Background: Regulation of NHE3 by ubiquitination has not been reported.
Results: Human NHE3, but not non-primates NHE3s, is ubiquitinated by Nedd4-2 and undergoes internalization at an increased rate.
Conclusion: Human NHE3 is uniquely regulated by ubiquitination.
Significance: This study provides a new mechanism of regulating NHE3 in human that may be relevant to diseases associated with increased Na+ and fluid absorption.
Keywords: Endocytosis, Hypertension, Protein-Protein Interaction, Sodium-Proton Exchange, Ubiquitin
Abstract
Na+/H+ exchanger NHE3 expressed in the intestine and kidney plays a major role in NaCl and HCO3− absorption that is closely linked to fluid absorption and blood pressure regulation. The Nedd4 family of E3 ubiquitin ligases interacts with a number of transporters and channels via PY motifs. A comparison of NHE3 sequences revealed the presence of PY motifs in NHE3s from human and several non-human primates but not in non-primate NHE3s. In this study we evaluated the differences between human and non-primate NHE3s in ubiquitination and interaction with Nedd4-2. We found that Nedd4-2 ubiquitinated human NHE3 (hNHE3) and altered its expression and activity. Surprisingly, rat NHE3 co-immunoprecipitated Nedd4-2, but its expression and activity were not altered by silencing of Nedd4-2. Ubiquitination by Nedd4-2 rendered hNHE3 to undergo internalization at a significantly greater rate than non-primate NHE3s without altering protein stability. Insertion of a PY motif in rabbit NHE3 recapitulated the interaction with Nedd4-2 and enhanced internalization. Thus, we propose a new model where disruption of Nedd4-2 interaction elevates hNHE3 expression and activity.
Introduction
Na+/H+ exchanger 3 (NHE3)2 is an integral protein expressed in the brush border membrane (BBM) of the epithelial cells in the small intestine, colon, and kidney where it conducts a bulk of Na+ and HCO3− absorption (1, 2). NHE3 is regulated by various intrinsic and extrinsic stimuli, including parathyroid hormone, glucocorticoids, serotonin, dopamine, angiotensin II, and bacterial toxin (3–7). NHE3 is present not only in BBM but also in intracellular vesicles so that the expression level of NHE3 in BBM is balanced by the rate of insertion from the intracellular pool and retrieval of NHE3 protein from the BBM. Inhibition of NHE3 induces endocytosis of NHE3, which is dependent on clathrin, dynamin, or lipid rafts (3, 8, 9). Similarly, simulation of NHE3 activity involves exocytic trafficking of NHE3 that increases NHE3 abundance at the plasma membrane (PM) (10–12). In addition, small GTPases, Na+/H+ exchanger regulatory factor 1 (NHERF1), NHERF2, megalin, and actin have been implicated in NHE3 trafficking (13–16).
Ubiquitination is a regulated post-translational modification that conjugates ubiquitin (Ub) to Lys residues of target proteins and controls their intracellular fate. The covalent ligation of the 76-amino acid peptide Ub to substrate proteins is a highly conserved process that occurs via sequential actions of three enzymes: Ub-activation enzyme E1, Ub-conjugating enzyme E2, and Ub ligase E3. E3 ligases play a pivotal role in ubiquitination because they recognize the acceptor proteins and, hence, dictate the high specificity of the reaction. The presence of multiple E3s in nature suggests their non-redundant roles (17). A protein can be mono-ubiquitinated, multi-ubiquitinated, or poly-ubiquitinated, and the type of ubiquitination determines the fate of the protein. The canonical role of ubiquitination is to mediate protein degradation, but ubiquitination modulates much broader and diverse functions involved in a myriad of cellular processes (17, 18).
Nedd4-1 (neural precursor cell expressed developmentally down-regulated 4-1) and Nedd4-2 belong to the Nedd4 family of E3 Ub ligases, which have an N-terminal C2 domain, three to four WW domains, and a C-terminal HECT domain (19, 20). Nedd4 proteins typically interact with protein substrates through a PY (PPXY) motif. Despite the similarity in their structures, Nedd4-1 and Nedd4-2 show distinct substrate preferences. Nedd4-1 preferentially targets tumor suppressors and membrane receptors, whereas Nedd4-2 ubiquitinates ion channels and transporters, including the epithelial Na+ channel (ENaC), cystic fibrosis transmembrane conductance regulator, and voltage-gated K+ channel (19–21). The importance of ubiquitination of a transport protein is underscored by a series of studies on ENaC. Nedd4-2 interacts with ENaC via the PY motifs in the C terminus of ENaC α, β, and γ subunits (22, 23). Mutations in PY motifs in ENaC block the interaction with Nedd4-2, preventing ubiquitination-dependent internalization of ENaC and thereby increasing ENaC expression in PM as found in Liddle's syndrome (22, 23). In keeping with this model, deletion of Nedd4-2 gene in mice resulted in elevated blood pressure and cardiac hypertrophy with increased ENaC expression (24).
Endocytic process is a crucial process for NHE3 regulation, but the status of NHE3 ubiquitination and whether NHE3 is regulated by ubiquitination have not been reported. In this study we show that NHE3s of human and several non-human primates, but not non-primate mammals, uniquely contain PY motifs. The PY motif in human NHE3 (hNHE3) mediates the interaction with Nedd4-2, which modulates cell surface expression and internalization of hNHE3.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
PS120 fibroblasts were cultured in DMEM supplemented with 1 mm sodium pyruvate, 50 units/ml penicillin, 50 μg/ml streptomycin, and 10% fetal bovine serum in a 5% CO2 humidified incubator at 37 °C. PS120 fibroblasts stably expressing rabbit (rb) NHE3 or opossum kidney NHE3 have been previously described (7). SK-CO15 and Caco-2bbe/rbNHE3V cells were cultured in DMEM supplemented with 10% FBS, penicillin, streptomycin, 1 mm sodium pyruvate, 15 mm HEPES, and 1× nonessential amino acids as previously described (25). Transfection was performed using Lipofectamine 2000 (Invitrogen) or the Neon Transfection System (Invitrogen).
Plasmids
The pcDNA3.1 constructs carrying hNHE3 or rbNHE3 fused at the C terminus with an antibody epitope derived from vesicular stomatitis virus glycoprotein (VSVG) have previously been described (15, 26). Rat NHE3 cDNA was kindly provided by John Orlowski (McGill University) (27). Triple HA tagged-rbNHE3 (3×HA-rbNHE3) and hNHE3 were gifts of Mark Donowitz (Johns Hopkins). Triple FLAG tag was cloned between amino acids 41 and 42 of hNHE3 by PCR to generate 3×FLAG-hNHE3. Point mutations in hNHE3 were made using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Englewood, CO). The presence of mutation was confirmed by nucleotide sequencing. C-terminal truncation constructs of hNHE3 were generated by PCR. Nedd4-1 and Nedd4-2 were gifts of Peter Snyder (University of Iowa). An HA tag was cloned at the N terminus to generate pcDNA/HA-Nedd4-1 and pcDNA/HA-Nedd4-2. Plasmid pMT123 encoding HA-Ub was a kind gift of Dirk Bohmann (University of Rochester). The lentiviral plasmid pLKO.1 containing short hairpin RNA (shRNA) targeting Nedd4-2, shNedd4-2, and non-targeting pLKO.1 containing a scrambled shRNA, control shRNA (shCon), were obtained from Sigma.
Antibodies
Rabbit polyclonal anti-NHE3 antibody EM450 and mouse monoclonal anti-VSVG P5D4 were described previously (25, 28). The following commercially obtained antibodies were used: mouse anti-HA (Santa Cruz, Dallas, TX), mouse anti-Ub P4D1 (Santa Cruz), mouse anti-FLAG (Sigma), mouse anti-β-actin (Cell Signaling, Danvers, MA), Alexa Fluor 488-conjugated mouse anti-HA and rabbit anti-FLAG antibodies (Cell Signaling), Alexa Fluor 647-conjugated wheat germ agglutinin (Invitrogen), and rabbit anti-β-tubulin (Abcam, Cambridge, MA).
Quantitative RT-PCR
Total RNA from PS120 or SK-CO15 cells was isolated using TRIzol (Invitrogen), and 2 μg of RNA was subsequently used to synthesize cDNA using the SuperScript III First Strand Synthesis kit as recommended by the manufacturer (Invitrogen). cDNA was amplified by using SYBR Green Supermix (Bio-Rad) on Mastercycler Realplex (Eppendorf, Hauppauge, NY). For a typical reaction, the reaction mixture consisted of 1 μl of cDNA, 10 μl of SYBR Green Supermix, and 0.5 μm target primers in a total volume of 20 μl. qPCR was initiated with pre-denaturation at 95 °C for 4 min followed by 40 cycles of amplification at 95 °C for 30 s (denaturation), 60 °C for 30 s (annealing), and 72 °C for 1 min (extension) as previously described (25). The primers used are as follows: Nedd4-2, human: 5′-GGGTGGTGAGGAACCAACG-3′ and 5′-TGATAGGTCGAGTCCAAGTTGT-3′; Nedd4-2, Chinese hamster PS120: 5′-CATGTGTGGTCTGGGAGATG-3′ and 5′-TGTGCCGGTAACAAATTGAA-3′. The amounts of Nedd4-2 mRNA were normalized to β-actin.
NHE3 Activity
The Na+-dependent changes in intracellular pH (pHi) by NHE3 was determined using the ratio-fluorometric, pH-sensitive dye 2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein acetoxymethyl ester (BCECF-AM) as described previously (7). Briefly, cells were washed in Na+ buffer (130 mm NaCl, 20 mm HEPES, 5 mm KCl, 1 mm tetramethylammonium (TMA)-PO4, 2 mm CaCl2, 1 mm MgSO4, and 25 mm glucose) and then dye-loaded by incubating with 6.5 μm BCECF-AM in the same solution for 10 min. The coverslips were mounted on a perfusion chamber mounted on an inverted microscope and superfused with NH4+ buffer (40 mm NH4Cl, 90 mm NaCl, 20 mm HEPES, 5 mm KCl, 1 mm TMA-PO4, 2 mm CaCl2, 1 mm MgSO4, and 25 mm glucose) for 5 min followed by perfusion with TMA+ buffer (130 mm TMA-Cl, 20 mm HEPES, 5 mm KCl, 1 mm TMA-PO4, 2 mm CaCl2, 1 mm MgSO4, and 25 mm glucose) for 2 min and subsequently with Na+ buffer. When necessary, Na+ buffer was supplemented with 50 μm HOE694 to inhibit NHE1 and NHE2 activities. The rate of Na+-dependent pH recovery was calculated by determining slopes along the early stage of pH recovery by linear least-squares analysis over a minimum of 9 s. Calibration of the fluorescence signal was performed using the K+/H+ ionophore nigericin as previously described (7). Comparisons of Na+/H+ exchange were made between measurements made on the same day. The microfluorometry was performed on an inverted fluorescence microscope, and photometric data were acquired using the Metafluor software (Molecular Devices, Sunnyvale, CA).
Detection of NHE3 Ubiquitination
Cells were lysed in cold lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm β-glycerophosphate, 2.5 mm sodium pyrophosphate, 1 mm Na2EDTA, 1 mm EGTA, 1 mm Na3VO4, 1 mm NaF, 10 mm leupeptin, 1% Triton X-100, protease inhibitors mixture, and 2.5 mm N-ethylmaleimide supplemented with 10 μm ALLN and 10 μm MG132 to inhibit proteasomal degradation. Protein concentration was determined by the bicinchoninic acid (BCA) assay (Sigma). Equal amounts of cell lysates (typically 300 μg) were incubated overnight with anti-VSVG, anti-FLAG, or EM450 antibody to immunoprecipitate NHE3. The immunocomplex was purified by incubating with 80 μl of protein A-Sepharose beads for 1 h followed by 3 washes in lysis buffer and 2 washes in PBS. NHE3-containing immunocomplex was eluted from the beads in 1× Laemmli buffer, resolved by SDS-PAGE, and immunoblotted with anti-HA or P4D1 antibody.
Surface Biotinylation
Surface biotinylation of NHE3 was performed as described (11). Briefly, cells were rinsed twice in PBS and incubated in borate buffer (154 mm NaCl, 7.2 mm KCl, 1.8 mm CaCl2, and 10 mm H3BO3, pH 9.0) for 10 min. Cells were then incubated for 40 min with 0.5 mg/ml sulfo-NHS-LC-biotin (Pierce) in borate buffer. Unbound sulfo-NHS-LC-biotin was quenched with Tris buffer (20 mm Tris, 120 mm NaCl, pH 7.4). Cells were lysed in lysis buffer and sonicated for 2 × 15 s. Lysate was agitated for 30 min and spun at 14,000 × g for 30 min at 4 °C to remove the insoluble cell debris. An aliquot was retained as the total fraction representing the total cellular NHE3. Protein concentration was determined, and 1 mg of lysate was then incubated with streptavidin-agarose beads (Pierce) for 2 h. The streptavidin-agarose beads were washed three times in lysis buffer and twice in PBS. All the above procedures were performed at 4 °C or on ice. Biotinylated surface proteins were then eluted by boiling the beads at 95 °C for 10 min. Dilutions of the total and surface NHE3 were resolved by SDS-PAGE and immunoblotted with an anti-VSVG antibody. Sequential cell surface biotinylation and immunoprecipitation were performed using monomeric avidin-agarose beads (Pierce), which allow elution of biotinylated proteins by excess free biotin as described (29). Densitometric analysis was performed using ImageJ software (National Institutes of Health).
NHE3 Internalization
Cells grown in their normal media were biotinylated using the membrane impermeable Sulfo-NHS-SS-Biotin (0.15 mg/ml, Pierce) in PBS for 10 min on ice. Labeled cells were washed 3 times with ice-cold PBS to remove unlabeled biotin and overlaid with DMEM preheated to 37 °C to initiate internalization. At each selected time point the cells were moved to 4 °C to halt internalization, and the remaining biotin on the cell surface was stripped with GSH buffer containing the membrane-impermeable reducing agent glutathione (GSH) (50 mm GSH, 75 mm NaOH, 75 mm NaCl, 1 mm EDTA, 0.1% BSA, pH 9) 2 times for 20 min each at 4 °C. GSH was neutralized with iodoacetamide (10 mm) in PBS. Cells were then rinsed with PBS, scraped, lysed in the lysis buffer described above, and sonicated for 2 × 15 s. Lysate was agitated for 30 min and spun at 14,000 × g for 30 min at 4 °C to remove insoluble cell debris. Supernatants containing equal amounts of protein were incubated with streptavidin beads to pull down the remaining biotinylated proteins. After extensive washes in lysis buffer, proteins were eluted from the streptavidin beads by boiling in reducing sample buffer. Eluted proteins were resolved by SDS-PAGE and immunoblotted as described (7).
NHE3 Half-life
SK-CO15 cells grown to 70–80% confluence were pretreated with 20 μm cycloheximide (Sigma). At each selected time point, cells were lysed in lysis buffer. Equal amounts of cell lysates were resolved by SDS-PAGE and immunoblotted with EM450 as described above. NHE3 expression was normalized to β-tubulin. Densitometry analysis was performed using ImageJ software (National Institutes of Health).
Immunofluorescence Imaging of Internalized NHE3
PS120 cells expressing 3×HA-rbNHE3, 3×FLAG-hNHE3, or truncated 3×FLAG-hNHE3 were infected with shNedd4-2 or shCon. Cells were rinsed twice with warm Hanks' balanced salt solution (HBSS) followed by rocking incubation with Alexa Fluor 488-conjugated mouse anti-HA or rabbit anti-FLAG antibody in HBSS at room temperature. After 30 min cells were rinsed and subjected to either direct fixation with 4% paraformaldehyde or kept at 37 °C for 3 h before fixation. Surfaces of the fixed cells were labeled with Alexa Fluor 647-conjugated wheat germ agglutinin. After three 10-min washes with PBS, the specimens were mounted with ProLong Gold Antifade Reagent (Invitrogen) and observed under a Zeiss LSM510 laser confocal microscope (Zeiss Microimaging, Thornwood, NY) coupled to a Zeiss Axioplan2e with ×60 Pan-Apochromat oil lenses.
Statistical Analysis
Statistical significance was determined by a paired t test. Data were expressed as the means ± S.E. A p value of <0.05 was considered significant.
RESULTS
Nedd4-2 Interacts with Human and Rat NHE3
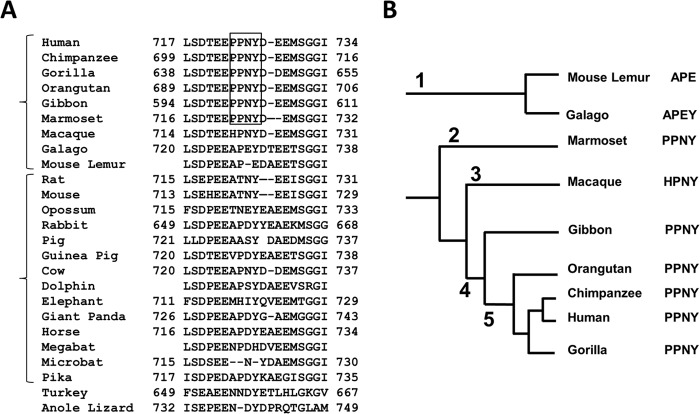
Because it was noted previously that hNHE3 has a PY motif (30), we aligned NHE3 sequences from 25 species (Fig. 1A). The analysis revealed that PY motifs are present in NHE3s of human and non-human primates but absent in NHE3s of 14 non-primate mammals, 1 bird, and 1 reptile. A phylogenetic tree of primates (Fig. 1B) shows that the primates of the Hominoidea family, which includes human, chimpanzee, gorilla, and closely related gibbon, have PY motifs in their NHE3s. The NHE3 of marmoset of the New World monkey family contains a PY motif. On the other hand, NHE3s of more distantly related mouse lemur and galago (also known as bushbabies) of the Prosimian suborder of primates and macaques of the Old World monkeys lack PY motifs.
FIGURE 1.
Alignment of NHE3 sequences. A, alignment of partial NHE3 sequences from 25 species is shown. PY motifs are enclosed by a square. The numbers indicate the positions of the amino acid residues. B, a phylogenetic tree of primates is shown. 1, Prosimians; 2, New World monkeys; 3, Old World monkeys; 4, Hominoidea; 5, Hominidae.
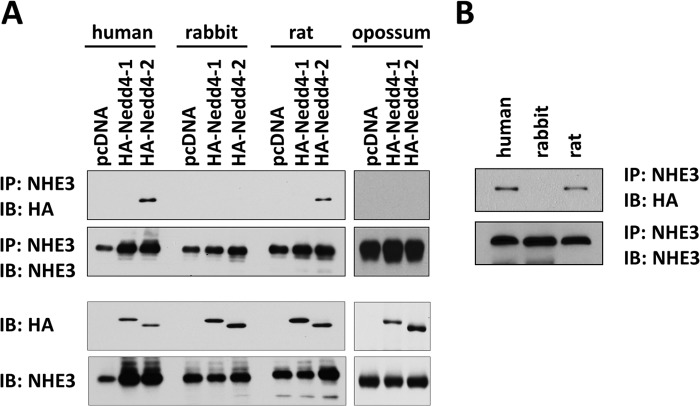
Because the presence of PY motif in NHE3 is limited to selected species, we compared whether there is a difference in the interaction with Nedd4 proteins among several NHE3s. We transiently co-expressed HA-Nedd4-1 or HA-Nedd4-2 and VSVG epitope-tagged NHE3 in HEK293 cells, and their interaction was assessed by immunoprecipitation of NHE3 followed by Western blot with anti-HA antibody. As illustrated in Fig. 2A, Nedd4-2 was co-immunoprecipitated with hNHE3 but not with rbNHE3 or opossum kidney NHE3. Surprisingly, rat NHE3 co-immunoprecipitated HA-Nedd4-2, although the association was reduced compared with hNHE3. None of the NHE3s interacted with Nedd4-1. To ensure the difference in Nedd4-2 interaction was not influenced by the use of a human cell line, co-immunoprecipitation of NHE3 and Nedd4-2 was performed in the Chinese hamster lung fibroblast PS120 cell line. Consistently, HA-Nedd4-2 was detected in anti-VSVG immunoprecipitates (Fig. 2B). Because ratNHE3 lacks a PY motif, these results could not assure the importance of PY motif in mediating interaction with Nedd4-2.
FIGURE 2.
Interaction of NHE3s with Nedd4 proteins. A, NHE3s containing a VSVG tag were transiently expressed in HEK293 cells along with HA-Nedd4-1 or HA-Nedd4-2. NHE3 was immunoprecipitated (IP) using VSVG antibody followed by immunoblotting (IB) using anti-HA antibody to detect co-immunoprecipitated HA-Nedd4-1 or HA-Nedd4-2. 300 μg of cell lysates were used for immunoprecipitation. The bottom two panels show relative expression levels of HA-Nedd4-1, HA-Nedd4-2, and NHE3 in the cell lysate. B, PS120 fibroblasts stably expressing human, rabbit, or rat NHE3 were transiently transected with pcDNA/HA-Nedd4-2. NHE3 was immunoprecipitated with anti-VSVG antibody, and co-immunoprecipitated Nedd4-2 was detected by anti-HA antibody. Representative figures from three independent experiments are shown.
Nedd4-2 Regulates hNHE3 Ubiquitination
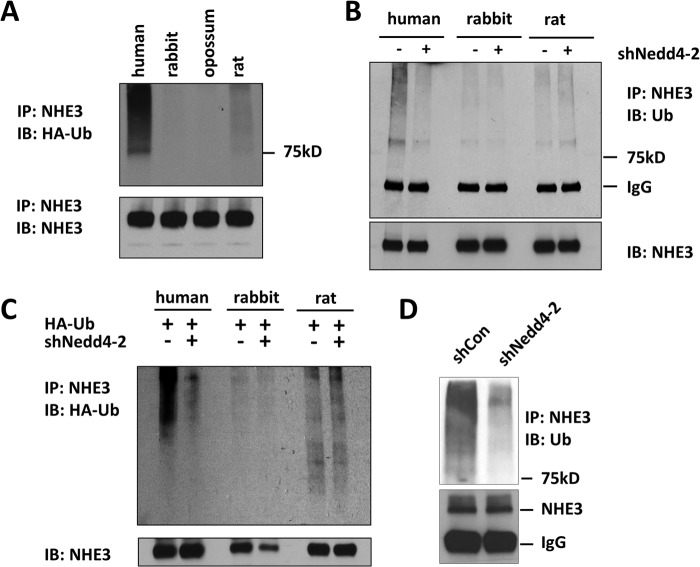
NHE3 constitutively recycles between PM and intracellular pools, but there is no convincing evidence that NHE3 is ubiquitinated. Hence, before determining the role of Nedd4-2 on NHE3 regulation, we sought to determine the ubiquitination status of NHE3 proteins. To this end, we expressed HA-Ub in NHE3-expressing PS120 fibroblasts. We did not transfect HA-Nedd4-2 in PS120 cells as these cells endogenously express Nedd4-2 mRNA. Fig. 3A shows that hNHE3 displayed constitutive ubiquitination that was detected as prominent high molecular mass species upward of 78 kDa, the observed molecular mass of NHE3. The ubiquitination level of ratNHE3 was significantly lower compared with hNHE3, whereas ubiquitination of rabbit and opossum NHE3 was visible only when the blot was overexposed.
FIGURE 3.
Nedd4-2 regulates ubiquitination of hNHE3. A, PS120 cells fibroblasts expressing human, rabbit, opossum, or rat NHE3 were transiently transfected with HA-Ub. NHE3 was immunoprecipitated (IP) using VSVG antibody, and the levels of NHE3 ubiquitination were determined by immunoblotting (IB) with a HA-Ub antibody. B, PS120/NHE3 cells were transfected with shCon or shNedd4-2. NHE3 was immunoprecipitated (lower panel) with anti-VSVG antibody followed by immunoblot using P4D1 to detect NHE3 ubiquitination (upper panel). Data are representative of at least three experiments. C, PS120/NHE3 cells were transiently transfected with HA-Ub and lentiviral shNedd4-2. shCon was used as a control. Ubiquitination of immunoprecipitated NHE3 was determined using anti-HA antibody. The bottom panel shows the total amounts of immunoprecipitated NHE3. D, ubiquitination of hNHE3 in SK-CO15 cells transfected with shCon or shNedd4-2 was determined.
To determine whether NHE3 ubiquitination is dependent on the presence of Nedd4-2, we silenced Nedd4-2 expression using lentiviral shRNA targeting Nedd4-2, shNedd4-2. Treatment of PS120 cells with shNedd4-2 decreased Nedd4-2 mRNA expression by 66% compared with shCon (not shown). Importantly, shNedd4-2 resulted in a significant decrease in hNHE3 ubiquitination levels detected by mouse anti-Ub antibody, P4D1, which recognizes both mono- and poly-Ub (Fig. 3B). On the contrary, shNedd4-2 did not significantly alter ubiquitination of ratNHE3 or rbNHE3. The lack of effect on ratNHE3 ubiquitination was surprising in light of its interaction with Nedd4-2. To confirm these unexpected results, a similar experiment was performed using cells transfected with HA-Ub. Consistent with the above results, ubiquitination level of hNHE3, but not that of rbNHE3 or ratNHE3, was decreased by silencing of Nedd4-2 (Fig. 3C). The lack of an effect in rbNHE3 and ratNHE3 was not due to underexposure of the gel as longer exposure of the gel did not reveal different results. To determine whether Nedd4-2 regulates ubiquitination of endogenously expressed hNHE3, we used the human colonic epithelial SK-CO15 cells that express NHE3 (25). Fig. 3D shows that shNedd4-2, which decreased the Nedd4-2 mRNA level by 75% (not shown), markedly lowered the hNHE3 ubiquitination level compared with shCon. Together, these results show that hNHE3 ubiquitination is uniquely regulated by Nedd4-2. Moreover, despite its apparent interaction with Nedd4-2, ratNHE3 ubiquitination is contrastingly lower than hNHE3 and is independent of the Nedd4-2 presence.
Silencing of Nedd4-2 Alters hNHE3 Activity
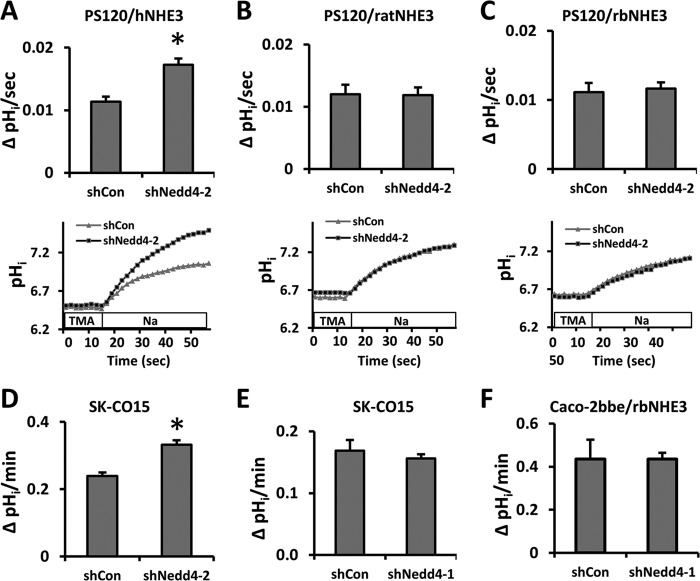
We next determined whether Nedd4-2 modulates NHE3 activity, which is defined as Na+-dependent pHi recovery. NHE3 activity was determined in PS120 cells treated with shCon or shNedd4-2. Nedd4-2 knockdown resulted in a significant increase in hNHE3 activity (Fig. 4A) but did not alter the activity of ratNHE3 or rbNHE3 compared with shCon-transfected cells (Fig. 4, B and C). The effect of Nedd4-2 knockdown on hNHE3 activity was confirmed in SK-CO15 cells where elevated NHE3 activity was observed (Fig. 4D).
FIGURE 4.
The activity of hNHE3 is stimulated by Nedd4-2 knockdown. The effect of Nedd4-2 knockdown on NHE3 activity was determined on PS120/hNHE3 (A), PS120/ratNHE3 (B), and PS120/rbNHE3 (C) cells. NHE3 activity was determined fluorometrically using the intracellular pH-sensitive dye BCECF-AM as described under “Experimental Procedures.” The cells were incubated in NH4+, washed with TMA, and Na+-dependent pH recovery was initiated by addition of Na+ buffer. NHE3 activity was expressed as the rate of Na+-dependent intracellular pH recovery (upper panel), and representative traces of Na-dependent pHi recovery are shown below. Results are presented as the means ± S.E. n ≥ 9. *, p < 0.01 compared with shCon. The rates of Na+-dependent pHi recovery were determined in SK-CO15 cells transfected with shNedd4-2 (D) or shNedd4-1 (E). F, NHE3 activity in Caco-2bbe/rbNHE3 cells transfected with shNedd4-1 or shCon was determined. n ≥ 9.
It was shown recently that NHE1 indirectly interacts with Nedd4-1 via β-arrestin (29). Hence, we examined the effect of silencing Nedd4-1 on NHE3 activity. Knockdown of Nedd4-1 did not affect hNHE3 activity in SK-CO15 cells (Fig. 4E). Similarly, we did not observe any effect of shNedd4-1 on rbNHE3 activity in Caco-2bbe cells (Fig. 4F) eliminating the possibility that rbNHE3 is indirectly regulated by Nedd4-1.
Knockdown of Nedd4-2 Alters hNHE3 Surface Expression and Ubiquitination
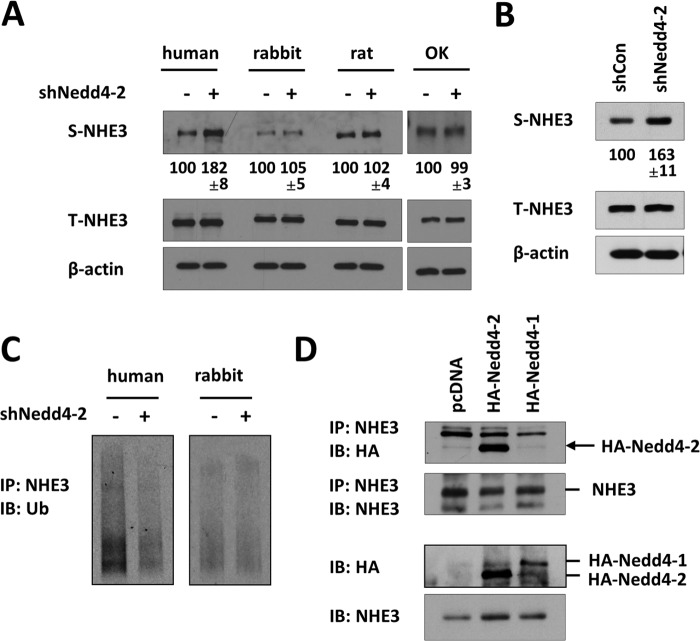
Ubiquitination of membrane proteins alters protein degradation or endocytosis. Hence, we sought to determine whether NHE3 expression is altered by Nedd4-2. ShNedd4-2 did not alter the total expression level of NHE3 expression in PS120 fibroblasts (Fig. 5A). However, a significant increase in surface abundance of hNHE3 was observed with shNedd4-2 compared with shCon. This observation was corroborated in SK-CO15 cells (Fig. 5B). In contrast, surface expression of rat, rabbit, or opossum NHE3 was not affected by shNedd4-2 (Fig. 5A).
FIGURE 5.
Nedd4-2 regulates hNHE3 surface membrane expression. A, surface biotinylation of PS120/NHE3 cells transfected with shCon (−) or shNedd4-2 (+) was performed as described under “Experimental Procedures.” NHE3 expression in the surface membrane (S-NHE3) and total lysate (T-NHE3) was determined by immunoblotting using VSVG antibody. β-Actin expression in lysates was used as a loading control. The expression levels of S-NHE3 were quantified relative to each species transfected with shCon. B, surface expression of NHE3 was determined in SK-CO15 cells transfected with shCon or shNedd4-2. EM450 was used for immunoprecipitation and subsequent immunoblotting for NHE3. C, ubiquitination of NHE3 in PS120/hNHE3 and PS120/rbNHE3 cells was determined by isolation of surface NHE3 by biotinylation followed by immunoblotting (IB) for Ub using P4D1. IP, immunoprecipitation. D, in PS120/hNHE3 expressing HA-Nedd4-1 or HA-Nedd4-2, surface membrane was separated by surface biotinylation. NHE3 was immunoprecipitated using anti-VSVG antibody from the surface fraction followed by immunoblotting with anti-HA antibody. Immunoprecipitated Nedd4-2 is indicated by an arrow. In all lanes a nonspecific band slightly smaller than HA-Nedd4-1 was visible. The bottom panels show the presence of HA-Nedd4-1, HA-Nedd4-2, and NHE3 in the surface fraction. Representative figures from three independent experiments are shown.
Because Nedd4-2 regulates hNHE3 surface expression, we investigated whether Nedd4-2 modulates ubiquitination of NHE3 at the cell surface. As shown in Fig. 5C, Nedd4-2 knockdown resulted in a marked reduction in ubiquitination levels of surface hNHE3. However, unlike human, the ubiquitination level of rbNHE3 on PM was not altered by shNedd4-2. The changes in surface hNHE3 ubiquitination suggested that Nedd4-2 might interact with hNHE3 at the cell surface. To test this possibility, we performed co-immunoprecipitation of hNHE3 and Nedd4-2 from the surface membrane fraction of PS120/hNHE3 cells. The presence of both Nedd4-1 and Nedd4-2 in the surface membrane fraction was observed, but surface hNHE3 co-immunoprecipitated Nedd4-2 but not Nedd4-1. These results indicate that Nedd4-2 interacts with hNHE3 at the surface membrane (Fig. 5D).
PY Motif Is Necessary for Nedd4-2 Interaction
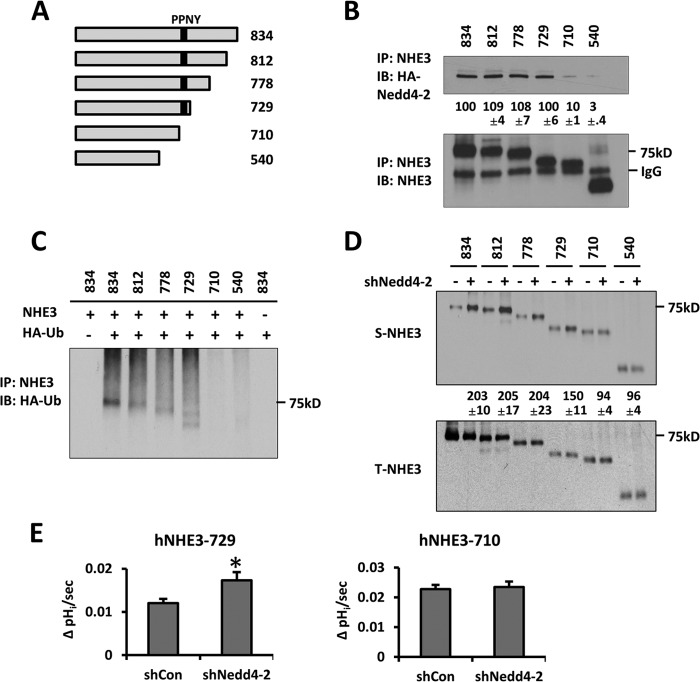
Our data thus far show that Nedd4-2 regulates human NHE3, which contains a PY motif. However, the observation that Nedd4-2 co-immunoprecipitated with ratNHE3 appears to imply that the PY motif might not be the determinant for Nedd4-2 recognition by NHE3. Therefore, we generated a series of constructs with truncation at the C terminus of hNHE3 (Fig. 6A). As illustrated in Fig. 6B, Nedd4-2 was present in the immunoprecipitates of full-length hNHE3 (834) and truncated hNHE3s, including 812, 778, and 729. However, co-immunoprecipitation of Nedd4-2 with hNHE3-710 or -545 was markedly decreased. It is noteworthy that a faint band of Nedd4-2 was detected with hNHE3-710 or -540, suggesting that either an additional Nedd4-2 interacting motif is present or the faint band corresponds to nonspecific interaction of Nedd4-2.
FIGURE 6.
Nedd4-2 binding is necessary for ubiquitination of hNHE3. A, the C-terminal truncation constructs of hNHE3 are shown. Numbers denote the amino acid position at the truncation site for each mutant. B, NHE3 variants were immunoprecipitated (IP, lower) followed by immunoblotting (IB) for HA-Nedd4-2 (upper). The relative amount of HA-Nedd4-2 co-immunoprecipitated was quantified with the interaction with full-length hNHE3-834 as 100%. C, cells expressing hNHE3 variants were transfected with HA-Ub, and NHE3 was immunoprecipitated. Ubiquitination levels are shown. D, surface expression of hNHE3 variants was determined by surface biotinylation. Cells were transfected with shNedd4-2 (+) or shCon (−) before surface biotinylation. Data are representative of three independent experiments. Quantification was done with the control transfected (−) lane of the respective samples as 100%. E, the effects of Nedd4-2 knockdown on Na+-dependent pHi recovery of hNHE3-729 and hNHE3-710 were determined. n = 9. *, p < 0.01 compared with shCon.
To determine whether the reduced interaction with Nedd4-2 is refractory to NHE3 ubiquitination, we compared ubiquitination of the hNHE3 truncation constructs in PS120 cells. Fig. 6C shows that hNHE3-834, -812, -778, and -729, which interacted with Nedd4-2, showed robust levels of ubiquitination as full-length hNHE3. In comparison, the ubiquitination levels of hNHE3-710 and -540 were significantly reduced. We showed above that silencing of Nedd4-2 altered hNHE3 expression at the cell surface. Consistently, shNedd4-2 resulted in increased surface abundance of hNHE3-834, -812, -778, and -729, whereas the surface abundance of hNHE3-710 and -540 was not affected (Fig. 6D). The changes in surface expression were accompanied by parallel changes in NHE3 activity. We observed that the activity of hNHE3-729 was significantly increased by shNedd4-2, whereas no effect on hNHE3-710 activity was noted (Fig. 6E). These data collectively demonstrate the importance of Nedd4-2 for hNHE3 ubiquitination and surface expression.
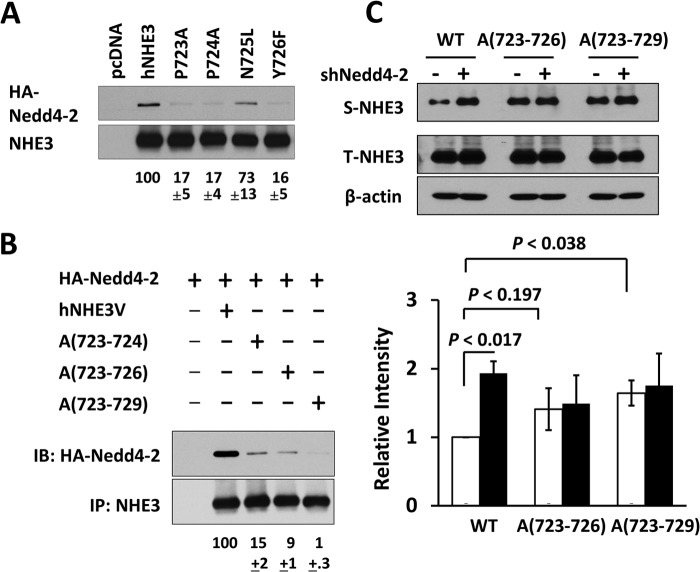
Because of the proximity of the residue 729 to the PY motif, we examined the importance of the PY motif in Nedd4-2 interaction by mutating the 723PPNY726 sequence. As illustrated in Fig. 7A, mutations in Pro-723, Pro-724, or Tyr-726 markedly reduced the Nedd4-2 binding efficiency to hNHE3. Altering Asn-725 showed a smaller but statistically significant decrease. Surprisingly, multiple mutations of PP to alanine (Ala-(723–724)) or PPNY to alanine (Ala-(723–726)) did not completely abolish the interaction with Nedd4-2. A study of the Nedd4-2 interaction with ENaC has shown that the optimal interaction with ENaC involves a conserved Leu adjacent to the PY motif (31). Human NHE3 lacks a Leu, but we mutated the DEE sequence posterior to the PY motif to alanine, Ala-(723–729). These additional mutations further decreased the interaction with Nedd4-2 (Fig. 7B), indicating a contribution by the DEE sequence. In addition, surface expression of Ala-(723–729) was increased compared with WT hNHE3 (Fig. 7C), consistent with the notion that Nedd4-2 decreases surface expression of NHE3. However, the change in the basal expression of Ala-(723–726) was small with p < 0.197, although we suspect that this could reach statistical significance with a larger sample number. Importantly, knockdown of Nedd4-2 did not alter surface expression of Ala-(723–726) or Ala-(723–729), further confirming the role of PY motif.
FIGURE 7.
The extended PY motif of hNHE3 mediates interaction with Nedd4-2. A, each of the amino acid residues in the PY motif in hNHE3 was changed to Ala, and the interaction with HA-Nedd4-2 was determined by co-immunoprecipitation in PS120 cells. The extent of interaction with Nedd4-2 was quantified relative to full-length hNHE3. B, the following mutations were made: PP of PPNY to Ala (A(723–724)); PPNY to Ala (Ala-(723–726)); PPNYDEE to Ala (Ala-(723–729)). The interaction with Nedd4-2 was determined as described above. IB, immunoblot. C, the effect of Nedd4-2 silencing was determined on surface expression of Ala-(723–726) and Ala-(723–729) in PS120 cells transfected with shNedd4-2 (+) or shCon (−). Representative figures from three independent experiments are shown. The bar graph shows quantification of S-NHE3/T-NHE3 in each sample normalized to WT transfected with shCon. Open bar, shCon; closed bar, shNedd4-2. n = 3.
Nedd4-2 Regulates hNHE3 Internalization
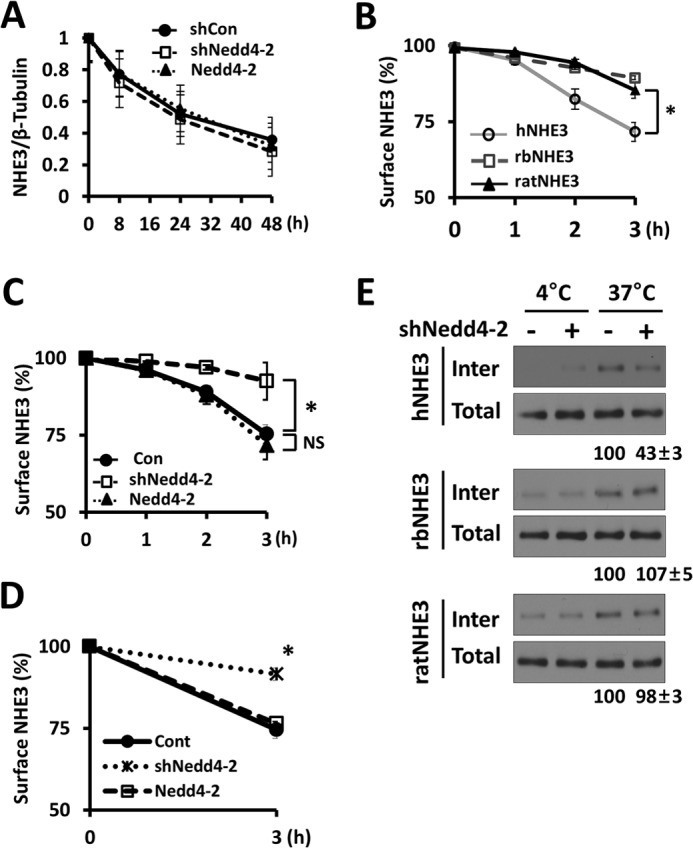
To determine the mechanism by which Nedd4-2 regulates hNHE3, we investigated whether Nedd4-2 alters hNHE3 protein stability. NHE3 expression in SK-CO15 cells was determined in the presence of cycloheximide to prevent translation. Fig. 8A shows that hNHE3 is a long-lived protein with a half-life of >20 h, and knockdown or overexpression of Nedd4-2 did not significantly alter NHE3 protein stability compared with control-transfected cells. Similarly, Nedd4-2 did not alter hNHE3 half-life in PS120 cells (not shown). These results indicate that a major effect of ubiquitination by Nedd4-2 is not degradation of NHE3.
FIGURE 8.
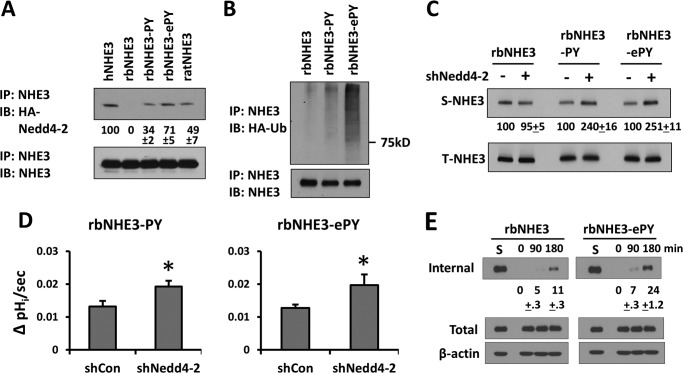
Nedd4-2 facilitates internalization of NHE3. A, SK-CO15 cells were transfected with shCon, shNedd4-2, or HA-Nedd4-2. The steady-state expression levels of NHE3 were determined in the presence of cycloheximide. The graph shows quantification of NHE3 protein bands is shown. n ≥ 3. B, internalization of hNHE3, rbNHE3, and ratNHE3 was determined in PS120 fibroblasts. The graph shows the amounts of internalized NHE3 quantified as fractions of NHE3 in the surface membrane at the start of the experiment. *, p < 0.01. C, internalization of hNHE3 in PS120 cells transfected with shCon, shNedd4-2, or Nedd4-2 was determined. *, p < 0.01. NS, not significant. D, internalization of endogenous hNHE3 was determined in SK-CO15 cells, which were pretransfected with shCon, shNedd4-2, or Nedd4-2. *, p < 0.01 versus shNedd4-2. E, PS120 fibroblasts expressing hNHE3, rbNHE3, or ratNHE3 were treated with shNedd4-2 or shCon. Western blots show internalized NHE3 3 h after the temperature shift to 37 °C, which initiated internalization. Controls were kept at 4 °C for 3 h. The amount of internalized NHE3 was determined relative to internalized NHE3 in control cells. Representative Western blots of three independent experiments are shown.
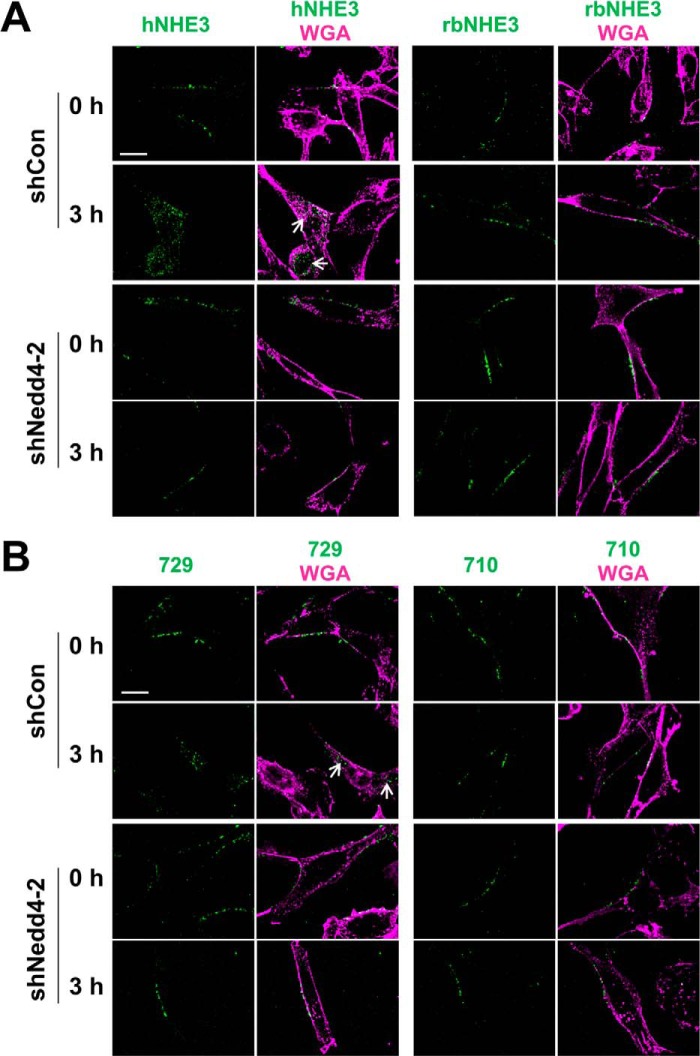
Ubiquitination of integral proteins can function as an internalization or endosomal signal (32, 33). Because NHE3 constitutively internalizes (34), we assessed the rate of hNHE3 internalization by biotinylating hNHE3 on the cell surface of PS120 cells. Cell surface hNHE3 was biotinylated using the membrane-impermeable, cleavable biotin analog Sulfo-NHS-SS-Biotin at 4 °C and then shifted to 37 °C to initiate internalization. At each selected time point, remaining biotin on the cell surface was removed, and biotinylated hNHE3 inside the cells were isolated. As illustrated in Fig. 8B, hNHE3 underwent constitutive endocytosis, leading to 32.6 ± 2.8% of surface NHE3 internalized at 3 h. The rates of internalization of ratNHE3 (14.5 ± 0.9%) and rbNHE3 (11.3 ± 1.7%) were, in comparison, significantly lower. Notably, knockdown of Nedd4-2 in PS120/hNHE3 cells reduced the rate of internalization of hNHE3 to the levels comparable to ratNHE3 or rbNHE3 (Fig. 8C). Consistent with the results from PS120 cells, knockdown of Nedd4-2 in SK-CO15 cells blocked internalization of BBM hNHE3 (Fig. 8D, 25 ± 3% with shCon versus 8 ± 2% with shNedd4-2). Overexpression of Nedd4-2 did not increase the rate of internalization in both PS120 and SK-CO15 cells, indicating that the expression level of Nedd4-2 is not a limiting factor (Fig. 8, C and D). In contrast to hNHE3, internalization of rbNHE3 or ratNHE3 was not altered by shNedd4-2 (Fig. 8E). Nedd4-2-dependent internalization of NHE3 was corroborated by immunofluorescence microscopic analysis. To this end, cells expressing exofacial tri-FLAG-tagged hNHE3, 3×FLAG-hNHE3, were labeled with anti-FLAG antibody conjugated with Alexa Fluor 488 without permeabilization. Fig. 9A, left panels, shows cross-sectional X-Y views of labeled PS120/3×FLAG-hNHE3 cells. After 3 h of initiating internalization by a temperature shift, fluorescence-labeled hNHE3 was localized inside the cell. In comparison, hNHE3 remained at the cell surface in shNedd4-2-transfected cells. Similarly, internalization of hNHE3-729 was blocked by knockdown of Nedd4-2 (Fig. 9B, left panels). However, 3×HA-rbNHE3 as well as hNHE3-710, which do not interact with Nedd4-2, did not exhibit a substantial level of endocytosis, and this was not altered by Nedd4-2 knockdown (Fig. 9, A and B, right panels).
FIGURE 9.
Internalization of hNHE3 is PY motif-dependent. A, PS120/3×FLAG-hNHE3 (left) and PS120/3×HA-rbNHE3 (right) were transfected with shCon (upper panels) or shNedd4-2 (lower panels) and labeled with Alexa Fluor 488-conjugated anti-FLAG or anti-HA antibody without permeabilization. Images of NHE3 (green) and wheat germ agglutinin (WGA; magenta) were captured at time 0 and 3 h of internalization. Arrows point to internalized NHE3. Scale bar, 10 μm. Representative figures from at least three independent experiments are shown. B, PS120 cells expressing 3×FLAG-hNHE3-729 or -710 in PS120 cells were transfected with shCon (top) or shNedd4-2 (bottom) and assayed for internalization as described above.
PY Motif Constitutes Nedd4-2-dependent Regulation of rbNHE3
Given the importance of the PY motif for Nedd4-2-dependent modulation of hNHE3, we questioned whether insertion of a PY motif in non-primate NHE3 can recapitulate Nedd4-2-dependent effects. Hence, we incorporated PY (PPNY) or ePY (PPNYDEE) motif in rbNHE3. The insertion of PY and ePY motifs enabled interaction with Nedd4-2 (Fig. 10A). Nedd4-2 binding efficiency of rbNHE3-PY and -ePY was slightly below and above ratNHE3, respectively, although hNHE3 interaction was strongest. Consistent with the Nedd4-2 interaction, ubiquitination levels of rbNHE3-PY and -ePY were greater than that of rbNHE3 (Fig. 10B). In addition, knockdown of Nedd4-2 significantly increased surface expression and Na+-dependent pHi recovery of rbNHE3-PY and -ePY (Fig. 10, C and D). Because Nedd4-2 facilitates internalization of hNHE3, we determined whether the rate of internalization of rbNHE3 was altered. Fig. 10E shows that rbNHE3-ePY internalized at an increased rate than rbNHE3. Together, these results show that PY motif is the primary determinant for Nedd4-2-dependent NHE3 surface expression and internalization.
FIGURE 10.
Extended PY motif constitutes Nedd4-2 dependence in rbNHE3. A, the interaction of Nedd4-2 with hNHE3, rbNHE3, rbNHE3-PY, rbNHE3-ePY, and ratNHE3 was compared by co-immunoprecipitation (IP) in PS120 cells. The interaction of HA-Nedd4-2 was quantified relative to hNHE3. IB, immunoblot. B, ubiquitination of rbNHE3, rbNHE3-PY, and rbNHE3-ePY was determined. The effect of Nedd4-2 knockdown on surface expression (C), and the rates of Na+-dependent pHi recovery of rbNHE3-PY and rbNHE3-ePY were determined (D). E, internalization of rbNHE3 and rbNHE3-ePY is shown. The amounts of internalized NHE3 as percent fraction of surface NHE3 (S) are shown. Data are representative of at least three independent experiments.
DISCUSSION
We report herein that the presence of PY motifs in NHE3 sequences is species-specific. NHE3s of primates, including human, great apes, and closely related gibbons, contain PY motifs. However, the macaque family of the Old World monkeys, which includes rhesus monkeys, lacks PY motifs. These results indicate that the presence of PY motifs in NHE3s is likely a recent evolutionary modification in human and non-human primates that belong to more modern branches of primate families.
A line of evidence shows that the Nedd4 family of E3 ligases interacts with and regulates a number of membrane proteins (19–21). Nedd4-1 is closely related to Rsp5 in Saccharomyces cerevisiae, which is the only Nedd4 family protein expressed in S. cerevisiae (19). In comparison, Nedd4-2 is distantly related to Rsp5, indicating that Nedd4-1 is likely the ancestral member of the family (19). Because previous studies have shown the preference of transporters and channels to interact with Nedd4-2 instead of Nedd4-1 (19, 20), we anticipated that hNHE3 selectively interacts with Nedd4-2. Indeed, we found that none of the NHE3s examined interacted with Nedd4-1. Human NHE3 showed robust interaction with Nedd4-2. On the other hand, the interaction between Nedd4-2 and ratNHE3 was not only surprising but conflicted with our hypothesis that the PY motif in hNHE3 governs Nedd4-2 interaction. It is noteworthy that previous studies have shown interaction with Nedd4 proteins in the absence of PY motifs (29, 35, 36). In the cases of NHE1 and β2-adrenergic receptor, ubiquitination and protein internalization by Nedd4-1 are mediated by the adaptor protein β-arrestin (29). Our unpublished study showed that β-arrestin interacted with all NHE3s included in the current study, suggesting the possibility of indirect interaction between NHE3 and Nedd4-2 via β-arrestin. However, we ruled out contribution by Nedd4-1 based on the observation that knockdown of Nedd4-1 had no effect on NHE3 activity in Caco-2bbe cells. Moreover, silencing of Nedd4-2 did not affect ratNHE3 or rbNHE3, hence excluding the involvement of Nedd4-2 in regulation of non-primate NHE3s. Although the interaction between ratNHE3 and Nedd4-2 remains perplexing, we suggest that Nedd4-1 and Nedd4-2 are not involved in regulation of non-primate NHE3s.
Because we found that ratNHE3 interacted Nedd4-2 in the absence of a PY motif, we used a set of hNHE3 truncations and point mutations to identify the Nedd4-2 binding motif. Ironically these efforts led to the PY motif as the core of the interaction. Deletion or mutations of the PY motif markedly decreased interaction with Nedd4-2, ubiquitination, and surface expression of hNHE3. A study of high resolution structural determination of Nedd4 and ENaC interaction showed that the C-terminal Leu residue that encompass the PPXYXXL sequence makes a contact with a WW domain of Nedd4-2 (37). Similarly, we found that mutations of the adjacent DEE sequence resulted in a further decrease in Nedd4-2 binding. Because of the polar nature of DEE sequence, whether DEE enhances Nedd4-2 interaction by strengthening the contact with a WW domain of Nedd4-2 is not clear.
The importance of interaction with Nedd4-2 via the PY motif was underscored by the observation that the insertion of a PY motif in rbNHE3 increased the rate of internalization and recapitulated Nedd4-2-dependent regulation of NHE3 expression and activity. However, the fact that Nedd4-2 did not regulate ratNHE3 despite its binding implies that the binding of Nedd4-2 does not warrant Nedd4-2-dependent regulation. Comparison of Nedd4-2 co-immunoprecipitation with different NHE3s (Fig. 10A) did not reveal a marked difference in Nedd4-2 binding between ratNHE3 and modified rbNHE3s. Hence, the lack of effect on ratNHE3 by Nedd4-2 does not appear to be the strength of Nedd4-2 interaction. However, in the case of ratNHE3, we cannot rule out the possibility that the interaction between ratNHE3 and Nedd4-2 occurs via a domain other than PY motif so that the spatial orientation of Nedd4-2 relative to ratNHE3 limits the accessibility of a specific site(s) for ubiquitination. Interestingly, we could not find distinct differences in the composition of Lys residues among various NHE3s that may help to identify potential sites of ubiquitination. Another aspect to note is that the current study evaluated basal, and not regulated, endocytosis of NHE3. It remains to be tested whether Nedd4-2 alters ubiquitination and endocytosis of ratNHE3 in response to a negative cue, such as forskolin or parathyroid hormone.
E3 Ub ligases function at both cell surface and intracellular locations (33). Previous studies on ENaC regulation by Nedd4-2 have shown that Nedd4-2 interacts with ENaC at the cell surface, inducing cell surface ENaC degradation (23, 38). Nedd4-2 colocalizes with amino acid transporter ATA2 on the plasma membrane in 3T3-L1 adipocytes (39). Our observation that Nedd4-2 catalyzed ubiquitination of surface hNHE3 suggests that Nedd4-2 interacts with hNHE3 at the cell surface.
NHE3 exists in multiple intracellular pools so that the primary means of NHE3 regulation involves the movement of NHE3 protein molecules between PM and the intracellular endosomal pools (14). We found that Nedd4-2 does not affect NHE3 protein stability. However, “cycloheximide chase” has an intrinsic limitation on determining degradation of long-lived proteins due to the potential effects of prolonged protein synthesis inhibition on overall cell function (40), and hence, we cannot completely rule out the role of ubiquitination in hNHE3 degradation based on the current results. While this work was being pursued, Armando et al. (41) reported that dopamine inhibited NHE3 in hRPTC human renal proximal tubule cells by inhibition of the deubiquitinating activity of Ub-specific ligase 48, USP48. Inhibition of USB48, which removes Ub from NHE3, decreased the half-life of NHE3 and enhanced NHE3 degradation. Whether the activity of USB48 or USB48-like deubiquitinating enzyme masks the effect of silencing Nedd4-2 is not known.
The salient finding of the current study is that Nedd4-2 mediates internalization of hNHE3 at a rate >2-fold greater than rabbit or rat NHE3. Several lines of evidence support the critical role of Nedd4-2 in enhanced internalization of NHE3: first, knockdown of Nedd4-2 reduced the rate of internalization; second, hNHE3-710 lacking the PY motif failed to internalize within 3 h; third, insertion of ePY motif markedly increased internalization of rbNHE3. So what is the potential significance of increased internalization of hNHE3? Studies by McDonough and co-workers (42, 43) have shown that acute hypertension mediates rapid inhibition of renal proximal tubule Na+ and fluid reabsorption by redistribution of NHE3 from BBM to the base of microvilli in rat proximal tubules. Hence, we speculate that the increased internalization of NHE3 might have given humans an adaptive trait that enables more efficient compensation for elevated Na+ reabsorption elsewhere in the nephron to maintain fluid and electrolyte homeostasis. However, further studies are needed to confirm this supposition and to decode evolutionary stress that could have resulted in the presence of PY motif in NHE3.
We have shown previously that serum and glucocorticoid-induced kinase 1 (SGK1) induced by glucocorticoids stimulates NHE3 (7, 15). SGK1 phosphorylates Nedd4-2, preventing its binding to PY motif (44). Because glucocorticoids stimulate both human and non-human NHE3s, whether Nedd4-2 plays a role in NHE3 regulation by glucocorticoid remains unclear (7).
Lines of evidence link NHE3 to the regulation of blood pressure and extracellular fluid volume (45, 46). NHE3-deficient mice exhibit lower blood pressure than WT mice, and NHE3 activity and expression are elevated in spontaneously hypertensive rats (47–49). However, a genetic linkage associating NHE3 with essential hypertension is yet to be reported, indicating that the association of NHE3 to hypertension is at the post-transcriptional level (50). Nedd4-2 is mapped to chromosome 18q, which has been identified as a possible locus influencing the systolic blood pressure in human (51). Despite the increasing interests in the functions of the Nedd4 family E3 Ub ligases, little is known about how Nedd4 proteins are regulated. Fouladkou et al. (52) reported the finding of several mutations of Nedd4-2 by screening patients with end-stage renal disease. One of these variants, Nedd4-2-P335L, inhibited ENaC less efficiently compared with WT Nedd4-2 due to increased basal phosphorylation that weakens its interaction with ENaC (53). It is not known yet whether these mutations in Nedd4-2 inflict similar effects on NHE3. However, we postulate that a cellular or genetic change, which impedes the interaction of Nedd4-2 with NHE3 in humans, could increase NHE3 expression and activity, contributing to salt-induced pathological conditions. Future studies are needed to validate this supposition.
Acknowledgment
The microscopy core was supported by National Institutes of Health Grant R24DK064399.
This work was supported, in whole or in part, by National Institutes of Health Grants DK061418 (to C. C. Y.) and T32DK007771 (to B. K. Y.). This work was also supported by American Heart Association Grant 13SDG1623001 (to P. H.).
- NHE3
- Na+/H+ exchanger 3
- Nedd4
- neural precursor cell expressed, developmentally down-regulated 4
- BBM
- brush border membrane
- Ub
- ubiquitin
- ENaC
- epithelial Na+ channel
- hNHE3
- human NHE3
- rbNHE3
- rabbit NHE3
- ratNHE3
- rat NHE3
- PM
- plasma membrane
- BCECF-AM
- 2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein acetoxymethyl ester
- shCon
- control shRNA
- VSVG
- vesicular stomatitis virus glycoprotein
- TMA
- tetramethylammonium.
REFERENCES
- 1. Bobulescu I. A., Moe O. W. (2009) Luminal Na+/H+ exchange in the proximal tubule. Pflugers Arch. 458, 5–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander R. T., Grinstein S. (2009) Tethering, recycling and activation of the epithelial sodium-proton exchanger, NHE3. J. Exp. Biol. 212, 1630–1637 [DOI] [PubMed] [Google Scholar]
- 3. Collazo R., Fan L., Hu M. C., Zhao H., Wiederkehr M. R., Moe O. W. (2000) Acute regulation of Na+/H+ exchanger NHE3 by parathyroid hormone via NHE3 phosphorylation and dynamin-dependent endocytosis. J. Biol. Chem. 275, 31601–31608 [DOI] [PubMed] [Google Scholar]
- 4. Hayashi H., Szászi K., Coady-Osberg N., Furuya W., Bretscher A. P., Orlowski J., Grinstein S. (2004) Inhibition and redistribution of NHE3, the apical Na+/H+ exchanger, by Clostridium difficile toxin B. J. Gen. Physiol. 123, 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gill R. K., Saksena S., Tyagi S., Alrefai W. A., Malakooti J., Sarwar Z., Turner J. R., Ramaswamy K., Dudeja P. K. (2005) Serotonin inhibits Na+/H+ exchange activity via 5-HT4 receptors and activation of PKCα in human intestinal epithelial cells. Gastroenterology 128, 962–974 [DOI] [PubMed] [Google Scholar]
- 6. Hu M. C., Fan L., Crowder L. A., Karim-Jimenez Z., Murer H., Moe O. W. (2001) Dopamine acutely stimulates Na/H exchanger (NHE3) endocytosis via clathrin-coated vesicles: dependence on protein kinase A-mediated NHE3 phosphorylation. J. Biol. Chem. 276, 26906–26915 [DOI] [PubMed] [Google Scholar]
- 7. He P., Lee S. J., Lin S., Seidler U., Lang F., Fejes-Toth G., Naray-Fejes-Toth A., Yun C. C. (2011) Serum- and glucocorticoid-induced kinase 3 in recycling endosomes mediates acute activation of Na+/H+ exchanger NHE3 by glucocorticoids. Mol. Biol. Cell 22, 3812–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chow C. W., Khurana S., Woodside M., Grinstein S., Orlowski J. (1999) The epithelial Na+/H+ exchanger, NHE3, is internalized through a clathrin-mediated pathway. J. Biol. Chem. 274, 37551–37558 [DOI] [PubMed] [Google Scholar]
- 9. Murtazina R., Kovbasnjuk O., Donowitz M., Li X. (2006) Na+/H+ exchanger NHE3 activity and trafficking are lipid raft-dependent. J. Biol. Chem. 281, 17845–17855 [DOI] [PubMed] [Google Scholar]
- 10. Fuster D. G., Bobulescu I. A., Zhang J., Wade J., Moe O. W. (2007) Characterization of the regulation of renal Na+/H+ exchanger NHE3 by insulin. Am. J. Physiol. Renal Physiol. 292, F577–F585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He P., Klein J., Yun C. C. (2010) Activation of Na+/H+ exchanger nhe3 by angiotensin II is mediated by inositol 1,4,5-triphosphate (IP3) receptor-binding protein released with IP3 (IRBIT) and Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 285, 27869–27878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang D., Sun H., Lang F., Yun C. C. (2005) Activation of NHE3 by dexamethasone requires phosphorylation of NHE3 at Ser-663 by SGK1. Am. J. Physiol. Cell Physiol 289, C802–C810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Biemesderfer D., Nagy T., DeGray B., Aronson P. S. (1999) Specific association of megalin and the Na+/H+ exchanger isoform NHE3 in the proximal tubule. J. Biol. Chem. 274, 17518–17524 [DOI] [PubMed] [Google Scholar]
- 14. Alexander R. T., Furuya W., Szászi K., Orlowski J., Grinstein S. (2005) Rho GTPases dictate the mobility of the Na/H exchanger NHE3 in epithelia: role in apical retention and targeting. Proc. Natl. Acad. Sci. U.S.A. 102, 12253–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yun C. C., Chen Y., Lang F. (2002) Glucocorticoid activation of Na+/H+ exchanger isoform 3 revisited. The roles of SGK1 and NHERF2. J. Biol. Chem. 277, 7676–7683 [DOI] [PubMed] [Google Scholar]
- 16. Weinman E. J., Steplock D., Shenolikar S. (2003) NHERF-1 uniquely transduces the cAMP signals that inhibit sodium-hydrogen exchange in mouse renal apical membranes. FEBS Lett. 536, 141–144 [DOI] [PubMed] [Google Scholar]
- 17. Piper R. C., Lehner P. J. (2011) Endosomal transport via ubiquitination. Trends Cell Biol. 21, 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hislop J. N., von Zastrow M. (2011) Role of ubiquitination in endocytic trafficking of g-protein-coupled receptors. Traffic 12, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang B., Kumar S. (2010) Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 17, 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rotin D., Staub O. (2011) Role of the ubiquitin system in regulating ion transport. Pfluegers Arch. 461, 1–21 [DOI] [PubMed] [Google Scholar]
- 21. Persaud A., Alberts P., Amsen E. M., Xiong X., Wasmuth J., Saadon Z., Fladd C., Parkinson J., Rotin D. (2009) Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol. Syst. Biol. 5, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Münster C., Chraïbi A., Pratt J. H., Horisberger J. D., Pearce D., Loffing J., Staub O. (2001) Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na+ channel cell surface expression. EMBO J. 20, 7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou R., Patel S. V., Snyder P. M. (2007) Nedd4-2 catalyzes ubiquitination and degradation of cell surface ENaC. J. Biol. Chem. 282, 20207–20212 [DOI] [PubMed] [Google Scholar]
- 24. Shi P. P., Cao X. R., Sweezer E. M., Kinney T. S., Williams N. R., Husted R. F., Nair R., Weiss R. M., Williamson R. A., Sigmund C. D., Snyder P. M., Staub O., Stokes J. B., Yang B. (2008) Salt-sensitive hypertension and cardiac hypertrophy in mice deficient in the ubiquitin ligase Nedd4-2. Am. J. Physiol. Renal Physiol. 295, F462–F470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoo B. K., Yanda M. K., No Y. R., Yun C. C. (2012) Human intestinal epithelial cell line SK-CO15 is a new model system to study Na+/H+ exchanger 3. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G180–G188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin S., Yeruva S., He P., Singh A. K., Zhang H., Chen M., Lamprecht G., de Jonge H. R., Tse M., Donowitz M., Hogema B. M., Chun J., Seidler U., Yun C. C. (2010) Lysophosphatidic acid stimulates the intestinal brush border Na+/H+ exchanger 3 and fluid absorption via LPA5 and NHERF2. Gastroenterology 138, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cabado A. G., Yu F. H., Kapus A., Lukacs G., Grinstein S., Orlowski J. (1996) Distinct structural domains confer cAMP sensitivity and ATP dependence to the Na+/H+ exchanger NHE3 isoform. J. Biol. Chem. 271, 3590–3599 [DOI] [PubMed] [Google Scholar]
- 28. Kreis T. E. (1986) Microinjected antibodies against the cytoplasmic domain of vesicular stomatis virus glycoprotein block its tranport to the cell surface. EMBO J. 5, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simonin A., Fuster D. (2010) Nedd4-1 and β-arrestin-1 are key regulators of Na+/H+ exchanger 1 ubiquitylation, endocytosis, and function. J. Biol. Chem. 285, 38293–38303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Itani O. A., Campbell J. R., Herrero J., Snyder P. M., Thomas C. P. (2003) Alternate promoters and variable splicing lead to hNedd4-2 isoforms with a C2 domain and varying number of WW domains. Am. J. Physiol. Renal Physiol. 285, F916–F929 [DOI] [PubMed] [Google Scholar]
- 31. Kanelis V., Farrow N. A., Kay L. E., Rotin D., Forman-Kay J. D. (1998) NMR studies of tandem WW domains of Nedd4 in complex with a PY motif-containing region of the epithelial sodium channel. Biochem. Cell Biol. 76, 341–350 [DOI] [PubMed] [Google Scholar]
- 32. d'Azzo A., Bongiovanni A., Nastasi T. (2005) E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6, 429–441 [DOI] [PubMed] [Google Scholar]
- 33. Kerscher O., Felberbaum R., Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 34. Kurashima K., Szabó E. Z., Lukacs G., Orlowski J., Grinstein S. (1998) Endosomal recycling of the Na+/H+ exchanger NHE3 isoform is regulated by the phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 273, 20828–20836 [DOI] [PubMed] [Google Scholar]
- 35. Persaud A., Alberts P., Hayes M., Guettler S., Clarke I., Sicheri F., Dirks P., Ciruna B., Rotin D. (2011) Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 30, 3259–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arroyo J. P., Lagnaz D., Ronzaud C., Vázquez N., Ko B. S., Moddes L., Ruffieux-Daidié D., Hausel P., Koesters R., Yang B., Stokes J. B., Hoover R. S., Gamba G., Staub O. (2011) Nedd4-2 modulates renal Na+-Cl− cotransporter via the aldosterone-SGK1-Nedd4-2 pathway. J. Am. Soc. Nephrol. 22, 1707–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kanelis V., Rotin D., Forman-Kay J. D. (2001) Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 8, 407–412 [DOI] [PubMed] [Google Scholar]
- 38. Kabra R., Knight K. K., Zhou R., Snyder P. M. (2008) Nedd4-2 induces endocytosis and degradation of proteolytically cleaved epithelial Na+ channels. J. Biol. Chem. 283, 6033–6039 [DOI] [PubMed] [Google Scholar]
- 39. Hatanaka T., Hatanaka Y., Setou M. (2006) Regulation of amino acid transporter ATA2 by ubiquitin ligase Nedd4-2. J. Biol. Chem. 281, 35922–35930 [DOI] [PubMed] [Google Scholar]
- 40. Yewdell J. W., Lacsina J. R., Rechsteiner M. C., Nicchitta C. V. (2011) Out with the old, in with the new? Comparing methods for measuring protein degradation. Cell Biol. Int. 35, 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Armando I., Villar V. A., Jones J. E., Lee H., Wang X., Asico L. D., Yu P., Yang J., Escano C. S., Jr., Pascua-Crusan A. M., Felder R. A., Jose P. A. (2014) Dopamine D3 receptor inhibits the ubiquitin-specific peptidase 48 to promote NHE3 degradation. FASEB J. 28, 1422–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yip K. P., Tse C. M., McDonough A. A., Marsh D. J. (1998) Redistribution of Na+/H+ exchanger isoform NHE3 in proximal tubules induced by acute and chronic hypertension. Am. J. Physiol. 275, F565–F575 [DOI] [PubMed] [Google Scholar]
- 43. Riquier A. D., Lee D. H., McDonough A. A. (2009) Renal NHE3 and NaPi2 partition into distinct membrane domains. Am. J. Physiol. Cell Physiol. 296, C900–C910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kamynina E., Staub O. (2002) Concerted action of ENaC, Nedd4-2, and Sgk1 in transepithelial Na+ transport. Am. J. Physiol. Renal Physiol 283, F377–F387 [DOI] [PubMed] [Google Scholar]
- 45. Bobulescu I. A., Di Sole F., Moe O. W. (2005) Na+/H+ exchangers: physiology and link to hypertension and organ ischemia. Curr. Opin. Nephrol. Hypertens. 14, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Girardi A. C., Di Sole F. (2012) Deciphering the mechanisms of the Na+/H+ exchanger-3 regulation in organ dysfunction. Am. J. Physiol. Cell Physiol. 302, C1569–C1587 [DOI] [PubMed] [Google Scholar]
- 47. Schultheis P. J., Clarke L. L., Meneton P., Miller M. L., Soleimani M., Gawenis L. R., Riddle T. M., Duffy J. J., Doetschman T., Wang T., Giebisch G., Aronson P. S., Lorenz J. N., Shull G. E. (1998) Renal and intestinal absorptive defects in mice lacking the NHE3 Na+/H+ exchanger. Nat. Genet. 19, 282–285 [DOI] [PubMed] [Google Scholar]
- 48. Noonan W. T., Woo A. L., Nieman M. L., Prasad V., Schultheis P. J., Shull G. E., Lorenz J. N. (2005) Blood pressure maintenance in NHE3-deficient mice with transgenic expression of NHE3 in small intestine. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R685–R691 [DOI] [PubMed] [Google Scholar]
- 49. LaPointe M. S., Sodhi C., Sahai A., Batlle D. (2002) Na+/H+ exchange activity and NHE-3 expression in renal tubules from the spontaneously hypertensive rat. Kidney Int. 62, 157–165 [DOI] [PubMed] [Google Scholar]
- 50. Lifton R. P., Hunt S. C., Williams R. R., Pouysségur J., Lalouel J.-M. (1991) Exclusion of the Na/H antiporter as a candidate gene in human essential hypretension. Hypertension 17, 8–14 [DOI] [PubMed] [Google Scholar]
- 51. Pankow J. S., Dunn D. M., Hunt S. C., Leppert M. F., Miller M. B., Rao D. C., Heiss G., Oberman A., Lalouel J. M., Weiss R. B. (2005) Further evidence of a quantitative trait locus on chromosome 18 influencing postural change in systolic blood pressure: the Hypertension Genetic Epidemiology Network (HyperGEN) Study. Am. J. Hypertens. 18, 672–678 [DOI] [PubMed] [Google Scholar]
- 52. Fouladkou F., Alikhani-Koopaei R., Vogt B., Flores S. Y., Malbert-Colas L., Lecomte M. C., Loffing J., Frey F. J., Frey B. M., Staub O. (2004) A naturally occurring human Nedd4-2 variant displays impaired ENaC regulation in Xenopus laevis oocytes. Am. J. Physiol. Renal Physiol. 287, F550–F561 [DOI] [PubMed] [Google Scholar]
- 53. Chandran S., Li H., Dong W., Krasinska K., Adams C., Alexandrova L., Chien A., Hallows K. R., Bhalla V. (2011) Neural precursor cell-expressed developmentally down-regulated protein 4–2 (Nedd4-2) regulation by 14-3-3 protein binding at canonical serum and glucocorticoid kinase 1 (SGK1) phosphorylation sites. J. Biol. Chem. 286, 37830–37840 [DOI] [PMC free article] [PubMed] [Google Scholar]