Abstract
Estimates of the total number of bacterial species1-3 suggest that existing DNA sequence databases carry only a tiny fraction of the total amount of DNA sequence space represented by this division of life. Indeed, environmental DNA samples have been shown to encode many previously unknown classes of proteins4 and RNAs5. Bioinformatics searches6-10 of genomic DNA from bacteria commonly identify novel noncoding RNAs (ncRNAs)10-12 such as riboswitches13,14. In rare instances, RNAs that exhibit more extensive sequence and structural conservation across a wide range of bacteria are encountered15,16. Given that large structured RNAs are known to carry out complex biochemical functions such as protein synthesis and RNA processing reactions, identifying more RNAs of great size and intricate structure is likely to reveal additional biochemical functions that can be achieved by RNA. We applied an updated computational pipeline17 to discover ncRNAs that rival the known large ribozymes in size and structural complexity or that are among the most abundant RNAs in bacteria that encode them. These RNAs would have been difficult or impossible to detect without examining environmental DNA sequences, suggesting that numerous RNAs with extraordinary size, structural complexity, or other exceptional characteristics remain to be discovered in unexplored sequence space.
Conserved secondary structures of novel RNAs can be identified by phylogenetic comparative sequence analysis18,19, whereby nucleotides and structures important for RNA function are revealed by identification of conserved sequences and nucleotide covariation (e.g. see Supplementary Fig. 1). We used this approach to identify over 75 new structured RNAs from bacteria or archaea. Among these are novel riboswitch classes that sense tetrahydrofolate, S-adenosylhomocysteine, and c-di-GMP, and other candidate cis-regulatory and noncoding RNAs (unpublished data). Based on available sequence data, several of these RNAs are present only in specific environments or in phyla with few available genome sequences (Supplementary Table 1). Here we report a special subset of new-found RNA structures that are extraordinary, either because they are extremely large and structurally complex or because they are produced in unusually high amounts.
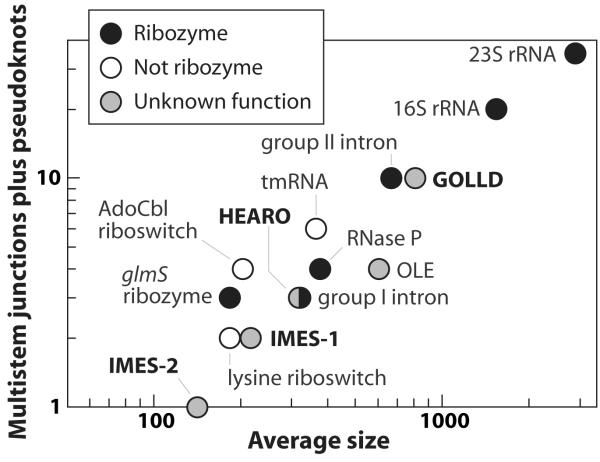
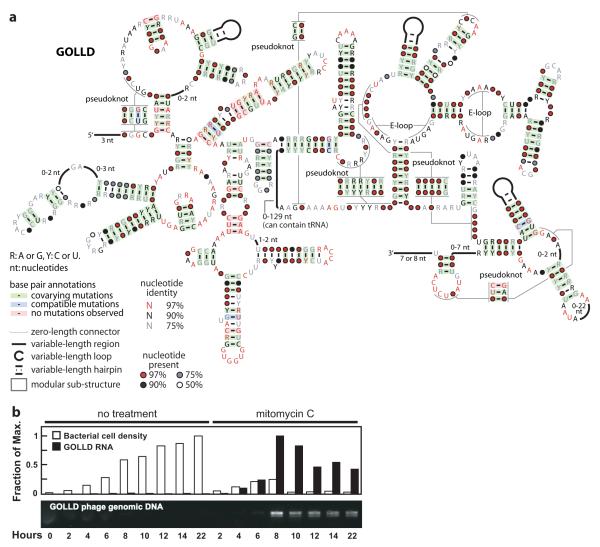
We identified two RNA structures (GOLLD and HEARO) that are among the largest complex bacterial ncRNAs known (Fig. 1). GOLLD (Giant, Ornate, Lake- and Lactobacillales-Derived) RNA is particularly striking because it represents the third-largest highly structured bacterial RNA discovered to date, ranking only behind 23S and 16S rRNAs. The structural complexity of GOLLD RNA (Fig. 2a), as quantified by the number of multistem junctions and pseudoknots, is similar to most self-splicing group II introns20. Also, as observed in many large ribozymes18-20, some regions of GOLLD RNA can adopt a diversity of complex folds (Supplementary Figure 2).
Figure 1. Size and structural complexity of new-found RNAs compared to the ten largest known bacterial ncRNAs with complex structures.
Structural complexity is represented by the number of multistem junctions plus pseudoknots (see full Methods for details). RNAs described in this report are in bold type. HEARO and Group I ribozyme symbols overlap. Narrowly distributed RNAs (present in only one bacterial class) are not included.
Figure 2. GOLLD RNAs.
a, Simplified consensus sequence and secondary structure model for the most common architecture of GOLLD RNAs. Annotated 5′ and 3′ ends reflect L. brevis transcripts observed by RACE experiments (Supplementary Fig. 3). b, Phage induction and expression of GOLLD RNA. Experimental details are presented in the full Methods.
We identified GOLLD RNAs by searching environmental sequences collected from Lake Gatún, Panama21, and representatives were subsequently identified in eight cultivated organisms distributed among three bacterial phyla. GOLLD RNAs are frequently located adjacent to tRNAs, and in three cases, a tRNA is predicted inside a variable region in GOLLD RNA itself (Fig. 2a and Supplementary Discussion).
In Lactobacillus brevis ATCC 367 and other organisms, GOLLD RNA resides in an apparent prophage. We therefore monitored GOLLD RNA transcription in L. brevis cultures grown with mitomycin C, an antibiotic that commonly induces prophages to lyse their hosts22. Increased GOLLD RNA expression correlates with bacteriophage particle production, and DNA corresponding to the GOLLD RNA gene is packaged into phage particles (Fig. 2b). Furthermore, most L. brevis GOLLD RNA transcripts made during bacteriophage production closely bracket the entire span of conserved sequences and structural elements as determined by mapping of the 5′ and 3′ termini (Supplementary Figure 3). Thus, expression of the entire noncoding RNA presumably is important for the bacteriophage lytic process.
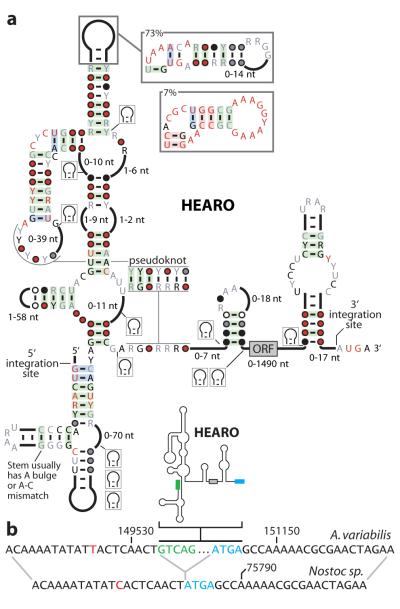
HEARO (HNH Endonuclease-Associated RNA and ORF) RNAs (Fig. 3a) often carry an embedded ORF that usually is predicted to code for an HNH endonuclease. This enzyme is commonly exploited by a variety of mobile genetic elements to achieve DNA transposition23. Thus HEARO RNA and its associated ORF together might constitute a mobile genetic element. The number of HEARO RNAs encoded by bacterial genomes varies widely. A total of 42 HEARO RNAs are predicted in Arthrospira maxima CS-328 (Supplementary Data), and most of these RNAs appear to represent recent duplications (Supplementary Fig. 4). When A. maxima HEARO sequences are aligned, it is apparent that the elements are highly conserved in sequence, while their flanking sequences show no conservation (Supplementary Fig. 5).
Figure 3. HEARO RNAs.
a, Consensus sequence and secondary structure model for HEARO RNAs. Annotations are as described in the legend to Fig. 3a. b, Typical sequence signature of HEARO genomic integration (see also Supplementary Fig. 6). (Top) HEARO element and flanking sequence in Anabaena variabilis ATCC 29413, plasmid C (NC_007412.1). Green text designates DNA corresponding to the first five nucleotides of conserved HEARO RNA. Blue text designates DNA corresponding to the conserved RUGA motif at each integration site. (Bottom) Homologous genome sequence lacking the HEARO element from Nostoc sp. PCC 7120, plasmid pCC7120beta (NC_003240.1). Red nucleotides identify positions that vary between the two genomes.
In some instances, homologs of the sequences flanking the consensus sequence can be identified in related bacterial species wherein the HEARO element is absent. These observations allow us to map putative integration events (Figure 3b, Supplementary Fig. 6), which are consistent with a requirement for integration immediately upstream of the sequence ATGA or GTGA. Self-splicing group I and group II introns frequently carry ORFs coding for endonucleases, and the combined action of the protein enzyme and ribozyme components permit transposition with a reduced chance for genetic disruption at the integration site23,24. The similarity in gene association between these RNAs suggests that HEARO RNAs may also process themselves. However, self-splicing could not be demonstrated using protein-free assays (unpublished data), and therefore HEARO may have a different function.
We observed expression of HEARO RNA from Exiguobacterium sibiricum (Supplementary Fig. 7), although we have not yet determined whether these RNAs undergo unusual processing in vivo. Structural probing experiments in vitro (Supplementary Fig. 8) reveal that an A. maxima HEARO RNA adopts most of the secondary structure features predicted from comparative sequence analysis data. Therefore, these RNAs may not require protein factors to form the folded state required for their biological function, just as some large ribozymes can form their active states without the obligate participation of proteins.
Four unusually abundant RNA structures, termed IMES-1 through IMES-4 (Supplementary Fig. 9), were identified in marine environmental sequences. The first three correspond to several noncoding RNA classes recently identified independently5, though our findings support different structural models (Supplementary Discussion). Expression of RNAs is often quantitated relative to 5S rRNA25, which is among the most abundant of bacterial RNAs. Remarkably, metatranscriptome sequences collected near Station ALOHA5,26 (Pacific Ocean) revealed that all IMES RNAs are exceptionally abundant (Supplementary Table 2). IMES-1 and IMES-2 RNAs are over five- and over two-fold more abundant than 5S rRNA, respectively.
Moreover, we find that IMES-1 RNA is also highly expressed in bacteria from another marine environment, in Block Island Sound (Atlantic Ocean), though not as abundantly as found in Station ALOHA samples (Supplementary Fig. 10). The high amounts of IMES-1 and IMES-2 RNAs are extremely rare for bacterial ncRNAs25, and only 6S RNA and total tRNAs are known to outnumber rRNAs27. Moreover, other than SprD28 and OxyS29 RNAs, all RNAs whose abundance is comparable to even the lower IMES-1 levels at Block Island Sound were reported by the early 1970s25,27.
Although we have identified numerous other noncoding RNAs in our searches (e.g. see Supplementary Table 1 and Supplementary Fig. 11), examples of ncRNAs with conserved sequence and structural complexity comparable to GOLLD and HEARO RNAs or with expression levels comparable to IMES RNAs are exceedingly rare. With few exceptions, these highly complex or abundant RNAs were discovered decades ago. One exception, OLE RNA16, is a complex-folded RNA recently discovered by conducting similar phylogenetic-comparative sequence analysis using DNA sequence data from cultured bacteria. This RNA is found in bacteria that can live under anaerobic conditions and that are commonly extremophilic. Thus GOLLD, HEARO, and OLE RNAs are members of a select group of large and complex-folded RNAs whose mysterious functions impact specialized groups of bacteria.
Only recently has sufficient DNA sequence data from cultured organisms been made available such that GOLLD and HEARO RNAs can be detected in a few disparate species, while IMES RNAs are not found at all within genome sequences derived from known bacteria. However, among the environmental sequences used to identify GOLLD and IMES RNAs, perhaps as many of 10 to 30 percent of bacterial cells in the relevant environment use these RNAs (Supplementary Table 3). Given that most bacterial species are extremely uncommon1-3, more RNAs with extraordinary characteristics likely remain undiscovered in rarer bacteria. Thus, improvements in sequencing technologies, cultivation methods, bioinformatics and experimental approaches are poised to yield a far greater spectrum of biochemical functions for large ncRNAs from bacterial, archaeal, and phage genomes.
METHODS SUMMARY
RNA motifs were discovered using a computational pipeline based on an early version of a method to cluster intergenic regions by sequence similarity17. The amounts of RNA expression in metatranscriptome data were established by the use of covariance model searches to identify IMES RNA and 5S RNA variants. Additional details on the sequence search and alignment methods are provided in the full Methods.
Information on oligonucleotides, bacterial cultures, and RNA analyses is detailed in the full Methods. GOLLD RNA expression was established by treating L. brevis cultures with mitomycin C (0.5 μg mL−1) to induce bacteriophage production. GOLLD RNA was detected by northern analysis and transcripts mapped by 5′-RLM-R A C E a n d 3′-RACE. Bacteriophages were detected from supernatant by PCR. IMES-1 RNA detection and quantitation was achieved using northern analysis of RNA samples isolated from bacteria collected by filtering ocean water. HEARO RNA was detected in vivo using RT-PCR of total RNAs isolated from cultured E. sibiricum cells.
METHODS
Detection and alignment of homologous RNA sequences
Novel classes of structured bacterial RNAs were identified using an updated method to cluster intergenic regions based on sequence similarity that is related to a recently published method17. Similar to our earlier method10, CMfinder30 was used to infer secondary structures from the clustered intergenic regions by simultaneously aligning based on conserved sequence and secondary structure features. The identified structures are subsequently used in covariance model-based homology searches31-33 to find additional examples that are used by CMfinder to refine its initial alignment. Motifs were scored using a variety of statistics as described previously10, and by inferring a phylogenetic tree using subroutines in Pfold34, then using pscore35. To find all homologs of the novel RNA classes, we used various homology search strategies that were previously developed11. The set of genome sequences searched were RefSeq36 version 32, and environmental sequences from acid mine drainage37, soil and whale fall38, human gut39,40, mouse gut41, gutless sea worms42, sludge communities43, termite hind gut44, marine samples at various depths near Station ALOHA45 and the global ocean survey21,46.
Protein-coding genes were annotated using several sources. For RefSeq sequences, we used the RefSeq annotation. For global ocean survey sequences, we used previously predicted proteins4. For certain sequences40,44,45, genes were predicted using the MetaGene program47 with default parameters. For the remainder, we used predictions downloaded from IMG/M48. Genes were classified into protein families based on the Conserved Domain Database49 version 2.08.
Multiple sequence alignments were developed using previously established methods11. Conservation statistics reflected in the consensus diagrams were calculated based on previously established protocols, and the following description is adapted from our previous report11. To reduce bias caused by nearly redundant sequences, sequences were weighted using Infernal’s implementation of the GSC algorithm. These weights were used to calculate nucleotide frequencies at each position in the alignment. Base pairs whose weighted frequency of non-canonical base pairs (neither Watson-Crick nor G-U) exceeded 10% were not classified as covarying, and are not shaded in consensus diagrams. Sequences in which both positions of the base pair are missing (deleted) or either nucleotide was uncertain (e.g., was the degenerate nucleotide code ‘N’) were not counted in the frequency. Base paired positions annotated as showing covariation (shaded green) meant that at least two Watson-Crick or G-U pairs were observed at the given base pair position and these two pairs differed at both nucleotides (e.g., A-U and C-G differ at both positions). If pairs were detected that differ at only one position (e.g., A-U and G-U), this is classified as compatible mutation. The method was varied for GOLLD RNA. GOLLD RNA is very large, and yet is mostly present in environmental sequence scaffolds that are relatively short. Thus, most of the detected GOLLD RNAs are fragmentary. We did not count parts of sequences that are not present in a particular sequence fragment in any of the statistics. Also, the diagram of GOLLD RNA in Figure 2a is based on the most common structure observed, but statistics for the highly conserved 3′ half (Supplementary Discussion) are based on all GOLLD RNAs, not just those with the most common structure.
RNA detection in metatranscriptome sequences
To determine if IMES RNAs are transcribed, we performed homology searches using accelerated covariance model searches implemented in RAVENNA31-33 on marine transcript sequences collected from Station ALOHA, in the open Pacific Ocean5,26. To accommodate the short transcript reads, we performed searches in “local” mode, which allows for partial matches to a query RNA model. To guard against false positives, we manually inspected predicted homologs, and used stringent E-value thresholds. Searches performed on known RNA motifs used as queries the relevant “seed” alignments in Rfam version 9.050.
Estimates of RNA distributions in genome and metagenome sequences
Note: For the HOT/ALOHA DNA samples in the “Subtropical Gyre” habitat (DeLong, et al.), we originally downloaded the GenBank files, which appear to be incorrectly annotated with metadata. (All GenBank sequences are annotated as being taken at 4000m depth.) Therefore, for the distribution analysis presented as Supplementary Table 1, the sequences were downloaded from the CAMERA web site (http://camera.calit2.net) and were searched in the same way as the metatranscriptome sequences, with an E-value cutoff of 10−4. When we aligned and inspected the hits, several contained long polynucleotide repeats, which are a common source of spurious low E-values; these hits were discarded. (As noted above, metatranscriptome hits were also inspected, but fit the expected consensus well, so no hits were discarded from those samples.)
Calculation of RNA class sizes and structural statistics
We enumerated bacterial ncRNAs with a complex structure (based on comparative or experimental data) that were present in more than one bacterial class. RNA classes were derived based on Rfam50 version 9.1 and NONCODE51. Sizes are the average reported by Rfam for the “seed” alignment, except as follows. All rRNA statistics are based on the E. coli model52. The RNase P size is the average of sequences in the two bacterial models in Rfam. Group II intron and HEARO RNA sizes were calculated as the average of RNA length minus their embedded ORF length. Because many HEARO RNAs are found in incomplete or environmental genomes, its ORFs are not well annotated. To avoid noise from misannotations (where typically the start codon is annotated upstream of the true start codon), we subtracted the entire variable-length region that can contain the ORF. Consequently, HEARO RNA sizes might be slightly underestimated. Group II intron and ORF sizes were derived from a previous study53. Conserved structures of known RNAs were taken from the literature for rRNAs as above52, group II introns54,55, OLE16, RNase P RNAs56, tmRNAs57, group I introns18 and riboswitches58. At least two consecutive Watson-Crick base pairs were required to define pseudoknots.
Although many quantitative definitions of structural complexity are possible, our use of multistem junctions and pseudoknots is equally applicable to a wide variety of RNAs for which comparative analysis or biochemical experiments are possible. Definitions based on other tertiary interactions, for example, would only be appropriate for RNAs that have been the subject of many detailed biochemical experiments.
Phylogenetic tree inference
The phylogenetic tree for HEARO RNAs was inferred as follows. First, from the HEARO multiple sequence alignment, we extracted the region 5′ to the point at which the ORF is sometimes inserted. This resulting alignment was converted to sequential PHYLIP-format using an in-house script, and used as input to PhyML59. PhyML version 3.0 was run with the command line:
phyml -i hearo-5prime.phylip --rand_start --n_rand_starts 10 -d nt -q -m GTR -f e -t e-v e -a e -s SPR -o tlr
Support for nodes in the resulting phylogenetic tree were calculated using the -b -4 option. The tree was drawn using the drawtree command from the PHYLIP package version 3.66, written by Joe Felsenstein.
Oligonucleotides
The sequences of oligonucleotides used in this study are given in the following table. Probes used for northern hybridizations of environmental RNA use IUPAC symbols for degenerate nucleotides.
| Description | Sequence |
|---|---|
| RT-PCR for HEARO RNA |
5′-ATCATACAGGTAGAGAATGGAAGGTGACAATG-3′ 5′-CGTCCGGTTGATAAACGATGTGACCAATC-3′ |
| PCR to generate template for Ama-1-29 RNA. Forward primer carries T7 RNA polymerase promoter. |
5′- CCAAGTAATACGACTCACTATAGGTCGTCGATAGTCAGCACCCCCGG -3′ 5′- CACGTAAAACTCCTGGGAGGGTTGG-3′ |
| Overlapping primers used to generate a fragment of Agrobacteriu m tumefaciens 5S rRNA as positive control in Northern hybridizatio n experiments. Forward primer carries T7 RNA polymerase promoter. |
5′-TAATACGACTCACTATAGGCGACCTGGTGGTCATCGCGGGGCGGCTGCAC CCGTTCCCTTTCCG-3′ 5′-CCTACTCTCCCGCGTCTTGAGACGAAGTACCATTGGCGCTGGGGCGTTTC ACGGCCGTGTTCGGAATGGGAACGGGT-3′ |
| Three overlapping primers used to generate synthetic IMES-1 RNA as positive control in Northern hybridizatio n experiments. Forward primer carries T7 RNA polymerase promoter. |
5′-TAATACGACTCACTATAGGTAATTTTCGACTAGTGACCAACTGCAGACGG AAGATCCTAGAGAAAAATTAAAGGAAGAGACCAAAGGGTGAAAGCAT-3′ 5′-GGAAGAGACCAAAGGGTGAAAGCATTTATAAGAGTCGATGATAAAAAACA GCTTATAAATCCACCAAGAATACAAGAGAAAGTATTCAAGGAG-3′ 5′-AAAACAGAGTCTAGCTCTGTTTCTTTAGTTCGAGGTCCTTAGGGGTCTAA AGTGAGATTTTCTTTAGCTCCTTGAATACTTTCTCTTG-3′ |
| Overlapping primers used to generate synthetic IMES-2 RNA as positive control in Northern hybridizatio n experiments. Forward primer carries T7 RNA polymerase promoter. |
5′-TAATACGACTCACTATAGGAAATGAATTAAGAGGCAACTCTTAACTGACC ATCTGGGGAAAAACCGAGAGGTTCAAGCCCAGAGGGCAGAAAACTCTAC-3′ |
| 5′-TTTTGGCACGTCGTTTTATGCGCTACCGCCATGCCTTCCACTCTACTATT TTAGCGCTACTCTGTAGAGTTTTCTGCCCTCTGGGC-3′ |
|
| Northern probe for IMES-1 RNA |
5′-ARGKGTCNDRAGTGAGATTYTCTTTAGCNCCTTGRNKDNTWTCTCTTNHN NYCTTGGTGG-3′ |
| Northern probe for IMES-2 RNA |
5′-TTTGGYWCGTCGTTKTANGCGCTACCG-3′ |
| Alternate Northern probe for IMES-3 RNA |
5′-ARYTSCGATCCAACYNRARRGTTGTGGACGATCTSA-3′ |
| Northern probe for IMES-4 RNA |
5′-AAWYTRMTTAYTAGGTTTGCGTGTAATAA-3′ |
| PCR of GOLLD gene in L. brevis. |
5′-GGTTAAAAAAAAGCCGCCT-3′ 5′-AGATTAACAGATTGAGAATACATCCG-3′ |
| PCR of non- phage DNA in L. brevis (negative control). |
5′-GACTGTAAAGATTGGTATTAATGGTTTC-3′ 5′-TTAGAGCGTTGCAAAGTGCA-3′ |
| PCR of phage DNA in L. brevis at a different locus from the GOLLD gene. |
5′-ATTCCCGCCGTGC-3′ 5′-CTGCTGCATCCATCTCA-3′ |
| Primers used to generate dsDNA template for in vitro transcriptio n of GOLLD 5′ RLM-RACE linker. |
5′- TTTCTACTCCTTCAGTCCATGTCAGTGTCCTCGTGCTCCAGTCGCCTATAGTGAGTCGTA TTA-3′ 5′-TAATACGACTCACTATAGG-3′ |
| Synthetic RNA linker used in GOLLD 5′ RLM RACE |
5′- CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′ |
| GOLLD 5′ RLM RACE reverse transcriptio n |
5′- CCGTTACCCGCGTTACGCTTAGACCAC-3′ |
| GOLLD 5′ RLM RACE PCR |
5′- CCGGTTCGTTTCCAGCTTAACGCCTTC-3′ 5′- GACTGGAGCACGAGGACACTGA-3′ |
| GOLLD 3′ RACE reverse transcriptio n |
5′- GCGGTCACGCTTACTTAGCCCTCACTGAAATTTTTTTTTTTTTTTTT-3′ |
| GOLLD 3′ RACE PCR |
5′- GCGGTCACGCTTACTTAGCCCTCACTGAA-3′ 5′- GAACGGGTGGAACTCCTTCCACCG-3′ |
In vitro-transcribed RNAs
RNAs corresponding to 5S rRNA, IMES-1 and IMES-2 were in vitro transcribed to use as standards or markers. The Template DNA for the in vitro transcription was assembled from overlapping oligonucleotides (see table above). The RNA sequences expected to result from the transcription reactions are as follows. Lowercase g’s represent G nucleotides that were added to improve transcription yield.
5S rRNA:
5′-ggCGACCUGGUGGUCAUCGCGGGGCGGCUGCACCCGUUCCCUUUCCGAACACG GCCGUGAAACGCCCCAGCGCCAAUGGUACUUCGUCUCAAGACGCGGGAGAGUA GG-3′
IMES-1 RNA
5′ -ggUAAUUUUCGACUAGUGACCAACUGCAGACGGAAGAUCCUAGAGAAAAAUUA AAGGAAGAGACCAAAGGGUGAAAGCAUUUAUAAGAGUCGAUGAUAAAAAACA GCUUAUAAAUCCACCAAGAAUACAAGAGAAAGUAUUCAAGGAGCUAAAGAAA AUCUCACUUUAGACCCCUAAGGACCUCGAACUAAAGAAACAGAGCUAGACUCUGUUUU-3′
IMES-2 RNA:
5′-ggAAAUGAAUUAAGAGGCAACUCUUAACUGACCAUCUGGGGAAAAACCGAGAG GUUCAAGCCCAGAGGGCAGAAAACUCUACAGAGUAGCGCUAAAAUAGUAGAG UGGAAGGCAUGGCGGUAGCGCAUAAAACGACGUGCCAAAA-3′
Bacteria and growth conditions
L. brevis ATCC 367 was grown in MRS broth (Becton-Dickinson) at 28°C. E. sibiricum 255-15 (gift of Debora Rodrigues) was grown in half-strength tryptic soy broth (Becton-Dickinson) at 37°C.
Phage induction experiments
L. brevis cultures were grown in MRS broth, and samples were taken at the indicated time points starting from cells in exponential phase at an OD600 of 0.15 to 0.2. For cultures treated with mitomycin C, the time points are relative to the addition of mitomycin C at 0.5 μg/mL. Each sample was centrifuged to isolate cells for RNA extraction and to recover supernatant for phage detection. RNA was isolated from pelleted cells using TRIzol LS (Invitrogen) in accordance with the manufacturer’s instructions. Supernatant was treated with DNase RQ1 (Promega) for 1 hour at 37°C to eliminate naked genomic DNA, followed by proteinase K (Roche) and EDTA treatment 30 minutes at 37°C followed by 30 minutes at 55°C to degrade DNase molecules and phage capsids. Proteinase K was heat inactivated at 96°C for 10 minutes. 1 μL of a 1/10 dilution was used to deliver phage DNA for PCR templates. To ensure phage identity of the DNA, two separate regions were amplified, and a bacterial genomic DNA region was used as a negative control against host DNA (see Oligonucleotides).
Collection of water samples and extraction of RNA
Shore water samples were collected in Long Island Sound at Lighthouse Point Park (41° 15′ 6.3” N, 72° 54′ 14.5” W) at 12 pm on April 26, 2009. Off-shore water samples were collected in Block Island Sound at coordinates 41° 19′ 17” N, 71° 32′ 11” W between 11:30 am and 12:00 pm on May 21, 2009. The off-shore sample was collected from 15-20 m depth (total depth was 27 m) using a sealed container whose seal was broken when it reached the specified depth. Since we did not have filtering equipment at the sampling sites, there was a delay of approximately 30 minutes for the shore sample, and 3 hours for the off-shore sample between collection and commencement of filtration. We obtained essentially identical results in Northern hybridization experiments using water that was filtered roughly 10 hours after collection as with water that was filtered 3 hours after collection.
Bacterial cells were collected from the water sample by vacuum filtration onto a 47 mm diameter, 0.22 μm pore durapore membrane filter (Millipore part #GVWP04700), with the use of a 37 mm diameter, 1.6 μm “APFA”-type glass-fiber prefilter (Millipore part #APFA03700). After filtration the filters were stored at -80°C. To extract RNA, lysozyme from chicken egg white (Sigma) at 1 mg/mL was applied directly to the filter, until the filter was covered (covering required 300 to 500 μL). The cells were subjected to three freeze/thaw cycles. TRIzol LS reagent was added directly to the filter and re-applied repeatedly to fully suspend cellular material. The TRIzol solution was collected and subsequent steps for RNA isolation followed the manufacturer’s instructions.
GOLLD and IMES-1 RNA analysis by Northern hybridization
Northern analysis of GOLLD RNA was conducted on RNA (2 μg per lane) separated using a denaturing (6% formaldehyde) 1% agarose gel electrophoresis in a running buffer of 20 mM MOPS (free acid), 8 mM sodium acetate, 1 mM EDTA (final pH of 7.0). Northern blots for IMES-1 were conducted on RNA (1 μg per lane) extracted from ocean water that was separated by denaturing (8 M urea) 6% polyacrylamide gel electrophoresis (PAGE). RNAs in the resulting gels were blotted by capillarity action to a Hybond-N+ membrane (Amersham Biosciences) and hybridization was conducted with 5′ radiolabeled oligonucleotides in Rapid-hyb buffer (GE Healthcare) with hybridization times ranging from 2 hours to 16 hours and 42°C to 45°C. Bands were quantified using a Storm PhosphorImager and ImageQuant software (Molecular Dynamics).
Standards for quantitation were created by probing an in-vitro transcribed IMES-1 RNA with an IMES-1-specific probe or a 5S rRNA-specific probe (see “In vitro-transcribed RNAs” section above for RNA sequences). The amount of in-vitro transcribed RNA applied to the gel ranged from 0.001 to 1 pmol. Total RNA isolated from ocean water samples was hybridized to the same membrane, and hybridized with the same probes.
Because the Standards and the Total RNA analysis lanes were part of the same gel and membrane, their intensities can be directly compared. Thus, for example, lane 7 of the IMES-1 analysis image (Supplementary Fig. 10) appears to contain ~0.01 pmol IMES-1 RNA. Our estimates assume that the in vitro-transcribed IMES-1 and 5S rRNA standards anneal to the probes with similar efficiencies to homologous RNAs found in environmental samples. The in-vitro transcribed 5S rRNA sequence is based on the α-proteobacterial A. tumefaciens sequence, since the species containing IMES-1 RNAs were predicted to be related to proteobacteria, and likely to α-proteobacteria. Sample 7 appears to contain ~0.2 pmol 5S rRNA.
GOLLD 5′-RLM RACE
A total of 10 μg RNA isolated from L. brevis eight hours after the addition of mitomycin C was treated with tobacco acid pyrophosphatase (Epicenter Biotechnologies) to remove any 5′ pyrophosphate or triphosphate in a total volume of 20 μL for 1 hour at 37 °C according to the manufacturer’s instructions. The reaction was terminated by phenol-chloroform extraction and the RNA was recovered by precipitation with ethanol.
The RNA was resuspended in deionized water and ligated using T4 RNA ligase 1 (New England Biolabs) to an RNA linker (0.25 μg, see Oligonucleotides) in a total volume of 20 μL incubated at 37°C for 1 hr according to the manufacturer’s instructions. The reaction was terminated and the RNA recovered as described above. The RNA linker was transcribed in vitro from a DNA template constructed by primer extension (see Oligonucleotides for primer sequences). The RNA was resuspended in deionized water and reverse transcription performed using a GOLLD specific primer (see Oligonucleotides) with Superscript-II reverse transcriptase (Invitrogen) in a total volume of 20 μL for 1.5 hrs at 42°C according to the manufacturer’s instructions. The reaction was terminated by heating at 65°C for 20 minutes and 1 μL used as template for PCR amplification with Taq polymerase (New England Biolabs) (see Oligonucleotides for primer sequences). PCR products were cloned using a TOPO-TA cloning kit (Invitrogen) and transformed into TOP10 cells (Invitrogen). Plasmids from 12 of the resulting colonies were purified (Qiagen) and sequenced (Keck DNA sequencing resource, Yale University).
The initial RLM-RACE experiments produced sequences with additional bases between the expected linker sequence and the genomic sequence from L. brevis resulting from run-on transcription of the in vitro transcribed RNA-linker60. Although the 5′ extents of the transcripts seemed clear, the entire RLM-RACE was repeated as above using a synthetic linker purchased from the Keck Biotechnology Resources Laboratory with a 2′ protecting TOM group, deprotected61 through treatment with 1 M tetrabutylammonium fluoride in tetrahydrofuran (Sigma) and subsequently purified by denaturing 6% PAGE. An additional eleven sequences were obtained and the combined results of both experiments are reported. Some significantly shorter sequences resulted from the second RACE experiment where the RNA was likely more degraded due to sample handling and additional freeze-thaw cycles between the two experiments. However, the dominant 5′ ends in the second experiment match the location determined from the first experiment.
GOLLD 3′ RACE
A total of 10 μg RNA isolated from L. brevis eight hours after the addition of mitomycin C was polyadenylated using E. coli poly(A) polymerase (New England Biolabs) according to the manufacturer’s instructions. The reaction was terminated and the RNA was recovered as described above. The polyadenylated RNA was resuspended in water and reverse transcription was conducted using Superscript II reverse transcriptase (Invitrogen) in a total volume of 20 μL at 42°C for 1.5 hours according to the manufacturer’s instructions. The reaction was terminated by heating at 65°C for 20 minutes and 1 μL was subsequently used as template for PCR using Taq polymerase (New England Biolabs) (see Oligonucleotides for primer sequences). PCR products were cloned and their DNA sequenced as described above.
RT-PCR analysis of HEARO RNA
E. sibiricum cultures were harvested during both stationary and logarithmic phase growth. The equivalent of 1 mL of cell culture at an OD600 of 3 were pelleted and resuspended in 100 μL of 3 mg/mL lysozyme in TE buffer (10 mM Tris-HCl [pH 7.5 at 23°C], 1 mM EDTA). Cell lysis was facilitated by multiple freeze-thaw cycles before isolating RNA with 1 mL of TRIzol using the manufacturer’s protocol. DNA was removed through digestion with DNase RQ1 (Promega) at 37°C for 1 hour. Approximately 2.5 μg of total RNA was used as template for reverse transcription at 55°C for 1.5 hours in a volume of 20 μL using SuperScript III (Invitrogen) reverse transcriptase primed by random DNA hexamers supplied by the vendor according to the manufacturer’s instructions. Negative control samples included for each analysis were prepared using identical conditions but without enzyme addition. 1 μL of each reverse transcription reaction was used to deliver cDNA templates for PCR.
In-line probing of HEARO (Ama-1-29 RNA)
DNA corresponding to the Ama-1-29 RNA was amplified from Arthrospira maxima genomic DNA by PCR, and the resulting DNA product was used as template for in vitro transcription using T7 polymerase to produce RNA (see Oligonucleotides for primer sequences). The RNA was purified by 6% denaturing PAGE, extracted from the gel slice in 200 mM NaCl, 10 mM Tris-HCl (pH 7.5 at 25°C), 1 mM EDTA and precipitated with ethanol. The RNA was resuspended in deionized water and separate aliquots of the RNA were 5′ or 3′ 32P-labeled.
For the 5′ labeling, 10 pmol of RNA was dephosphorylated using rAPid alkaline phosphatase (Roche) according to the manufacturer’s instructions. The phosphatase reaction was terminated by heating at 95°C for three minutes and the dephosphorylated RNA was subsequently 5′ 32P-labeled in 5 mM MgCl2, 25 mM CHES (pH 9.0 at 25°C), 3 mM DTT using 40 μCi of γ-32P ATP and 20 U T4 polynucleotide kinase (New England Biolabs) and incubated at 37 °C for 1 hr. The RNA was purified by denaturing PAGE and recovered from the gel as described above.
For the 3′ labeling, 50 pmol of RNA was incubated in 50 mM Tris-HCl (pH 7.8 at 25°C), 10 mM MgCl2, 10 mM DTT, 2 mM ATP, 10% DMSO with 150 μCi of pCp [5′-32P], and 50 U of T4 RNA ligase 1 (New England Biolabs) at 4°C for 40 hours. RNA was purified and recovered as described above. In-line probing reactions were prepared with radiolabeled RNAs as noted and analyzed by denaturing 10% PAGE essentially as described previously62.
Supplementary Material
Acknowledgements
We thank Nick Carriero and Rob Bjornson for assisting our use of the Yale Life Sciences High Performance Computing Center (NIH grant RR19895-02), Capt. Thad Gruczka for advice and assistance in ocean water collection, Jingying Yang for assistance with the analysis of the dct-1 motif, Debora Rodrigues for E. sibiricum, Donald Bryant for A. maxima genomic DNA and Patrick O’Donoghue, Ming Hammond, Narasimhan Sudarsan, Sanshu Li, Jeffrey Barrick, Zizhen Yao, Larry Ruzzo, and Elizabeth Tseng for helpful advice. J.P. and M.M.M. were supported by postdoctoral fellowships from the Canadian Institutes of Health Research and National Institutes of Health, respectively. R.R.B is a Howard Hughes Medical Institute Investigator.
Footnotes
Author Contributions Z.W. and R.R.B conceived of the study and R.R.B supervised the research. Z.W. created bioinformatics scripts and prepared RNA sequence alignments. J.P. conducted GOLLD and IMES RNA experiments. M.M. conducted GOLLD RACE and HEARO RNA experiments. Z.W. and R.R.B. wrote the manuscript, and all authors participated in editing.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA. 95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA. 99:10234–10236. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bent SJ, Forney LJ. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2008;2:689–695. doi: 10.1038/ismej.2008.44. [DOI] [PubMed] [Google Scholar]
- 4.Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature. 2009;459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- 6.Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- 7.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrick JE, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U S A. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao Z, Barrick J, Weinberg Z, Neph S, Breaker R, Tompa M, Ruzzo WL. A computational pipeline for high-throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput. Biol. 2007;3:e126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg Z, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer M, et al. Identification of candidate structured RNAs in the marine organism ‘Candidatus Pelagibacter ubique’. BMC Genomics. 10:268. doi: 10.1186/1471-2164-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montange RK, Batey RT. Riboswitches: emerging theses in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 14.Roth A, Breaker RR. The structure and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puerta-Fernandez E, Barrick JE, Roth A, Breaker RR. Identification of a large noncoding RNA in extremophilic eubacteria. Proc. Natl. Acad. Sci. USA. 2006;103:19490–19495. doi: 10.1073/pnas.0607493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng HH, Weinberg Z, Gore J, Breaker RR, Ruzzo WL. Finding non-coding RNAs through genome-scale clustering. J Bioinform Comput Biol. 2009;7:373–88. doi: 10.1142/s0219720009004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 19.Pace NR, Thomas BC, Woese CR. Probing RNA structure, function, and history by comparative analysis. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. pp. 113–141. [Google Scholar]
- 20.Toor N, Keating KS, Pyle AM. Structural insights into RNA splicing. Curr. Opin. Struct. Biol. 2009;19:1–7. doi: 10.1016/j.sbi.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raya RR, Hébert EM. In: Bacteriophages: methods and protocols. Clokie MRJ, editor. Vol. 1. Humana Press; New York: 2009. [Google Scholar]
- 23.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 24.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu. Rev. Genetics. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 25.Wassarman KM, Zhang A, Storz G. Small RNAs in Escherichia coli. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 26.Frias-Lopez J, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci U S A. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wassarman KM. 6S RNA: a regulator of transcription. Mol. Microbiol. 2007;65:1425–1431. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- 28.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A Small, Stable RNA Induced by Oxidative Stress: Role as a Pleiotropic Regulator and Antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 30.Yao Z, Weinberg Z, Ruzzo WL. CMfinder – a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 31.Weinberg Z, Ruzzo WL. Sequence-based heuristics for faster annotation of non-coding RNA families. Bioinformatics. 2006;22:35–39. doi: 10.1093/bioinformatics/bti743. [DOI] [PubMed] [Google Scholar]
- 32.Eddy SR, Durbin R. RNA Sequence Analysis Using Covariance Models. Nucleic Acids Res. 1994;22:2079–88. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein RJ, Eddy SR. RSEARCH: finding homologs of single structured RNA sequences. BMC Bioinformatics. 2003;4:44. doi: 10.1186/1471-2105-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao Z. Dissertation. University of Washington; Seattle, WA: 2008. Genome scale search of noncoding RNAs: bacteria to vertebrates. [Google Scholar]
- 36.Pruitt K, Tatusova T, Maglott D. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 38.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 39.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 42.Woyke T, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–5. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 43.Garcia Martin H, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006;24:1263–9. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- 44.Warnecke F, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–5. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 45.DeLong EF, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 46.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 47.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–30. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markowitz VM, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–8. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:192–196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardner PP, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–40. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C, et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–5. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai L, Zimmerly S. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 2002;30:1091–102. doi: 10.1093/nar/30.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boudvillain M, Pyle AM. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO. J. 1998;17:7091–104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–52. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haas ES, Brown JW, Pitulle C, Pace NR. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl. Acad. Sci. USA. 1994;91:2527–31. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zwieb C, Wower I, Wower J. Comparative sequence analysis of tmRNA. Nucleic Acids Res. 1999;27:2063–71. doi: 10.1093/nar/27.10.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 60.Cazenave C, Uhlenbeck OC. RNA-template-directed RNA synthesis by T7 polymerase Proc. Natl. Acad. Sci. USA. 1994;91:6972–6976. doi: 10.1073/pnas.91.15.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu T, Ogilvie TT, Pon RT. Prevention of chain cleavage in the chemical synthesis of 2′ silylated oligoribonucleotides. Nucleic Acids Res. 1989;17:3501–3517. doi: 10.1093/nar/17.9.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. In: Wilusz J, editor. Methods in Molecular Biology vol 419: Post-Transcriptional Gene Regulation. Humana Press; Totowa, NJ: 2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.