Background: Myosin-19 is strongly associated with mitochondria and plays a role in the transport of mitochondria.
Results: The light chains of myosin-19 are the regulatory light chains of myosin-2. ADP release is rate-limiting for acto-Myo19 ATPase and ADP strongly inhibits myosin-19 motor function.
Conclusion: Myosin-19 is a plus-end-directed, high-duty ratio molecular motor.
Significance: Myosin-19 functions as a molecular motor.
Keywords: Actin, ATPase, Intracellular Trafficking, Mitochondria, Myosin
Abstract
Class XIX myosin (Myo19) is a vertebrate-specific unconventional myosin, responsible for the transport of mitochondria. To characterize biochemical properties of Myo19, we prepared recombinant mouse Myo19-truncated constructs containing the motor domain and the IQ motifs using the baculovirus/Sf9 expression system. We identified regulatory light chain (RLC) of smooth muscle/non-muscle myosin-2 as the light chain of Myo19. The actin-activated ATPase activity and the actin-gliding velocity of Myo19-truncated constructs were about one-third and one-sixth as those of myosin-5a, respectively. The apparent affinity of Myo19 to actin was about the same as that of myosin-5a. The RLCs bound to Myo19 could be phosphorylated by myosin light chain kinase, but this phosphorylation had little effect on the actin-activated ATPase activity and the actin-gliding activity of Myo19-truncated constructs. Using dual fluorescence-labeled actin filaments, we determined that Myo19 is a plus-end-directed molecular motor. We found that, similar to that of the high-duty ratio myosin, such as myosin-5a, ADP release rate was comparable with the maximal actin-activated ATPase activity of Myo19, indicating that ADP release is a rate-limiting step for the ATPase cycle of acto-Myo19. ADP strongly inhibited the actin-activated ATPase activity and actin-gliding activity of Myo19-truncated constructs. Based on the above results, we concluded that Myo19 is a high-duty ratio molecular motor moving to the plus-end of the actin filament.
Introduction
Myosins are actin-based molecular motors that play an essential role in various cell processes such as muscle contraction, cytokinesis, endocytosis, adhesion, and organelle transport. So far all identified myosins contain three distinct domains, i.e. the motor domain, the neck region or lever arm, and the tail. The motor domain is responsible for actin binding and ATP hydrolysis. The neck region consists of IQ motifs and the bound calmodulin (CaM)2 or CaM-like light chains. The tail domain is largely responsible for targeting of the motor to its subcellular localization and may be involved in the regulation of motor activity (1–3). Based on the motor domain sequence, the myosin superfamily is currently comprised of more than 35 classes (4, 5).
Class XIX Myosin (Myo19) is a vertebrate-specific unconventional myosin and is one of the least studied myosins among the 25 unconventional myosins identified in human. Based upon the deduced amino acid sequence, it was predicted that Myo19 consists of a motor domain, a neck region containing three IQ motifs, and a short tail domain (Fig. 1A) (6). Cell biology experiments revealed that Myo19 is strongly associated with mitochondria and plays a role in the transport of mitochondria along actin filament in human cells (7). Recently, Adikes and co-workers (8) expressed human Myo19-truncated constructs in Sf9 insect cells and characterized the motor properties of Myo19. They found that Myo19 displayed actin-activated ATPase activity and in vitro actin-gliding activity, indicating that Myo19 is a functional motor (8). However, several important issues regarding the motor function of Myo19 have not been solved.
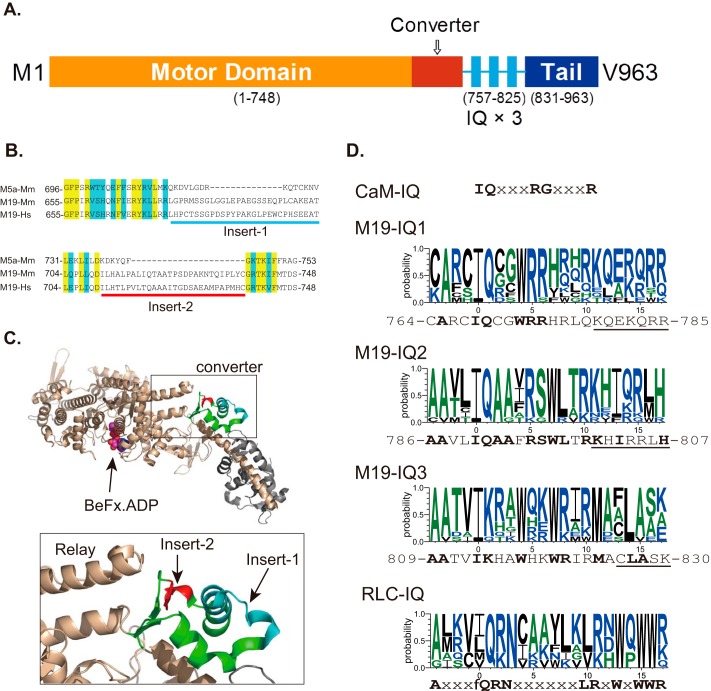
FIGURE 1.
Diagram of Myo19 structure and sequence alignment of Myo19. A, predicted structure of Myo19. The diagram was not drawn to scale. B, sequence alignment of the converter of myosin-5a and Myo19 reveals two inserts in the converter of Myo19. Yellow and blue shades indicate the identical and conserved residues, respectively. Myo5a-Mm, mouse myosin-5a (GI: 115511052); M19-Mm, mouse Myo19 (GI: 254939539); M19-Hs, human Myo19 (GI: 254939537). C, location of the two Myo19-specific inserts in the crystal structure of myosin-5a (PDB code 1W7J). Upper, the head domain of myosin-5a; lower, the expanded view of the converter. The converter is colored green with insert 1 in cyan and insert 2 in red. Insert 1 and 2 are adjacent to each other and insert 2 is adjacent to the tip of the relay helix. D, consensus sequences of CaM-binding IQ motif, IQ motifs of Myo19, and RLC-binding IQ motif of myosin-2 and -18. CaM-IQ, consensus sequence of the six IQ motifs of mouse myosin-5a (GI: 115511052). M19-IQ1, -IQ2, and -IQ3, sequence logo of the consensus sequence of the three IQ motifs of 11 class XIX myosins, including mouse (GI: 254939539), rat (GI: 254939541), human (GI: 254939537), cattle (GI: 254939551), elephant (GI: 344285323), rabbit (GI: 291405654), cat (GI: 410980571), whale (GI: 466018728), chicken (GI: 513216545), Xenopus laevis (GI: 82178330), and zebrafish (GI: 189519181). The sequence shown at the bottom is mouse Myo19. Conserved residues are shown in bold. Underlined residues are not essential for RLC binding (for details see ”Discussion“). RLC-IQ, sequence logo of the consensus sequence of IQ2 motifs of five muscle myosin-2 and two myosin-18, including scallop muscle myosin (GI: 5611), human non-muscle myosin-2a (GI: 47678583), -2b (GI: 219841954), -2c (GI: 116284394), smooth muscle myosin (GI: 46486992), myosin-18a (GI: 24660442), and myosin-18b (GI: 219841774). Sequence logos were generated by an online program, WebLOGO.
First, the identity of the light chain associated with Myo19 IQ motifs remains to be determined. CaM is a common light chain bound to the IQ motif of unconventional myosin identified so far. Adikes and co-workers (8) coexpressed Myo19 with CaM and found stoichiometric association of CaM with Myo19. However, the consensus sequences of Myo19 IQ motifs (IQ1, AXXIQXXWRR; IQ2, AAXXIQAAXRSWLXRKXIXXXH; and IQ3, AAXXiKXXWXXWRXXMXXLA) are quite different from the consensus sequence of CaM-bound IQ motif in myosin, i.e. IQXXXRGXXXR (Fig. 1D). So, it is necessary to examine if light chain other than CaM serves as endogenous light chains of Myo19.
Second, the directionality of the Myo19 motor moving along the actin filament is not known. Compared to other myosins, Myo19 contains two unique inserts in a region of the motor domain known as the converter (Fig. 1B). In the crystal structure of myosin-5a (PDB 1W7J), the two inserts of Myo19 are adjacent to one another and the second one is adjacent to the tip of the relay helix (Fig. 1C). The converter is the major component connecting the core of the motor domain with the lever arm. During the ATP hydrolysis cycle, the converter amplifies and directs the lever arm movements resulting from the conformational change in the core of the motor domain (9). It is well established that the minus-end-directed movement of class VI myosin is dictated by a unique insert of ∼38 amino acids located between the converter and lever arm (10, 11). Although the insert of Myo19 is located within the converter instead of between the converter and the lever arm, it is potentially critically for the direction of the lever arm movement of Myo19 during the ATP hydrolysis cycle, thus affecting the direction of Myo19 movement along actin filaments.
Third, it remains to be determined if Myo19 is a high-duty ratio motor. During each ATP hydrolysis cycle, myosin motors cyclically attach to and detach from actin filaments. Duty ratio is the fraction of the cycle time that myosin motor attaches to actin filament. Although myosin motor domains are highly conserved, their motor duty ratios are very different from one to another. Most members of class II myosin (myosin-2 or conventional myosin) are low-duty ratio motors that spend most of the time during the ATP hydrolysis cycle detached from actin filaments. An advantage of the low-duty ratio myosin motor is that many molecules can simultaneously interact with one actin filament thus producing a large force without interfering with each other. In contrast, a number of unconventional myosins, including myosin-5, -6, -7, and -10, are high-duty ratio motors that spend most of the time during the ATP hydrolysis cycle attached to the actin filament (12–15). Upon proper dimerization, those high-duty ratio motors can move individually along the actin filament, thus becoming a processive motor and suitable for intracellular transport (16–20).
The kinetic of high-duty ratio motor are quite different from low-duty ratio motor. In general, ADP release is the rate-limiting step for the high-duty ratio myosin (12–13, 20). Because myosin·ADP strongly binds to actin, these myosins spend most of their kinetic cycle strongly bound to actin. In contrast, the ADP release rate for the low-duty ratio myosins is 1–2 orders of magnitude faster than the steady-state actin-activated ATPase rate (21). Therefore, it is critical to measure the ADP release rate to determine whether Myo19 is a high-duty ratio motor.
In the present study, we identified the regulatory light chain of smooth muscle/non-muscle myosin as the light chain of Myo19. We found that, similar to vertebrate myosin-5a, Myo19 moves to the plus-end of the actin filament. In addition, we found that the ADP release rate of Myo19 is comparable with the maximal steady-state actin-activated ATPase rate, suggesting that ADP release is a rate-limiting step for acto-Myo19 and Myo19 is a high-duty ratio motor.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes and DNA modifying enzymes were purchased from New England BioLabs (Beverly, MA), unless indicated otherwise. Anti-FLAG M2 antibody, anti-FLAG M2-agarose, phosphoenol pyruvate, 2,4-dinitrophenyl-hydrazine, pyruvate kinase, ADP, malachite green oxalate, glucose oxidase, casein, and sodium molybdate dihydrate were from Sigma. Nickel-nitrilotriacetic acid-agarose was from Qiagen (Germany). ATP was purchased from Amresco. Rhodamine-phalloidin and Alexa 488-phalloidin were from Invitrogen. Catalase was from Worthington Biomedical Co. FLAG peptide (DYKDDDDK) was synthesized by Augct Co. (Beijing, China). Oligonucleotides were synthesized by Invitrogen. Actin, CaM, and MLCK (myosin light chain kinase) were prepared as described previously (22, 23). Two mouse myosin-5a constructs, i.e. Myo5a-1IQ (containing the motor domain and 1st IQ, amino acids 1–791) and Myo5a-ΔT (myosin-5a with the globular tail domain deleted), were prepared as described (23, 24). SkM (skeletal muscle myosin) was prepared from rabbit muscle according to a published method (25). SkM-S1 (skeletal muscle myosin subfragment-1) was prepared from SkM by chymotrypsin digestion as described (26).
Expression and Purification of Myo19 and Myo5a
The cDNA of Myo19 (GI: 254939538) was reversely transcribed from mouse kidney total RNA using AccuScript Reverse Transcriptase (Stratagene), amplified by Pfu-Ultra DNA polymerase (Stratagene) with the following primers: ACTAGGATCCATGCTCCAGCAGGTGAATG (underlined, BamHI) and ACGTCTCGAGTCACACTTGGACTGGC (underlined, XhoI), and cloned into the BamHI site and XhoI site of pFastBacHFTb (a modified baculovirus transfer vector pFastHTb, Life Technology as described previously (27)). Myo19-truncated constructs were created by introducing a stop codon at the indicated positions. All Myo19 constructs contain an N-terminal FLAG tag (DYKDDDDK) for purification and an Avi-tag (GLNDIFEAQKIEWHE) to facilitate actin-gliding assay was attached to the C terminus of Myo19 constructs. Recombinant baculoviruses were prepared using the Bac-To-Bac system (Invitrogen) as described previously (24).
To express Myo19, Sf9 cells were infected with the recombinant viruses encoding Myo19 with or without the recombinant virus encoding the light chains. The expressed Myo19 was purified by anti-FLAG M2 affinity chromatography as described previously (24). The concentrations of the purified Myo19 were determined by SDS-PAGE and Coomassie Blue staining with a known concentration of smooth muscle myosin protein as standard.
RLC Expression and Purification
The cDNA of RLC9 and RLC12b was obtained by RT-PCR using mouse kidney RNA and cloned into pT7-7 bacterial expression vector. RLC9 and RLC12b were expressed in BL21(DE3) and purified as described by Ikebe et al. (28). The concentrations of purified RLC9 and RLC12b were determined by the absorbance at 280 nm (A280 equals 5.01 mg/ml of RLC9 and 3.77 mg/ml of RLC12b). Approximately 48 mg of RLC9 and 35 mg of RLC12b were obtained from 1 liter of culture, respectively.
To prepare recombinant baculovirus encoding RLC, cDNA of RLC9 or RLC12b was subcloned into pFastBac baculovirus transfer vector (Invitrogen). Recombinant baculoviruses were prepared using a Bac-To-Bac system (Invitrogen) as described previously (24).
Identification of the Light Chains of Myo19
We first prepared a crude extract of myosin light chains from mouse kidney. About 1 g of adult mouse kidneys was homogenized in 10 ml of lysis buffer (30 mm Tris-HCl (pH 7.5), 8 m urea, 5 mm DTT, and 10 μg/ml of leupeptin). The homogenization was clarified by centrifugation at 25,000 × g for 1 h at 4 °C. The proteins in the supernatant were denatured by 5% TCA and then collected by centrifugation at 25,000 × g for 30 min at 4 °C. The precipitation was dissolved with 8 m urea (adjust pH to 7.0 with 1 m Tris base), and dialyzed three times (6 h each) against 1 liter of 30 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 1 mm DTT. The dialyzed sample was clarified by centrifugation at 13,000 × g for 10 min at 4 °C and the supernatant was used as crude extract of myosin light chains.
To identify the light chains associated with the Myo19 heavy chain, we co-purified the M19-2IQ with crude extract of the myosin light chain from mouse kidney. FLAG-tagged M19-2IQ was expressed in Sf9 cells and absorbed onto anti-FLAG M2-agarose. About 0.5 ml of anti-FLAG M2-agarose containing ∼0.2 mg of M19-2IQ was incubated with 1 ml of light chain extract in Binding Buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm EGTA, and 1 mm DTT) at 4 °C for 2 h. After washing away the unbound proteins with Binding Buffer, the bound protein was eluted with 0.2 mg/ml of FLAG peptide in Binding Buffer. The eluted proteins were subjected to SDS-PAGE (4–20%) and Coomassie Blue staining. A control experiment was performed similarly except omitting M19-2IQ or light chain extract. The bands in SDS-PAGE specifically associated with M19-2IQ were excised and subjected to trypsin digestion and LC-MS/MS analysis.
ATPase Assay
Unless otherwise indicated, the steady-state ATPase activity of Myo19 was measured in a plate-based, ATP regeneration system as described previously (29). The steady-state ATPase activities were measured in a solution containing ∼50 nm Myo19 construct, 20 mm MOPS-KOH (pH 7.0), 50 mm NaCl, 1 mm MgCl2, 1 mm DTT, 0.25 mg/ml of BSA, 0.5 mm ATP, 2.5 mm phosphoenol pyruvate, 20 units/ml of pyruvate kinase, 4 μm RLC, 1 mm EGTA, and actin up to 80 μm at 25 °C. The reaction was stopped at various times between 4 to 60 min by adding 25 μl of reaction solution to a well of a 96-well plate (flat bottom) containing 100 μl of 0.36 mm 2,4-dinitrophenyl hydrazine (Sigma) and 0.4 m HCl. After incubation at 37 °C for 15 min, 50 μl of 2.5 m NaOH and 0.1 m EDTA were added to each well and the absorptions at 450 nm were recorded in a microplate reader. To examine the effects of ADP on the actin-activated ATPase activity of Myo19, we performed an ATPase assay in the absence of the ATP regeneration system, and the liberated phosphate was determined by the malachite green method (30).
Stopped-flow Measurements of ADP Release Rate
Transient kinetic measurements were performed by using a Chirascan SF3 stopped-flow instrument (Applied Photophysics, Surrey, UK) having a 1.2-ms dead time at 25 °C in a buffer containing 20 mm MOPS-KOH (pH 7.0), 100 mm NaCl, 1 mm MgCl2, 1 mm EGTA, and 1 mm DTT. Mant-ADP fluorescence was excited at 360 nm, and emission was selected using a 395-nm long pass filter. Mant-ADP fluorescence was recorded in the stopped flow after mixing 1 mm ATP with 1 μm mant-ADP and 1 μm M19-1IQ in the absence or presence of 1.5 μm phalloidin-stabilized actin filaments (1.5 μm actin and 1.5 μm phalloidin). Data fitting was done with KaleidaGraph (Synergy Software, Reading, PA).
Phosphorylation of RLC of Myo19 by MLCK
To examine the phosphorylation of RLC of Myo19 by MLCK, the purified Myo19·RLC complex (∼2 μm) was incubated with MLCK (0.03 mg/ml) in a solution containing 30 mm Tris-HCl (pH 7.5), 50 mm KCl, 1 mm DTT, 1 mm MgCl2, 10 μm CaM, 0.1 mm CaCl2, and 1 mm ATP at 25 °C. At the indicated time, 110 μl of reaction solution was mixed with 10% TCA (final concentration) and the denatured protein was precipitated by centrifugation at 12,000 × g for 5 min. The precipitated protein was solubilized in 20 μl of sample solution (8 m urea, 10 mm DTT, and 0.005% bromphenol blue) and analyzed by urea/glycerol PAGE as described previously (22).
Biotinylation of Myosin by BirA
cDNA of Escherichia coli biotin ligase (BirA) was amplified from BL21(DE3) DNA by PCR and cloned into pET30a (Novagen). To express BirA, BL21(DE3) was transformed with BirA/pET30a and induced by isopropyl 1-thio-β-d-galactopyranoside. The expressed BirA was purified by nickel-nitrilotriacetic acid-agarose chromatography according to standard procedures. About 10 mg of BirA was obtained from 200 ml of culture. Myosin construct containing an Avi-tag was biotinylated by BirA as described by Cull and Schatz (31) with minor modifications. Briefly, myosin construct containing a C-terminal Avi-tag (2–6 μm) was incubated with 20 μg/ml of BirA in a solution containing 50 mm Bicine buffer (pH 8.3), 5 mm ATP, 10 mm magnesium acetate, 50 μm d-biotin at 30 °C for 30 min. The extent of biotinylation of the target protein was examined by Western blot using anti-biotin HRP-linked antibody (Cell Signaling Co.). The biotinylated myosin was dialyzed against 10 mm imidazole (pH 7.5), 100 mm KCl, 1 mm EGTA, 1 mm DTT, and 10% glycerol was extensively to remove free biotin and used immediately or aliquoted and stored in −80 °C.
In Vitro Actin-gliding Assay
The actin-gliding activity of biotinylated Myo19 (see above) was performed as follows. About 1 mg/ml of Biotin-LC-BSA (catalog number 29130, Thermo) in Rigor solution (25 mm imidazole (pH 7.5), 100 mm KCl, 5 mm MgCl2, and 1 mm EGTA) was absorbed on a nitrocellulose-coated flow chamber and incubated on ice for 5 min. After blocking with 1 mg/ml of casein in Rigor solution, the flow chamber was perfused with 0.2 mg/ml of streptavidin (Roche Applied Science) in Rigor solution and incubated on ice for 5 min. After washing away the unbound streptavidin with 1 mg/ml of casein, biotinylated Myo19 (20–200 nm in Rigor solution) was applied to the flow chamber and incubated on ice for 10 min. After washing away the unbound myosin with 1 mg/ml of casein in Rigor solution, rhodamine-phalloidin-labeled F-actin (7 nm in Rigor solution) was applied to the flow chamber and incubated on ice for 5 min. The unbound F-actin was washed away by Motility buffer I (Rigor solution plus 100 mm β-mercaptethanol, 2.5 mg/ml of glucose, 2 units/μl of catalase, 40 units/ml of glucose oxidase). Before observation, the flow chamber was perfused with Motility buffer II (Motility buffer I plus 0.5% methylcellulose, 6 μm RLC9, 6 μm RLC12b, and 0.5–2 mm ATP). The movement of F-actin was recorded at 25 °C under an Olympus IX71 inverted microscope. The velocity of F-actin movement was determined with in-house written Matlab (Mathworks, Inc.) program (32). In case that many actin filaments did not move smoothly or did not move, we pre-spun myosin proteins with F-actin in the presence of 1 mm ATP before application to the coverslip and washed the coverslip with unlabeled F-actin before application of labeled F-actin.
Actin-gliding activities of myosin-5a and skeletal muscle myosin were measured similarly with the following changes. For myosin-5a, myosin protein (10–100 nm in Rigor solution) was attached to the glass surface directly instead of via biotinylated-BSA/streptavidin and motility buffer II contained 12 μm CaM instead of 6 μm RLC9 and 6 μm RLC12b. For skeletal muscle myosin, myosin protein in high salt buffer (0.5–2 mg/ml in 0.6 m KCl, 1 mm EDTA, 1 mm DTT, and 10 mm potassium phosphate, pH 6.5) was attached to the glass surface directly and motility buffer II contains 25 mm KCl.
To determine the directionality of myosin on F-actin, dual fluorescence-labeled actin filaments were used for actin-gliding assay. Dual fluorescence-labeled actin filaments were prepared according to Homma et al. (15).
RESULTS
Identification of the Light Chains Associated with Myo19
To investigate the motor function of Myo19, we produced a number of Myo19-truncated constructs containing the motor domain and various numbers of IQ motifs (Fig. 2A). To facilitate purification, a FLAG tag was attached to the N terminus of each constructs.
FIGURE 2.
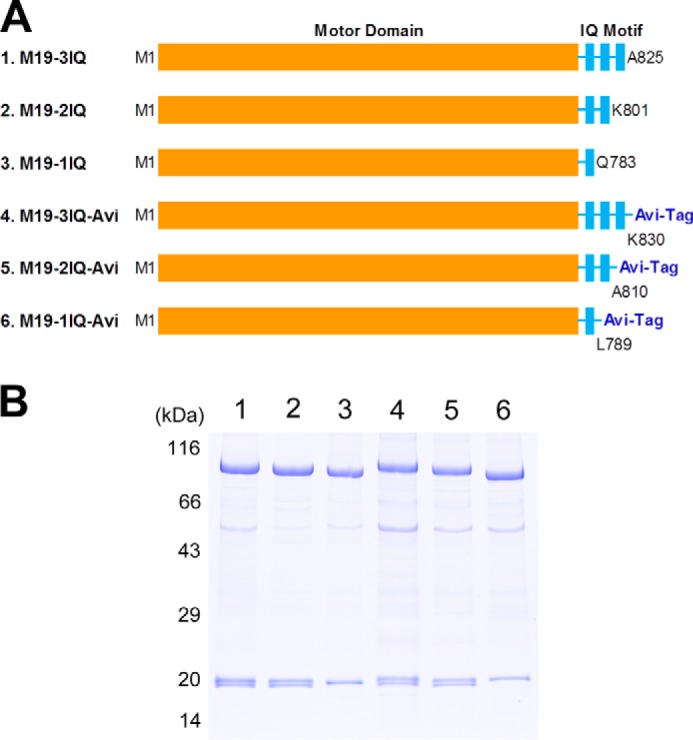
Myo19 truncated constructs. A, schematic diagrams of Myo19 constructs used in this study. To facilitate purification, a FLAG tag was attached to the N terminus of each construct. To facilitate actin-gliding assay, an Avi-tag was attached to the C terminus of the last three constructs. Amino acid numbers of the constructs are indicated. B, SDS-PAGE (4–20%) of purified Myo19-truncated constructs. Myo19-truncated construct was coexpressed with RLC9 and RLC12b in Sf9 cells and purified by anti-FLAG affinity agarose. Lane 1, M19-3IQ; 2, M19-2IQ; 3, M19-1IQ; 4, M19-3IQ-Avi; 5, M19-2IQ-Avi; 6, M19-1IQ-Avi.
Because CaM is a common light chain bound to the IQ motif of the unconventional myosin identified so far, we co-expressed Myo19-truncated constructs with CaM in Sf9 cells and purified Myo19 with anti-FLAG-agarose. We found that CaM copurified with M19-1IQ was barely detectable and the CaM copurified with M19-2IQ or M19-3IQ was far below stoichiometry (Fig. 3A). The lower than expected amount of CaM associated with Myo19 was not due to insufficient CaM in Sf9 cells, as a similar amount of baculovirus encoding CaM was sufficient to saturate six IQ motifs of myosin-5a (data not shown). Thus, we expected that the light chain other than CaM binds to the IQ motifs of Myo19.
FIGURE 3.
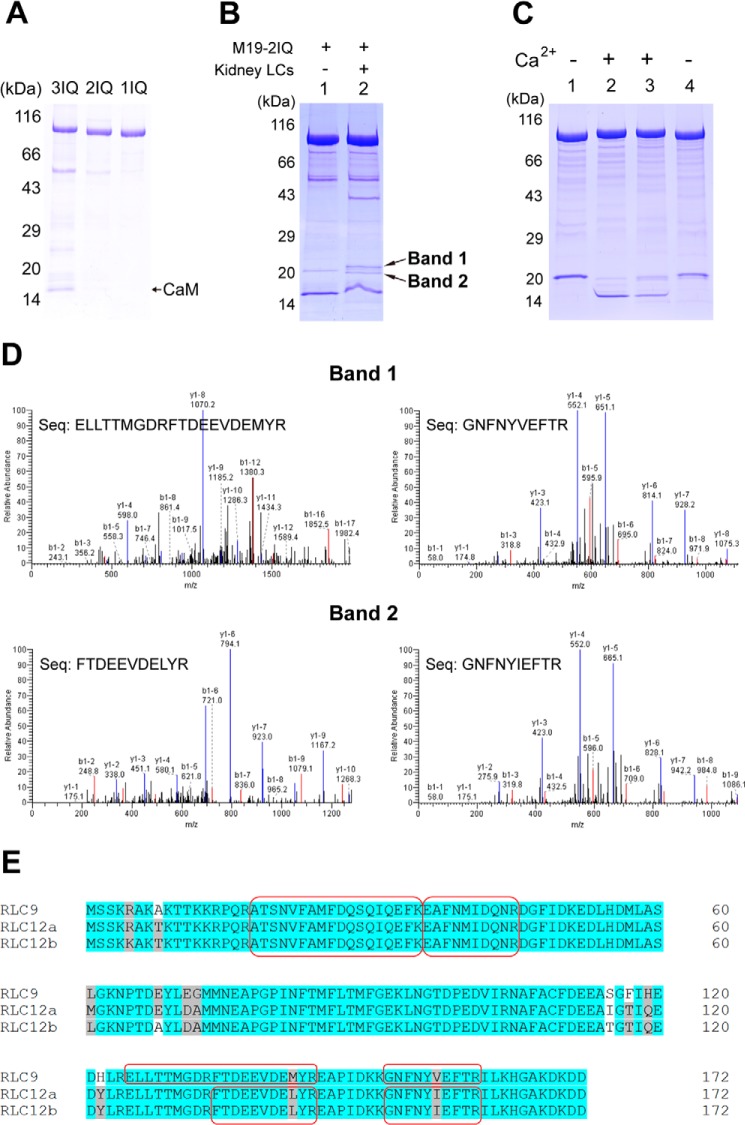
Identification of the light chain of Myo19. A, SDS-PAGE (4–20%) of the purified Myo19-truncated constructs coexpressed with CaM in Sf9 cells. B, SDS-PAGE (4–20%) of M19-2IQ co-purified with crude extract of myosin light chain from mouse kidney. Lane 1, purified M19-2IQ expressed in Sf9 cells; 2, re-purified M19-2IQ after incubated with crude extract of myosin light chain from mouse kidney. Band-1 and band-2 indicate the two specific ∼20-kDa bands co-purified with M19-2IQ. C, CaM gel shift assay of the light chain co-purified with M19-2IQ. Lanes 1 and 2, purified M19-2IQ expressed in Sf9 cells; lanes 3 and 4, re-purified M19-2IQ after incubation with crude extract of myosin light chain from mouse kidney. Lanes 1 and 4 were run under EGTA conditions, lanes 2 and 3 were run under Ca2+ conditions. For details, see ”Experimental Procedures.“ D, MS/MS analysis of band-1 and band-2 in lane 2, the two specific ∼20-kDa bands co-purified with M19-2IQ. Two peptides corresponding to RLC9 and RLC12b were detected in band-1 and band-2, respectively. The sequences of the peptides were shown. E, amino acid sequence alignment of three isoforms of mouse RLC. The sequences shaded in gray are conserved but not identical residues, and the ones in white are non-conserved residues. The peptide sequences identified in MS/MS are boxed.
To identify the light chains associated with Myo19, we first prepared the crude extract of myosin light chains from mouse kidney. We expected that mouse kidney contains the light chains for Myo19, as Northern blot shows that Myo19 is expressed at a high level in multiple human tissues, including kidney (7). We mixed the purified M19-2IQ with the crude extract of myosin light chains from kidney, and re-purified M19-2IQ with anti-FLAG-agarose. The re-purified M19-2IQ contains two distinct ∼20-kDa bands in addition to the ∼17-kDa band (Fig. 3B).
We suspected the ∼17-kDa band is CaM. Therefore, we performed CaM gel shift assay. As expected, the ∼17-kDa band displayed a motility shift in response to Ca2+ during SDS-PAGE, indicating that the ∼17-kDa band was CaM (Fig. 3C). To determine the identities of the two ∼20-kDa bands, we performed mass spectrometry analysis. The amino acid sequence of the upper ∼20-kDa band matched mouse RLC9 and the lower ∼20-kDa band matched mouse RLC12a or RLC12b (Fig. 3D).
The mouse express three highly conserved RLCs for smooth muscle/non-muscle myosin-2, i.e. RLC9, RLC12a, and RLC12b (33). RLC12a and RLC12b are highly homologous with only 4 of 172 amino acid residue differences. RLC9 shares ∼95% identity with RLC12a and RLC12b (Fig. 3E). Although RLC9 is expressed at a high level in smooth muscle (thus named smooth muscle myosin RLC) and RLC12b is absent in striate muscle, all three RLCs are expressed in non-muscle tissues (33).
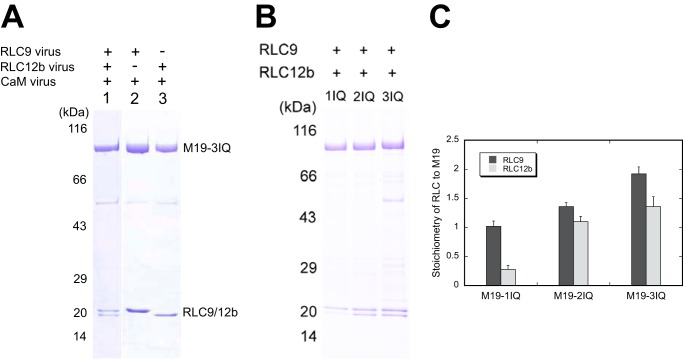
To determine whether RLC associates with Myo19, we co-expressed M19-3IQ with RLC9, RLC12b, and CaM in Sf9 cells and purified M19-3IQ with anti-FLAG-agarose. The purified M19-3IQ strongly associated with RLC9 and/or RLC12b, but not with CaM (Fig. 4A), suggesting that the light chain of Myo19 is RLC but not CaM. To determine the binding site of RLC in Myo19, we co-expressed M19-1IQ, -2IQ, or -3IQ with RLC9 and RLC12b in Sf9 cells. The purified M19-1IQ stoichiometrically associated with RLC9, whereas M19-2IQ associated with RLC9 and RLC12b in approximately equal stoichiometry (Fig. 4B), indicating that the light chain bound to the first IQ motif is RLC9 and that to the second IQ motif is RLC12b. On the other hand, M19-3IQ associated with approximate 2 m RLC9 and 1 m RLC12b, indicating that the light chain bound to the third IQ motif is RLC9.
FIGURE 4.
Identification of the light chain binding sites in Myo19. Myo19-truncated constructs were co-expressed with different groups of light chains. The purified Myo19 samples were subjected to SDS-PAGE (4–20%) and Coomassie Blue staining. A, SDS-PAGE of purified M19-3IQ coexpressed with the different light chains (indicated at the top of the gel) in Sf9 cells. B, SDS-PAGE of purified M19-1IQ, -2IQ, and -3IQ coexpressed with RLC9 and RLC12b. C, stoichiometry of RLC9 and RLC12b to Myo19 heavy chain. Quantifications of the light chain and heavy chain were done with ImageJ software. The molar ratio of RLC versus the heavy chain was calculated based on the corresponding molecular weight masses. Values are mean ± S.D. from three independent assays.
Therefore, we coexpressed Myo19-truncated constructs with RLC9 and RLC12b in Sf9 cells and purified the expressed protein with anti-FLAG affinity chromatography. All purified Myo19-truncated constructs contained stoichiometric RLC9 and/or RLC12b (Fig. 2B).
The Actin-activated ATPase Activity of Myo19
One basic property of myosin is the ability to hydrolyze ATP. Most characterized myosins have ATPase activity, which is enhanced by actin. The motor domain of Myo19 shares 30–40% homology with myosin-5a, myosin-6, and myosin-7a/b, and contains the highly conserved sequences in the ATP-binding site, including P-loop, Switch-I, and Switch-II, suggesting that Myo19 should have ATPase activity.
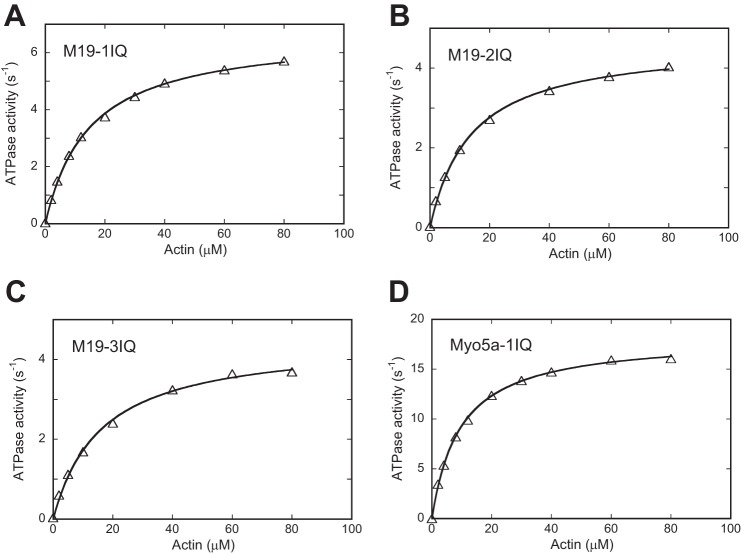
We measured the steady-state ATPase activities of M19-1IQ, -2IQ, and -3IQ, using an ATP-regenerating system in the presence of various concentrations of actin. All three M19 constructs exhibited low ATPase activity in the absence of actin of less than 0.04 s−1 and actin markedly enhanced the ATPase activity of all three M19 constructs (Table 1). The actin dependence exhibited a hyperbolic saturation curve versus the increasing actin concentration, defining Vmax and Kactin values of 6.75 ± 0.33 s−1 and 15.2 ± 4.0 μm for M19-1IQ (Fig. 5A and Table 1). The Vmax was approximately one-third of Myo5a-1IQ, a truncated myosin-5a containing the motor domain and the 1st IQ motif. The Kactin of M19-1IQ was similar to that of Myo5a-1IQ (9.6 μm) under the same assay conditions (Fig. 5, A and D, and Table 1), indicating that, similar to myosin-5a, Myo19 has a high affinity to actin during ATP hydrolysis cycle. Similar results were obtained for M19-2IQ and -3IQ (Fig. 5, B and C, and Table 1).
TABLE 1.
Summary of actin-activated ATPase activity and actin-gliding activity of Myo19 truncated constructs
Actin-activated ATPase activities were measured as described in the legend to Fig. 5 except with the indicated concentration of NaCl. Velocity of actin filaments in actin-gliding activity were measured as described in the legend to Fig 7. All data, except that of Myo5a-1IQ, are the mean ± S.D. of 2–6 independent assays of at least 2 independent preparations.
| Conditions | V0 | Vmax | Kactin | Velocity | |
|---|---|---|---|---|---|
| s−1 | μm | nm/s | |||
| M19-1IQ | 25 mm NaCl | 5.73 ± 0.04 (2) | 10.3 ± 1.3 (2) | 24.5 ± 1.0 (2) | |
| 50 mm NaCl | 0.032 ± 0.001 (3) | 6.75 ± 0.33 (6) | 15.2 ± 4.0 (6) | 27.1 ± 1.2 (2) | |
| 100 mm NaCl | 7.62 ± 0.22 (2) | 24.8 ± 2.3 (2) | 34.5 ± 2.7 (6) | ||
| 150 mm NaCl | 8.77 ± 0.04 (2) | 45.6 ± 5.2 (2) | |||
| M19-2IQ | 50 mm NaCl | 0.035 ± 0.002 (6) | 4.44 ± 0.65 (3) | 17.2 ± 4.9 (3) | 42.4 ± 2.4 (6) |
| M19-3IQ | 50 mm NaCl | 0.035 ± 0.001 (6) | 4.20 ± 0.72 (3) | 20.0 ± 6.8 (3) | 46.8 ± 4.7 (6) |
| Myo5a-1IQ | 50 mm NaCl | 18.21 | 9.60 |
FIGURE 5.
Actin-activated ATPase activities of Myo19-truncated constructs. ATPase activities of M19-1IQ (A), M19-2IQ (B), and M19-3IQ (C) were measured in a solution containing 50 mm NaCl, 20 mm MOPS (pH 7.0), 1 mm MgCl2, 1 mm EGTA, 0.25 mg/ml of BSA, 1 mm DTT, 0.5 mm ATP, 2.5 mm phosphoenol pyruvate, 20 units/ml of pyruvate kinase, 4 μm RLC9, 4 μm RLC12b, and 0–80 μm actin. ATPase activity of Myo5a-1IQ (D) was measured similarly except that RLCs was substituted with 12 μm CaM. Curves are the least squares fits of the data points based upon the equation, V = (Vmax × [actin]/Kactin + [actin]). Although multiple independent assays of at least 2 preparations have been performed, data from a single assay are presented. Vmax and Kactin from multiple assays are summarized in Table 1.
The actin-activated ATPase activity of M19 was moderately sensitive to ionic strength. With the increase of NaCl from 25 to 150 mm, KActin of M19-1IQ was increased from 10.3 to 45.6 μm and Vmax was moderately increased from 5.73 to 8.77 s−1 (Table 1).
Although addition of exogenous RLCs had no effect on the actin-activated ATPase activities of M19-1IQ and -2IQ, exogenous RLCs (4 μm RLC9 and 4 μm RLC12b) nearly double the actin-activated ATPase activity of M19-3IQ (Fig. 6A). These results suggest that the affinity between RLCs and the IQ3 motif is significantly lower than that between RLCs and IQ1/2. The relatively lower affinity between RLC and M19-3IQ is not due to impropriate truncation of IQ3, because the actin-activated ATPase activity of M19-3IQ with an extended C terminus (residues of 1–830) showed a similar RLC dependence (data not shown). To quantify the affinity between light chain and IQ3, we measured the actin-activated ATPase activity of M19-3IQ in the presence of exogenous light chains. As shown in Fig. 6B, actin-activated ATPase activity of M19-3IQ was markedly activated by exogenous light chain with a hyperbolic saturation curve, defining apparent Kd (dissociation constant) of 0.076, 0.300, 0.202, and 0.328 μm for RLC9, RLC12a, RLC12b, and CaM, respectively. Among the four light chains, RLC9 exhibited the lowest Kd, consistent with the conclusion that the light chain bound to IQ3 is RLC9 (Fig. 4B).
FIGURE 6.
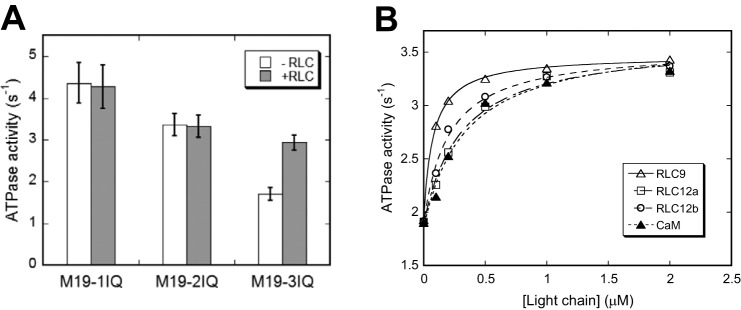
Exogenous light chain enhances actin-activated ATPase activity of M19–3IQ. A, actin-activated ATPase activity of Myo19-truncated constructs in the presence or absence of 4 μm RLC9 and RLC12b. Values are mean ± S.D. from two to four independent assays. B, effects of exogenous light chain on actin-activated ATPase activity of M19-3IQ. ATPase assay was performed in 50 mm NaCl, 20 mm MOPS (pH 7.0), 1 mm MgCl2, 1 mm EGTA, 0.25 mg/ml of BSA, 1 mm DTT, 0.5 mm ATP, 2.5 mm phosphoenol pyruvate, 20 units/ml of pyruvate kinase, 40 μm actin, and 0–2 μm light chain. The stimulation of ATPase activities by exogenous light chain was fitted with a hyperbolic equation: V = V0 + Vmax × [LC]/(Kd + [LC]), where V0, the activity in the absence of exogenous light chain; Vmax, the activity stimulated by exogenous light chain; [LC], the concentration of exogenous light chain; Kd, the apparent affinity between the light chain and M19-IQ3. The apparent Kd values are 0.076 ± 0.027, 0.321 ± 0.076, 0.223 ± 0.068, and 0.335 ± 0.049 μm for RLC9, RLC12a, RLC12b, and CaM, respectively. Data were average of two independent assays.
In Vitro Actin-gliding Activity of Myo19
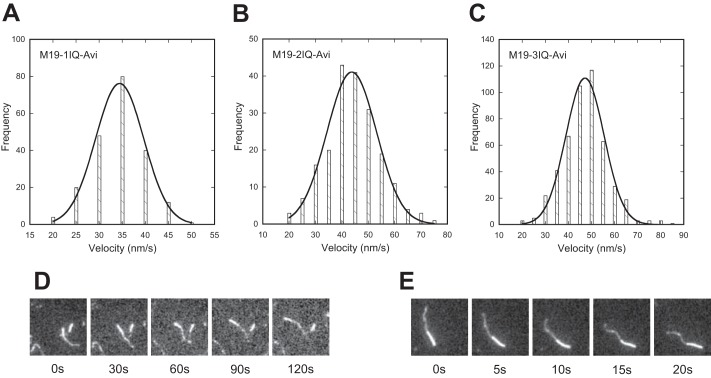
It has been shown that overexpression of Myo19 in mammalian cells enhances mitochondrial movement (7), suggesting that Myo19 functions as a molecular motor in vivo. Here we characterized the molecular motor function of Myo19 constructs using an in vitro actin-gliding assay. To facilitate the attachment of Myo19 on the nitrocellulose-coated glass surface, we fused an Avi-tag to the C terminus of Myo19 constructs, producing M19–1IQ-Avi, M19–2IQ-Avi, and M19–3IQ-Avi (Fig. 2). The purified Myo19-truncated construct with the C-terminal Avi-tag was biotinylated by BirA ligase in vitro and then attached on the glass surface via biotinylated BSA and streptavidin. To ensure that all IQ motifs of Myo19 constructs were full occupied, we added exogenous RLC9 and RLC12b in the actin-gliding assay system. All three Myo19 constructs supported robust movement of actin filaments (supplemental Movies S1–S3). The velocities of actin filaments glided by M19-1IQ-Avi, M19-2IQ-Avi, and M19-3IQ-Avi were 34.5 ± 2.7, 42.4 ± 2.4, and 46.8 ± 4.7 nm/s, respectively (Fig. 7, A–C). The actin-gliding velocity of Myo19 was moderately sensitive to ionic strength. With the increase of NaCl from 25 to 100 mm, the actin-gliding velocity of M19-1IQ was moderately increased from 24.5 to 34.5 nm/s (Table 1).
FIGURE 7.
Actin-gliding activity of Myo19. Biotinylated Myo19-truncated constructs were attached to a coverslip via biotinylated BSA and streptavidin, and the movements of rhodamine-labeled actin filaments were recorded with an Olympus IX71 inverted microscope at room temperature (about 25 °C). The velocity was analyzed by MATLAB. The solid line shows a fit to a single Gaussian curve. For details, see ”Experimental Procedures.“ A–C, histogram of multimolecular actin-gliding velocity of Myo19-truncated constructs, i.e. M19-1IQ-Avi (A), M19-2IQ-Avi (B), and M19-3IQ-Avi (C). D and E, direction of the actin filament translocated by M19-1IQ-Avi (D) and Myo5a-ΔT. E, actin filaments were dual fluorescence-labeled. Plus-end was labeled with Alexa 488 (dim) and minus end with rhodamine (bright). Actin filaments were moved toward the minus-end by M19-1IQ-Avi and Myo5a-ΔT.
We noticed that the actin-gliding velocity of Myo19 was significantly lower than that of myosin-5a. Under the same assay conditions, the velocity of Myo5a-ΔT (a mouse myosin-5a construct with the GTD deleted) (24) was 287 nm/s (supplemental Movie S4), similar to the published value (34, 35). It is noteworthy that, similar to myosin-5a, Myo19 constructs at a low density (∼25 nm) were sufficient to support the movement of actin filaments and Myo19 constructs at a relatively high density (>50 nm) to rapidly shred actin filaments into small fragments, suggesting that M19 is a high-duty ratio motor.
Myo19 Is a Plus-end-directed Motor
It is of interest to determine the directionality of Myo19 movement. Sequence alignment revealed that, compared with myosin-5a, Myo19 contains two unique inserts in a region of the motor domain known as the converter (Fig. 1B). In the crystal structure of myosin-5a (PDB 1W7J), the two inserts are adjacent to one another and the second one is adjacent to the tip of the relay helix (Fig. 1C). It is known that the minus-end-directed movement of class VI myosin is dictated by a unique insert of ∼38 amino acids located between the converter and the lever arm (10, 11).
To determine the directionality of Myo19 movement, we performed a actin-gliding assay using dual fluorescence-labeled actin filaments. We found that most of the filaments (>95%) in the presence of M19-1IQ exhibited plus-end-directed movements, similar to that of myosin-5a, a known plus-end-directed motor (Fig. 7, D and E). The few actin filaments (<5%) that appeared to support minus-end-directed movements were likely due to mislabeling of the fluorescence probes. These results indicate that Myo19 is a plus-end-directed motor and the unique insert in the converter of Myo19 plays a role other than reverse direction of movement of the lever arm.
ADP Release Is a Rate-limiting Step for ATPase Cycle of Acto-Myo19
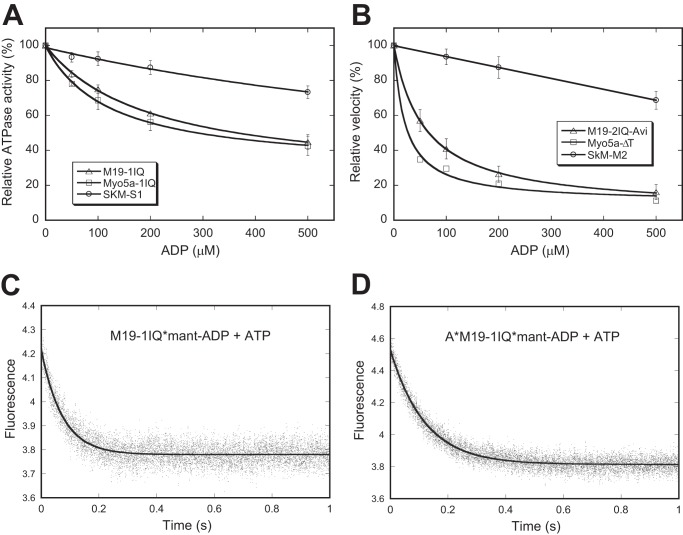
ADP release from actomyosin is the rate-limiting step for the ATP hydrolysis cycle in high-duty ratio motors, such as myosin-5a and myosin-6 (12, 13). It has been shown that the steady-state ATPase activity of myosin-5a and myosin-6 are strongly inhibited by ADP, whereas the low-duty ratio motor such as skeletal muscle myosin is only slightly inhibited by ADP (13, 36). To examine the effects of ADP on the steady-state ATPase activity of Myo19, we measured the actin-activated ATPase activity of M19-1IQ in the absence of the ATP regeneration system and in the presence of various initial ADP concentrations (0 to 500 μm) (Fig. 8A). As controls, the same experiments were conducted with Myo5a-1IQ (a myosin-5a-truncated construct containing the motor domain and the 1st IQ motif) and skeletal muscle myosin S1. The activity of skeletal muscle myosin S1 was inhibited <25% by 500 μm ADP, whereas that of Myo5a-1IQ was strongly inhibited by ADP (about 60% in the presence of 500 μm ADP) (Fig. 8A). Similar to that of Myo5a-1IQ, the actin-activated ATPase activity of M19-1IQ was strongly inhibited by ADP (about 60% in the presence of 500 μm ADP).
FIGURE 8.
ADP-off is the rate-limiting step for the motor activity of Myo19. A, effects of ADP on actin-activated ATPase activity of M19-1IQ, Myo5a-1IQ, and SkM-S1. Actin-activated ATPase activity was measured in the presence of 40 μm actin, 500 μm ATP, and 0–500 μm ADP and the released phosphate was determined by the malachite green method. The inhibition of ATPase activities by ADP was fitted with a hypophobic equation. B, effects of ADP on actin-gliding activity of M19–1IQ, Myo5a-ΔT, and skeletal muscle myosin (SkM-M2). Actin-gliding activity was measured in the presence of 500 μm ATP and 0–500 μm ADP. In the absence of ADP, the actin-gliding activities of M19-2IQ-Avi, Myo5a-ΔT, and skeletal muscle myosin were 40, 280, and 3.64 μm/s, respectively. C and D, dissociation of mant-ADP from M19-1IQ (C) and acto-M19-1IQ (D). The fluorescence intensity of mant-ADP was recorded in a stopped flow after mixing 1 mm ATP with and 1 μm mant-ADP and 1 μm M19–1IQ in the absence (C) or presence (D) of 1.5 μm phalloidin-stabilized actin filaments. The smooth line is the fit to single exponential kinetics, with Kobs of 13.65 s−1 (C) and 8.37 s−1 (D).
To examine the effects of ADP on actin-gliding activity of Myo19, we measured the rate of actin-gliding activity of M19-2IQ-Avi in the presence of a constant initial ATP concentration of 500 μm and various initial ADP concentrations (0 to 500 μm). As shown in Fig. 8B, ADP strongly inhibited the actin-gliding activities of M19-2IQ-Avi and Myo5a-ΔT, but only slightly inhibited that of skeletal muscle myosin.
These results suggest that, similar to that of myosin-5a, ADP release is likely a rate-limiting step in the ATP hydrolysis cycle of acto-Myo19. To test this possibility, we directly measured the ADP release rate by following the fluorescence decrease of mant-ADP from acto-M19-1IQ upon ATP binding in a stopped-flow instrument. The transient was best fit to single-exponential kinetics, and the rate of mant-ADP dissociation was ∼8 s−1 (Fig. 8D), which was comparable with the Vmax of the steady-state ATPase activity, indicating that ADP release is a rate-limiting step in the ATP hydrolysis cycle of acto-M19-1IQ.
Interestingly, the rate constant for dissociation of mant-ADP from M19-1IQ (in the absence of actin) was ∼14 s−1 (Fig. 8C), indicating the ADP release is not a rate-limiting step for the M19-1IQ ATPase cycle in the absence of actin and actin does not greatly affect the ADP release rate from M19-1IQ. A similar effect of actin on ADP release has been found for the myosin-6 ATPase cycle, where the dissociation rate constants of mant-ADP in the absence and presence of actin are virtually equal (13). Taken together, the above results indicate that ADP release is the rate-limiting step of the ATPase cycle for acto-Myo19, suggesting that Myo19 is a molecular motor with high-duty ratio.
Phosphorylation of RLC by MLCK Does Not Affect the Motor Activity of Myo19-truncated Constructs
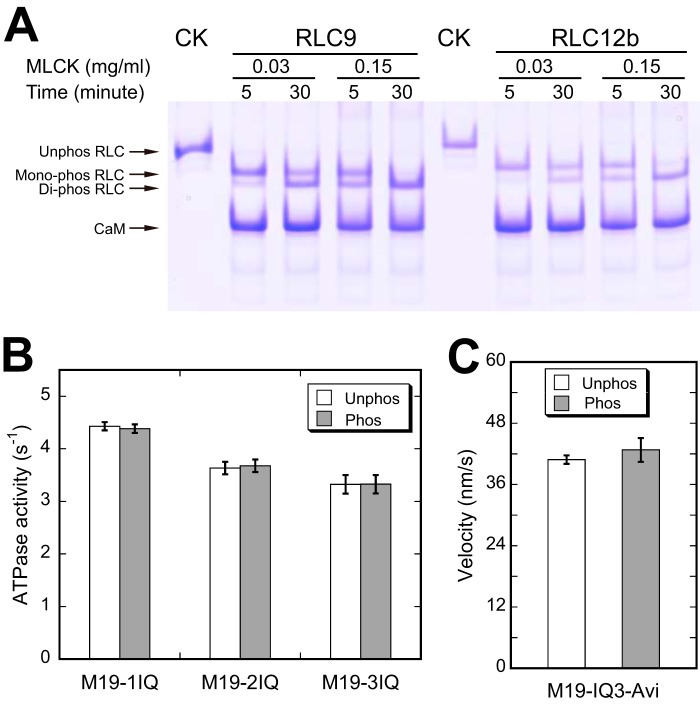
It is well documented that the light chains bound on the IQ motifs of myosin heavy chain generally play a crucial role in the regulation of motor activity. One of the best studied examples is the activation of smooth muscle myosin motor function by phosphorylation of RLC9 catalyzed by MLCK (37). Because the light chain bound to the IQ motifs of Myo19 is RLC, it is possible that phosphorylation of RLC affects the motor activity of Myo19.
To determine whether the RLC bound to Myo19 heavy chain could be phosphorylated by MLCK, we incubated the M19-3IQ·RLC complex with MLCK and examined the phosphorylation of RLC by urea/glycerol PAGE. Under conditions similar to ATPase assay, both RLC9 and RLC12b in M19-3IQ were completely phosphorylated by MLCK within 5 min (Fig. 9A). To investigate the effects of RLC phosphorylation by MLCK on the ATPase activity of Myo19, we measured the actin-activated ATPase activity of Myo19-truncated constructs in the absence or presence of MLCK and CaM. As shown in Fig. 9B, phosphorylation of RLC9 and RLC12b by MLCK did not significantly change the actin-activated ATPase activity of Myo19-truncated constructs. Similarly, phosphorylation of RLC9 and RLC12b did not significantly change the actin-gliding velocities of Myo19-truncated constructs (Fig. 9C).
FIGURE 9.
Effects of RLC phosphorylation on the motor function of Myo19. A, phosphorylation of the RLC of M19-3IQ by MLCK. M19-3IQ was co-expressed with RLC9 or RLC12b in Sf9 cells and purified by anti-FLAG affinity chromatography. The purified M19-3IQ/RLC9 or M19-3IQ/RLC12b was incubated with MLCK (0.03 mg/ml) in phosphorylation solution (30 mm Tris-HCl, pH 7.5, 50 mm KCl, 1 mm DTT, 1 mm MgCl2, 10 μm CaM, 0.1 mm CaCl2, and 1 mm ATP) at 25 °C for the indicated times. Phosphorylation of RLC was determined by urea/glycerol PAGE and visualized by Coomassie Blue staining. CK represents the unphosphorylated control, which was not treated with MLCK. B, effects of RLC phosphorylation on the actin-activated ATPase activity of Myo19. ATPase assay was performed in 50 mm KCl, 25 mm Tris-HCl (pH 7.5), 1 mm MgCl2, 0.25 mg/ml of BSA, 1 mm DTT, 0.5 mm ATP, 2.5 mm phosphoenol pyruvate, 20 units/ml of pyruvate kinase, 40 μm actin, 0.1 mm CaCl2, 2 μm CaM with 0.03 mg/mg of MLCK (phosphorylated conditions) or without MLCK (unphosphorylated conditions) at 25 °C. The reaction was stopped at various times between 4 and 60 min as described under “Experimental Procedures.” C, effects of RLC phosphorylation on in vitro actin-gliding activity of Myo19-3IQ-Avi. Prior to actin-gliding assay, biotinylated M19-3IQ-Avi was phosphorylated by 0.15 mg/ml of MLCK in phosphorylation solution at 25 °C for 5 min. Actin-gliding assay was performed as described in the legend of Fig. 7.
DISCUSSION
In the present study, we analyzed the ATPase activity and the actin-gliding activity of Myo19. Several lines of evidence obtained here show that the enzymatic properties of Myo19 are similar to that of high-duty ratio motors such as myosin-5a. First, the ADP release rate is comparable with the steady-state actin-activated ATPase activity of Myo19, indicating that ADP release is rate-limiting for acto-Myo19. It is well established that ADP release from actomyosin is the rate-limiting step for high-duty ratio motors, such as myosin-5a, -6, and -7, but not for low-duty ratio motors, such as smooth muscle myosin-2 (12, 13, 21, 36). Because myosin·ADP strongly binds to actin, these myosins spend most of their kinetic cycle strongly bound to actin, thus being a high-duty ratio motor. Second, similar to those of high-duty ratio myosin, actin-activated ATPase activity and the actin-gliding activity of Myo19 ADP were strongly inhibited by ADP. Third, the Kactin values of Myo19-truncated constructs are similar to that of myosin-5a, indicating that Myo19 has high affinity to actin during the ATPase hydrolysis cycle. Fourth, similar to myosin-5a, Myo19 at low-density is sufficient to support actin gliding and at high density shred actin filaments into short fragments.
High-duty ratio myosins, including myosin-5a, -6, -7, and -10, can move processively along the actin filament upon proper dimerization (16–18, 20). Several forms of dimerization of myosin motor domain have so far been identified, including parallel coiled-coil (such as myosin-5a) (38), antiparallel coiled-coil (such as myosin-10 (39)), and cargo-binding protein (such as myosin-6 (40)). Sequence analysis of the Myo19 tail reveals neither parallel coiled-coil (7) nor antiparallel coiled-coil. It remains to be determined if Myo19 forms a dimer upon binding to its cargo and functions as a processive motor in vivo.
It is intriguing that Myo19 IQ motifs bind to RLCs of smooth muscle and nonmuscle myosin-2, but not CaM. Both CaM and RLC belong to the EF-hand protein family. Both have amino- and carboxyl-terminal lobes separated by a flexible helix and each lobe contains a pair of EF-hands. Each EF-hand in CaM is able to bind one Ca2+ ions, whereas that in RLC lacks the ability to bind Ca2+ due to the absence of key residues for Ca2+ binding (41). Although CaM has broad roles in cells and binds to diversified targets including myosin, RLC is restricted to myosin.
It is known that RLCs bind to the IQ2 motif of smooth muscle and nonmuscle myosin-2 (42, 43). The consensus sequence of the RLC binding IQ motif is AXXXφQRN(X)6VKXWXWWR, where φ is Ile, Leu, Val, or Met, X is any amino acid (Fig. 1D). In the crystal structure of the scallop myosin-2 regulatory domain (IQ1/2 in complex with essential light chain and RLC), the core residues (φQRN) of IQ2 form strong interactions with the C-lobe of RLC and the last consensus residues (WXWWR) form a sharp bend and interact with the N-lobe of RLC (44). The RLC-binding IQ motif was also identified in myosin-18 (45). Guzik-Lendrum et al. (46) recently demonstrated that RLC indeed binds to myosin-18.
Alignment of 11 Myo19 revealed three non-canonical RLC-binding IQ motifs, i.e. AXXIQXXWRR (Myo19-IQ1), AAXXIQAAXRSWLXRKXIXXXH (Myo19-IQ2), and AAXXIKXXWXXWRXXMXXLA (Myo19-IQ3), where X is any amino acid, underlined residues are not essential for RLC binding (Fig. 1D). The C-terminal 8 residues (RKXIXXXH) in Myo19-IQ2 and the C-terminal 5 residues (MXXLA) in Myo19-IQ3 are not essential for RLC binding, because our M19-2IQ and M19-3IQ constructs do not contain those residues but still stoichiometrically associate with RLCs. The three non-canonical RLC-binding IQ motifs of Myo19 are quite different from the canonical RLC-binding IQ motifs of myosin-2 and myosin-18 (Fig. 1D). The most discernible difference is the absence of the WXWWR sequence in Myo19 IQ motifs. Also, the core consensus residues of IQRN in the canonical RLC-binding IQ motif is different from that in Myo19 IQ motifs. In addition, the RLC-binding motifs of Myo19 are much shorter than the canonical RLC-binding IQ motif. Thus, we expect that the conformation of RLC bound to the IQ motifs of Myo19 is likely different from that bound to the IQ2 of myosin-2 and -18.
It is well established that MLCK-catalyzed phosphorylation of RLC9 in smooth muscle myosin (37, 47) and RLC12b in non-muscle myosin-2 (48) stimulates the motor activity. We found that both RLC12b and RLC9 in Myo19 can be phosphorylated by MLCK, but this phosphorylation has little effect on the motor activity of Myo19-truncated constructs. It is known that phosphorylation of RLC by MLCK regulates the motor activity of smooth muscle myosin but not the truncated smooth muscle myosin having the coiled-coil tail completely deleted (22). Thus, it is possible that MLCK regulates the motor activity of full-length Myo19, but not the truncated Myo19. Unfortunately, we could not obtain a high quality full-length Myo19 protein to test this possibility. Another possibility is that Myo19 is regulated by kinases other than MLCK. Although MLCK phosphorylates RLC at Ser-19 and Thr-18 (37), other kinases, such as Cdc2 kinase (49) and PKC (50), phosphorylate RLC at Ser-1, Ser-2, and Thr-9. More experiments are required for understanding the regulation mechanism of Myo19 by RLC phosphorylation.
While the present study was being conducted, Adikes et al. (8) reported a biochemical and bioinformatic analysis of the Myo19 motor domain. They also concluded that Myo19 is a molecular motor with high-duty ratio. However, they only provided indirect evidence, i.e. MYO19 constructs displayed high actin affinity in the presence of ATP in actin co-sedimentation assays. In the present study, we performed kinetic analysis and found that the ADP release rate is comparable with the maximal steady-state actin-activated ATPase activity of Myo19, indicating that ADP release is rate-limiting for acto-Myo19 and Myo19 is a high-duty ratio motor. All characterized high-duty ratio myosins so far have ADP release as the rate-limiting step.
In addition, there are several major differences between this study and Adikes et al. (8). First, we identified RLCs as the light chain of Myo19 and prepared the recombinant Myo19-truncated constructs with RLCs, whereas Adikes et al. (8) used CaM as the light chain of Myo19. Our study clearly shows that the affinity of RLC to Myo19 is much higher than that of CaM to Myo19. Second, our Myo19 constructs have about twice higher actin-activated ATPase activity than that reported by Adikes et al. (8). One possible reason for the discrepancy is that different protein quantification methods were used in those two studies. To obtain the accurate steady-state ATPase activity of myosin, it is essential to measure myosin concentration properly. Unfortunately, Adikes et al. (8) did not indicate the method for Myo19 quantification. In our study, the Myo19 concentration was determined by SDS-PAGE and Coomassie Blue staining with a known concentration of smooth muscle myosin protein as standard. We constantly found that the Myo19 concentration determined by this method was substantially lower than that estimated by absorbance at A280. Third, our Myo19 constructs have about three times lower actin-gliding velocity than that reported by Adikes et al. (8). At present the reason of this apparent discrepancy is obscure. In our assays, we used a well characterized myosin, i.e. mouse myosin-5a, as control and obtained the velocity of myosin-5a consistent with the published values. We found that the actin-gliding velocities of Myo19-truncated constructs were about one-sixth that of myosin-5a.
In summary, we identified RLCs of smooth and non-muscle myosin-2 as the light chains of Myo19, demonstrated that Myo19 is a functional motor moving to the plus-end of actin filament, and found that ADP release is rate-limiting for the ATP hydrolysis cycle of acto-Myo19. Based upon these results, we concluded that Myo19 is a plus-end-directed, high-duty ratio molecular motor.
Supplementary Material
Acknowledgment
We thank Dr. Howard White (Eastern Virginia Medical School) for reading the manuscript.
This work was supported by National Basic Research Program of China Grant 2013CB932802 and National Natural Science Foundation of China Grants 31201071 and 31171367.

This article contains supplemental Movies S1–S4.
- CaM
- calmodulin
- Myo19
- myosin-19
- MLCK
- myosin light chain kinase
- RLC
- regulatory light chain
- SkM
- skeletal muscle myosin
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1. Hartman M. A., Finan D., Sivaramakrishnan S., Spudich J. A. (2011) Principles of unconventional myosin function and targeting. Annu. Rev. Cell Dev. Biol. 27, 133–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hammer J. A., 3rd, Sellers J. R. (2012) Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13, 13–26 [DOI] [PubMed] [Google Scholar]
- 3. Akhmanova A., Hammer J. A., 3rd (2010) Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 22, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Foth B. J., Goedecke M. C., Soldati D. (2006) From the cover: new insights into myosin evolution and classification. Proc. Natl. Acad. Sci. U.S.A. 103, 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Odronitz F., Kollmar M. (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berg J. S., Powell B. C., Cheney R. E. (2001) A millennial myosin census. Mol. Biol. Cell 12, 780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quintero O. A., DiVito M. M., Adikes R. C., Kortan M. B., Case L. B., Lier A. J., Panaretos N. S., Slater S. Q., Rengarajan M., Feliu M., Cheney R. E. (2009) Human Myo19 is a novel myosin that associates with mitochondria. Curr. Biol. 19, 2008–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adikes R. C., Unrath W. C., Yengo C. M., Quintero O. A. (2013) Biochemical and bioinformatic analysis of the myosin-XIX motor domain. Cytoskeleton 70, 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dominguez R., Freyzon Y., Trybus K. M., Cohen C. (1998) Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell 94, 559–571 [DOI] [PubMed] [Google Scholar]
- 10. Ménétrey J., Bahloul A., Wells A. L., Yengo C. M., Morris C. A., Sweeney H. L., Houdusse A. (2005) The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature 435, 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park H., Li A., Chen L. Q., Houdusse A., Selvin P. R., Sweeney H. L. (2007) The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc. Natl. Acad. Sci. U.S.A. 104, 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De La Cruz E. M., Wells A. L., Rosenfeld S. S., Ostap E. M., Sweeney H. L. (1999) The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. U.S.A. 96, 13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De La Cruz E. M., Ostap E. M., Sweeney H. L. (2001) Kinetic mechanism and regulation of myosin VI. J. Biol. Chem. 276, 32373–32381 [DOI] [PubMed] [Google Scholar]
- 14. Yang Y., Kovács M., Xu Q., Anderson J. B., Sellers J. R. (2005) Myosin VIIB from Drosophila is a high duty ratio motor. J. Biol. Chem. 280, 32061–32068 [DOI] [PubMed] [Google Scholar]
- 15. Homma K., Saito J., Ikebe R., Ikebe M. (2001) Motor function and regulation of myosin X. J. Biol. Chem. 276, 34348–34354 [DOI] [PubMed] [Google Scholar]
- 16. Mehta A. D., Rock R. S., Rief M., Spudich J. A., Mooseker M. S., Cheney R. E. (1999) Myosin-V is a processive actin-based motor. Nature 400, 590–593 [DOI] [PubMed] [Google Scholar]
- 17. Rock R. S., Rice S. E., Wells A. L., Purcell T. J., Spudich J. A., Sweeney H. L. (2001) Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. U.S.A. 98, 13655–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun Y., Sato O., Ruhnow F., Arsenault M. E., Ikebe M., Goldman Y. E. (2010) Single-molecule stepping and structural dynamics of myosin X. Nat. Struct. Mol. Biol. 17, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nagy S., Ricca B. L., Norstrom M. F., Courson D. S., Brawley C. M., Smithback P. A., Rock R. S. (2008) A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. U.S.A. 105, 9616–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Y., Kovács M., Sakamoto T., Zhang F., Kiehart D. P., Sellers J. R. (2006) Dimerized Drosophila myosin VIIa: a processive motor. Proc. Natl. Acad. Sci. U.S.A. 103, 5746–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cremo C. R., Geeves M. A. (1998) Interaction of actin and ADP with the head domain of smooth muscle myosin: implications for strain-dependent ADP release in smooth muscle. Biochemistry 37, 1969–1978 [DOI] [PubMed] [Google Scholar]
- 22. Ma R. N., Mabuchi K., Li J., Lu Z., Wang C. L., Li X. D. (2013) Cooperation between the two heads of smooth muscle myosin is essential for full activation of the motor function by phosphorylation. Biochemistry 52, 6240–6248 [DOI] [PubMed] [Google Scholar]
- 23. Lu Z., Shen M., Cao Y., Zhang H. M., Yao L. L., Li X. D. (2012) Calmodulin bound to the first IQ motif is responsible for calcium-dependent regulation of myosin 5a. J. Biol. Chem. 287, 16530–16540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X. D., Jung H. S., Mabuchi K., Craig R., Ikebe M. (2006) The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J. Biol. Chem. 281, 21789–21798 [DOI] [PubMed] [Google Scholar]
- 25. Pollard T. D. (1982) Myosin purification and characterization. Methods Cell Biol. 24, 333–371 [DOI] [PubMed] [Google Scholar]
- 26. Weeds A. G., Taylor R. S. (1975) Separation of subfragment-1 isoenzymes from rabbit skeletal muscle myosin. Nature 257, 54–56 [DOI] [PubMed] [Google Scholar]
- 27. Wang Z., Edwards J. G., Riley N., Provance D. W., Jr., Karcher R., Li X. D., Davison I. G., Ikebe M., Mercer J. A., Kauer J. A., Ehlers M. D. (2008) Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell 135, 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ikebe M., Reardon S., Schwonek J. P., Sanders C. R., 2nd, Ikebe R. (1994) Structural requirement of the regulatory light chain of smooth muscle myosin as a substrate for myosin light chain kinase. J. Biol. Chem. 269, 28165–28172 [PubMed] [Google Scholar]
- 29. Li X. D., Jung H. S., Wang Q., Ikebe R., Craig R., Ikebe M. (2008) The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proc. Natl. Acad. Sci. U.S.A. 105, 1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kodama T., Fukui K., Kometani K. (1986) The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J. Biochem. 99, 1465–1472 [DOI] [PubMed] [Google Scholar]
- 31. Cull M. G., Schatz P. J. (2000) Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 326, 430–440 [DOI] [PubMed] [Google Scholar]
- 32. Nitzsche B., Ruhnow F., Diez S. (2008) Quantum-dot-assisted characterization of microtubule rotations during cargo transport. Nat. Nanotechnol. 3, 552–556 [DOI] [PubMed] [Google Scholar]
- 33. Park I., Han C., Jin S., Lee B., Choi H., Kwon J. T., Kim D., Kim J., Lifirsu E., Park W. J., Park Z. Y., Kim do H., Cho C. (2011) Myosin regulatory light chains are required to maintain the stability of myosin II and cellular integrity. Biochem. J. 434, 171–180 [DOI] [PubMed] [Google Scholar]
- 34. Homma K., Saito J., Ikebe R., Ikebe M. (2000) Ca2+-dependent regulation of the motor activity of myosin V. J. Biol. Chem. 275, 34766–34771 [DOI] [PubMed] [Google Scholar]
- 35. Wang F., Chen L., Arcucci O., Harvey E. V., Bowers B., Xu Y., Hammer J. A., 3rd, Sellers J. R. (2000) Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J. Biol. Chem. 275, 4329–4335 [DOI] [PubMed] [Google Scholar]
- 36. De La Cruz E. M., Sweeney H. L., Ostap E. M. (2000) ADP inhibition of myosin V ATPase activity. Biophys. J. 79, 1524–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kamm K. E., Stull J. T. (1989) Regulation of smooth muscle contractile elements by second messengers. Annu. Rev. Physiol. 51, 299–313 [DOI] [PubMed] [Google Scholar]
- 38. Cheney R. E., O'Shea M. K., Heuser J. E., Coelho M. V., Wolenski J. S., Espreafico E. M., Forscher P., Larson R. E., Mooseker M. S. (1993) Brain myosin-V is a two-headed unconventional myosin with motor activity.[comment]. Cell 75, 13–23 [DOI] [PubMed] [Google Scholar]
- 39. Lu Q., Ye F., Wei Z., Wen Z., Zhang M. (2012) Antiparallel coiled-coil-mediated dimerization of myosin X. Proc. Natl. Acad. Sci. U.S.A. 109, 17388–17393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu C., Feng W., Wei Z., Miyanoiri Y., Wen W., Zhao Y., Zhang M. (2009) Myosin VI undergoes cargo-mediated dimerization. Cell 138, 537–548 [DOI] [PubMed] [Google Scholar]
- 41. Moncrief N. D., Kretsinger R. H., Goodman M. (1990) Evolution of EF-hand calcium-modulated proteins: I. relationships based on amino acid sequences. J. Mol. Evol. 30, 522–562 [DOI] [PubMed] [Google Scholar]
- 42. Li X. D., Saito J., Ikebe R., Mabuchi K., Ikebe M. (2000) The interaction between the regulatory light chain domains on two heads is critical for regulation of smooth muscle myosin. Biochemistry 39, 2254–2260 [DOI] [PubMed] [Google Scholar]
- 43. Cremo C. R., Wang F., Facemyer K., Sellers J. R. (2001) Phosphorylation-dependent regulation is absent in a nonmuscle heavy meromyosin construct with one complete head and one head lacking the motor domain. J. Biol. Chem. 276, 41465–41472 [DOI] [PubMed] [Google Scholar]
- 44. Houdusse A., Cohen C. (1996) Structure of the regulatory domain of scallop myosin at 2-Å resolution: implications for regulation. Structure 4, 21–32 [DOI] [PubMed] [Google Scholar]
- 45. Tan I., Yong J., Dong J. M., Lim L., Leung T. (2008) A tripartite complex containing MRCK modulates lamellar actomyosin retrograde flow. Cell 135, 123–136 [DOI] [PubMed] [Google Scholar]
- 46. Guzik-Lendrum S., Heissler S. M., Billington N., Takagi Y., Yang Y., Knight P. J., Homsher E., Sellers J. R. (2013) Mammalian myosin-18A, a highly divergent myosin. J. Biol. Chem. 288, 9532–9548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cremo C. R., Hartshorne D. J. (2008) Smooth-muscle myosin II. in Proteins and Cell Regulation, pp. 125, 169, Springer, New York [Google Scholar]
- 48. Hu A., Wang F., Sellers J. R. (2002) Mutations in human nonmuscle myosin IIA found in patients with May-Hegglin anomaly and Fechtner syndrome result in impaired enzymatic function. J. Biol. Chem. 277, 46512–46517 [DOI] [PubMed] [Google Scholar]
- 49. Satterwhite L. L., Lohka M. J., Wilson K. L., Scherson T. Y., Cisek L. J., Corden J. L., Pollard T. D. (1992) Phosphorylation of myosin-II regulatory light chain by cyclin-p34cdc2: a mechanism for the timing of cytokinesis. J. Cell Biol. 118, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ikebe M., Hartshorne D. J., Elzinga M. (1987) Phosphorylation of the 20,000-dalton light chain of smooth muscle myosin by the calcium-activated, phospholipid-dependent protein kinase. Phosphorylation sites and effects of phosphorylation. J. Biol. Chem. 262, 9569–9573 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.