Background: Unnatural amino acids can be genetically incorporated into 7-transmembrane receptors.
Results: A photoreactive amino acid introduced into the neurokinin-1 receptor cross-links substance P to the N-terminal and extracellular loop II domains of the receptor.
Conclusion: The extracellular domain of the neurokinin-1 receptor possesses multiple potential binding sites for substance P.
Significance: A photocross-linking methodology reveals novel interaction sites in the neurokinin-1-receptor-substance P complex.
Keywords: G Protein-coupled Receptor (GPCR), Homology Modeling, Membrane Protein, Protein Cross-linking, Structural Biology, Amber Codon Suppression, Photoactive Unnatural Amino Acid
Abstract
Substance P (SP) is a neuropeptide that mediates numerous physiological responses, including transmission of pain and inflammation through the neurokinin-1 (NK1) receptor, a G protein-coupled receptor. Previous mutagenesis studies and photoaffinity labeling using ligand analogues suggested that the binding site for SP includes multiple domains in the N-terminal (Nt) segment and the second extracellular loop (ECLII) of NK1. To map precisely the NK1 residues that interact with SP, we applied a novel receptor-based targeted photocross-linking approach. We used amber codon suppression to introduce the photoreactive unnatural amino acid p-benzoyl-l-phenylalanine (BzF) at 11 selected individual positions in the Nt tail (residues 11–21) and 23 positions in the ECLII (residues 170(C-10)–193(C+13)) of NK1. The 34 NK1 variants were expressed in mammalian HEK293 cells and retained the ability to interact with a fluorescently labeled SP analog. Notably, 10 of the receptor variants with BzF in the Nt tail and 4 of those with BzF in ECLII cross-linked efficiently to SP, indicating that these 14 sites are juxtaposed to SP in the ligand-bound receptor. These results show that two distinct regions of the NK1 receptor possess multiple determinants for SP binding and demonstrate the utility of genetically encoded photocross-linking to map complex multitopic binding sites on G protein-coupled receptors in a cell-based assay format.
Introduction
The neurokinin (NK)2 receptors belong to the rhodopsin-like class A 7-transmembrane receptors and are divided in three subtypes; NK1, NK2, and NK3. NK1 is widely expressed in both the central and the peripheral nervous systems, whereas NK2 is preferentially expressed in the peripheral system (1). The main endogenous ligand for the NK1 receptor is the excitatory neuropeptide substance P (SP), which is known to play roles in pain perception, inflammation, and vasodilation (1). SP is a member of the tachykinin peptide family, which also includes neurokinin-A (NKA) and neurokinin-B (NKB). The tachykinin neuropeptides have a characteristic C-terminal motif (FXGLM-NH2) that is believed to be crucial for activity, whereas their N-terminal (Nt) tails contribute to selectivity for the specific receptor subtypes (2–4). SP was originally thought to be the sole ligand for the NK1 receptor, and the related peptides, NKA and NKB, appeared to be agonists for the NK2 and NK3 receptors, respectively. This three ligand-three receptor hypothesis was primarily based on competition binding studies, which demonstrated that NKA and NKB competed very poorly against SP. However, this view has been questioned by homologous competition binding studies showing that the NKA and NKB peptides also have high affinity for NK1 and activate the receptor with nanomolar potencies (5, 6). It is believed that SP and NKA/NKB bind to different active receptor conformations rather than to distinct receptor sites (3). In this regard SP binding to NK1 was reported to be “biphasic” and to suggest an “SP-preferring” and a “general tachykinin” conformation of the receptor (3). Moreover, these states of NK1 might represent the two distinctive conformations that are involved in interaction with two different Gα proteins, Gs and Gq (7).
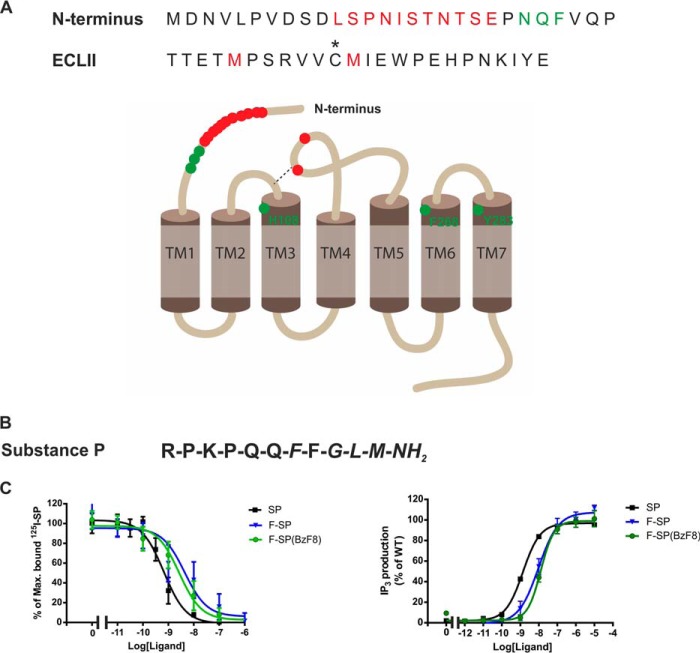
Mutational analysis of the extracellular part of NK1 indicated that residues 23–25 in the Nt tail along with residues His-108, Phe-268, and Tyr-287 were important for the binding of SP (Fig. 1, highlighted in green) (8–11). A photocross-linking approach using synthetic artificial SP analogues containing an unnatural amino acid, p-benzoyl-l-phenylalanine (BzF), was also employed to investigate how SP interacts with the NK1 receptor (12–19). To identify the cross-linking site of the SP analog on the NK1 receptor, this method required that receptor residues or fragments that cross-linked to the SP analog were determined using proteolytic digestion and/or mass spectrometry of the cross-linked complex. The results showed that SP(BzF3), where Lys-3 in SP is substituted with BzF, could cross-link to the Nt tail of the receptor between amino acids 11 and 21 as well as to a segment (residues 173(C-7)–177(C-3))3 located in the extra cellular loop II (ECLII) (14, 16). Furthermore, it has also been reported that SP with BzF substitutions at Pro-4 and Gln-5 cross-link to Met-174(C-6) of ECLII (18, 20). In addition, the substitution of Phe-8 of SP, SP(BzF8), resulted in cross-linking to Met-181(C+1) and possibly Met-174(C-6) (highlighted in red, Fig. 1A) (15, 17, 19, 21). Taken together, the previous ligand binding studies indicated that the SP peptide presumably interacts with epitopes presented on the outer part of the extracellular portion of the NK1 receptor, but the overall results have not led to a satisfactory refined model of how SP binds to the NK1 receptor.
FIGURE 1.
Interaction between substance P and the NK1 receptor. A, amino acid sequences for part of the N terminus and the ECLII of the NK1 receptor. Residues marked in green are found to be important for SP binding by mutational analysis. Residues colored in red are residues or fragments of NK1 that cross-linked to photoreactive analogues of SP. The asterisk represents one of the conserved cysteines bridging ECLII to TM-III. B, sequence of the endogenous peptide substance P, the conserved amidated C-terminal, FXGLM-NH2, is shown in italics. C, binding properties of SP (black), F-SP (blue), and F-SP(BzF8) (green) to NK11D4, measured in competition binding against 125I-labeled Lys-3]-SP. Functionality of the NK11D4 receptor, measured as SP (black), F-SP (blue), and F-SP(BzF8) (green), induced IP3 production in HEK293T cells transiently expressing the NK11D4 receptor.
To test and refine the model proposed for NK1-SP interaction, we employed a recently developed amber suppression technology for genetically introducing the photoreactive BzF directly into expressed G protein-coupled receptors (22). The site-specific incorporation of BzF relies on the expression of an orthogonal pair of a suppressor tRNA and an engineered aminoacyl-tRNA synthetase (23). We characterized the capacity for 34 BzF-NK1 mutants to interact with and cross-link to a fluorescent SP analog in a cell-based assay. We then carried out UV activation of the NKI-SP complexes to induce cross-links where the BzF was in close proximity to the bound SP. Our cross-linking data confirm that SP has multiple interaction sites within the 11–21 region of the Nt tail of NK1 and also identifies multiple interactions of SP with residues in ECLII. In addition, the study demonstrates that our novel approach is generally and readily applicable for investigating ligand binding in expressed G protein-coupled receptors.
EXPERIMENTAL PROCEDURES
Materials and Ligands
SP was purchased from Bachem. 125I-Labeled Lys-3]-SP was purchased from PerkinElmer Life Sciences. Fluorescein-labeled SP (F-SP) and F-SP(BzF8) that contains BzF at residue 8 were synthesized at the Proteomics Resource Center at Rockefeller University. The fluorescein was conjugated to the N terminus of each of the peptides with an aminohexanoyl linker.
Molecular Biology
The gene encoding the NK1 receptor was cloned into the eukaryotic expression vector pcDNA3.1+ (Invitrogen). The NK1 gene contained an N-terminal FLAG epitope and a C-terminal 1D4 epitope (TETSQVAPA), which is recognized by the 1D4 monoclonal antibody (1D4 mAb). Mutations were constructed by the use of the QuikChange lightning site-directed mutagenesis kit according to the manufacturer's protocol (Agilent Technologies). Amber suppressor tRNATyr (Bst-Yam) and tRNA synthetase from Escherichia coli were engineered to solely recognize BzF as previously described (23).
Cell Culture and Transfection
HEK293T cells were grown in Dulbecco's modified Eagle's Glutamax media (DMEM/Q, Invitrogen) supplied with 10% fetal bovine serum at 37 °C in a 5% CO2 atmosphere. HEK293T cells were transfected using Lipofectamine and Plus reagent (Invitrogen). Transfections were performed in 6-well plates using 0.09 μg of wild-type NK1 (WT-NK1) or 0.9 μg of NK1 amber variants, 0.9 μg of amber suppression tRNA-Tyr from B. stearothermophilis and 0.09 μg of tRNA synthetase (BzRS) per well with 2.5 μl of Plus reagent and 5 μl of Lipofectamine. After 3–5 h of transfection, each well was supplemented with DMEM/Q to a final concentration of 10% FBS and 0.5 mm BzF. For competition binding and IP3 assays, transfections were performed in 96-well plates using 1/30 of the amount of DNA plasmids and transfection reagents.
Competition Binding Assay
HEK293T cells were plated in poly-d-lysine-coated 96-well plates at a density of 500–10,000 cells/well aiming at 5–10% binding of the radioactive ligand. The 4th day binding experiments were performed for 3 h at 4 °C using ∼25 pm 125I-labeled Lys-3]-SP (PerkinElmer Life Sciences). Binding assays were performed in 0.1 ml of a HEPES buffer (pH 7.4, supplemented with 1 mm CaCl2, 5 mm MgCl2, 0.1% (w/v) bovine serum albumin, and 40 μg/ml bacitracin). Nonspecific binding was determined as the binding in the presence of 1 μm unlabeled SP. After 2 washes in cold buffer, lysis buffer (200 mm NaOH, 1% SDS) was added for 30 min, and radioactivity was counted.
IP3 Assay
The day after transfection cells were harvested, and 25,000 cells/well were plated in 96-well plates in DMEM/Q with 10% FBS and 5 μl of myo-[2-3H]inositol (American Radiolabeled Chemicals)/ml of DMEM media (final concentration 20 mCi/mmol). The following day the cells were washed twice with PBS and incubated in Hanks' balanced salt solution with 10 mm LiCl for 30 min at 37 °C. Subsequently, ligand was added and incubated for an additional 45 min at 37 °C. After removal of the ligand 50 μl of 10 mm cold formic acid was added to each well, and the cells were placed on ice for 30 min. Cell extract (20 μl) was added to an opaque white 96-well plate, and 80 μl of YSi SPA (scintillation proximity assay) beads (diluted 1:8 with double distilled H2O to a final concentration of 1 mg of YSi SPA beads/well, purchased from PerkinElmer Life Sciences) was added to each well. The plate was shaken for 2–4 h and spun down for 5 min. The signal was counted using the Trilux plate reader (PerkinElmer Life Sciences).
Photocross-linking Experiment
HEK293T cells expressing WT-NK1 or BzF-NK1 mutants were incubated with 100 nm concentrations of ligand in Hanks' balanced salt solution (pH 7.5) containing 0.1% BSA for 10 min at 37 °C in a 5% CO2 atmosphere. The cells were placed 5.5 inches from Maxima ML-3500S UV-A light (Spectronics Corp.) and exposed to UV light 15 min in a 4 °C cold room. The cells were subsequently harvested using PBS with aprotinin and PMSF.
Immunoblot Analysis
The harvested cells were lysed in 1% (w/v) CHAPSO buffer (50 mm Tris-HCl, 250 mm NaCl, 1 mm EDTA, 10% glycerol, 0.1 mm PMSF, pH 8) with a protease inhibitor mixture (Roche Applied Science) for 45 min at 4 °C on a nutator. NK1 receptors were immunopurified from the lysate using Sepharose beads conjugated to 1D4 mAb overnight at 4 °C. The following day beads were washed 3 times with CHAPSO buffer, and the NK1-ligand complex was mixed in 2× LDS loading buffer (Invitrogen) and released from the beads by incubation at 37 °C for 1 h. The eluted samples were resolved by SDS-PAGE and transferred into Immobilon membrane (Millipore). The membrane was incubated with either 1D4 (National Cell Culture Center, 1:3000 dilution) or anti-fluorescein antibody (Abcam, 1:3000 dilution) and subsequently probed with horseradish peroxidase-conjugated secondary antibodies (KLP Inc., goat anti-mouse, 1:20,000 dilution or goat anti-rabbit 1:20,000 dilution, respectively). Signals were visualized by enhanced chemiluminescence substrate (Thermo Scientific) and exposed to HyBlot CL Autoradiography film (Denville Scientific, Inc.)
Homology Modeling of the NK1 Receptor
Comparative modeling of the complex was performed with the automodel routine of the software package Modeler Version 9.8 (24) using the full-length human NTSR1 receptor crystal structure (25) as a template (UniProt: P30989). The sequence alignment provided as input for automodel was calculated with ClustalW (26) and manually refined in MacVector 12.5 (MacVector, Inc.). Modeling of SP utilized the neurotensin (NTS) residues 8–13 that are resolved in the crystal structure (PDB code 4grv) as a template. A set of 100 structures was generated. The structures were aligned to minimize the root mean square deviation of the receptor residues 23–315 using Vmd 1.9.1 (27).
RESULTS
Previous studies using mutational analysis and photocross-linking of peptide agonist analogues have mapped out potential sites within the Nt domain and the ECLII of NK1 to be important for NK1-SP interaction (8, 9, 18–20). To provide a more complete description of this ligand-receptor interaction, we have employed an approach that enables in vivo incorporation of BzF at single sites within NK1. The method involves the expression of BzF mutants of NK1, incubation with the agonist SP, UV irradiation, and determination of positive cross-links by immunoblot analysis. Our approach is ultrasensitive as the benzophenone group of BzF only reacts with properly oriented C-H bonds within a distance of about 3 Å (28). To detect cross-linked NK1-SP complexes, fluorescein conjugated to the N terminus of SP or to SP with BzF at residue 8 (F-SP(BzF8)) served as an antibody recognition tag. Moreover, the carboxyl end of the NK1 receptor gene was equipped with a sequence encoding the 1D4 epitope to create a tagged fusion NK11D4 receptor.
Functionality of the NK11D4 Receptor and the Modified SP Analogues
The recombinant NK11D4 receptor with a C-terminal 1D4 epitope bound SP with high affinity (Fig. 1C, Table 1) as previously described (7). Moreover, the fluorescently labeled SP analogues (F-SP and F-SP(BzF8)) only displayed a small decrease in affinity (10-fold) for the receptor (Fig. 1C, Table 1). The NK11D4 construct was functionally active as measured in SP-induced IP3 accumulation assay, showing a very similar EC50 value to that of the wild-type receptor (29) (Fig. 1C), whereas the SP analogues showed a small shift in potency comparable to the change in affinity for these ligands (Fig. 1C, Table 1). However, the modified ligands (i.e. F-SP and F-SP(BzF8)) were capable of activating the NK11D4 receptor to the same extent as the unmodified ligand as judged by Emax values (Fig. 1C, Table 1).
TABLE 1.
Ligand binding properties of BzF-substituted NK11D4 receptor mutants in positions 11–21 of the Nt and 172(C-8)–183(C+3) of the ECLII region using 125I-labeled Lys3]-SP as a radioligand
The constructs were expressed in transiently transfected HEK293T cells. Values are shown ± S.E.
| Construct (N terminus) | n | Kd | Bmax | Construct (ECLII) | n | Kd | Bmax |
|---|---|---|---|---|---|---|---|
| nm | fmol/105 cell | nm | fmol/105 cell | ||||
| WT NK1 (SP) | 5 | 0,53 ± 0.09 | 768 ± 68 | ||||
| WT NK1 (F-SP) | 4 | 7.90 ± 6.44 | |||||
| WT NK1 (F-SP8) | 4 | 3.13 ± 1.58 | |||||
| Leu-11BzF | 5 | 0.32 ± 0.06 | 207 ± 31 | Glu-172BzF (C-8) | 4 | 0.29 ± 0.05 | 93 ± 14 |
| Ser-12BzF | 6 | 0.46 ± 0.10 | 401 ± 74 | Thr-173BzF (C-7) | 4 | 0.23 ± 0.04 | 18 ± 3 |
| Pro-13BzF | 5 | 0.53 ± 0.09 | 281 ± 94 | Met-174BzF (C-6) | 4 | 0.20 ± 0.03 | 62 ± 17 |
| Asn-14BzF | 5 | 0.38 ± 0.06 | 129 ± 7 | Pro-175BzF (C-5) | 4 | 0.27 ± 0.04 | 14 ± 4 |
| Ile-15BzF | 6 | 0.83 ± 0.61 | 184 ± 41 | Ser-176BzF (C-4) | 4 | 0.13 ± 0.009 | 6 ± 3 |
| Ser-16BzF | 5 | 0.24 ± 0.08 | 141 ± 65 | Arg-177BzF (C-3) | 4 | 0.33 ± 0.10 | 12 ± 3 |
| Thr-17BzF | 6 | 0.40 ± 0.11 | 315 ± 103 | Val-178BzF (C-2) | 3 | 0.17 ± 0.05 | 15 ± 6 |
| Asn-18BzF | 5 | 0.84 ± 0.38 | 312 ± 103 | Val-179BzF (C-1) | 4 | 1.23 ± 0.43 | 27 ± 14 |
| Thr-19BzF | 5 | 0.34 ± 0.06 | 155 ± 41 | Met-181BzF (C + 1) | 4 | 0.14 ± 0.02 | 19 ± 4 |
| Ser-20BzF | 5 | 0.57 ± 0.22 | 307 ± 17 | Ile-182BzF (C + 2) | 4 | 0.63 ± 0.42 | 8 ± 2 |
| Glu-21BzF | 5 | 0.55 ± 0.08 | 439 ± 140 | Glu-183Bzf (C + 3) | 4 | 0.16 ± 0.01 | 9 ± 1 |
F-SP(BzF8) Selectively Labels the NK11D4 Receptor
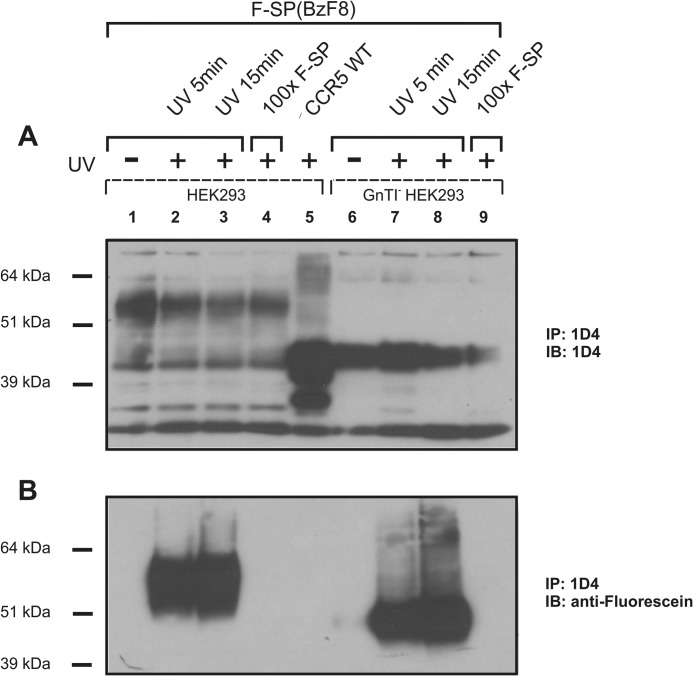
In positive control experiments, we employed ligand-based cross-linking followed by immunoprecipitation. When expressed in normal HEK293T cells, the main NK1-positive bands correspond to receptors with molecular masses of about 55 and 46 kDa (Fig. 2, lanes 1–4). However, when the NK11D4 receptor was expressed in a N-acetylglucosaminyltransferase I-negative HEK293 cell line incapable of forming N-glycosylations (30), only the 46-kDa band corresponding to the calculated molecular mass of NK1 (Fig. 2, lanes 6–9) was detected. Based on our observation, we conclude that the 46-kDa band corresponds to the unmodified NK1 receptor, whereas the 55-kDa band constitutes heavily N-glycosylated forms of the NK1 receptor, possibly modified at two potential N-glycosylation sites in the N terminus (i.e. residues 14 and 18).
FIGURE 2.
Expression and labeling of the NK11D4 receptor. Western blot analysis of cell lysate from HEK293T cells expressing the NK11D4 receptor. NK11D4 receptor was incubated for 10 min with 100 nm F-SP(BzF8) unexposed (lane 1) or exposed (lanes 2 and 3) to UV light. UV exposure was also performed in the presence of a 100-fold excess of unlabeled SP (F-SP) (lane 4). No unspecific binding was observed in lane 5 where CCR51D4 was stimulated with 100 nm F-SP(BzF8) and subsequently exposed to UV light for 15 min. Lanes 6–9 are equivalent to lanes 1–4 but display NK11D4 receptor expressed in an N-acetylglucosaminyltransferase I-negative (GnTI−) HEK293 cell line that prevents formation of complex N-glycans. IP, immunoprecipitate; IB, immunoblot.
It has previously been shown that the photoreactive SP analog SP(BzF8) was capable of cross-linking to WT-NK1 (12). In our experiments we utilized a fluorescein-labeled version of SP, F-SP(BzF8), thereby enabling the detection of the ligand-receptor complex using an anti-fluorescein antibody. As shown in Fig. 2, an anti-fluorescein-positive band corresponding to N-glycosylated NK1 (55-kDa band) was only observed when F-SP(BzF8) was present and the complex was exposed to UV light (compare lanes 1 and 2). Moreover, the complex formation between receptor and ligand could be prevented by competing with a 100-fold excess of F-SP (lane 4), indicating that the photocross-linked complexes are F-SP-specific. In addition, no cross-linking between F-SP(BzF8) and another 7-transmembrane family member, CCR51D4, was detected upon ligand stimulation and UV exposure (Fig. 2, lane 5), demonstrating the specificity of the ligand. A slight increase in cross-linking was seen if the UV exposure time was extended from 5 to 15 min (lanes 2 and 3). Taken together, these experiments revealed high specificity in classical ligand-based cross-linking to the NK11D4 receptor.
Detection of Interaction Sites between F-SP and Bzf Incorporated into the Nt Tail of NK1
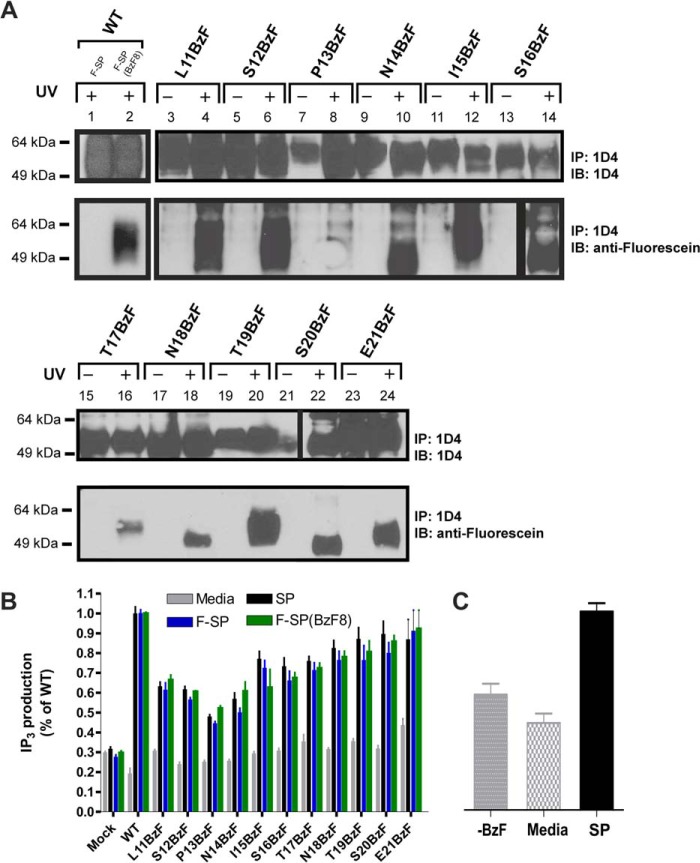
To incorporate BzF into the NK11D4 receptor, the amber stop codon (UAG) was introduced at distinct positions in the receptor gene using site-directed mutagenesis. Efficiency of BzF incorporation was detected by measuring the level of full-length receptor using antibody against the 1D4 epitope. Previous mutational analyses showed that residues 23–25 in the Nt tail of NK1 were crucial for binding of SP to the receptor (9). Furthermore, the photoreactive SP analog, SP(BzF3), cross-linked upon UV irradiation to the N terminus of NK1 (14). We re-examined ligand interaction to this portion of the receptor by generating receptor variants, in which a single residue was substituted with BzF for each of the positions 11–21. It is worth noting that substitution of the two potential N-glycosylation sites Asn-14 or Asn-18 of NK1 does not seem to interfere with binding of SP (8).
The incorporation of BzF in the N terminus of the receptor did not interfere with the binding of SP, as the affinity of the ligand was comparable with the WT receptor (Table 1), and furthermore complex formation between the NK11D4-BzF mutants and F-SP(BzF8) was detected in all sites (data not shown). The insertion of BzF at various positions in the N terminus of the receptor was verified by 1D4 detection (Fig. 3, panel A), and cross-linking between BzF-substituted NK1 variants and F-SP was detected by the immunoblots presented in Fig. 3, panel A. Various incorporation levels of BzF were detected for each of the Nt receptor mutants (Fig. 3). Differences in immunoreactivity for the Pro-13BzF and Ile-15BzF mutants comparing UV and non-exposed samples are most likely a result of loading uncertainty. In the case of Ser-20BzF mutant, transfer insufficiency most likely is the cause of the low immunoreactivity of the non-exposed Ser-20BzF sample. The increased gel mobility of complexes formed by the substitution of Asn-14, Ser-16, Asn-18, and Ser-20 is thought to be the consequence of removing the potential N-glycosylation motifs (NX(S/T)) as shown in Fig. 2. Strong cross-linking to F-SP could be detected with the Leu-11BzF, Asn-14BzF, Ile-15BzF, Ser-16BzF, Thr-17BzF, Asn-18BzF, Thr-19BzF, Ser-20BzF, and Glu-21BzF substitutions. A slightly weaker cross-link was observed when BzF was incorporated at position 12 in the NK11D4 receptor, whereas very little if any cross-linking could be detected with the Pro-13BzF substitution (Fig. 3, panel A). Importantly, the immunoblot showed that cross-linking between F-SP and the BzF-substituted receptors was strictly UV-dependent. Performing quantitative analysis of the labeling efficiency of F-SP to each of the mutants is difficult because the expression levels among the mutant receptors vary significantly and the efficiency of photocross-linking is very low even under ideal conditions. This limitation of the method due to the photochemistry of the cross-linkers has been discussed previously (31, 32).
FIGURE 3.
Interaction between F-SP and N-terminal NK11D4-BzF mutants. A, HEK293 cells transiently expressed NK11D4-BZF mutants in positions 11–21 of the N terminus. NK11D4 WT receptor was either stimulated with 100 nm F-SP (lane 1) or F-SP(BzF8) (lane 2) for 10 min before 15 min of UV exposure. All NK11D4-BZF mutants were incubated with 100 nm F-SP before 15 min of UV exposure. B, functional analysis of the NK11D4-BZF mutants of the N terminus as measured by IP3 accumulation without stimulation (Media) or after 45 min incubation with either SP, F-SP, or F-SP(BzF8). C, functional analysis of the S20BzF NK11D4 mutant in the absence of BzF but stimulated with 100 nm SP (−BzF), with BzF present but without ligand stimulation (Media), or with both BzF present in the media and stimulation of 100 mm SP. IP, immunoprecipitate; IB, immunoblot.
All the NK11D4-BzF mutants were tested for their ability to stimulate IP3 production when exposed to 100 nm ligand (SP, F-SP, and F-SP (BzF8)). Specifically, Leu-11BzF, Ser-12BzF, Pro-13BzF and Asn-14BzF-NK1 variants showed a somewhat lower signaling efficacy (≈50% of WT-NK1), whereas the receptors substituted at positions 18–21 exhibited a response close to that of WT-NK1 (Fig. 3, panel B). Importantly, receptor signaling was dependent on the presence of BzF in the growth medium and in turn to the synthesis of full-length NK11D4 as shown for the receptor mutant Ser-20BzF. Thus, a similar low level of IP3 accumulation was observed for ligand-stimulated cells that have been cultivated without BzF and for non-stimulated cells expressing the Ser-20BzF-substituted receptor (Fig. 3, panel C).
Detection of New Interaction Sites between F-SP and Bzf Incorporated into the ECLII of NK1
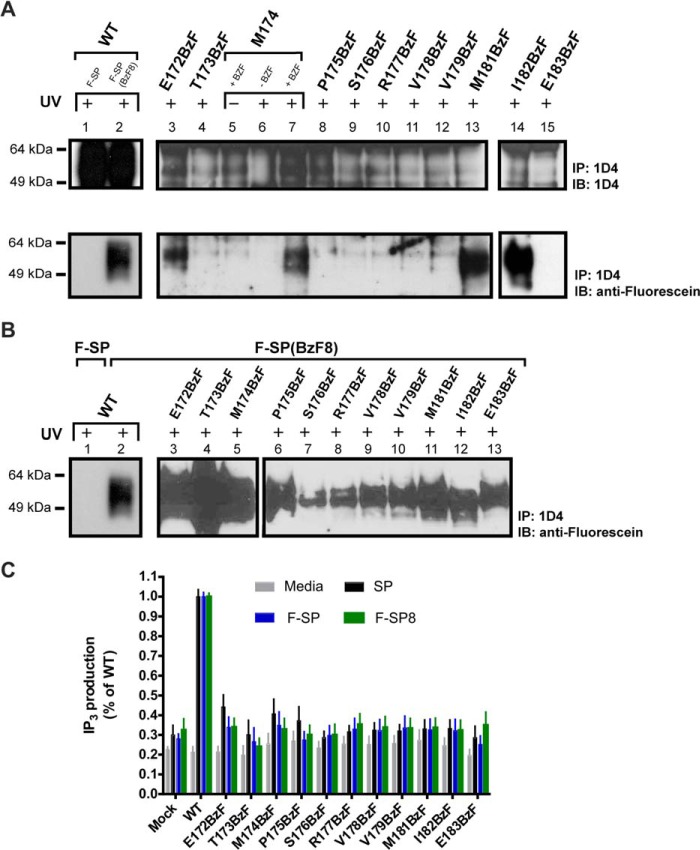
Because ligand-based cross-linking to Met-174(C-6) and Met-181(C+1) in the ECLII of NK1 had previously been suggested (15, 17, 19, 21), we decided to further investigate the ECLII region. It has been established that Cys-180 forms an intramolecular disulfide bridge with Cys-105 that is essential for SP binding (33). Hence, Cys-180 in the ECLII was excluded from BzF incorporation.
All BzF-substituted mutants were revealed to bind SP with high affinity (Table 1), and all variants were capable of cross-linking with the photoreactive ligand F-SP(BzF8) (Fig. 4, panel B, only residues 172(C-8)–183(C+3) are shown). In contrast, the number of binding sites at the cell surface, Bmax, appeared drastically lowered compared with both WT receptor and N-terminal-substituted mutants (Table 1). Although Western blot analysis also suggested that expression of receptors with ECLII substitution was considerably lower compared with the receptors substituted at the N terminus (Fig. 4, panel A, only residues 172(C-8)–183(C+3) are shown for clarity), we were able to identify four potential photocross-linking sites among the 23 tested residues in ECLII (i.e. positions 172(C-8), 174(C-6), 181(C+1), and 182(C+2)), (Fig. 4, panel A, only residues 172(C-8) to 183(C+3) are shown). In addition, controls showed that no fluorescein positive bands were observed when cells were grown in the absence of BzF or the UV exposure step was omitted (Fig. 4, lanes 5 and 6). However, when the BzF-substituted ECLII-receptors were tested for their ability to accumulate IP3 upon ligand stimulation by SP, F-SP or F-SP(BzF8), no significant ligand-dependent responses were detected (Fig. 4, panel C). Because the relatively low expression level of all ECLII receptor variants were confirmed by both immunoblot and ligand binding analysis, it seemed most likely that lack of response was due to the low abundance of the modified NK11D4 receptors rather than a defect in their signaling ability.
FIGURE 4.
Interaction between F-SP/F-SP(BzF8) and ECLII NK11D4-BzF mutants. HEK293 cells were transiently expressing NK11D4-variants in position 172(C-8) to 183(C+3) of the ECLII. A, WT receptor was either stimulated with 100 nm F-SP (lane 1) or F-SP(BzF8) (lane 2) for 10 min before 15 min of UV exposure. All NK11D4-BZF mutants were incubated with 100 nm F-SP before 15 min of UV exposure. IP, immunoprecipitate; IB, immunoblot. B, WT NK11D4 was either stimulated with F-SP (lane 1) or F-SP(BzF8) (lane 2) for 10 min before 15 min of UV exposure. The NK11D4-BZF mutants were incubated for 10 min with F-SP(BzF8) before 15 min of UV exposure. C, functional analysis of the NK11D4-BZF mutants of the ECLII as measured by IP3 accumulation without stimulation or after 45 min of stimulation with either SP, F-SP, or F-SP(BzF8).
Together, the present cross-linking study of the ECLII region in NK1 not only confirmed previous findings that residues Met-174(C-6) and Met-181(C+1) are located in close proximity to the ligand in NK1-SP complexes but also yielded two additional neighboring residues that potentially interact with SP, Glu172(C-8) and Ile-182(C+2).
Modeling of the Extracellular Region of the NK1 Receptor
To date, no x-ray structure of the NK1 receptor has been reported. However, using the recently characterized neurotensin receptor, belonging to the same subgroup of G protein-coupled receptors as a template, we prepared a three-dimensional model of the NK1 receptor in complex with SP and generated 100 likely lowest energy structural conformations (Fig. 5). The N-terminal part of NTSR1 (residue 1–19) is unresolved, and the corresponding segment is consequently also excluded from our proposed model of NK1. The C-terminal six amino acids of SP are well ordered and appear to dock into a cleft on the surface of the receptor. The rest of the N-terminal portion of SP moves radially to sweep out a cone-shaped volume that might facilitate contacts with multiple residues of the Nt tail of NK1 (Fig. 5).
FIGURE 5.
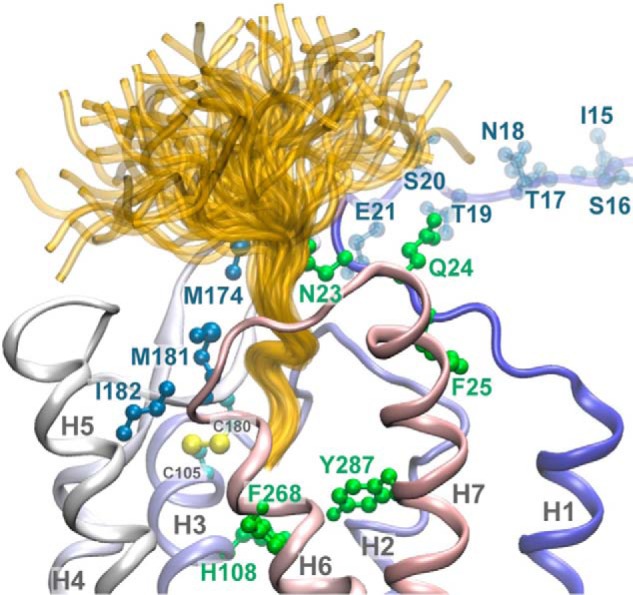
Structural model of the NK1 receptor in complex with the SP ligand. SP is shown in yellow. All 100 SP structural conformations resulting from the modeling exercise are shown. NK1 residues highlighted in shades of blue indicate the sites of cross-linking between NK1 and SP using site-specific introduction of BzF. Residues shown in green represent sites that when mutated caused loss of SP binding. The N-terminal residues 15–21 of the receptor are not structurally defined due to a lack of homology with the template structure and are rendered in a transparent representation. The Ct six amino acid residues of SP are well ordered and bind in a pocket on the extracellular surface of the receptor. The rest of SP is not well constrained, so multiple conformations are possible. The high mobility of both the amino portion of SP and the Nt tail of NK1 explains the observation that multiple sites on the Nt tail of NK1 photocross-link to bound SP.
Because of the large challenges in modeling of loop regions and the N-terminal segments of G protein-coupled receptors (34), the present model of NK1 is only a first structure-based attempt to probe for the potential proximity of the identified receptor residues to a bound SP molecule. However, the proposed three-dimensional model of NK1 clearly shows that the residues identified in the cross-linking experiments above are located in close proximity and constitutes a binding pocket for SP (Fig. 5). Furthermore, some residues previously identified as binding determinants for SP are also closely clustered in the presumed binding pocket for SP (Fig. 5) (8–11).
DISCUSSION
Numerous studies have aimed at identifying regions in the NK1 receptor involved in the binding of its ligands (2, 4, 8, 9, 17, 18, 35–38). This receptor is of particular interest because it can bind three different endogenous peptides as well as a number of synthetic ligands, one of which is an antiemetic drug on the market (3, 5, 39, 40). Furthermore, NK1 has the ability to signal through different pathways, and specific substitutions in the TM segment are known to introduce biased signaling (7), presumably by stabilizing a receptor conformation that has a preference for one particular signaling pathway. In addition, bias in signaling can be introduced by modifying the main endogenous ligand SP (41). Thus, further studies of NK1 and its ligands are essential for a full understanding of structure-function relationships and hence for development of new drugs.
Genetic and biochemical evidence indicate that the N terminus of NK1 acts as an important determinant for ligand binding (8, 9). Specifically, mutational analysis has verified that residues 23–25 are strongly required for proper binding of SP. Of these residues, Asn-23 and Gln-24 are only retained in the NK3 receptor, whereas Phe-25 is conserved in all three receptors (9). Furthermore, the photoreactive ligand SP(BzF3) cross-links to a specific portion of the Nt tail consisting of residues 11–21 (14, 16). In this study we generated NK11D4 receptor variants with substitutions of specific residues with BzF. These mutants enabled us to investigate the localization of residues 11–21 relative to ligand in binary complexes of NK1 and fluorescein-tagged SP. We first provided evidence that each of the mono-BzF-substituted receptors retained the ability to interact with the ligand, SP, as well as the ability to be stimulated by the ligands. Furthermore, we demonstrated that all substitutions, with the one exception of Pro-13BzF, could cross-link to F-SP, albeit with different efficiencies. Thus, our data indicate that residues 11–21 are in close proximity to F-SP in binary NK11D4-ligand complexes. Together, our results suggest that several side chains in this region of NK1 may interact with SP and that the N terminus of the receptor may play a key role in formation of the binding pocket for this ligand.
In particular, our data indicate that almost all of the residues probed in the N-terminal segment between positions 11 and 21 are in close enough proximity to the NK1-bound F-SP for the BzF side chain to cross-link the ligand. Ligand-based cross-linking has previously identified a similar fragment of the N terminus to be in proximity to the ligand (14, 16). We did not probe residue numbers 23–25 as they previously have been shown by mutational analysis to be essential for SP binding (8, 9). The high resolution x-ray structure of the closely related neurotensin receptor has shown that the corresponding peptide ligand interacts with a large number of residues in the receptor, including Leu-55 and Asp-56 in the Nt segment (25). However, our interpretation of the results from the present study would be that the membrane proximal Nt segment of the NK1 receptor is sufficiently flexible and dynamic to allow for efficient cross-linking at multiple sites, although probably only residues at some of these positions directly interact with the ligand in the final complex. It should also be taken into account that the ligand may have several intermediate binding modes on its way into the most high affinity binding site.
Moreover, we also employed the cross-linking approach to define residues in ECLII that are close to F-SP in binary receptor-ligand complexes. This extracellular loop is known to be important for ligand recognition and signaling of several 7-transmembrane receptors (42, 43) and in crystal structures of 7-transmembrane receptors ECLII takes up very different conformations, indicating that the loop may be important for ligand specificity (44–47). Specifically the structure of the agonist-bound neurotensin receptor reveals that the ECLII for this related receptor also forms part of the ligand binding pocket (25). It is well known that the highly conserved cysteine bridge between ECLII and TM-III (Fig. 1) is critical for NK1-SP interaction (33). Furthermore, the two loop residues Met-174(C-6) and Met-181(C+1) are close to the ligand in NK1-SP complexes (15, 17, 19, 21). Specifically, the conserved disulfide bridge positions Met-181(C+1) approximately one helical turn above His-108 (colored green in Fig. 1), which also has been found to be important for SP binding to NK1 (9).
Our investigation of the ECLII residues with BzF-encoded receptors, which were all proficient in SP binding, demonstrated cross-linking to the F-SP ligand in only 4 of 23 tested ECLII residues. Of these Met-174(C-6) and Met-181(C+1) have been identified previously by ligand-based cross-linking as being involved in SP binding (15, 17, 19, 21). However our amber-codon-induced, receptor-based cross-linking approach identified two additional interaction partners: Glu-172(C-8) and Ile-182 (C+2). There is a striking difference between the cross-linking pattern in the Nt segment where almost all residues were capable of cross-linking to the ligand and ECLII where only 4 of 23 tested residues were positive. Most likely this difference is due to a much more conformationally constrained structure of ECLII as compared with the Nt segment. Thus probably only residues that are directly involved in SP binding in the final receptor-ligand complex are able to form the chemical link originating from the BzF side chain. Thus the four identified positions will be particularly important fix points in future molecular modeling attempts designed to understand ligand-receptor interactions.
Due to the poor expression of some of the ECLII-substituted receptors, no significant functional IP3 response could be detected upon SP stimulation. However, the BzF-encoded receptors are capable of binding SP and also cross-linking with the photoactive ligand, SP(BzF8). This is a very interesting observation indicating differences in detection limit based on the employed method.
Collectively, our cross-linking experiments provide evidence that the SP ligand is bound near the edge of the extracellular domain of the NK1 receptor. This localization is consistent with data from studies of the solvent accessibility of the ligand in the receptor-bound complex. Thus, both the C- and N-terminal parts of SP were found to be fully accessible in binary complexes (40). Moreover, NMR data suggested that the C terminus of SP was inserted farther into the membrane of bicelles compared with its N terminus (48).
Furthermore, our results support a proposed binding model of SP to the NK1 receptor (35). The model, which is mainly based on NMR data for the ECLII loop and photoaffinity labeling studies, projects the N terminus of SP toward TM-I and the N terminus of NK1 and the amidated C terminus of SP toward the extracellular end of TM-III and TM-VI. The models proposed by Pellegrini et al. (35) suggest slightly different binding modes of the N terminus of SP depending on whether the Lys-3 residue of SP interacts with the N terminus (i.e. within residues 11–21) and/or with the ECLII (i.e. within residues 172(C-8)–177(C-3)) of NK1. In agreement with our presented data here, the model where Lys-3 makes interactions with both regions of NK1 demonstrates that the binding pocket is capped by the N terminus of NK1, thus resulting in multiple interactions with SP. Specifically, the models predict that Phe-7 of SP is crucial for ligand binding. Thus, in the proposed binding model, Phe-7 interacts with multiple residues in NK1; among them several of the residues are located in the 11–21 region of the N terminus. Finally, the model proposes that Phe-8 of SP, besides interacting with Met-181(C+1), also interacts with Ile-182(C+2) (35).
Because no crystal structure of the NK1 receptor is available, the structural information gained from the x-ray structure of an agonist-bound NTSR1 receptor was used to construct a three-dimensional model of the structure of NK1. Importantly and in agreement with our results, this three-dimensional model shows that the sites identified by photocross-linking are in close proximity and constitutes parts of the ligand binding pocket for SP (Fig. 5). Residues previously recognized as binding determinants for SP are also found to be located in the binding pocket of NK1 (8–11). This supports our assertion that the proposed three-dimensional model of NK1 represents an accurate interaction between the receptor and SP.
In summary, we have adapted a photocross-linking approach using unnatural amino acid mutagenesis to investigate the complex between the NK1 receptor and its endogenous ligand, SP. Using this method, we identified >12 side chains in the N terminus and ECLII of NK1 that are in close proximity to the ligand in binary complexes, indicating that these two regions of the receptor possess multiple determinants for SP binding. The generated three-dimensional model of NK1 further validates these residues as potential important sites for the interaction between SP and the NK1 receptor. Interestingly, these sites had not been identified in previous studies using conventional mutagenesis techniques (8–11). Thus, by applying the photocross-linking technique, we are able to describe a more comprehensive representation of the interactions between the NK1 receptor and its ligand SP.
Acknowledgments
We thank the Proteomic Resource Center at the Rockefeller University for the synthesis of the F-SP and F-SP(BzF8) peptide analogues. The Novo Nordisk Foundation Center for Basic Metabolic Research is supported by an unconditional grant from the Novo Nordisk Foundation to the University of Copenhagen.
This work was supported by an International Research Alliance between the Novo Nordisk Foundation Center for Basic Metabolic Research and Rockefeller University.
Residues in ECLII are numbered based on their proximity to the highly conserved Cys-180. For example, Met-174 is depicted as residue ECLII C-6.
- NK
- neurokinin
- NK1
- neurokinin-1
- NKA
- neurokinin A
- NKB
- neurokinin B
- NTSR1
- neurotensin receptor
- BzF
- p-benzoyl-phenylalanine
- ECLII
- extracellular loop II
- IP3
- inositol triphosphate
- Nt
- N-terminal
- SP
- substance P
- CHAPSO
- 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonic acid.
REFERENCES
- 1. Pennefather J. N., Lecci A., Candenas M. L., Patak E., Pinto F. M., Maggi C. A. (2004) Tachykinins and tachykinin receptors: a growing family. Life Sci. 74, 1445–1463 [DOI] [PubMed] [Google Scholar]
- 2. Regoli D., Boudon A., Fauchére J. L. (1994) Receptors and antagonists for substance P and related peptides. Pharmacol. Rev. 46, 551–599 [PubMed] [Google Scholar]
- 3. Maggi C. A., Schwartz T. W. (1997) The dual nature of the tachykinin NK1 receptor. Trends Pharmacol. Sci. 18, 351–355 [DOI] [PubMed] [Google Scholar]
- 4. Pradier L., Ménager J., Le Guern J., Bock M. D., Heuillet E., Fardin V., Garret C., Doble A., Mayaux J. F. (1994) Septide: an agonist for the NK1 receptor acting at a site distinct from substance P. Mol. Pharmacol. 45, 287–293 [PubMed] [Google Scholar]
- 5. Hastrup H., Schwartz T. W. (1996) Septide and neurokinin A are high-affinity ligands on the NK-1 receptor: evidence from homologous versus heterologous binding analysis. FEBS Lett. 399, 264–266 [DOI] [PubMed] [Google Scholar]
- 6. Maggi C. A. (2000) Principles of tachykininergic co-transmission in the peripheral and enteric nervous system. Regul. Pept. 93, 53–64 [DOI] [PubMed] [Google Scholar]
- 7. Holst B., Hastrup H., Raffetseder U., Martini L., Schwartz T. W. (2001) Two active molecular phenotypes of the tachykinin NK1 receptor revealed by G-protein fusions and mutagenesis. J. Biol. Chem. 276, 19793–19799 [DOI] [PubMed] [Google Scholar]
- 8. Fong T. M., Yu H., Huang R. R., Strader C. D. (1992) The extracellular domain of the neurokinin-1 receptor is required for high-affinity binding of peptides. Biochemistry 31, 11806–11811 [DOI] [PubMed] [Google Scholar]
- 9. Fong T. M., Huang R. R., Yu H., Strader C. D. (1993) Mapping the ligand binding site of the NK-1 receptor. Regul. Pept. 46, 43–48 [DOI] [PubMed] [Google Scholar]
- 10. Holst B., Zoffmann S., Elling C. E., Hjorth S. A., Schwartz T. W. (1998) Steric hindrance mutagenesis versus alanine scan in mapping of ligand binding sites in the tachykinin NK1 receptor. Mol. Pharmacol. 53, 166–175 [DOI] [PubMed] [Google Scholar]
- 11. Huang R. R., Yu H., Strader C. D., Fong T. M. (1994) Interaction of substance P with the second and seventh transmembrane domains of the neurokinin-1 receptor. Biochemistry 33, 3007–3013 [DOI] [PubMed] [Google Scholar]
- 12. Boyd N. D., Kage R., Dumas J. J., Silberman S. C., Krause J. E., Leeman S. E. (1995) Localization of the peptide binding domain of the NK-1 tachykinin receptor using photoreactive analogues of substance P. Ann. N.Y. Acad. Sci. 757, 405–409 [DOI] [PubMed] [Google Scholar]
- 13. Boyd N. D., Kage R., Dumas J. J., Krause J. E., Leeman S. E. (1996) The peptide binding site of the substance P (NK-1) receptor localized by a photoreactive analogue of substance P: presence of a disulfide bond. Proc. Natl. Acad. Sci. U.S.A. 93, 433–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bremer A. A., Leeman S. E., Boyd N. D. (2001) Evidence for spatial proximity of two distinct receptor regions in the substance P (SP)*neurokinin-1 receptor (NK-1R) complex obtained by photolabeling the NK-1R with p-benzoylphenylalanine3-SP. J. Biol. Chem. 276, 22857–22861 [DOI] [PubMed] [Google Scholar]
- 15. Girault S., Sagan S., Bolbach G., Lavielle S., Chassaing G. (1996) The use of photolabelled peptides to localize the substance-P-binding site in the human neurokinin-1 tachykinin receptor. Eur. J. Biochem. 240, 215–222 [DOI] [PubMed] [Google Scholar]
- 16. Li Y. M., Marnerakis M., Stimson E. R., Maggio J. E. (1995) Mapping peptide-binding domains of the substance P (NK-1) receptor from P388D1 cells with photolabile agonists. J. Biol. Chem. 270, 1213–1220 [DOI] [PubMed] [Google Scholar]
- 17. Macdonald D., Mierke D. F., Li H., Pellegrini M., Sachais B., Krause J. E., Leeman S. E., Boyd N. D. (2001) Photoaffinity labeling of mutant neurokinin-1 receptors reveals additional structural features of the substance P/NK-1 receptor complex. Biochemistry 40, 2530–2539 [DOI] [PubMed] [Google Scholar]
- 18. Sachon E., Bolbach G., Chassaing G., Lavielle S., Sagan S. (2002) Cgamma H2 of Met-174 side chain is the site of covalent attachment of a substance P analog photoactivable in position 5. J. Biol. Chem. 277, 50409–50414 [DOI] [PubMed] [Google Scholar]
- 19. Lequin O., Bolbach G., Frank F., Convert O., Girault-Lagrange S., Chassaing G., Lavielle S., Sagan S. (2002) Involvement of the second extracellular loop (E2) of the neurokinin-1 receptor in the binding of substance P. Photoaffinity labeling and modeling studies. J. Biol. Chem. 277, 22386–22394 [DOI] [PubMed] [Google Scholar]
- 20. Li H., Macdonald D. M., Hronowski X., Costello C. E., Leeman S. E., Boyd N. D. (2001) Further definition of the substance P (SP)/neurokinin-1 receptor complex. MET-174 is the site of photoinsertion p-benzoylphenylalanine4 SP. J. Biol. Chem. 276, 10589–10593 [DOI] [PubMed] [Google Scholar]
- 21. Kage R., Leeman S. E., Krause J. E., Costello C. E., Boyd N. D. (1996) Identification of methionine as the site of covalent attachment of a p-benzoyl-phenylalanine-containing analogue of substance P on the substance P (NK-1) receptor. J. Biol. Chem. 271, 25797–25800 [DOI] [PubMed] [Google Scholar]
- 22. Grunbeck A., Huber T., Sakmar T. P. (2013) Mapping a ligand binding site using genetically encoded photoactivatable cross-linkers. Methods Enzymol. 520, 307–322 [DOI] [PubMed] [Google Scholar]
- 23. Ye S., Köhrer C., Huber T., Kazmi M., Sachdev P., Yan E. C., Bhagat A., RajBhandary U. L., Sakmar T. P. (2008) Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem. 283, 1525–1533 [DOI] [PubMed] [Google Scholar]
- 24. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 25. White J. F., Noinaj N., Shibata Y., Love J., Kloss B., Xu F., Gvozdenovic-Jeremic J., Shah P., Shiloach J., Tate C. G., Grisshammer R. (2012) Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 27. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 28. Sato S., Mimasu S., Sato A., Hino N., Sakamoto K., Umehara T., Yokoyama S. (2011) Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid. Biochemistry 50, 250–257 [DOI] [PubMed] [Google Scholar]
- 29. Holst B., Nygaard R., Valentin-Hansen L., Bach A., Engelstoft M. S., Petersen P. S., Frimurer T. M., Schwartz T. W. (2010) A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J. Biol. Chem. 285, 3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reeves P. J., Callewaert N., Contreras R., Khorana H. G. (2002) Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grunbeck A., Huber T., Abrol R., Trzaskowski B., Goddard W. A., 3rd, Sakmar T. P. (2012) Genetically encoded photocross-linkers map the binding site of an allosteric drug on a G protein-coupled receptor. ACS Chem. Biol. 7, 967–972 [DOI] [PubMed] [Google Scholar]
- 32. Grunbeck A., Sakmar T. P. (2013) Probing G protein-coupled receptor-ligand interactions with targeted photoactivatable cross-linkers. Biochemistry 52, 8625–8632 [DOI] [PubMed] [Google Scholar]
- 33. Elling C. E., Raffetseder U., Nielsen S. M., Schwartz T. W. (2000) Disulfide bridge engineering in the tachykinin NK1 receptor. Biochemistry 39, 667–675 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen E. D., Norn C., Frimurer T. M., Meiler J. (2013) Assessment and challenges of ligand docking into comparative models of G-protein coupled receptors. PLoS ONE 8, e67302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pellegrini M., Bremer A. A., Ulfers A. L., Boyd N. D., Mierke D. F. (2001) Molecular characterization of the substance P*neurokinin-1 receptor complex: development of an experimentally based model. J. Biol. Chem. 276, 22862–22867 [DOI] [PubMed] [Google Scholar]
- 36. Riitano D., Werge T. M., Costa T. (1997) A mutation changes ligand selectivity and transmembrane signaling preference of the neurokinin-1 receptor. J. Biol. Chem. 272, 7646–7655 [DOI] [PubMed] [Google Scholar]
- 37. Rosenkilde M. M., Cahir M., Gether U., Hjorth S. A., Schwartz T. W. (1994) Mutations along transmembrane segment II of the NK-1 receptor affect substance P competition with non-peptide antagonists but not substance P binding. J. Biol. Chem. 269, 28160–28164 [PubMed] [Google Scholar]
- 38. Wijkhuisen A., Sagot M. A., Frobert Y., Créminon C., Grassi J., Boquet D., Couraud J. Y. (1999) Identification in the NK1 tachykinin receptor of a domain involved in recognition of neurokinin A and septide but not of substance P. FEBS Lett. 447, 155–159 [DOI] [PubMed] [Google Scholar]
- 39. Fong T. M., Cascieri M. A., Yu H., Bansal A., Swain C., Strader C. D. (1993) Amino-aromatic interaction between histidine 197 of the neurokinin-1 receptor and CP 96345. Nature 362, 350–353 [DOI] [PubMed] [Google Scholar]
- 40. Turcatti G., Zoffmann S., Lowe J. A., 3rd, Drozda S. E., Chassaing G., Schwartz T. W., Chollet A. (1997) Characterization of non-peptide antagonist and peptide agonist binding sites of the NK1 receptor with fluorescent ligands. J. Biol. Chem. 272, 21167–21175 [DOI] [PubMed] [Google Scholar]
- 41. Sachon E., Girault-Lagrange S., Chassaing G., Lavielle S., Sagan S. (2002) Analogs of Substance P modified at the C terminus which are both agonist and antagonist of the NK-1 receptor depending on the second messenger pathway. J. Pept. Res. 59, 232–240 [DOI] [PubMed] [Google Scholar]
- 42. Peeters M. C., van Westen G. J., Li Q., IJzerman A. P. (2011) Importance of the extracellular loops in G protein-coupled receptors for ligand recognition and receptor activation. Trends Pharmacol. Sci. 32, 35–42 [DOI] [PubMed] [Google Scholar]
- 43. Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 44. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Katritch V., Cherezov V., Stevens R. C. (2012) Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol. Sci. 33, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 47. Rasmussen S. G., Choi H. J., Rosenbaum D. M., Kobilka T. S., Thian F. S., Edwards P. C., Burghammer M., Ratnala V. R., Sanishvili R., Fischetti R. F., Schertler G. F., Weis W. I., Kobilka B. K. (2007) Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature 450, 383–387 [DOI] [PubMed] [Google Scholar]
- 48. Gayen A., Goswami S. K., Mukhopadhyay C. (2011) NMR evidence of GM1-induced conformational change of substance P using isotropic bicelles. Biochim. Biophys. Acta 1808, 127–139 [DOI] [PubMed] [Google Scholar]