Abstract
Web-based social media is increasingly being used across different settings in the health care industry. The increased frequency in the use of the Internet via computer or mobile devices provides an opportunity for social media to be the medium through which people can be provided with valuable health information quickly and directly. While traditional methods of detection relied predominately on hierarchical or bureaucratic lines of communication, these often failed to yield timely and accurate epidemiological intelligence. New web-based platforms promise increased opportunities for a more timely and accurate spreading of information and analysis. This article aims to provide an overview and discussion of the availability of timely and accurate information. It is especially useful for the rapid identification of an outbreak of an infectious disease that is necessary to promptly and effectively develop public health responses. These web-based platforms include search queries, data mining of web and social media, process and analysis of blogs containing epidemic key words, text mining, and geographical information system data analyses. These new sources of analysis and information are intended to complement traditional sources of epidemic intelligence. Despite the attractiveness of these new approaches, further study is needed to determine the accuracy of blogger statements, as increases in public participation may not necessarily mean the information provided is more accurate.
Key words: social media, epidemiological intelligence, informatics, flu, infectious disease.
Significance for public health
The shift in human interaction and media consumption to online communication has changed the way people seek information. The increased frequency in the use of the Internet via computer or mobile devices provides an opportunity for social media to be the means of providing people with valuable health information directly and quickly. The development of web-based social media analytics has given public health officers and physicians an earlier insight into the emergence and spread of infectious diseases. Availability of timely and accurate information is especially useful for the rapid identification of an outbreak of an infectious disease necessary to promptly and effectively develop public health responses.
Introduction
Social networking enables the health care industry to interact with and educate patients through social media. Social media platforms such as Twitter and Facebook are the open forum spaces in which social networks are created and maintained. Another term for a social networking platform is a social networking site, or SNS. As a forum, an SNS generally allows users to upload commentary and content onto online open space.
From 2005 to 2009, SNS usage quadrupled thus increasing connectivity and prompting efforts to identify new opportunities of using social media to impact population health.1 Timely, accessible, and credible health information is critical for improving public health outcomes, taking action during an outbreak, and preventing illness.2 The increased frequency in the use of the Internet via computer or mobile devices provides an opportunity for social media to be the medium through which users can be provided with valuable health information quickly and directly. Social media platforms and web searches provide access to emerging health trends, which decreases the interim period before health events occur.
Web-based social media is increasingly being used across different settings in the health care industry. Large hospital networks are using social media platforms to disseminate information to patient support groups by using both video and live chats.3 Facebook, Twitter, and YouTube are being used to transmit information at health conferences while health institutes have created outlets to create a dialogue with communities. Social media provides another communication channel for patients and providers to exchange information.
The shift in human interaction and media consumption to online communication has changed the way people seek information. Search engines and social media have become the key sources for enhancing communication of health information among community members to prevent and understand illness. The content entered into search engines or posted on social media accounts provide valuable information about the detection of infectious disease. This previously relied on doctors and health care workers reporting cases of a disease. With the emergence of novel web technologies, analysing web searches to provide an early warning about an impending epidemic in the human population can minimise the time that elapses between onset and detection.
The increase in public participation in online blogs and forums allows for more accessible and transparent information. Information relating to the emergence or spread of an infectious disease can be accessed from individuals who report personal cases in online forums such as Twitter. The capability to cheaply aggregate, process, and analyse this kind of intelligence is now possible with web-based analytics developed by companies such as Google and Yahoo.4
The development of web-based social media analytics has given public health officers and physicians an earlier insight into the emergence and spread of infectious diseases. While traditional methods of detection relied predominately on hierarchical or bureaucratic lines of communication, these often failed to yield timely and accurate epidemiological intelligence.4 Web-based analytics and information processing gives provide health practitioners with the opportunity of consolidating previously inaccessible and disparate information sources. These new sources of analysis and information are intended to complement traditional sources of intelligence about epidemics.
Analysis of user searches for infectious disease intelligence and surveillance
The rapid identification of an outbreak of an infectious disease is necessary to more promptly and effectively develop public health responses. The Canadians were among the first to pioneer the development of web-based surveillance technologies. In the 1990s, Health Canada created the Global Public Health Intelligence Network.5 The network uses autonomous news feed aggregators based on established search queries to collect articles containing relevant information about the possibility of a public health emergency.5
The Canadian network was particularly useful during the 2002 outbreak of severe acute respiratory syndrome (SARS) in the Guangdong Province of China, as well as the 2008 Canadian listeriosis outbreak. In the case of SARS in China, the network identified the outbreak more than two months before the World Health Organization publically published details on cases of the new respiratory illness.5 With regards to the listeriosis outbreak in Canada, the network indicated a statistically significant spike in the pattern of listeriosis-related searches nearly a month before the declaration of the outbreak.5 In both cases, the network-based intelligence predicted a public health crisis before it was officially declared. The data the network obtained was derived from the searches conducted by individuals accessing social media on the Internet.
When used to undertake infectious disease surveillance, social media-based models can track infectious disease trends in real time in order to predict, observe, and minimise the harm caused by outbreak events.5
A 2006 study by Eysenbach tracked influenza (flu)-related searches on the web for syndromic surveillance to determine whether an automated analysis of trends in Internet searches could be useful to predict outbreaks such as influenza epidemics.6 Eysenbach developed a model for predicting a flu outbreak on the basis of changes in Internet search activity for flu-related information.6 The model was evaluated against a traditional surveillance method which uses sentinel physicians who manually report encounters with sick patients demonstrating flu-like illness (ILI) to a public health agency.6
To obtain statistics on the prevalence of searches, Eysenbach created an advertising campaign on Google Adsense that appeared for Canadian searchers only who entered flu or flu symptoms into Google. The ad read, Do you have the flu? Fever, Chest discomfort, Weakness, Aches, Headache, Cough? and provided a link to a generic patient education website. Eysenbach aggregated daily statistics on impressions and clicks provided by Google to match the time periods of the weekly national FluWatch reports. The number of advertising clicks correlated better with flu events than flu-like illness (ILI) reports from sentinel physicians. Internet clicks also were a timelier marker than these reports in that they performed better in predicting the flu events of the following week. Correlation coefficients in the reports from sentinel physicians were better for the status of the current week than for predicting the following week. A study conducted by Polgreen et al.7 also examined the occurrence of ILI and flu as related to information collated by an Internet search engine. They used Yahoo to predict an increase in culture positive for flu 1-3 weeks in advance.7 Their model was predicated on the theory that the pattern of how and when people search may provide clues or early indications about future health-related concerns and expectations.7 For instance, when someone believes himself to be sick, he will first search on the internet for disease-related symptoms related to his symptoms before checking in with a public health professional or physician. In developing their model, Polgreen et al. determined whether there was a relationship between the web-based search terms related to a disease and the actual cases of disease.7 In order to relate the search data to a measure of the actual occurrence of flu, the investigators used reports generated by clinical laboratories (members of the National Respiratory and Enteric Virus Surveillance System) that compiled the total number of respiratory specimens that were positive for the disease.7 The web-based search data were compiled at national level and included queries that contained information pertinent to seasonal flu events. Specific census regions were studied that identified the geographical location of origin of the Internet protocol addresses conducting the searches.7 The two sets of data were then correlated and compared. A positive relationship was found between the fraction of flu-related queries and rates of cultures positive for flu two weeks later.7
Despite discovering a relationship between search queries and new flu cases, the study was limited due to the relatively short period of time in which it was conducted (four years), as well as false positives associated with some searches, i.e. searches conducted due to events unrelated to the actual occurrence of flu in a population. These limitations may be corrected by extending the duration of future studies and restricting data sources to websites dedicated solely to medical information, such as WebMD and MD Consult.7 While future studies may refine existing techniques, such refinements do not address the fact that data collected on searches might not have always represented an actual geographical location as per a specific census region.7 In order to address this issue, search data must be analysed with a greater degree of specificity. However, this analytical specificity might represent a breach in search user privacy. Therefore, additional studies might only use aggregated search volumes representing larger geographical regions for surveillance purposes.7
Ginsberg et al.8 conducted such a study on aggregated search volumes towards the early detection of disease activity. Millions of Google search queries were monitored for health-seeking behaviour. These query trends were then correlated to the percentage of physician visits in which a patient displays ILI.8 Similar to previously mentioned studies, Ginsberg et al. asserted that search engine trends on flu could be used to predict the occurrence of actual flu events. Their system built upon previous attempts by using an automated method of discovering flu-related search queries.8 Billions of individual searches over five years of Google search logs were used to inform more comprehensive models for use in flu surveillance.8
Instead of correlating search data to the occurrence of positive flu cultures, Ginsberg et al. examined the probability that a random physician visit in a particular region would be related to a random search query submitted from the same region.8 The investigators examined a larger aggregated search query database due to the fact that such correlations are only meaningful across large populations.8 The model was able to obtain a good association with Centres for Disease Control and Prevention (CDC)-reported ILI percentages with a mean correlation of 0.97.8 The mean indicated a strong relationship between the occurrence of a physician visit and an ILI-related search query.
The web-based search query data accurately predicted the increase in ILI percentages. Such data can be acquired, processed, and analysed quickly. As a result, the ILI estimates found by Ginsberg et al. were consistently 1-2 weeks ahead of CDC ILI surveillance reports.8 Search query-based predictive models may give public health professionals a greater advantage in developing earlier responses to seasonal outbreaks of flu. Due to the widespread utilisation and popularity of the Google search engine, Google-based search query analytics is one of the most timely and broad-reaching flu monitoring systems available today. Like other web-based surveillance systems, however, Ginsberg et al.’s system remains susceptible to false alerts resulting from ILI inquiries unrelated to the actual occurrence of flu in any one population group. Therefore, human analysis and interpretation of search query data remains necessary.
While promising as predictive instruments, the intelligence gathering tools used by the four studies are limited by their data collection methodologies. In each study, the data were procured from a particular population, namely wired Internet users. Thus, the studies limited sampling to a specific population. While Ginsberg et al. attempted to correlate searches made in a particular region to physician visits in that same region, none of the studies made any attempt to relate the actual number of Internet users in a region to the actual population of that region. For instance, in some rural areas of the United States, close to a quarter of a region’s population may be without an Internet connection.9
Future studies should incorporate statistics that can illustrate the percentage of wired versus non-wired segments of the populations. In addition, an attempt should be made to determine what proportion of the wired population participated in the reporting of ILI.
Identifying potential networks of disease communicability within wired social networks
Social media and web searches help identify individuals within a network who should be targeted for vaccinations to prevent the spread of disease. Analysing information related to disease from web searches provides public health officials with a more accurate and timely disease surveillance system.
During the H1N1 pandemic in the fall and winter of 2009, Christakis of Harvard University used his social networking methodology to analyse the spread of flu among a group of college students.10 The results from researchers who watched for flu reports among the highly connected people predicted the peak of the flu epidemic 16 days ahead of time.10 Christakis predicted the spread of the flu through social networks; social media and web searches have the ability to make this prediction more immediate.
Social networks determine patterns of disease progression among the entire population. In order to arrest the progression of an infectious disease, often very small quantities of vaccine must be administered to a large at-risk population. The highest priority groups identified in an outbreak may be best for individual protection, but may not, however, be optimal to protect the entire population.11 Highest priority groups for the H1N1 vaccine were health care workers and people who were at risk of severe complications if infected. This included pregnant women, young children, people who lived with or cared for children under six months of age, and children aged 5-18 years with chronic medical conditions. A long-standing policy for seasonal flu includes priority vaccination of the elderly, even though school children and working adults transfer disease at a higher rate due to their having a higher contact rate.
A recent study conducted by researchers at Yale University School of Medicine and Clemson University found that consideration of transmission is an important factor when developing a vaccination policy.11 Furthermore, the study concluded that previous and new CDC recommendations are suboptimal based on five outcome measures: total infections averted, total deaths averted, years of life lost, contingent valuation (an assumption of life value based on age), and economic costs. The controversy over who should be eligible to receive a vaccine when the supply is limited is not one that is easily resolved. New technologies, such as real-time surveillance, have provided access to unprecedented resources that can be used to fight the spread of infectious diseases. The ability to quickly and efficiently disseminate information plays a vital role in preventing an outbreak from getting out of control.11
Analysis of user-generated content (blogs) towards predicting outbreaks
Blogs enable users to upload a linked series of postings about a particular topic in a forum setting. The blogosphere is the aggregation of these postings. Blogs may be considered a form of social networking.12 Data mining of blogs for the purpose of flu surveillance gives a uniform voice to the web-connected public for the purpose of disease reporting. Previously, those who contributed public health-related information to blogs and other online social media outlets only did so to communicate with other individuals who were using the same information interfaces. Data mining of these sources now allows these contributions to be aggregated and studied. What was previously an inchoate and fragmented forum becomes a single voice when contributions to online forums are collated and analysed.13
Previous efforts aimed at estimating the flu prevalence in a population relied solely upon extrapolation of formally diagnosed cases.13 Blogs provide additional information to better inform traditional epidemiological models. Incorporating these sources into their work, Corley et al. developed a system that identifies blog communities that share flu-related postings.13 Trends in postings were correlated to CDC ILI patient reporting at sentinel healthcare providers.13 Patterns in flurelated postings were then further discerned via graph-based data mining to identify structural anomalies in the flu blogosphere that correspond to increases in ILI.13
Spinn3r was used to automatically process and analyse the content of thousands of blogs. Spinn3r is a web and social media (WSM) indexing service that conducts real-time indexing with the throughput power of 100,000 new blogs per hour.13 Spinn3r collected, processed, and discriminated the blogs containing flu keywords. Those selected blogs were then further analysed by text mining. Text mining is the process of discovering information in large text collections and automatically identifying interesting patterns and relationships in textual data.13 To further identify nuanced interrelationships between blogs, such as influence of a particular blogger on flu-related material, the Subdue system was developed. Subdue was devised for general purpose automated discovery, concept learning, and hierarchical clustering.13 Whereas a human being might miss a larger pattern in aggregate data which might indicate the beginning of a flu outbreak, Subdue will be able to immediately recognise and flag these less visible phenomena in a larger data set. Public health officials could then analyse the aggregated and tamed data to determine the relevance of computer-indicated trends (Figure 1).
Figure 1.
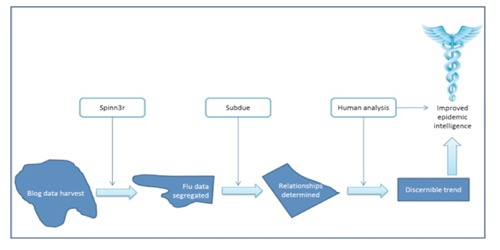
Blog content analysis process.
To determine whether CDC ILI surveillance was correlated to WSM, Corley et al. compared the two data series with Pearson’s correlation statistic.13 The CDC ILI reports and the WSM were shown to have correlated strongly with a Pearson statistic of r=0.545 with 95% confidence.13 Despite the appeal of such a comprehensive WSM collation system, it has two limitations; i) sample bias; and ii) the reliability of blogger statements. Those who post on blogs generally tend to have access to higher levels of technology and education. Thus, only the literate and wired segments of society would be tapped as sources of flu intelligence. Further study must be conducted to determine whether this skews response variety. Secondly, the verisimilitude of blogger statements must also be verified. It is possible that bloggers may intentionally create false alarms for the purpose of garnering increased following and popularity.
While critics of blogosphere mining may indicate problems associated with the veracity of the data compiled under this approach, they fail to note that more traditional epidemiological studies also often rely upon limited samples as well. Furthermore, traditional studies are also subject to issues related to response validity. Until empirical evidence demonstrates to the contrary, it must be assumed that blogger posts related to ILI are no more or less accurate than the responses generated by respondents in more traditional surveys.
Data analysis of blogosphere sources and traditional epidemic intelligence sources may be undertaken in conjunction with one another in order to develop a more comprehensive tracking of outbreaks. Incorporating an analysis of the blogosphere would serve to complement and bolster more traditional studies. Similar to the meta-analysis that is often applied in the social sciences, a holistic approach to disease tracking could incorporate emerging sources of web-based intelligence into the traditional methodologies currently in use by epidemiologists.
The potential benefit of WSM-based flu surveillance far outweighs the potential risk. Before the advent of WSM and blogging, the general public’s participation in flu reporting only went as far as calling a public health official (if he or she felt inclined to do so) or visiting a physician who would report ILI to the CDC. Now, if someone has neither the time nor desire to meet directly with a public health official or physician, he or she could, instead, just blog or tweet a statement similar to I’m coming down with the flu. A web-based analytical system such as Subdue will then quickly register the posting.
The Internet has made disease reporting and surveillance much more accessible and timely. It has also provided a greater opportunity to the wired public to participate in tracking the emergence of flu outbreaks. This increased participation might also benefit international public health initiatives. For instance, if a repressive country’s government is reluctant to disclose the outbreak of disease for political reasons, those connected to the Internet will still be able to disseminate the truth to the rest of the world.
Mapping disease outbreaks in real time
A geographical information system (GIS) is a technological tool for understanding geographical issues and making intelligent decisions.14 During an outbreak, GIS provides tools that speed the collection of accurate field data. Complex statistical and other analyses applied with GIS technology provide relevant information to support sound decisions. GIS analysis can, for example, locate a potential disease hot spot and calculate a nearby hospital’s ability to handle the expected increase in service demand if an outbreak should occur. Public health emergencies are often protracted events. Effects of a disease outbreak (or an environmental disaster such as a chemical spill) have the potential for a long-term impact on the health and wellbeing of a community. Public health organisations rely on GIS analysis tools to assess data collected in the process of monitoring long-term health effects.15
To give public health officials an intuitive, spatially-based and collaborative online resource with which to analyse the emergence and spread of infectious disease, multiple web-based programmes must be used in combination. Previously, web-based programmes that housed different data sources and display methods had to be used independently of one another. Advances in information technology now allow for the creation of mashups, or web sites or services that weave data from different sources into a new and consolidated data source or service.16 Mashup programmes such as My Maps and Mapplets allow the layman to integrate disparate data sources.
Geocoded health data displayed by a mashed-up GIS make it possible to visualise and track diseases in a simulated environment. These programmes combine geocoded epidemiological data from public health reports with programmes such as Google Maps and Second Life to offer the user virtual travel through a GIS-based environment. A mashed-up avatar-interface programme like Second Life also allows multiple users in different locations to access the GIS-based health data simulation simultaneously and interact with each other.16
Mashups are made possible by data formatters, connectors, visualisation, sharing, and web Application Programming Interfaces (API). These web-based technologies are the various building blocks of a mashup. Data formatters allow the user to visually map the web content to a particular structure. This makes content extraction and formatting over the web easier.16 Data connectors allow users to geographically create a pipe or workflow for the connection and integration of different formats of data.16 Data visualisation takes the data that has been standardised by the data connectors and visualises it with a programme such as Google Maps or Google Earth.14 The data can then be shared through collaboration forums in which users can view and utilise the work of others.15 Finally, web API gives the mashup developers the ability to further add to and edit existing mashups.
Mashups integrate numerical and spatial data for public health decision support.16 These programmes give public health officials the ability to combine intuitive GIS displays with geocoded health data. They can also be continuously up-dated and changed at any time and in any location through the use of web API. In short, this technology allows for increased levels of collaboration, particularly across great distances, as well as an improved epidemiology intelligence platform.
Conclusions
Online forums and advances in information technology have resulted in new methods and opportunities for understanding the spread of infectious disease. Whereas previous public health systems suffered from the information lag associated with reliance upon bureaucratic hierarchies, new web-based platforms promise increased opportunities for more timely and accurate information dissemination and analysis. Public health officials will increasingly rely upon intuitive GIS-generated models based upon aggregated data from the web to analyse the spread of infectious disease. Further studies are needed to determine the accuracy of blogger statements as increases in public participation may not necessarily mean the information provided is more accurate.
As society becomes increasingly wired, it becomes more important to analyse Internet-based sources of information. Online participation has exploded over the course of the past few decades and is expected to expand still further. Public health officials must be aware of these new trends and understand them in order to adjust how they gather information. Web searches, blog postings, social networks, and geographical information systems should not be thought of as replacements for traditional epidemic intelligence tools. Rather, they should be considered useful supplements to existing methodologies. Like the electronic database for the librarian, data taken from the Internet can be an invaluable tool to the epidemiologist. Careful study and analysis will ensure the appropriate application of these new web-based tools.
References
- 1.Chou WS, Hunt Y, Beckjord E, et al. Social media use in the United States: implications for health communication. J Med Internet Res 2009;11:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNab C. What social media offers to health professionals and citizens. Bull World Health Organ 2009;87:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thaker SI, Nowacki AS, Mehta NB, Edwards AR. How U.S. hospitals use social media. Ann Intern Med 2001;154:707-8 [DOI] [PubMed] [Google Scholar]
- 4.Dukic V, Lopes H, Polson N.Tracking flu epidemics using Google flu trends and particle learning. Social Science Research Network. 2009. Available from: http://papers.ssrn.com/sol3/papers.cfm?abstract_id=1513705
- 5.Wilson K, Browstein J. Early detection of disease outbreaks using the Internet. CMAJ 2009;180:829-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eysenbach G. Infodemiology: tracking flu-related searches on the web for syndromic surveillance. AMIA Annu Symp Proc 2006;244-8 [PMC free article] [PubMed] [Google Scholar]
- 7.Polgreen P, Chen Y, Pennock D, Nelson F. Using internet searches for influenza surveillance. Clin Infect Dis 2008;47:1443-7 [DOI] [PubMed] [Google Scholar]
- 8.Ginsberg J, Monhebbi M, Patel R, et al. Detecting influenza epidemics using search engine query data. Nature 2008;457:1012-4 [DOI] [PubMed] [Google Scholar]
- 9.Kim G.Three out of four Americans have access to the internet, according to Nielsen//NetRatings. Available from: http://www.nielsen-online.com/pr/pr_040318.pdf
- 10.Christakis N, Fowler J. Social network sensors for early detection of contagious outbreaks. PLoS One 2010;5:e12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medlock J, Galvani AP. Optimizing influenza vaccine distribution. Science 2009;3258:1705-8 [DOI] [PubMed] [Google Scholar]
- 12.Gaudeul A, Peroni C. Reciprocal attention and norm of reciprocity in blogging networks. Available from: http://papers.ssrn.com/sol3/papers.cfm?abstract_id=1577062
- 13.Corley C, Cook D, Mikler A, Singh K. Text and structural data mining of influenza mentions in web and social media. Int J Environ Res Public Health 2010;7:597-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geographic Information System GIS. Introduction. Collegial centre for educational materials development; 2008. Available from: http://www.ccdmd.qc.ca/en/gis/before.html
- 15.ESRI. Redlands: ESRI; GIS best practices early detection and response to infectious disease; [about 37 screens]. Available from: http://www.esri.com/library/bestpractices/early-detection.pdf
- 16.Boulos M, Scotch M, Cheung K, Burden D. Web GIS in practice VI: a demo playlist of geo-mashups for public health neogeographers. Int J Health Geogr 2008;7:2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]


