains and can activate Cdc42 in the vicinity of ligand-bound integrins. Upon ligand binding, integrins undergo a conformational change rendering their β subunit cytoplasmic domains accessible for binding to Talin (31, 32) or to Filamin A (Fig. 6, 3) (33), thus with the actin cytoskeleton. Our study and others (28, 34, 35) show that both Talin-1/2 and Filamin A are required for podosome and sealing zone formation. These proteins compete for the same binding site of the integrin β subunit cytoplasmic domain to allow the spatial temporal organization of different integrin-linked F-actin meshworks (36). Thus, FGD6 may regulate the interactions of integrins with different F-actin meshworks. Another FGD6 interactor is IQGAP1, which binds Cdc42 (37) and has barbed end capping activity of actin filaments (38). IQGAP1 contains an atypical phosphoinositide binding domain at its C terminus allowing its binding to PI(3,4,5)P3 (39, 40), consistent with its localization to podosomes/sealing zones of osteoclasts. Cdc42 binding to IQGAP1 induces a change in IQGAP1 conformation thereby allowing its interaction with N-WASP and the Arp2/3 complex, thus triggering actin polymerization (37). FGD6 activating Cdc42 could therefore control IQGAP1-dependent actin polymerization at podosomes. Whether IQGAP1 forms complexes with Talin-1/2 or Filamin A remains to be determined.
FGD6 binds via its FYVE domain to PI(3)P-rich early endosomes and/or transcytotic vesicles (Fig. 6, 4). FGD6 forms a complex with the actin nucleation-promoting factor WASH, a Cdc42 interactor. WASH is known to sustain the sorting activity of the retromer complex needed for recycling of endocytosed membrane components (22, 41). Both WASH and retromer are essential to maintain osteoclast polarity. We show that transcytotic vesicles are surrounded by early endosomes and these structures can fuse with each other. This may allow exchange of membrane components, as seen during phagocytosis (42), required for retrieval of membrane components to the ruffled border. Such a process could allow retrieval of lysosomal-associated membrane proteins (Lamps), vacuolar ATPases, or other proteins to maintain the integrity and function of the ruffled border. To interact with PI(3)P-rich early endosomes, transcytotic vesicles may undergo PIP turnover as seen during endocytosis (30). This PIP maturation of transcytotic vesicles may allow FGD6 to remain bound to membranes by switching from PI(3,4,5)P3, PI(4,5)P2, or PI(3,4,)P2 binding states to a PI(3)P binding state during membrane maturation. FGD6 binding to maturing trancytotic vesicles and early endosomes could regulate Cdc42- and WASH-dependent actin polymerization to sustain the retromer sorting function. Thus, endosomal membrane recycling would be essential for maintaining osteoclast polarity. Interestingly, the maintenance of cell polarity in Drosophila is also linked with the retromer-dependent membrane recycling of Crumbs (43).
FGD6 is a member of the FGD family (7, 44–46). Our MS-based proteomic approaches coupled to functional studies, as used here for FGD6, may provide an experimental pipeline for understanding the precise function of the different FGD members in bone biology and homeostasis of other tissues.
Supplementary Material
Acknowledgments
We thank the different lab members for helpful discussions and T. Pursche, J. Klemm, D. Thiel, C. Gütte, S. Lenhard, A. Engelbrecher, M. Bergert, and J. Lehmann for technical assistance. We are grateful to Alexis Gautreau for the anti-WASH1 antibodies.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants TRR 83/1-2010, TRR 83/2-2014, HO 2584/1-1, HO 2584/2-1, HO 2584/6-1, HO 2584/8-1, HO 2584/9-1, and TU-Dresden.

This article contains supplemental Table S1 and Videos S1–S10.
- PH
- pleckstrin homology
- GEF
- guanine nucleotide exchange factor
- FYVE
- domain present in Fab1, YotB, Vac1p, and EEA1
- OD
- osteologic disc
- RANKL
- receptor activator of NF-κB ligand
- PIPs
- phosphatidylinositol phosphates
- PI
- phosphatidylinositol
- PI(3)P
- phosphatidylinositol 3-phosphate
- PI(4)P
- phosphatidylinositol 4-phosphate
- PI(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PI(3,4,5)P3
- phosphatidylinositol −3,4,5-trisphosphate
- aa
- amino acid(s)
- ANOVA
- analysis of variance
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- WASH
- Wiskott-Aldrich Syndrome Protein and SCAR Homolog.
REFERENCES
- 1. Crockett J. C., Rogers M. J., Coxon F. P., Hocking L. J., Helfrich M. H. (2011) Bone remodelling at a glance. J. Cell Sci. 124, 991–998 [DOI] [PubMed] [Google Scholar]
- 2. Teitelbaum S. L. (2011) The osteoclast and its unique cytoskeleton. Ann. N.Y. Acad. Sci. 1240, 14–17 [DOI] [PubMed] [Google Scholar]
- 3. Stenbeck G., Horton M. A. (2004) Endocytic trafficking in actively resorbing osteoclasts. J. Cell Sci. 117, 827–836 [DOI] [PubMed] [Google Scholar]
- 4. Lowell C. A., Soriano P. (1996) Knockouts of Src-family kinases: stiff bones, wimpy T cells, and bad memories. Genes Dev. 10, 1845–1857 [DOI] [PubMed] [Google Scholar]
- 5. Czupalla C., Mansukoski H., Pursche T., Krause E., Hoflack B. (2005) Comparative study of protein and mRNA expression during osteoclastogenesis. Proteomics 5, 3868–3875 [DOI] [PubMed] [Google Scholar]
- 6. Heckel T., Czupalla C., Expirto Santo A. I., Anitei M., Arantzazu Sanchez-Fernandez M., Mosch K., Krause E., Hoflack B. (2009) Src-dependent repression of ARF6 is required to maintain podosome-rich sealing zones in bone-digesting osteoclasts. Proc. Natl. Acad. Sci. U.S.A. 106, 1451–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Egorov M. V., Capestrano M., Vorontsova O. A., Di Pentima A., Egorova A. V., Mariggiò S., Ayala M. I., Tetè S., Gorski J. L., Luini A., Buccione R., Polishchuk R. S. (2009) Faciogenital dysplasia protein (FGD1) regulates export of cargo proteins from the golgi complex via Cdc42 activation. Mol. Biol. Cell 20, 2413–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Estrada L., Caron E., Gorski J. L. (2001) Fgd1, the Cdc42 guanine nucleotide exchange factor responsible for faciogenital dysplasia, is localized to the subcortical actin cytoskeleton and Golgi membrane. Hum. Mol. Genet. 10, 485–495 [DOI] [PubMed] [Google Scholar]
- 9. Orrico A., Galli L., Cavaliere M. L., Garavelli L., Fryns J. P., Crushell E., Rinaldi M. M., Medeira A., Sorrentino V. (2004) Phenotypic and molecular characterisation of the Aarskog-Scott syndrome: a survey of the clinical variability in light of FGD1 mutation analysis in 46 patients. Eur. J. Hum. Genet. 12, 16–23 [DOI] [PubMed] [Google Scholar]
- 10. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo J., Deng Z. L., Luo X., Tang N., Song W. X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., He T. C. (2007) A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 12. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−[Delta][Delta] CT) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 13. Yamaki M., Nakamura H., Takahashi N., Udagawa N., Ozawa H. (2005) Transcytosis of calcium from bone by osteoclast-like cells evidenced by direct visualization of calcium in cells. Arch. Biochem. Biophys. 440, 10–17 [DOI] [PubMed] [Google Scholar]
- 14. Kartner N., Yao Y., Li K., Crasto G. J., Datti A., Manolson M. F. (2010) Inhibition of osteoclast bone resorption by disrupting vacuolar H+-ATPase a3-B2 subunit interaction. J. Biol. Chem. 285, 37476–37490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pellegrin S., Mellor H. (2008) Rho GTPase activation assays. Curr. Protoc. Cell Biol. Chapter 14, Unit 14.8 [DOI] [PubMed] [Google Scholar]
- 16. Czupalla C., Mansukoski H., Riedl T., Thiel D., Krause E., Hoflack B. (2006) Proteomic analysis of lysosomal acid hydrolases secreted by osteoclasts: implications for lytic enzyme transport and bone metabolism. Mol. Cell Proteomics 5, 134–143 [DOI] [PubMed] [Google Scholar]
- 17. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 18. Boyle W. J., Simonet W. S., Lacey D. L. (2003) Osteoclast differentiation and activation. Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 19. Luxenburg C., Winograd-Katz S., Addadi L., Geiger B. (2012) Involvement of actin polymerization in podosome dynamics. J. Cell Sci. 125, 1666–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakamura I., Takahashi N., Sasaki T., Tanaka S., Udagawa N., Murakami H., Kimura K., Kabuyama Y., Kurokawa T., Suda T. (1995) Wortmannin, a specific inhibitor of phosphatidylinositol-3 kinase, blocks osteoclastic bone resorption. FEBS Lett. 361, 79–84 [DOI] [PubMed] [Google Scholar]
- 21. Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D., Gautreau A. (2009) The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 [DOI] [PubMed] [Google Scholar]
- 22. Gomez T. S., Billadeau D. D. (2009) A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vives V., Laurin M., Cres G., Larrousse P., Morichaud Z., Noel D., Côté J. F., Blangy A. (2011) The Rac1 exchange factor Dock5 is essential for bone resorption by osteoclasts. J. Bone Miner. Res. 26, 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ito Y., Teitelbaum S. L., Zou W., Zheng Y., Johnson J. F., Chappel J., Ross F. P., Zhao H. (2010) Cdc42 regulates bone modeling and remodeling in mice by modulating RANKL/M-CSF signaling and osteoclast polarization. J. Clin. Invest. 120, 1981–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ren X. R., Du Q. S., Huang Y. Z., Ao S. Z., Mei L., Xiong W. C. (2001) Regulation of CDC42 GTPase by proline-rich tyrosine kinase 2 interacting with PSGAP, a novel pleckstrin homology and Src homology 3 domain containing rhoGAP protein. J. Cell Biol. 152, 971–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zou W., Teitelbaum S. L. (2010) Integrins, growth factors, and the osteoclast cytoskeleton. Ann. N.Y. Acad. Sci. 1192, 27–31 [DOI] [PubMed] [Google Scholar]
- 27. McHugh K. P., Hodivala-Dilke K., Zheng M. H., Namba N., Lam J., Novack D., Feng X., Ross F. P., Hynes R. O., Teitelbaum S. L. (2000) Mice lacking β3 integrins are osteosclerotic because of dysfunctional osteoclasts. J. Clin. Invest. 105, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chellaiah M. A., Biswas R. S., Yuen D., Alvarez U. M., Hruska K. A. (2001) Phosphatidylinositol 3,4,5-trisphosphate directs association of Src homology 2-containing signaling proteins with gelsolin. J. Biol. Chem. 276, 47434–47444 [DOI] [PubMed] [Google Scholar]
- 29. Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A. (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 30. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 31. Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., Ginsberg M. H., Calderwood D. A. (2003) Talin binding to integrin β tails: a final common step in integrin activation. Science 302, 103–106 [DOI] [PubMed] [Google Scholar]
- 32. Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A. A., McLean M. A., Sligar S. G., Taylor K. A., Ginsberg M. H. (2010) Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 188, 157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim H., McCulloch C. A. (2011) Filamin A mediates interactions between cytoskeletal proteins that control cell adhesion. FEBS Lett. 585, 18–22 [DOI] [PubMed] [Google Scholar]
- 34. Leung R., Wang Y., Cuddy K., Sun C., Magalhaes J., Grynpas M., Glogauer M. (2010) Filamin A regulates monocyte migration through Rho small GTPases during osteoclastogenesis. J. Bone Miner. Res. 25, 1077–1091 [DOI] [PubMed] [Google Scholar]
- 35. Marzia M., Chiusaroli R., Neff L., Kim N. Y., Chishti A. H., Baron R., Horne W. C. (2006) Calpain is required for normal osteoclast function and is down-regulated by calcitonin. J. Biol. Chem. 281, 9745–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kiema T., Lad Y., Jiang P., Oxley C. L., Baldassarre M., Wegener K. L., Campbell I. D., Ylänne J., Calderwood D. A. (2006) The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell 21, 337–347 [DOI] [PubMed] [Google Scholar]
- 37. Brandt D. T., Grosse R. (2007) Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep. 8, 1019–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pelikan-Conchaudron A., Le Clainche C., Didry D., Carlier M. F. (2011) The IQGAP1 protein is a calmodulin-regulated barbed end capper of actin filaments: possible implications in its function in cell migration. J. Biol. Chem. 286, 35119–35128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dixon M. J., Gray A., Schenning M., Agacan M., Tempel W., Tong Y., Nedyalkova L., Park H. W., Leslie N. R., van Aalten D. M., Downes C. P., Batty I. H. (2012) IQGAP proteins reveal an atypical phosphoinositide (aPI) binding domain with a pseudo C2 domain fold. J. Biol. Chem. 287, 22483–22496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dixon M. J., Gray A., Boisvert F. M., Agacan M., Morrice N. A., Gourlay R., Leslie N. R., Downes C. P., Batty I. H. (2011) A screen for novel phosphoinositide 3-kinase effector proteins. Mol. Cell. Proteomics 10.1074/mcp.M110.003178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Duleh S. N., Welch M. D. (2010) WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton 67, 193–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fairn G. D., Grinstein S. (2012) How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 33, 397–405 [DOI] [PubMed] [Google Scholar]
- 43. Pocha S. M., Wassmer T., Niehage C., Hoflack B., Knust E. (2011) Retromer controls epithelial cell polarity by trafficking the apical determinant Crumbs. Curr. Biol. 21, 1111–1117 [DOI] [PubMed] [Google Scholar]
- 44. Huber C., Mårtensson A., Bokoch G. M., Nemazee D., Gavin A. L. (2008) FGD2, a CDC42-specific exchange factor expressed by antigen-presenting cells, localizes to early endosomes and active membrane ruffles. J. Biol. Chem. 283, 34002–34012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ono Y., Nakanishi H., Nishimura M., Kakizaki M., Takahashi K., Miyahara M., Satoh-Horikawa K., Mandai K., Takai Y. (2000) Two actions of frabin: direct activation of Cdc42 and indirect activation of Rac. Oncogene 19, 3050–3058 [DOI] [PubMed] [Google Scholar]
- 46. Pasteris N. G., Nagata K., Hall A., Gorski J. L. (2000) Isolation, characterization, and mapping of the mouse Fgd3 gene, a new faciogenital dysplasia (FGD1; Aarskog Syndrome) gene homologue. Gene 242, 237–247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.