Background: A polysaccharide lyase (Smlt1473) from Stenotrophomonas maltophilia degrades multiple anionic polysaccharides in a pH-regulated manner.
Results: Mutation of predicted substrate-binding residues significantly increased activity and specificity toward poly-β-d-mannuronic acid or poly-β-d-glucuronic acid.
Conclusion: Substrate specificity is highly sensitive to conserved, charged, and aromatic residues flanking the active site.
Significance: Smlt1473 may serve as a platform for the design of robust, highly specific polysaccharide lyases.
Keywords: Carbohydrate Processing, Enzyme Mechanism, Enzyme Mutation, Polysaccharide, Protein Design, Lyase, Substrate Specificity
Abstract
Anionic polysaccharides are of growing interest in the biotechnology industry due to their potential pharmaceutical applications in drug delivery and wound treatment. Chemical composition and polymer length strongly influence the physical and biological properties of the polysaccharide and thus its potential industrial and medical applications. One promising approach to determining monomer composition and controlling the degree of polymerization involves the use of polysaccharide lyases, which catalyze the depolymerization of anionic polysaccharides via a β-elimination mechanism. Utilization of these enzymes for the production of custom-made oligosaccharides requires a high degree of control over substrate specificity. Previously, we characterized a polysaccharide lyase (Smlt1473) from Stenotrophomonas maltophilia k279a, which exhibited significant activity against hyaluronan (HA), poly-β-d-glucuronic acid (poly-GlcUA), and poly-β-d-mannuronic acid (poly-ManA) in a pH-regulated manner. Here, we utilize a sequence structure guided approach based on a homology model of Smlt1473 to identify nine putative substrate-binding residues and examine their effect on substrate specificity via site-directed mutagenesis. Interestingly, single point mutations H221F and R312L resulted in increased activity and specificity toward poly-ManA and poly-GlcUA, respectively. Furthermore, a W171A mutant nearly eliminated HA activity, while increasing poly-ManA and poly-GlcUA activity by at least 35%. The effect of these mutations was analyzed by comparison with the high resolution structure of Sphingomonas sp. A1-III alginate lyase in complex with poly-ManA tetrasaccharide and by taking into account the structural differences between HA, poly-GlcUA, and poly-ManA. Overall, our results demonstrate that even minor changes in active site architecture have a significant effect on the substrate specificity of Smlt1473, whose structural plasticity could be applied to the design of highly active and specific polysaccharide lyases.
Introduction
Polysaccharide lyases (PLs)2 are a class of carbohydrate-modifying enzymes that catalyze the degradation of polyuronides via a β-elimination mechanism, cleaving the glycosidic bond between two adjacent sugar residues and generating an unsaturated hexenuronic acid at the nonreducing end of the oligosaccharide product (1). Three chemical steps occur during the β-elimination mechanism as follows: (i) neutralization of the negatively charged C5 carboxylate group on the +1 sugar ring, reducing the pKa of the C5 proton; (ii) abstraction of the C5 proton by a Brønsted base; and (iii) electron transfer resulting in double bond formation between C4 and C5 of the +1 sugar ring and donation of a proton by a Brønsted acid to the glycosidic oxygen resulting in the cleavage of the C4–O-1 glycosidic bond (1). Despite diverse substrate specificity, secondary structure content, and tertiary folds, this three-step mechanism is highly conserved across the 23 different polysaccharide lyase families (PL-1 to PL-23) (2, 3) and can be divided into two general categories as follows: metal ion-assisted and metal ion-independent lyases, the latter of which utilize nearby arginine, asparagine, or glutamine residues for substrate neutralization with a highly conserved tyrosine or histidine acting as the base and tyrosine as the acid (3). A consensus has not been met regarding the specific roles of the catalytic histidine and tyrosine, namely whether tyrosine acts as proton acceptor and donor with histidine stabilizing an intermediate, the mechanism proposed for alginate lyases (4) and xanthan lyases (5), or whether histidine acts as the proton acceptor and tyrosine as the proton donor, the mechanism proposed for hyaluronan lyases (6, 7). Furthermore, Shaya et al. (8) proposed a third mechanism for heparinase II in which the residue responsible for accepting the C5 proton is dependent on the structure of the substrate itself. Thus, although the catalytically active residues of PLs are highly conserved, delineating the individual roles of each residue in the mechanism of catalysis remains an area of active research. Additional studies suggest that variations in the polysaccharide structure itself and interactions between the substrate and other noncatalytic active site residues are important in the precise positioning of the cleavable glycosidic bond with respect to the catalytic tyrosine and histidine and therefore may dictate what role each plays (8–10).
The crystal structures of PLs reveal that despite different secondary structure and tertiary folds, the lyases contain a long deep cleft to accommodate at least five sugar rings of the polysaccharide substrate (4, 8, 10–14). Furthermore, the residues along this cleft form hydrogen bonds, salt bridges, and van der Waals contacts with not only the +1 and −1 sugar residues between which cleavage takes place but also the sugar rings located two or three residues away from the site of cleavage (3). Instead of participating directly in catalysis, these substrate-binding residues are thought to initially attract and then precisely orient the substrate in a geometry that facilitates catalysis (10, 15). Therefore, substrate specificity is derived from shaping the active site cleft to precisely fit a given polysaccharide structure and form the appropriate bonds with the various substituents of the sugar rings. Given that all PLs utilize the β-elimination mechanism and yet lyases from different families exhibit vastly different substrate specificity, understanding how the substrate-binding residues dictate specificity would allow for engineering of highly specialized lyases with numerous industrial and medical applications. These applications include polysaccharide sequence determination by cleavage with a set of well characterized lyases and subsequent analysis of oligosaccharide products (16), saccharification of alginic acid and poly-β-d-glucuronic acid (poly-GlcUA) found abundantly in brown and green algae for use in biofuels (17, 18), and utilization of enzymes highly active against polysaccharides secreted by bacteria during biofilm formation as an adjuvant in the treatment of chronic bacterial infection (19).
Previously, we characterized a putative alginate lyase (Smlt1473) from the opportunistic pathogen Stenotrophomonas maltophilia strain k279a (9). Smlt1473 exhibited broad, yet pH-regulated substrate specificity toward both bacterial and mammalian substrates, including poly-β-d-mannuronic acid (poly-ManA), poly-GlcUA, and HA, with optimal poly-ManA activity observed at pH 9, poly-GlcUA at pH 7, and HA at pH 5. To understand the basis for pH-regulated substrate specificity of Smlt1473 and to determine roles for noncatalytic residues in substrate specificity, we identified nine putative substrate-binding residues (Lys42, Tyr115, Lys162, Arg163, Trp171, Arg218, His221, Tyr225, and Arg312) and expressed, purified, and characterized nine single mutants (K42L, Y115F, K162L, R163L, W171A, R218L, H221F, Y225F, and R312L). In particular, we find several of these mutations play key roles in altering and, in some cases, increasing both substrate specificity and overall activity. For example, H221F increases the poly-ManA activity 2-fold and shifts the ratio of poly-ManA/poly-GlcUA turnover from 0.1 to 2.0. Conversely, R312L increases poly-GlcUA activity 2.5-fold and shifts the ratio of poly-GlcUA/poly-ManA turnover from 9.6 to 340.8. Calculation of enzyme efficiency (kcat/Km) revealed that mutants H221F and Y225F exhibited a 2.4-fold higher efficiency toward poly-ManA cleavage compared with wild-type Smlt1473. Collectively, our results indicate that the noncovalent interactions between noncatalytic residues and substrate are critical for not only overall enzymatic activity but also substrate specificity and optimal reaction conditions, such as pH, and can be grouped into categories based on their positioning within the active site cleft. Furthermore, the apparent structural plasticity of Smlt1473 offers possibilities for selection of lyases with enhanced specificity toward a particular polysaccharide substrate (9).
EXPERIMENTAL PROCEDURES
Subcloning and Site-directed Mutagenesis
Unless otherwise stated, standard molecular biology techniques were used for subcloning and site-directed mutagenesis (20). An Escherichia coli codon-optimized nucleotide sequence of smlt1473 (corresponding to GenBankTM accession number CAQ45011) was subcloned into pET28a(+) (Invitrogen) as an NcoI-XhoI insert with no stop codon, resulting in a C-terminal His6 tag. Mutagenic primers were designed via PrimerX, and amino acid substitutions were generated via the QuikChange II site-directed mutagenesis kit (Agilent Technologies). Nucleotide sequences containing mutations were confirmed by DNA sequencing (GeneWiz).
Expression and Purification
For expression and purification, methods similar to those described previously (9) were used with the following modifications. Soluble cell lysate was passed over a column containing 15 ml of Ni2+-bound chelating Sepharose Fast Flow resin (GE Healthcare) pre-equilibrated in IMAC Buffer A (20 mm HEPES, 500 mm NaCl, 10% v/v glycerol, 10 mm imidazole) at a flow rate of 2 ml/min via a BioLogic LP chromatography system (Bio-Rad) with fraction collector. The column was washed for 70 min with IMAC Buffer A to remove any unbound proteins before applying a gradient from 0 to 30% IMAC Buffer B (20 mm HEPES, 500 mm NaCl, 10% v/v glycerol, 500 mm imidazole) over the course of 80 min to wash away any nonspecifically bound proteins, and finally Smlt1473 was eluted from the column by running IMAC Buffer B for 20 min. Fractions were assayed for protein content via Bradford reagent (Bio-Rad), and samples containing purified Smlt1473 were pooled together and dialyzed against 4 liters of 20 mm HEPES, 100 mm NaCl, 5% v/v glycerol, 20 mm imidazole for 20 h at 4 °C and then 25 mm sodium phosphate buffer, pH 8, for 40 h at 4 °C with one buffer exchange. Protein concentration was determined via absorbance measurements at 280 nm, and molar extinction coefficients were estimated from primary amino acid sequences (21).
Polysaccharide Substrates
Sodium alginate, medium viscosity, and hyaluronic acid, potassium salt from human umbilical cord, were obtained from MP Biomedicals. Chondroitin sulfate and poly-α-d-galacturonic acid (poly-GalA) were obtained from Alfa Aesar. Heparin and heparan sulfate were obtained from Sigma and Sagent Pharmaceuticals, respectively. Poly-GlcUA was prepared from Avicel PH-105 NF (FMC Biopolymer), and its structure was confirmed by 1H NMR as described previously (9). Poly-ManA, poly-GulA, and poly-MG block structures were prepared from sodium alginate by partial acid hydrolysis, and its structure was confirmed by 1H NMR as described previously (9). Poly-GlcUA, poly-ManA, poly-GulA, and poly-MG samples were dialyzed against 4 liters of double distilled H2O for 40 h at 4 °C with one buffer exchange and then lyophilized. The resulting powders were stored at 4 °C until needed.
Enzyme Activity Assays
The β-elimination mechanism employed by polysaccharide lyases generates a double bond between C4 and C5 of the sugar ring at the newly formed nonreducing end that can be monitored as a change in absorbance at 235 nm (22). Absorbance measurements were taken in 1-s intervals over the course of at least 5 min via an Ultrospec 3300 pro UV-visible spectrophotometer with a detection limit of 0.001 absorbance units at 235 nm/min. One unit of enzyme activity was defined as an increase in absorbance at 235 nm of 1.0/min at 25 °C (1 unit = 1 ΔA235 nm min−1) (9, 16). In general, 14 μg of purified wild-type and mutant Smlt1473 was added to a solution containing 1 mg/ml substrate in a final volume of 350 μl, with the exception of poly-GlcUA, in which 1.4 μg of enzyme was added to each reaction due to higher activity against poly-GlcUA. The pH of all reactions was maintained via specific buffers for a given pH range at a total ionic strength of 30 mm (acetate for pH 4–6, phosphate for pH 5–8, Tris for pH 6–9, and glycine for pH 9–10). Additionally, the Michaelis constant (Km) and turnover number (kcat) of wild-type and mutant Smlt1473 against poly-GlcUA and poly-ManA were determined by varying polysaccharide concentrations. An extinction coefficient of 6150 m−1 cm−1 at 235 nm was used to convert absorbance to product concentration (23). Initial rates (vi) were fit to the Michaelis-Menten equation, v = kcatE0S/(Km + S), via the generalized reduced gradient (GRG2) nonlinear optimization program (24). The R2 and correlation values for each substrate were greater than 0.943 and 0.944, respectively. All reactions were carried out in triplicate, and error is reported as standard deviation.
Lyase activity was independently determined via a thiobarbituric acid assay in which the unsaturated product is converted to a pre-chromogen by oxidation with periodic acid and then reacted with thiobarbituric acid to form a pink chromogen with an absorbance maximum of 550 nm (25). For these experiments, 4 μg of purified enzyme (0.4 μg for poly-GlcUA) was mixed to solutions containing 1 mg/ml substrate in a total reaction volume of 100 μl. Reactions were allowed to proceed for 5 min before heating the samples at 100 °C for 10 min to degrade the enzyme and stop the reaction.
Sequence Alignment
According to the Carbohydrate Active Enzymes (CAZy) database, Smlt1473 belongs to polysaccharide lyase family 5 (PL-5), which currently contains a total of 105 lyases (2). The primary amino acid sequence of each PL-5 lyase was downloaded and parsed to remove any entry that shared a greater than 95% pairwise sequence identity with another entry in an effort to reduce redundancy in the database. The remaining 50 lyases, including Spingomonas sp. A1 (PDB code 1QAZ) (11), were aligned with Smlt1473 using Clustal Omega (26).
Homology Modeling
A homology model of Smlt1473 was constructed using Swiss-Model (27–29) based on the alginate lyase A1-III (PDB code 1QAZ) from Spingomonas sp. (11), as described previously (9). All images of the resultant model were generated via PyMOL (30).
RESULTS
Identification of Putative Substrate-binding Residues in Smlt1473 via Homology Modeling and Sequence Alignment
As stated previously, high resolution structures of various PLs have indicated that residues located inside the active site cleft yet not directly involved in the β-elimination mechanism form numerous noncovalent bonds with the substrate, including hydrogen bonds, salt bridges, and van der Waals contacts (4, 8, 10–14). The types of residues fall into three general categories as follows: (i) positively charged residues such as arginine or lysine that facilitate substrate binding and positioning by forming salt bridges with the negatively charged carboxylic acid groups present in the substrate (4, 14). Additionally, HA lyases from Streptococcus pneumoniae and Streptococcus agalactiae contain positive patches at the entrance of the cleft thought to assist in drawing the substrate into the cleft (6, 31). (ii) Aromatic residues such as tryptophan and phenylalanine undergo C–H/π interactions with the sugar rings of the substrate (32), thus optimizing its position with respect to the catalytically active residues to promote enzymatic activity (10). (iii) Polar residues such as tyrosine and asparagine form hydrogen bonds with various substituents on the substrate, further assisting in binding and positioning (4, 12, 14).
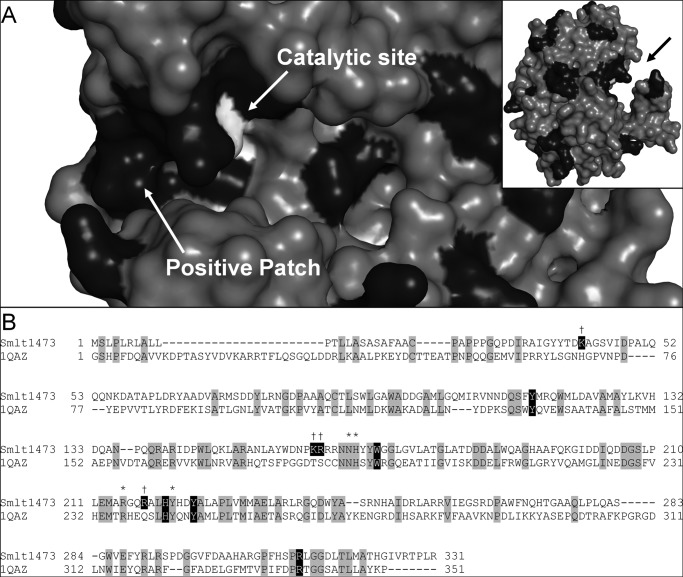
In an effort to identify putative residues that participate in substrate binding, a homology model for Smlt1473 was constructed with Swiss-Model using alginate lyase A1-III (PDB code 1QAZ) from Spingomonas sp. (11). In addition to sharing the greatest sequence identity with Smlt1473 among all solved lyase crystal structures (30%), A1-III lyase is the only PL-5 member whose high resolution structure has been determined (2). From the Smlt1473 model, the predicted catalytic residues (Asn167, His168, Arg215, and Tyr 222), highlighted in white in Fig. 1A, are clustered in a deep trench-like cleft (9). A cluster of basic residues (Lys42, Lys162, Arg163, and Arg218) form a positive patch at the entrance of the active site, highlighted in black in Fig. 1A, consistent with the HA lyases of S. pneumoniae and S. agalactiae (6, 31). Additionally, a sequence alignment with 1QAZ (Fig. 1B) revealed that the cleft contained a conserved tryptophan residue (Trp171) and several conserved polar/charged residues (Tyr115, His221, Tyr225, and Arg312) that undergo noncovalent interactions with the alginate substrate in A1-III lyase (4, 14). Thus, nine putative substrate-binding residues were identified based on homology modeling and sequence alignment with 1QAZ (Lys42, Tyr115, Lys162, Arg163, Trp171, Arg218, His221, Tyr225, and Arg312). The above amino acids were conservatively mutated in an effort to test specific properties of the given residue. For example, lysine and arginine were predicted to interact with the substrate via electrostatic attraction; therefore, these residues were mutated to leucine to remove the positive charge while minimizing side chain size variation. Tyrosine and histidine were predicted to interact with the substrate via hydrogen bonding; therefore, these residues were mutated to phenylalanine to remove the polar groups while maintaining side chain ring structure. Tryptophan was predicted to interact with the substrate via C–H/π bonds and other hydrophobic interactions; therefore, this residue was mutated to alanine to remove the aromatic ring and reduce its hydrophobicity. These mutations provide a structural basis for probing their effects on pH-dependent lyase activity against a broad range of polysaccharides, including poly-ManA, poly-GlcUA, and HA.
FIGURE 1.
Identification of putative substrate-binding residues in Smlt1473. A, surface homology model of Smlt1473 constructed with Swiss-Model and Protein Data Bank file 1QAZ as a template. A deep trench-like cleft was identified (black arrow, inset). Catalytically active residues are highlighted in white. Positively charged residues are highlighted in black. A positive patch was identified at the entrance of the active site, immediately adjacent to the catalytically active residues. B, sequence alignment of Smlt1473 with Sphingomonas sp. A1-III lyase (PDB code 1QAZ) (11). Identical residues are highlighted in gray. Residues predicted to be located in the active site cleft and participate in substrate binding are highlighted in black. Residues predicted to participate in the catalytic mechanism of Smlt1473 are marked by asterisks. Residues located in positive patch are marked by daggers.
Enzymatic Activity Screening of Mutant Lyases Revealed Diverse Substrate Specificity
The lyase activity of purified wild-type and mutant Smlt1473 was tested against the following 10 polyuronides: alginate (MP Biomedicals); poly-ManA, poly-GulA, poly-MG, poly-GlcUA, poly-GalA (Alfa Aesar); HA (MP Biomedicals); heparin (Sigma); heparan sulfate (Sagent Pharmaceuticals); and chondroitin sulfate (Alfa Aesar). The supplemental Table S1 provides the specific activity of each mutant lyase against each substrate (1 mg/ml) at the optimal pH values determined previously for wild-type Smlt1473 (9). Note that no detectable activity was measured for any of the mutant lyases against poly-GalA, heparin, and heparan sulfate at pH 5, 7, and 9. Fig. 2 displays the specific activity of each mutant against each of the three main substrates (poly-ManA, poly-GlcUA, and HA). All mutant lyases exhibited diminished activity toward HA, the most inactive being the W171A mutant with a specific HA activity of 0.4 ± 0.1 units/mg, compared with wild-type at 41.8 ± 1.5 units/mg (Table S1 and Fig. 2).
FIGURE 2.
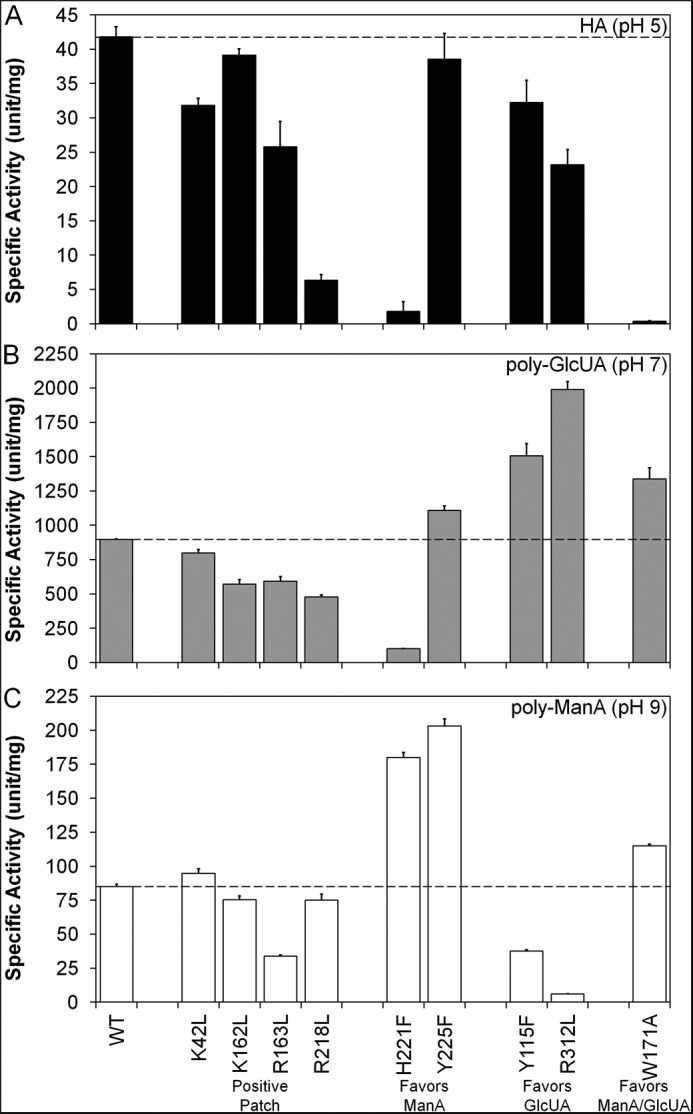
Specific activity of wild-type and mutant Smlt1473 against HA (A), poly-GlcUA (B), and poly-ManA (C). Purified wild-type and mutant Smlt1473 (14 μg) was mixed with 1 mg/ml HA in 30 mm sodium acetate, pH 5, or poly-ManA in 30 mm Tris, pH 9, in a total reaction volume of 350 μl. For poly-GlcUA, the amount of enzyme added to 1 mg/ml poly-GlcUA in 30 mm Tris, pH 7, was reduced to 1.4 μg due to higher activity. Enzymatic activity was monitored by absorbance at 235 nm. One unit of activity was defined as an increase in absorbance at 235 nm of 1.0 per min at 25 °C. Dashed lines indicate wild-type activity for comparison with other mutants. All reactions were performed in triplicate, and error is reported as standard deviation.
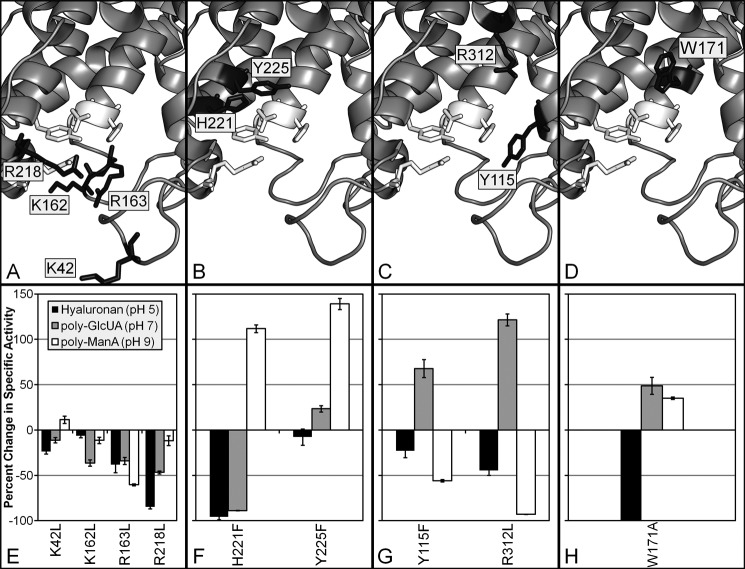
In contrast to HA, however, the Smlt1473 mutant lyases displayed significant changes in activity toward poly-GlcUA at pH 7 and poly-ManA at pH 9 and in some cases with specific mutants exhibiting opposite effects on activity for poly-ManA versus poly-GlcUA (supplemental Table S1 and Fig. 2). For example, the H221F mutant was 2.1 times more active than wild-type against poly-ManA, yet it retained only 11% activity toward poly-GlcUA. Conversely, the R312L mutant was 2.2 times more active than wild-type against poly-GlcUA, yet it retained only 7% activity toward poly-ManA (supplemental Table S1 and Fig. 2). To better organize and understand the effect each mutation has on lyase activity, the mutants were subdivided into four groups as follows: mutation of residues located in the positive patch flanking the active site (K42L, K162L, R163L, and R218L; Fig. 3, A and E) mutations that exhibited increased activity and specificity toward poly-ManA (H221 and Y225F; Fig. 3, B and F); mutations that exhibited increased activity and specificity toward poly-GlcUA (Y115F and R312L; Fig. 3, C and G), and mutations that exhibited increased activity and specificity toward poly-ManA and poly-GlcUA (W171A; Fig. 3, D and H).
FIGURE 3.
Location of putative substrate-binding residues in Smlt1473. A–D, homology model of Smlt1473 built with Swiss-Model and Protein Data Bank code 1QAZ. Images were generated in PyMOL. Predicted catalytic residues (Asn167, His168, Arg215, and Tyr222) are highlighted in white. Putative substrate-binding residues of interest are highlighted in black. E–H, percent change in specific activity of mutant lyases from wild-type Smlt1473 for HA at pH 5, poly-GlcUA at pH 7, and poly-ManA at pH 9. Enzymatic activity was monitored by change in absorbance at 235 nm. All reactions were performed in triplicate, and error is reported as standard deviation. Residues are organized into four groups as follows: residues located in the positive patch (Lys42, Lys162, Arg163, and Arg218; A and E) and residues whose mutation favors poly-ManA cleavage (His221, Tyr225; B and F), poly-GlcUA cleavage (Tyr115, Arg312; C and G), and poly-ManA/poly-GlcUA cleavage (Trp171; D and H).
Mutation of Residues Located in Positive Patch
Homology modeling of Smlt1473 using 1QAZ (11) as a template revealed a cluster of basic residues (Lys42, Lys162, Arg163, and Arg312) located near the entrance of the active site and immediately adjacent to the catalytic residues (Figs. 1A and 3A). Similar positive patches were observed in the crystal structures of HA lyases from S. pneumoniae and S. agalactiae. In both structures, the positive patch residues were predicted to play a role in the initial drawing of the negatively charged substrate into the active site via electrostatic attraction (6, 31). Thus, we expected that mutation of the homologous basic residues located in the equivalent positive patch of Smlt1473 (Lys42, Lys162, Arg163, and Arg218) would also result in decreased lyase activity due to a diminished ability to attract the acidic polysaccharide to the active site. The loss of activity by removing positive charges from this patch is also expected to be substrate-independent due to each substrate containing the negatively charged C5 carboxylic acid groups. As shown in Fig. 3E, mutation of each of the four basic residues to nonpolar leucine (K42L, K162L, R163L, and R218L) resulted in diminished activity toward HA at pH 5, poly-GlcUA at pH 7, and poly-ManA at pH 9, with the exception of K42L, which exhibited a modest (11%) increase in activity toward poly-ManA compared with wild type. The K42L mutant also displays decreases of less than 25% in activity toward HA and poly-GlcUA (Fig. 3E) and is located distal to the catalytic tetrad and other three basic residues (Fig. 3A), indicating it may play a less significant role in substrate attraction. Overall, we see loss of activity against all three main substrates (poly-ManA, poly-GlcUA, and HA) when replacing positive charges in the predicted positive patch with hydrophobic residues (K42L, K162L, R163L, and R218L), which is consistent with our expected result that these play a general role in initial substrate binding due to the net negative charge of the polysaccharides.
Mutations That Exhibited the Greatest Change in Activity with Respect to Wild Type for Poly-ManA
In contrast to the cluster of positive residues involved in initial substrate binding, mutations to predicted substrate-binding residues within the active site cleft (H221F and Y225F; Fig. 3B) resulted in significantly increased activity and specificity toward poly-ManA (Fig. 3F). The H221F mutant exhibited a 111% increase in poly-ManA activity, as well as a 96 and 89% decrease in HA and poly-GlcUA activity, respectively (Fig. 3F), making it highly selective for poly-ManA. The specific activity ratios of poly-ManA/poly-GlcUA and poly-ManA/HA were shifted 10- and 50-fold relative to wild type, respectively, indicating a significant increase in poly-ManA specificity (supplemental Table S1 and Fig. 2). The Y225F mutant exhibited similar trends to H221F, with a 139% increase in poly-ManA activity, as well as a 23% increase and 7% decrease in poly-GlcUA and HA activity, respectively (Fig. 3F). Likewise, the specific activity ratios of poly-ManA/poly-GlcUA and poly-ManA/HA were both shifted by more than a factor of 2 in favor of poly-ManA (supplemental Table S1 and Fig. 2). Taken together, the H221F and Y225F mutants both exhibited significant increases in both activity and specificity toward poly-ManA (Fig. 3F). Finally, whereas wild-type Smlt1473 exhibited the highest specific activity toward poly-GlcUA (898.2 ± 3.2 units/mg) versus poly-ManA (85.0 ± 1.7 units/mg) and HA (41.8 ± 1.5 units/mg), the H221F mutant switched to having the highest specific activity toward poly-ManA (180.0 ± 3.6) versus poly-GlcUA (100.7 ± 2.8 units/mg) and HA (1.8 ± 1.4 units/mg) (supplemental Table S1 and Fig. 2). Thus, both predicted substrate-binding residues (His221 and Tyr225) play key roles in negative regulation of poly-ManA specificity, with mutations to either causing a significant increase in selectivity and activity toward poly-ManA.
Mutations That Exhibited the Greatest Change in Activity with Respect to Wild Type for Poly-GlcUA
As with poly-ManA-selective point mutations, point mutations to additional predicted substrate-binding residues (Y115F and R312L; Fig. 3C) resulted in significantly increased activity and specificity toward poly-GlcUA (Fig. 3G). The R312L mutant exhibited a 121% increase in poly-GlcUA activity, as well as a 45 and 93% decrease in HA and poly-ManA activity, respectively (Fig. 3G), making it highly selective for poly-GlcUA. The specific activity ratios of poly-GlcUA/poly-ManA and poly-GlcUA/HA were shifted more than 30- and 4-fold in favor of poly-GlcUA, respectively (supplemental Table S1 and Fig. 2). The Y115F mutant exhibited similar trends, with the specific activity ratios shifting nearly 4-fold in favor of poly-GlcUA versus poly-ManA and more than 2-fold in favor of poly-GlcUA versus HA (supplemental Table S1 and Fig. 2). Taken together, the R312L and Y115F mutants exhibit increases in both activity and specificity toward poly-GlcUA (Fig. 3G), indicating these residues play a key role in negative regulation of poly-GlcUA activity.
Mutations That Exhibited the Greatest Change in Activity with Respect to Wild Type for Poly-GlcUA and Poly-ManA
Mutation of the highly conserved Trp171 located in the center of the active site cleft (Fig. 3D) resulted in significantly increased activity and specificity toward poly-ManA and poly-GlcUA (Fig. 3H). The W171A mutant exhibited a 49% increase in poly-GlcUA activity, 35% increase in poly-ManA activity, and was almost inactive against HA, shifting both the poly-GlcUA/HA and poly-ManA/HA specific activity ratios over 160-fold in favor of poly-GlcUA and poly-ManA (Fig. 3H).
Changes in Michaelis Constant and Enzyme Efficiency
To differentiate the effect each mutation had on substrate binding and product turnover, the Michaelis constant (Km), which reflects the substrate concentration necessary for catalysis to occur, and turnover number (kcat) were determined for each mutant against poly-GlcUA at pH 7 and poly-ManA at pH 9 (Table 1). Kinetic parameters were not determined for HA due to all nine mutants exhibiting a diminished activity toward that substrate. Given that required substrate concentration for catalysis and turnover rate are dependent on one another, the ratio of the two parameters (kcat/Km) is considered a measure of enzyme efficiency toward a given substrate (10, 14). Fig. 4 depicts the percent change of enzyme efficiency with respect to wild type for either poly-GlcUA at pH 7 or poly-ManA at pH 9. Based on the equation derived by Kelly et al. (33) for polysaccharide lyase catalysis, a change in kcat/Km indicates the mutant enzyme is binding the substrate with a different affinity. Furthermore, although Km may approximate substrate affinity, it is also affected by other rate and equilibrium constants, and therefore kcat/Km is used as a descriptive parameter for substrate binding (33).
TABLE 1.
Kinetic parameters of Smlt1473 and mutants against poly-GlcA and poly-ManA
Purified wild-type and mutant Smlt1473 was added to 16 different solutions containing 7.8 to 3000 μg/ml (22.2 to 487.8 mm) of either poly-GlcUA at pH 7 or poly-ManA at pH 9. Enzymatic activity was monitored by absorbance at 235 nm. All reactions were preformed in triplicate. Turnover rate (kcat) was calculated using an extinction coefficient of 6150 m−1 cm−1 for the unsaturated product (23). Km values are based upon an effective molecular weight of 352 for the disaccharide sugar unit, the smallest product formed by Smlt1473 during catalysis (9, 44).
| Poly-GlcUA 30 mm Tris, pH 7 |
Poly-ManA 30 mm Tris, pH 9 |
|||
|---|---|---|---|---|
| kcat (s−1) | Km (mm) | kcat (s−1) | Km (mm) | |
| Wild type | 31.9 | 0.17 | 3.3 | 0.35 |
| Positive patch | ||||
| K42L | 29.0 | 0.18 | 3.7 | 0.46 |
| K162L | 20.3 | 0.10 | 2.8 | 0.33 |
| R163L | 21.3 | 0.18 | 1.4 | 0.67 |
| R218L | 17.8 | 0.19 | 2.8 | 0.36 |
| Favors ManA | ||||
| H221F | 3.4 | 0.06 | 7.0 | 0.31 |
| Y225F | 39.1 | 0.21 | 7.9 | 0.35 |
| Favors GlcUA | ||||
| Y115F | 59.6 | 0.38 | 1.3 | 0.15 |
| R312L | 80.7 | 0.59 | 0.2 | 0.52 |
| Favors ManA/GlcUA | ||||
| W171A | 49.4 | 0.27 | 4.9 | 0.67 |
FIGURE 4.
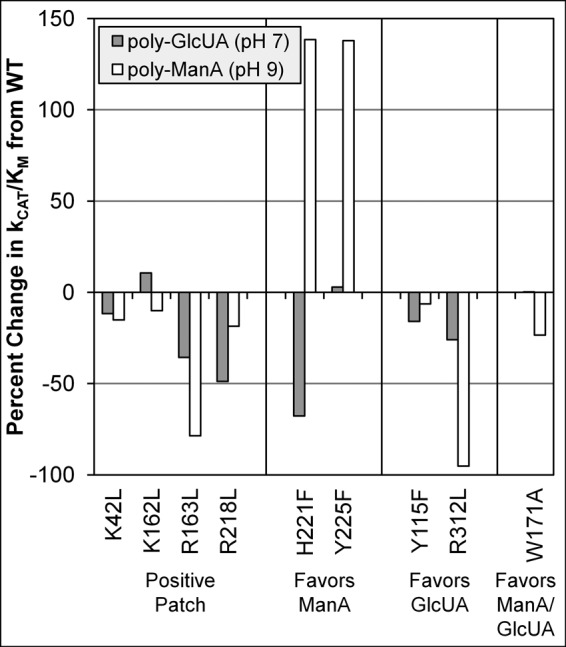
Percent change in enzyme efficiency (kcat/Km) of Smlt1473 mutants from wild type for poly-GlcUA at pH 7 and poly-ManA at pH 9.
In general, mutations of residues in the positive patch (K42L, K162L, R163L, and R218L) resulted in a decrease in enzyme efficiency for both substrates, due to either a decrease in kcat, an increase in Km, or in most cases both (Table 1 and Fig. 4). The one exception was the K162L mutant against poly-GlcUA, which displayed a 10% increase in efficiency (Fig. 4) due to the reduction in Km from 0.17 to 0.10 mm between wild-type and K162L (Table 1). Although this increase in efficiency is unexpected, it is important to recall that the positive patch is only partially disrupted by the removal of one of the charged groups, and other subtle changes in active site architecture may result in more efficient binding of a particular substrate (see under “Discussion”). Overall, mutations to either arginine (R163L and R218L) appeared to have a greater detrimental effect on enzyme efficiency than lysine mutations (K42L and K162L).
For substrate-binding residues H221F and Y225F, both exhibited a 138% increase in enzyme efficiency toward poly-ManA, due to significant increases in kcat and decreases in Km values, indicating greater overall enzyme specificity for poly-ManA for both mutants. Additionally, the H221F mutant exhibited a 68% decrease in enzyme efficiency toward poly-GlcUA, emphasizing its significantly increased activity and specificity for poly-ManA (Table 1 and Fig. 4). Similarly, the substrate-binding residues Y115F, W171A, and R312L demonstrated increases in Km as well as kcat for poly-GlcUA compared with wild type, and thus none of these mutants exhibited a significant increase in enzyme efficiency for poly-GlcUA. However, R312L, the most active mutant toward poly-GlcUA (kcat of 80.7 s−1 versus 31.9 s−1 for wild type), displayed a 95% decrease in enzyme efficiency for poly-ManA, further indicating this mutant's unique and significantly increased activity and specificity for poly-GlcUA (Table 1 and Fig. 4). Overall, these results further reinforce the idea that the positive patch residues (Lys42, Lys162, Arg163, and Arg218) play a general role in substrate binding, as evidenced by uniform decreases in enzyme efficiency for mutations to any of these residues versus interior substrate-binding residues, where major increases in enzyme efficiency are observed for poly-ManA (H221F and Y225F).
Heat Map of Changes in Substrate Specificity Caused by Mutation of Putative Substrate-binding Residues
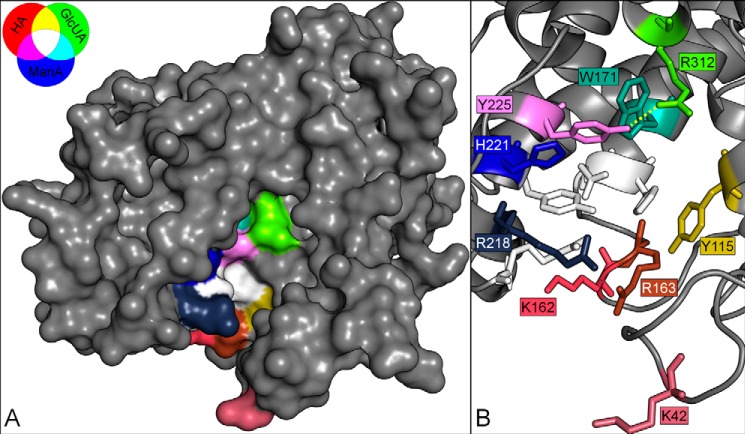
In an effort to summarize the effect of each mutation on substrate specificity, the putative substrate-binding residues were colored according to the effect of mutating each residue on activity toward HA, poly-GlcUA, and poly-ManA. More precisely, the specific activity of each mutant against poly-GlcUA was scaled to the color green with values ranging from 0, corresponding to the mutant with the lowest poly-GlcUA activity (H221F), to 255, corresponding to the mutant with the highest specific activity (R312L). A similar approach was taken for HA with red and poly-ManA with blue (Fig. 5). Interestingly, mutations with similar effects on substrate specificity clustered together in the homology model. For example, H221F and Y225F both favored poly-ManA cleavage and are localized to the left side of the active site, whereas Y115F and R312L both favored poly-GlcUA cleavage and are localized to the right side. Finally, W171A favored both poly-ManA and poly-GlcUA cleavage and is located in the middle of the active site (Fig. 5).
FIGURE 5.
Heat map of putative substrate-binding residues with respect to substrate specificity. Surface model (A) and stick model (B) of Smlt1473 built with Swiss-Model and Protein Data Bank code 1QAZ. Images were generated in PyMOL. Predicted catalytic residues (Asn167, His168, Arg215, and Tyr222) are highlighted in white. Each putative substrate-binding residue is colored according to the effect of mutating each residue on activity toward HA (red), poly-GlcUA (green), and poly-ManA (blue). The specific activity of the mutants against each substrate was scaled to the intensity of the corresponding color. The hydrogen bond between Tyr225 and Arg312 is highlighted by a yellow dashed line.
Changes in Optimal pH of R312L Mutant
Our previous work concluded Smlt1473 was a multifunctional lyase that exhibited broad but pH-regulated substrate specificity, with optimal HA activity at pH 5, poly-GlcUA activity at pH 7, and poly-ManA activity at pH 9 (9). Given the highly pH-sensitive nature of wild-type Smlt1473, it was important to differentiate whether the nine mutations changed the specific activity or the optimal pH of the enzyme against a given substrate. To that end, the pH optimum of each of the nine mutants was determined by measuring the activity at 1 pH unit above and below the optimal pH of wild-type Smlt1473 (pH 4–6 for HA, pH 6–8 for poly-GlcUA, and pH 8–10 poly-ManA). All mutants were found to exhibit optimal pH values identical to wild type with the exception of R312L against poly-GlcUA and poly-ManA, where the optimal pH was 1 unit less than that of wild-type Smlt1473 (pH 6 for poly-GlcUA and pH 8 for poly-ManA) (data not shown). Interactions between catalytic residues and nearby charged residues have been shown to significantly influence the apparent pKa value of the catalytic residues (34). Based on the homology model of Smlt1473, the side chain of Arg312 lies ∼8 Å from the catalytic His168 and Tyr222 (Figs. 3C and 5), and it lies 3 Å from the hydroxyl group of Tyr225, which in turn lies 3 Å from the catalytic Tyr222 (Figs. 3B and 5). Disruption of the interaction between Tyr225 and Arg312 was estimated by PROPKA 3.1 (35–38) to increase the pKa of Tyr225 by 1.24. A similar shift of 1.46 was estimated upon mutation of the corresponding arginine in 1QAZ (11). The hydrogen bond between the side chains of Tyr225 and Arg312 is highlighted in Fig. 5 by a yellow dashed line. Given the close proximity of Tyr225 to Arg312 and the catalytic Tyr222, it is possible that Arg312 influences catalysis indirectly via Tyr225 (Fig. 5).
DISCUSSION
The homology model of Smlt1473 revealed a cluster of basic residues (Lys42, Lys162, Arg163, and Arg218) located at the entrance of the active site cleft immediately adjacent to the catalytic tetrad (Fig. 1). Similar positive patches observed in the crystal structures of HA lyases from S. pneumoniae and S. agalactiae are thought to participate in substrate binding by drawing the negatively charged polysaccharide toward the active site via electrostatic attraction (6, 31). Therefore, we predicted that mutation of residues located in the positive patch of Smlt1473 would result in an overall reduction of enzymatic activity regardless of substrate. As shown in Fig. 3E, mutants K42L, K162L, R163L, and R218L resulted in diminished activity toward each substrate with the exception of K42L, which exhibited a modest (11%) increase in activity toward poly-ManA. Furthermore, each positive patch mutant displayed reduced enzymatic efficiency toward poly-GlcUA and poly-ManA, with the exception of K162L, which exhibited a modest (10%) increase in efficiency toward poly-GlcUA (Fig. 4). In general, arginine mutations (R163L and R218L) appeared to have a greater detrimental effect on activity and enzyme efficiency than lysine mutations (K42L and K162L), implying the arginine residues play a more important role in substrate attraction and neutralization, perhaps due to the ability of the guanidinium group to interact with the C5 carboxylate of the substrate in three different orientations and form a larger number of electrostatic interactions, such as salt bridges and hydrogen bonds, compared with lysine (39, 40).
Multiple sequence alignment of Smlt1473 with the PL-5 lyases listed in the CAZy database (2) revealed that Trp171 was conserved across all members of this family. Given the high degree of conservation and predicted location in the center of the active site cleft (Fig. 3D), we anticipated a significant role for Trp171 in regulation of enzyme activity. Further evidence in favor of a role for conserved tryptophan in regulation of lyase activity comes from previous studies on the effect of mutating an analogous, conserved tryptophan residue (W292A) in the PL-8 HA lyase from S. pneumoniae, which retained 4% activity compared with wild type (10). Interestingly, crystal structures of the W292A mutant revealed that the HA substrate was still bound to the active site but was misaligned with respect to the catalytic residues (10). Consistent with the role of tryptophan aligning HA substrates within the active site, the analogous W171A in Smlt1473 resulted in essentially complete elimination of HA activity (Fig. 3H). However, in unexpected contrast, W171A exhibited increased activity toward poly-ManA and poly-GlcUA (Fig. 3H), implying this residue was not essential for the optimal positioning of these additional substrates, but rather it was specific to HA. One possible explanation for this contrast in activities for different substrates is that HA is composed of repeating heterodisaccharide units of GlcUA and N-acetylglucosamine, whereas poly-GlcUA and poly-ManA are both homopolysaccharides. The β-elimination mechanism of PLs requires that a uronic acid be present in the +1 subsite; therefore, the heterodisaccharide repeat of HA could require additional, unique positioning in the active site conferred by Trp171. Thus, Trp171 plays a key role in Smlt1473 in conferring activity for HA and is analogous to other tryptophan residues in related HA lyases where it is essential for activity by appropriately positioning the heterodisaccharide repeat within the active site for catalysis.
The remaining four substrate-binding mutants can be split into two groups with opposite effects on substrate specificity; H221F and Y225F exhibited increased activity and specificity for poly-ManA (Fig. 3F), and Y115F and R312L exhibited increased activity and specificity for poly-GlcUA (Fig. 3G). Based on the model proposed by Shaya et al. (8), it is interesting to consider the structural differences between poly-ManA and poly-GlcUA and how those differences dictate which residues in the active site undergo substrate binding and effect the overall positioning of the substrate with respect to the catalytic residues. In their model for heparinase II, the residue responsible for proton abstraction during the β-elimination mechanism is dependent on the structure of the substrate itself. For heparin cleavage, a histidine residue accepts the proton, whereas for heparan sulfate cleavage, a tyrosine located on the opposite side of the substrate accepts the proton. This model is based on the fact that heparin contains mostly GlcUA, whereas heparan sulfate contains mostly iduronic acid, the C5 epimer of GlcUA. The H5 proton abstracted during catalysis points in opposite directions for each substrate and therefore can only be abstracted by the catalytic residue it faces. Similarly, GlcUA and ManA are C2 epimers of each other, with the second hydroxyl group pointing down in the equatorial position for GlcUA and up in the axial position for ManA (Fig. 6, top). Computational studies and x-ray crystallography have shown that this subtle difference has a dramatic effect on the polymeric structure (41, 42), namely poly-GlcUA favors a 21 helix (similar to cellulose) with 180° turns between each subsequent residue, whereas poly-ManA favors a 32 helix with 120° turns between each residue. This results in poly-GlcUA having a planar shape with the substituents of every other residue pointing in the same direction (41). Alternatively, poly-ManA has a more rod-like shape with the sugar rings of three subsequent residues lying in three separate planes (Fig. 6, bottom) (42). Crystal structures of poly-ManA in complex with A1-III alginate lyase from Spingomonas sp. show that bound substrate retains the 32 confirmation with minor distortions between the −1 and +1 residues (4, 14). There is currently no crystal structure of a lyase in complex with poly-GlcUA, but because poly-ManA retains its native structure upon binding, one might consider that poly-GlcUA would do the same and therefore lead to dramatic changes in relative activity dependent on orientation of the substrates in the active site due to their unique structures (Fig. 6). These structural differences likely account for the opposite effects of a single mutant on poly-ManA and poly-GlcUA activity and specificity. Further confirmation of this idea will require more detailed, high resolution structures of poly-GlcUA-specific lyases with bound substrate to compare against poly-ManA-specific lyases.
FIGURE 6.
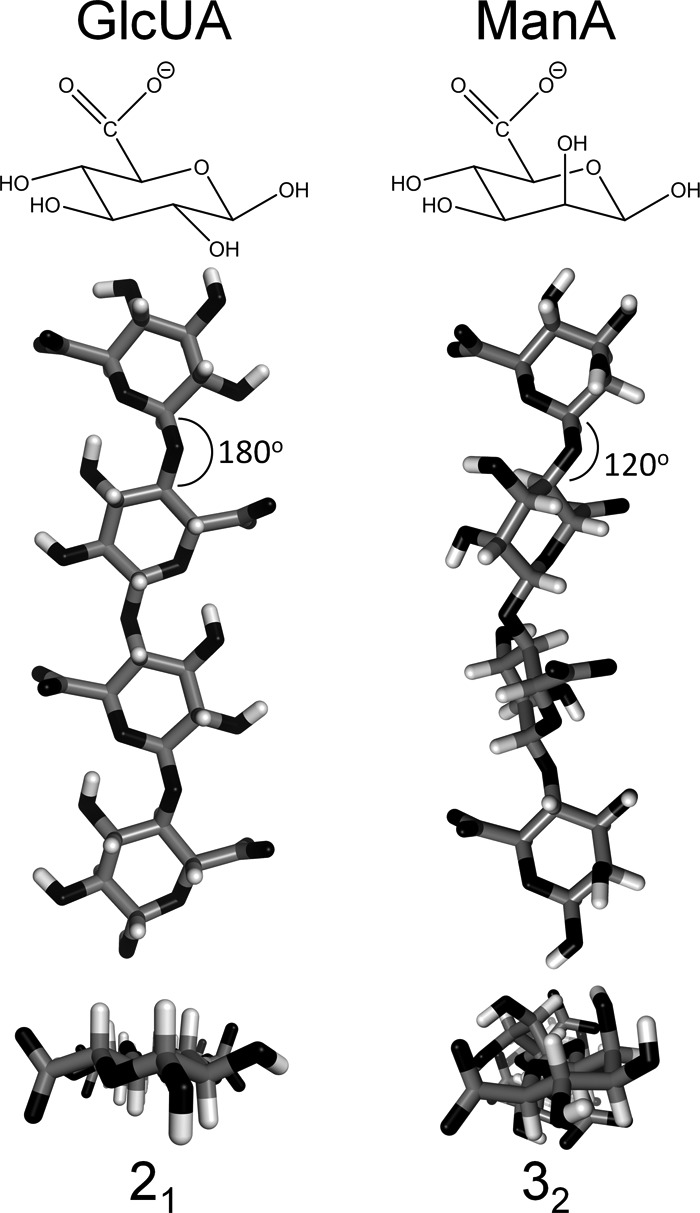
Structural differences between poly-ManA and poly-GlcUA. Top, chair diagram of GlcUA and ManA, which are C2 epimers of each other. The C2 hydroxyl group is pointing down in the equatorial position for GlcUA and pointing up in the axial position for ManA. Bottom, three-dimensional structure of poly-GlcUA in a 21 helix (41) and poly-ManA in 32 helix (42). Structures were downloaded from PolySac3Db and rendered in PyMOL.
To better understand the effect of mutations H221F, Y225F, Y115F, and R312L on substrate specificity, the corresponding residues were located in the solved crystal structure of A1-III alginate lyase in complex with poly-ManA tetrasaccharide (PDB code 4F13) (14) and analyzed for protein-protein and ligand/protein interactions (43) in an effort to explain the increased or decreased poly-ManA activity exhibited by the above mutants. However, these interactions must be evaluated with caution, because most mutations will lead to subtle structural changes that are difficult to predict. As stated previously, there are currently no solved crystal structures of a lyase in complex with poly-GlcUA; therefore, an equivalent analysis for poly-GlcUA activity could not be performed. The H221F mutant exhibited increased poly-ManA activity (Fig. 3F) and substrate affinity (Table 1). The equivalent His245 forms a hydrogen bond with O3 of the −1 sugar ring. Although mutation to phenylalanine results in the loss of that hydrogen bond, the larger six-carbon ring was predicted to double the surface area between this residue and the −1 sugar ring (43), perhaps resulting in a stronger C–H/π interaction and therefore increased substrate binding. Interestingly, the Nϵ2 group of His245 lies 3.2 Å away from the C2 equatorial position of the −1 sugar ring, which is where the C2 hydroxyl group of poly-GlcUA would be located, implying the histidine would be capable of hydrogen bonding with both O3 and O2 of poly-GlcUA. The loss of this additional hydrogen bond may account for the decrease in poly-GlcUA activity observed for the H221F mutant. The Y225F mutant exhibited increased poly-ManA activity (Fig. 3F) with little change in Km (Table 1), implying this mutation influences catalytic activity instead of substrate binding. The equivalent Tyr249 forms a partially stacked interaction with the −1 ManA sugar ring and a hydrogen bound with the side chain of Arg342 (equivalent to Arg312 in Smlt1473). Mutation to phenylalanine would preserve the C–H/π interaction with the substrate but eliminate the hydrogen bond with arginine. Disruption of the interaction between Tyr225 and Arg312 in R312L also influenced the optimal pH of Smlt1473 (see “Results”), implying this bond plays an indirect role in catalysis.
The Y115F mutant exhibited reduced poly-ManA activity (Fig. 3G), yet increased substrate affinity (Table 1). The equivalent Tyr137 forms a partially stacked interaction with the +1 sugar ring. Mutation to phenylalanine may increase the C–H/π interaction between the residue and sugar ring and prevent release of the unsaturated product, therefore resulting in a decrease in overall activity but an increase in substrate affinity. The R312L mutant exhibited reduced poly-ManA activity (Fig. 3G) and reduced substrate affinity (Table 1). The equivalent Arg342 forms a salt bridge with the carboxylic acid group of the −1 sugar ring. Electrostatic attraction between positively charged amino acids and the negatively charged substrate is a key factor in polysaccharide binding, and therefore it is not surprising that a mutation to leucine results in a decrease in substrate binding and activity. With regard to the increased poly-GlcUA activity of R312L, Arg312 may not be involved in neutralizing the carboxyl group of the −1 GlcUA sugar ring due to the structural differences between poly-ManA and poly-GlcUA (Fig. 6). Removal of the arginine side chain may trigger a conformational change in the active site cleft that facilitates poly-GlcUA cleavage. Clearly, further structural analysis of Smlt1473 is required to fully elucidate the effect of each mutation on enzymatic activity and substrate specificity.
Overall, our results also point to the utility of a sequence structure guided approach in the identification of key residues responsible for regulating poly-ManA, poly-GlcUA, and HA activity in Smlt1473. By utilizing a homology model based on 1QAZ (Fig. 1), we were able to identify nine key residues within the predicted active site, and we found point mutations that led to dramatic increases in activity as well as selectivity for poly-GlcUA and poly-ManA. The ability to engineer a high degree of selectivity yet retain potent activity for a given substrate is of increasing interest in processing naturally occurring polysaccharides such as alginate and glucuronan for a diverse array of applications, including biomass pre-processing for fermentation-based biofuel production and generating defined chemical compositions and sizes of polysaccharides as drug delivery systems. Previous studies have used random mutagenesis coupled to selection to identify poly-α-l-guluronic (poly-GulA)-specific alginate lyases by randomly mutating AlyA from Klebsiella pneumoniae, which natively exhibited both high poly-MG and poly-GulA activity. Although effective in identifying poly-GulA-specific lyases, the increase in selectivity came at the expense of significantly reduced overall activity (16). In contrast, our sequence-structure guided approach was able to focus our initial search to a narrow set of nine conserved residues, with point mutations giving rise to highly active and specific poly-ManA and poly-GlcUA lyases. Moreover, our results highlight the structural plasticity of the Smlt1473 active site, with key residues important for conferring activity to HA (Trp171), poly-GlcUA (His221 and Tyr225), and poly-ManA (Tyr115 and Arg312), enabling engineering of selectivity for a particular substrate. Future studies are focused on probing the structural specificity responsible for poly-GlcUA, which will enable comparison of substrate structure with active site binding and catalysis to determine what role substrate structure plays in enzyme activity, selectivity, and catalysis.

This article contains supplemental Table S1.
- PL
- polysaccharide lyase
- GlcUA
- d-glucuronic acid
- ManA
- d-mannuronic acid
- HA
- hyaluronan
- PL-5
- polysaccharide lyase family 5
- GulA
- l-guluronic acid
- MG
- mannuronic acid and guluronic acid.
REFERENCES
- 1. Gacesa P. (1987) Alginate-modifying enzymes: A proposed unified mechanism of action for the lyases and epimerases. FEBS Lett. 212, 199–202 [Google Scholar]
- 2. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garron M. L., Cygler M. (2010) Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20, 1547–1573 [DOI] [PubMed] [Google Scholar]
- 4. Yoon H. J., Hashimoto W., Miyake O., Murata K., Mikami B. (2001) Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution. J. Mol. Biol. 307, 9–16 [DOI] [PubMed] [Google Scholar]
- 5. Maruyama Y., Hashimoto W., Mikami B., Murata K. (2005) Crystal structure of Bacillus sp. GL1 xanthan lyase complexed with a substrate: insights into the enzyme reaction mechanism. J. Mol. Biol. 350, 974–986 [DOI] [PubMed] [Google Scholar]
- 6. Jedrzejas M. J., Mello L. V., de Groot B. L., Li S. (2002) Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate. J. Biol. Chem. 277, 28287–28297 [DOI] [PubMed] [Google Scholar]
- 7. Rigden D. J., Jedrzejas M. J. (2003) Structures of Streptococcus pneumoniae hyaluronate lyase in complex with chondroitin and chondroitin sulfate disaccharides. Insights into specificity and mechanism of action. J. Biol. Chem. 278, 50596–50606 [DOI] [PubMed] [Google Scholar]
- 8. Shaya D., Zhao W., Garron M. L., Xiao Z., Cui Q., Zhang Z., Sulea T., Linhardt R. J., Cygler M. (2010) Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate. J. Biol. Chem. 285, 20051–20061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. MacDonald L. C., Berger B. W. (2014) A polysaccharide lyase from Stenotrophomonas maltophilia with a unique, pH-regulated substrate specificity. J. Biol. Chem. 289, 312–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nukui M., Taylor K. B., McPherson D. T., Shigenaga M. K., Jedrzejas M. J. (2003) The function of hydrophobic residues in the catalytic cleft of Streptococcus pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms. J. Biol. Chem. 278, 3079–3088 [DOI] [PubMed] [Google Scholar]
- 11. Yoon H. J., Mikami B., Hashimoto W., Murata K. (1999) Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 A resolution. J. Mol. Biol. 290, 505–514 [DOI] [PubMed] [Google Scholar]
- 12. Lunin V. V., Li Y., Linhardt R. J., Miyazono H., Kyogashima M., Kaneko T., Bell A. W., Cygler M. (2004) High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism. J. Mol. Biol. 337, 367–386 [DOI] [PubMed] [Google Scholar]
- 13. Elmabrouk Z. H., Vincent F., Zhang M., Smith N. L., Turkenburg J. P., Charnock S. J., Black G. W., Taylor E. J. (2011) Crystal structures of a family 8 polysaccharide lyase reveal open and highly occluded substrate-binding cleft conformations. Proteins 79, 965–974 [DOI] [PubMed] [Google Scholar]
- 14. Mikami B., Ban M., Suzuki S., Yoon H. J., Miyake O., Yamasaki M., Ogura K., Maruyama Y., Hashimoto W., Murata K. (2012) Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III. Acta Crystallogr. D Biol. Crystallogr. 68, 1207–1216 [DOI] [PubMed] [Google Scholar]
- 15. Wong T. Y., Preston L. A., Schiller N. L. (2000) ALGINATE LYASE: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54, 289–340 [DOI] [PubMed] [Google Scholar]
- 16. Tøndervik A., Klinkenberg G., Aarstad O. A., Drabløs F., Ertesvåg H., Ellingsen T. E., Skjåk-Bræk G., Valla S., Sletta H. (2010) Isolation of mutant alginate lyases with cleavage specificity for di-guluronic acid linkages. J. Biol. Chem. 285, 35284–35292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S. I., Choi S. H., Lee E. Y., Kim H. S. (2012) Molecular cloning, purification, and characterization of a novel poly-MG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2. Appl. Microbiol. Biotechnol. 95, 1643–1653 [DOI] [PubMed] [Google Scholar]
- 18. Elboutachfaiti R., Delattre C., Petit E., Michaud P. (2011) Polyglucuronic acids: structures, functions and degrading enzymes. Carbohydrate Polymers 84, 1–13 [Google Scholar]
- 19. Hamza A., Piao Y. L., Kim M. S., Choi C. H., Zhan C. G., Cho H. (2011) Insight into the binding of the wild type and mutated alginate lyase (AlyVI) with its substrate: a computational and experimental study. Biochim. Biophys. Acta 1814, 1739–1747 [DOI] [PubMed] [Google Scholar]
- 20. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3 Ed., pp. 1.1–1.162, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Wilkins M. R., Gasteiger E., Bairoch A., Sanchez J. C., Williams K. L., Appel R. D., Hochstrasser D. F. (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552 [DOI] [PubMed] [Google Scholar]
- 22. Preiss J., Ashwell G. (1962) Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-l-erythro-5-hexoseulose uronic acid. J. Biol. Chem. 237, 309–316 [PubMed] [Google Scholar]
- 23. Farrell E. K., Tipton P. A. (2012) Functional characterization of AlgL, an alginate lyase from Pseudomonas aeruginosa. Biochemistry 51, 10259–10266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fylstra D., Lasdon L., Watson J., Waren A. (1998) Design and use of the Microsoft Excel Solver. Interfaces 28, 29–55 [Google Scholar]
- 25. Weissbach A., Hurwitz J. (1959) The formation of 2-keto-3-deoxyheptonic acid in extracts of Escherichia coli B. I. Identification. J. Biol. Chem. 234, 705–709 [PubMed] [Google Scholar]
- 26. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The Swiss-Model workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 28. Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. (2009) Protein structure homology modeling using Swiss-Model workspace. Nat. Protoc. 4, 1–13 [DOI] [PubMed] [Google Scholar]
- 29. Bordoli L., Schwede T. (2012) Automated protein structure modeling with SWISS-MODEL workspace and the protein model portal. Methods Mol. Biol. 857, 107–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 31. Mello L. V., De Groot B. L., Li S., Jedrzejas M. J. (2002) Structure and flexibility of Streptococcus agalactiae hyaluronate lyase complex with its substrate. Insights into the mechanism of processive degradation of hyaluronan. J. Biol. Chem. 277, 36678–36688 [DOI] [PubMed] [Google Scholar]
- 32. Asensio J. L., Ardá A., Cañada F. J., Jiménez-Barbero J. (2013) Carbohydrate-aromatic interactions. Acc. Chem. Res. 46, 946–954 [DOI] [PubMed] [Google Scholar]
- 33. Kelly S. J., Taylor K. B., Li S., Jedrzejas M. J. (2001) Kinetic properties of Streptococcus pneumoniae hyaluronate lyase. Glycobiology 11, 297–304 [DOI] [PubMed] [Google Scholar]
- 34. Harris T. K., Turner G. J. (2002) Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life 53, 85–98 [DOI] [PubMed] [Google Scholar]
- 35. Li H., Robertson A. D., Jensen J. H. (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 36. Bas D. C., Rogers D. M., Jensen J. H. (2008) Very fast prediction and rationalization of pKa values for protein–ligand complexes. Proteins 73, 765–783 [DOI] [PubMed] [Google Scholar]
- 37. Olsson M. H., Søndergaard C. R., Rostkowski M., Jensen J. H. (2011) PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 [DOI] [PubMed] [Google Scholar]
- 38. Søndergaard C. R., Olsson M. H. M., Rostkowski M., Jensen J. H. (2011) Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 [DOI] [PubMed] [Google Scholar]
- 39. Wigley D. B., Lyall A., Hart K. W., Holbrook J. J. (1987) The greater strength of arginine: Carboxylate over lysine carboxylate ion pairs implications for the design of novel enzymes and drugs. Biochem. Biophys. Res. Commun. 149, 927–929 [DOI] [PubMed] [Google Scholar]
- 40. Barlow D. J., Thornton J. M. (1983) Ion-pairs in proteins. J. Mol. Biol. 168, 867–885 [DOI] [PubMed] [Google Scholar]
- 41. Braccini I., Grasso R. P., Pérez S. (1999) Conformational and configurational features of acidic polysaccharides and their interactions with calcium ions: a molecular modeling investigation. Carbohydr. Res. 317, 119–130 [DOI] [PubMed] [Google Scholar]
- 42. Atkins E. D., Nieduszynski I. A., Mackie W., Parker K. D., Smolko E. E. (1973) Structural components of alginic acid. I. The crystalline structure of poly-β-d-mannuronic acid. Results of x-ray diffraction and polarized infrared studies. Biopolymers 12, 1865–1878 [DOI] [PubMed] [Google Scholar]
- 43. Sobolev V., Sorokine A., Prilusky J., Abola E. E., Edelman M. (1999) Automated analysis of interatomic contacts in proteins. Bioinformatics 15, 327–332 [DOI] [PubMed] [Google Scholar]
- 44. Rye C. S., Matte A., Cygler M., Withers S. G. (2006) An atypical approach identifies Tyr234 as the key base catalyst in chondroitin AC lyase. ChemBioChem 7, 631–637 [DOI] [PubMed] [Google Scholar]