Abstract
An expanding number of metabolite-binding riboswitch classes are being discovered in the non-coding portions of bacterial genomes. Findings over the last decade indicate that bacteria commonly use these RNA genetic elements as regulators of metabolic pathways and as mediators of changes in cell physiology. Some riboswitches are surprisingly complex, and they rival protein factors in their structural and functional sophistication. Each new riboswitch discovery expands our knowledge of the biochemical capabilities of RNA and some give rise to new questions that require additional study to be addressed. Some of the greatest prospects for riboswitch research and some of the more interesting mysteries are discussed in this review.
Ten Years of Riboswitch Research
The first experimental proofs that some messenger RNAs carry metabolite-sensing domains to control gene expression were published nearly a decade ago (Mironov et al., 2002; Nahvi et al., 2002; Winkler et al., 2002a; 2002b). The name “riboswitch” was chosen to reflect the fact that each of these RNA motifs functions as a genetic switch (Nahvi et al., 2002). These initial reports and many subsequent studies confirmed earlier speculation (Gold et al., 1997a; 1997b; Gelfand et al., 1999; Nou and Kadner, 2000; Miranda-Rios et al., 2001; Stormo and Ji, 2001) that certain mRNAs might directly sense and respond to changing concentrations of small molecules. The list of experimentally validated riboswitch classes has grown to 24, and there are many hundreds or even thousands of representatives for some of these classes in the current DNA sequence databases.
The known collection of riboswitch classes probably constitute only a tiny portion of the total that exist in the biosphere, and so many new classes are likely to be discovered in the future (Ames and Breaker, 2010; Breaker, 2011). Some of the characteristics established in published studies on riboswitches are sure to be very general, whereas other properties are noteworthy for their rarity and for the interesting capabilities they offer to the cells that carry these unique riboswitches. One of the most understandable features of a riboswitch is that the ligand-binding “aptamer” domain is its most conserved component. This makes sense because the aptamer is formed from only four types of nucleotides and these must fold to form a precise sensor for a ligand (Figure 1A, Top) that never changes through evolution. In contrast, the sequence and structure of the regulatory “expression platform” of each riboswitch can vary greatly (Barrick and Breaker, 2007) because there are far more ways in which simple RNA structures can influence such processes as transcription, translation, and RNA processing (Figure 1A, Bottom).
Figure 1. Riboswitch ligands and mechanisms.
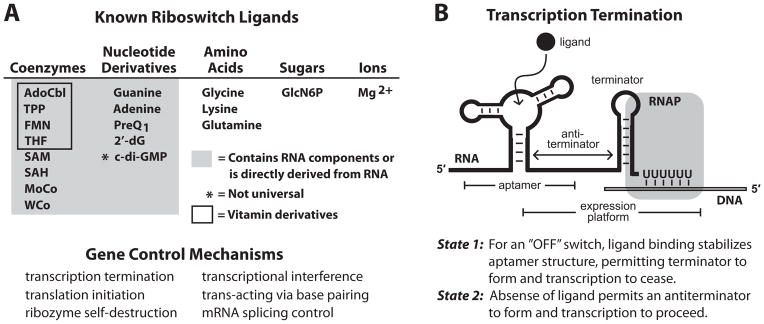
(A) (Top) List of the riboswitch ligands with biochemical and or genetic validation. (Bottom) Demonstrated mechanisms for riboswitch-mediated gene control.
(B) Schematic representation of the most common form of riboswitch-mediated gene regulation: transcription termination. Image depicts RNA polymerase (RNAP) in the act of transcribing the U-rich portion of an intrinsic transcription terminator stem in “State 1”, wherein ligand has been bound by the aptamer and transcription will terminate. Alternatively, in “State 2” (not shown), the absence of ligand allows nucleotides from the aptamer to form a competing anti-terminator stem that allows transcription to pass beyond the U-rich termination site. Other riboswitch mechanisms are described elsewhere (Breaker, 2011).
Another readily understood feature of bacterial riboswitches is that they are almost exclusively located in the 5′ untranslated regions (5′ UTRs) of the mRNAs whose expression they control. This arrangement permits the riboswitch to be synthesized first and gives time for the riboswitch to respond to metabolite binding before the full-length mRNA (or entire operon) is produced. The foremost mechanism for riboswitch-mediated gene control appears to be transcription termination (Figure 1B). Bacteria favor this arrangement probably to save time and to avoid wasting energy producing full-length mRNAs that are not needed. Thus, RNA structural diversity, the relative ease of regulating mRNA expression, and the requirement for biochemical efficiency all influence the structures, genomic locations, and mechanisms of riboswitches.
These and many other common characteristics of metabolite-sensing riboswitches (Dambach and Winkler, 2009; Roth and Breaker, 2009; Serganov, 2009) have been revealed over the last decade. However, there are some very interesting puzzles and paradoxes that lie where knowledge becomes sparse. While the strange characteristics of some riboswitches are believed to have been resolved, others remain mysterious and await future resolution. In this manuscript, I describe some of the more interesting aspects of riboswitches that are sure to hold new surprises and reveal new insights into the functional complexity of these noncoding RNA elements and their utility in many organisms.
When is an RNA Called a Riboswitch?
The term riboswitch was originally coined (Nahvi et al., 2002) to designate an RNA genetic switch that directly binds metabolites without the obligate involvement of proteins. This name and its definition were inspired by the name and function of ribozymes. The term ribozyme designates an enzyme-like RNA that can promote a chemical transformation without the obligate involvement of proteins, although the activities of many ribozymes are enhanced by protein partners. As with ribozymes, proofs of riboswitch activity in protein-free assays along with confirmatory genetic data were necessary before claims of their existence were widely accepted by the scientific community (Mironov et al., 2002; Nahvi et al., 2002; Winkler et al., 2002a; 2002b). Also like most ribozymes, each riboswitch must harness the limited chemical diversity of RNA nucleotides alone to form a selective ligand binding pocket akin to the active site of a ribozyme. This means that most riboswitch classes have conserved sequence and structural characteristics that are similar in complexity to some of the smaller ribozyme classes.
A number of bacterial gene control discoveries that intimately involve special RNA structures preceded the discovery of metabolite-binding riboswitches, and all these systems have at least one or sometimes several features in common (Table 1). These similarities have motivated some to retroactively rename previously-known systems as riboswitches, leading to some confusion. Gene control by bacterial attenuation mechanisms (Yanofsky, 1981) involves ribosome stalling under low amino acid concentrations to cause alternative folding of the mRNA located upstream of the open reading frame (ORF). Such classic attenuation mechanisms for bacterial gene control make use of intrinsic terminator stems (Yarnell and Roberts, 1999; Gusarov and Nudler, 1999), which are also commonly used by riboswitches. However, attenuation systems react to changing amino acid concentrations by sensing the availability of aminoacyl-tRNAs, which correlates with the concentrations of their cognate amino acid. In other words, the ligand sensor is the aminoacyl-tRNA synthase, and eventually the ribosome, and not the mRNA itself as is the case with metabolite-binding riboswitches.
Table 1. Features of various bacterial gene regulation systems involving RNA.
For a review of bacterial gene regulation systems that rely on RNA components, see Waters and Storz, 2009.
| RNA System | Sensory Targets | Sensor Component | Expression Platform |
|---|---|---|---|
| Riboswitches | Metabolites Inorganic Ions | Aptamer | Intrinsic Terminators RBS Occlusion Stems Alternative Splicing Others |
| Attenuation (uORFs) | Amino Acids via Charged tRNAs | Ribosomes and Amino-acyl tRNA synthases | Intrinsic Terminators |
| T boxes | Amino Acids via Uncharged tRNAs | mRNA Base Pairing and Other Contacts | Intrinsic Terminators |
| Thermosensors | Temperature | Thermosensitive RNA Structure | RBS Occlusion Stems |
| Riboregulators | Metabolites and Other Signals | Protein Factor or Unknown | RBS Occlusion Stems or Direct RBS Occlusion |
| Protein-binding RNAs | Metabolites, Proteins, and Other Signals | Protein Factor | Intrinsic Terminators |
Other gene control systems involving mRNA structures also exist with similarities to riboswitches. In the 1980s and 1990s, several groups reported the existence of metabolite-sensing proteins that bind to mRNA 5′ UTRs and control mRNA folding and expression, as exemplified by the TRAP (Shimotsu et al., 1986) and HutP (Oda et al., 1992) proteins. These proteins typically bind short spans of nucleotides or small structural domains within an mRNA, and this association is mediated by small molecule binding to the protein. Likewise, three other RNA-based regulatory systems, RNA Thermosensors, T boxes, and riboregulators, do not have any obligate need for protein factors. RNA “thermosensors” (Klinert and Narberhaus, 2009) usually form simple thermo-responsive base-paired structures that control access to the ribosome binding site of an mRNA. T box RNAs (Grundy and Henkin, 1993) bind non-aminoacylated tRNAs to regulate gene expression under low amino acid concentrations. T box RNAs do not directly bind the amino acid whose concentration they respond to, but rather respond indirectly to low amino acid concentration changes by binding non-aminoacylated tRNAs. Binding of the tRNA is mediated in part by short antisense Watson-Crick pairing between the anticodon nucleotides and nucleotides of the T box, and so this class of regulatory elements also has characteristics of small riboregulators (Gottesman, 2005) that use simple Watson-Crick base pairing to the mRNAs they control.
All these systems frequently use alternative RNA structures to control gene expression that are similar to the expression platform domains found with many metabolite-binding riboswitches. However, none of these systems use direct metabolite binding by RNA to measure the concentration of the small molecule target as do riboswitches. This need for the riboswitch to form the direct sensor of the small-molecule ligand places enormous requirements on the sequence and structure of the aptamer domain. Indeed, riboswitch aptamers appear to be among the most highly-conserved biopolymer domains in all of biology, and such conservation of sequence and structure provides a useful way to classify riboswitch representatives.
Even armed with these basic distinctions between the various types of bacterial regulatory systems involving RNA, it is not always possible to draw solid lines dividing all types. A definition that encompasses the vast majority of one type of regulatory RNA sometimes includes one or more motifs that blurs the lines and could be classified as another type. Although this causes frustration for those seeking perfect definitions, the source of this frustration – the great structural and function diversity of RNA – is wonderfully harnessed by cells to build truly striking RNA sensors and switches. In subsequent sections, I highlight a variety of additional aspects of metabolite-binding riboswitches that suggest there is far more diversity and complexity to these RNA devices that remain to be revealed.
How Many Riboswitches Exist?
Common questions that emerged soon after the first riboswitch validation studies were published centered on whether riboswitches were biochemical oddities or whether those sensing adenosylcobalamin (AdoCbl) (Nahvi et al., 2002), thiamin pyrophosphate (TPP) (Mironov et al., 2002; Winkler et al., 2002b), and flavin mononucleotide (FMN) (Mironov et al., 2002; Winkler et al., 2002a) represented just part of a large undiscovered collection of metabolite-sensing RNAs. With the subsequent discovery and validation of about two dozen riboswitch classes, we can now confidently say that bacterial genomes will carry far more classes with distinct structural architectures, additional ligand specificities, and novel gene control mechanisms (Ames and Breaker, 2010; Breaker, 2011). For some organisms such as Bacillus subtilis, the number of known riboswitch classes is greater than the number of validated protein factors that directly bind metabolites and control gene expression. Although this may be hard to believe, researchers have much more validation and accounting work to do before declaring with confidence which biopolymer dominates metabolite sensing and gene regulation in even the simplest organisms.
The fact that the AdoCbl, TPP and FMN riboswitches were the first experimentally validated classes is not at all surprising. These three classes, along with subsequently-reported riboswitch classes for S-adenosylmethionine (SAM) (Epshtein et al., 2003; McDaniel et al., 2003, Winkler et al., 2003), lysine (Grundy et al., 2003; Sudarsan et al., 2003a), guanine/adenine (Mandal et al., 2003; 2004b), glycine (Mandal et al., 2004a), and the bacterial second messenger c-di-GMP (Sudarsan et al., 2008), are among the ten most-common riboswitch classes known to exist. Their high frequency of occurrence in the genomes of bacteria certainly increased the chances that researchers using search methods based either on genetics or bioinformatics would have quickly encountered them. Interestingly, two of the top ten most common classes are riboswitch candidates called yybP (Barrick et al., 2004) and ydaO (Block et al, 2010). These “orphan riboswitches” and others that lack assigned ligands will be discussed in greater detail below.
There is an intriguing pattern to the frequency of occurrence of validated riboswitch classes in the genomes of sequenced organisms (Ames and Breaker, 2010; Breaker, 2011). Specifically, there are an ever increasing number of ever more rare riboswitch classes, and this pattern is reminiscent of data that fits a power law distribution. If this trend persists, then there could be hundreds of undiscovered classes encoded among the first 1000 bacterial genomes that have been sequenced. Indeed, orders of magnitude more classes may remain to be discovered among the great diversity of microbes on the planet that have yet to be sequenced. Unfortunately, many of these classes will be exceedingly rare and difficult or even impossible to discover and catalog by using existing genetics and bioinformatics approaches.
Although we may not have a complete set of the natural riboswitch classes anytime soon, we can speculate on the characteristics that many of these RNAs are likely to exhibit. Given the collection of known classes, we can see that bacterial cells care very much about sensing and responding to changing concentrations of key coenzymes and metabolites. Sometimes a riboswitch class is exceedingly well conserved in sequence and architecture across all species, such as is found with the TPP riboswitch class (Barrick and Breaker, 2007). In contrast, several other ubiquitous metabolites have two or more distinct riboswitch classes that sense them. For example, there are two different ways that RNA folds to bind the natural guanine analog called prequeuosine 1 (PreQ1) (Roth et al., 2007; Meyer et al., 2008). The smallest PreQ1-I class is formed by as few as 34 nucleotides, and perhaps its small size and simpler architecture explains why it is more common than the larger PreQ1-II class.
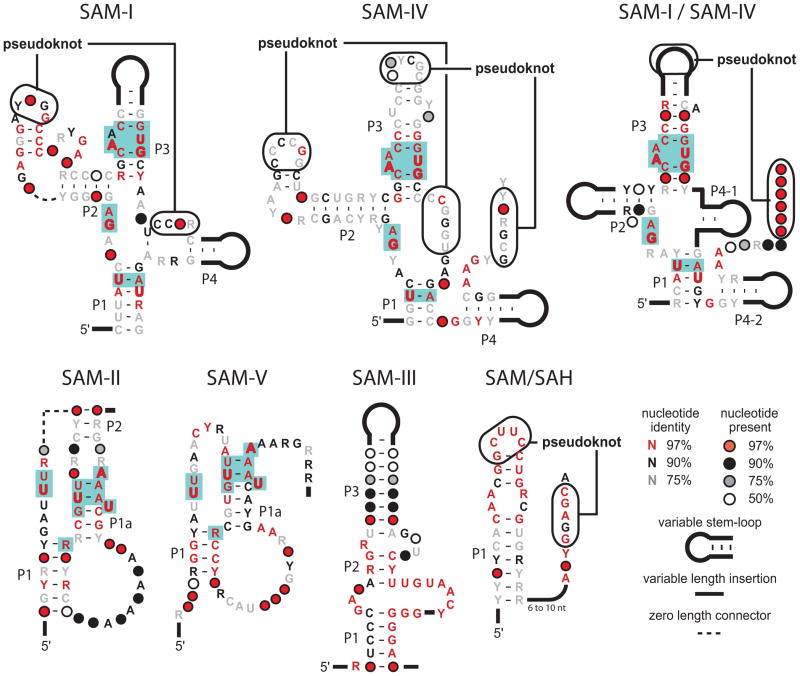
Likewise, there are at least two distinct riboswitch classes for the important bacterial second messenger c-di-GMP (Sudarsan et al., 2008; Lee et al., 2010). Both c-di-GMP-I and c-di-GMP-II classes involve the formation of conserved secondary and tertiary sub-structures interspersed with highly-conserved nucleotides. Globally, these structures have nothing in common, and yet they form precise binding pockets for the same second messenger molecule. Perhaps most striking is the fact that at least five major classes of RNAs have been found that sense SAM or its metabolic derivative S-adenosylhomocysteine (SAH) (Figure 2) (Wang and Breaker, 2008; Weinberg et al., 2008). Thus, among the many new classes of riboswitches that are likely to be discovered in the future, both novel motifs for additional metabolites and novel motifs for metabolites that already have validated riboswitch classes are likely to be encountered. Undoubtedly, the full spectrum of RNA structural and functional potential will be on display.
Figure 2. The collection of riboswitch classes that sense SAM and/or SAH.
Depicted are the consensus sequence and structural models for four major families of riboswitches that respond to SAM. The SAM-I family is comprised of representatives classified as SAM-I (or S box), SAM-IV and SAM-I/IV riboswitches. Note that nucleotides shaded in turquoise are identical in these three consensus models, and these nucleotides are intimately involved informing the SAM-binding site (Montange and Batey, 2006). Similarly, the SAM-II family is comprised of SAM-II and SAM-V riboswitches that share nucleotides that form the ligand-binding pocket. The remaining two families are represented by SAM-III (or SMK box) and SAM-SAH riboswitch classes. A fifth riboswitch class that rejects SAM and selectively binds its metabolic derivative S-adenosylhomocysteine (SAH) (Wang et al., 2008) is not shown. P1 and related notations indicate base paired substructures.
Why are Some Riboswitch Types so Rare?
One question that emerges when reviewing the distributions of riboswitch representatives is that some classes are extremely rare, despite the fact that they appear to serve as very effective metabolite sensors and gene control systems. In some instances, the reason for the rarity of a riboswitch class is obvious: it has stiff competition for its job from other riboswitch classes with near-identical functional characteristics. This is most evident by examining the collection of riboswitch classes noted above that sense SAM (Figure 2). This collection includes the very common SAM-I class (Epshtein et al., 2003; McDaniel et al., 2003, Winkler et al., 2003) and its close variants SAM-IV (Weinberg et al., 2008) and SAM-I/IV (Weinberg et al., 2010), the more rare SAM-II (Corbino et al., 2005) and SAM-V variants (Poiata et al., 2009), SAM-III or SMK class (Fuchs et al., 2006), and the exceedingly rare SAM/SAH class (Weinberg et al., 2010). Organisms clearly have a diversity of SAM riboswitch classes to choose from, which erodes the dominance of any single class.
Some competing SAM riboswitch classes such as the SAM/SAH motif are so simple that they might easily emerge through evolution on multiple occasions, and be more readily propagated. Despite this advantage, representatives of the SAM-SAH class are few in number, which might be due to the fact that it has relatively poor affinity and specificity for SAM compared to its competitor riboswitch classes (Weinberg et al., 2010). The rarity of other riboswitch representatives, particularly of the purine-sensing family is far more puzzling as noted below.
The collection of purine-sensing riboswitches (Kim and Breaker, 2008) together rank as the 7th most common family of validated riboswitches (Ames and Breaker, 2010; Breaker, 2011). Members of this family are recognized by their architecture consisting of a three-stem junction wherein the loops of two stems associate through tertiary interactions (Figure 3) (Mandal et al., 2003; Batey et al., 2004; Serganov et al., 2004). Representatives can selectively bind guanine (Mandal et al., 2003), adenine (Mandal et al., 2004b), or 2′-deoxyguanosine (Kim et al., 2007), depending on the identities of nucleotides within the regions joining the stems (J1-2, J2-3 and J3-1). Guanine and adenine riboswitches vary at one key nucleotide in J3-1 that directly base pairs to the ligand (nucleotide 74). In contrast, the riboswitches that sense 2′-deoxyguanosine accommodate the extra 2′-deoxyribose moiety by exploiting several key mutations in the J2-3 region (Edwards and Batey, 2009).
Figure 3. Purine riboswitch variants and their ligand specificities.
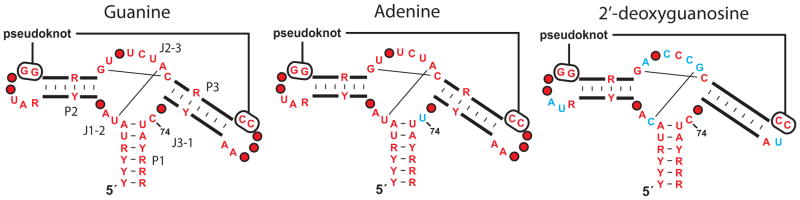
Depicted are the consensus sequence and secondary structure models for riboswitch aptamers that selectively respond to guanine, adenine or 2′-deoxyguanosine. Red nucleotides are present in greater than 90% of the guanine riboswitch representatives. Blue nucleotides in the adenine and 2′-deoxyguanosine aptamers differ from the guanine consensus. Other annotations are as described in the legend to Figure 2.
Intriguingly, there are only two known examples of 2′-deoxyguanosine riboswitches, and these are present in only one bacterial species, Mesoplasma florum. It is very perplexing why 2′-deoxyguanosine-sensing riboswitches that are only a few mutations away from one of the most common riboswitch types (guanine-sensing) are present in only one organism. The ligand, 2′-deoxyguanosine, is present in all organisms that make DNA, and it seems reasonable to assume that more organisms may find it beneficial to monitor the concentration of this nucleotide. Furthermore, since a single point mutation can convert a guanine riboswitch into one that is selective for adenine (Mandal et al., 2004b), why are there no analogous natural variants of the 2′-deoxyguanosine aptamer that sense the related universal metabolite, 2′-deoxyadenosine?
Perhaps organisms have little to gain by carefully monitoring the concentrations of these DNA nucleotides, which could explain why a riboswitch that should be easy to access through evolution may be so rare. Alternatively, other species that do keep tabs on the concentrations of their DNA monomers might use different sensors based on protein factors or perhaps by exploiting a different riboswitch class. Numerous additional questions like these are likely to emerge as researchers try to more comprehensively understand the regulation of bacterial metabolic networks.
The Difficult Task of Matching Newfound Riboswitches with Ligands
The list of metabolites known to be sensed by riboswitches so far is a near perfect subset of compounds that are universal to modern life. This collection includes eight coenzymes or coenzyme derivatives, three of the 20 standard amino acids, four purine nucleobases or their derivatives, and the essential amino sugar glucosamine-6-phosphate (Figure 1A). Perhaps the validated riboswitch ligand with the most restricted phylogenetic distribution is c-di-GMP, which none-the-less serves as a near universal second messenger in bacteria (Jenal and Malone, 2006).
The fact that the known riboswitch ligands include such prevalent metabolites simply could be due to the high probability of researchers encountering widespread riboswitches for widespread ligands. However, the list of compounds sensed by riboswitches is surely to expand greatly, even just by solving the ligand identities of the approximately two dozen candidate or orphan riboswitch classes that have been published to date (Barrick et al., 2004; Corbino et al., 2005; Weinberg et al., 2007). The ligand for most newfound classes is inferred from the functions of the proteins produced from genes associated with the riboswitch. However, this approach begins to fail for rare riboswitches because of the sparse and imperfect assignment of protein functions. Even some widespread riboswitch candidates remain unresolved because they are associated with many genes encoding proteins of unknown function.
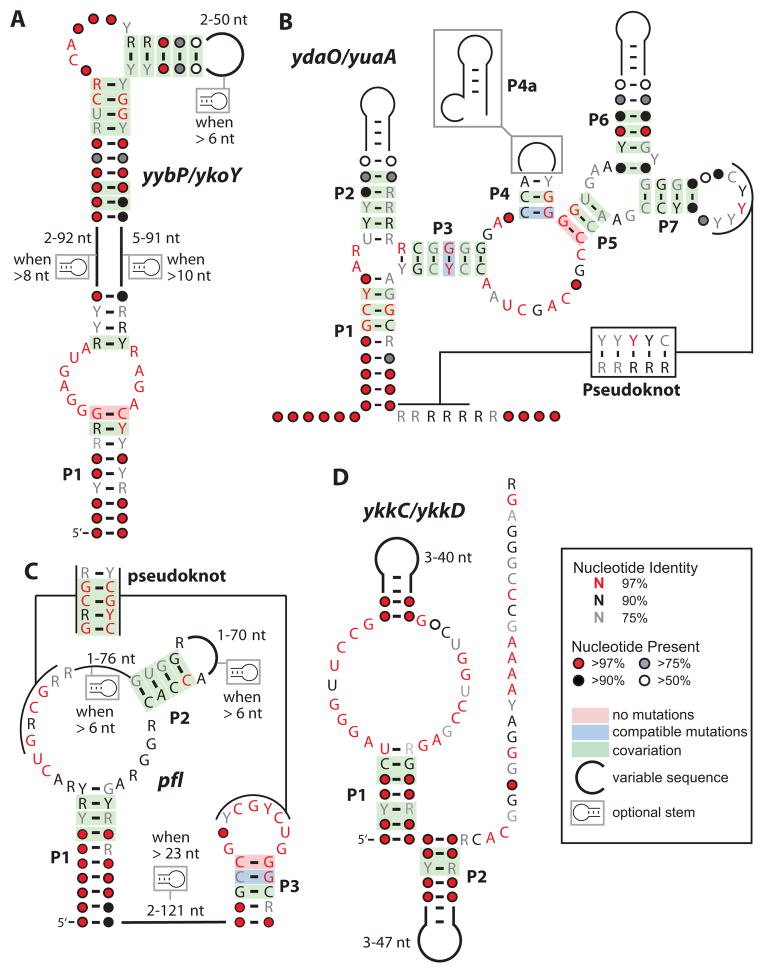
The problems of ligand assignment are showcased by four of the most numerous orphans (Block et al., 2010; Meyer et al., 2011) (Figure 4). If experimentally confirmed, yybP motif RNAs (Barrick et al, 2004) would constitute the fourth most common riboswitch class in bacteria. Its relatively simple structure is widely associated with genes involved in cellular pH control, suggesting the RNA could be a direct sensor of protons or hydroxide ions. Intriguingly, evidence of alkaline-dependent transcription activation has been reported (Nechooshtan et al., 2009) for an RNA carrying a yybP motif representative from E. coli.
Figure 4. The most abundant orphan riboswitch classes.
The consensus sequences and secondary structure models are depicted for the four most common classes of RNA elements believed to be riboswitch aptamers (Block et al., 2010; Meyer et al., 2011).
(A) The yybP/ykoK motif RNAs are frequently associated with genes whose protein products mediate pH stress responses.
(B) The ydaO/yuaA motif RNAs are frequently associated with genes involved in osmotic stress responses.
(C) The pfl motif RNAs control genes centered on folate metabolism.
(D) The ykkC/ykkD motif RNAs are associated with genes for transporters and for purine metabolism.
If yybP representatives are direct pH sensors, it still seems puzzling why this single RNA class is used so frequently when many pH-responsive RNA structures presumably could easily have emerged in evolution. At extreme pH values (less than 5 and more than 9), RNA structures are substantially affected due to protonation or deprotonation of nucleobases, which disrupts Watson-Crick base pairing. However, some RNA tertiary structures can shift the pKa values of functional groups, and this effect is observed for some ribozymes (Bevilacqua et al., 2004). A direct pH sensor made of RNA would need to create one or more pKa shifts to provide a structure that responds to pH changes near the physiological norm. Perhaps such a specialized adaptation explains why the yybP RNA motif is so widespread and well conserved.
It seems likely that many riboswitch candidates discovered in the future will be increasingly difficult to assign ligand specificity. Ever more rare classes will offer ever fewer gene associations that provide clues to the identity of the target ligand. Furthermore, new riboswitch candidates may be specialized in some bacterial lineages or individual species to respond to compounds that are present only in these organisms. In other words, the lack of information on gene function and the fact that many compounds sensed by riboswitches currently might be unknown to science will be major challenges facing riboswitch discoverers.
Where are the NAD and CoA Riboswitches?
It has been proposed (White, 1976) that many coenzymes, particularly those derived from or carrying fragments of nucleotides, are molecular relics from the RNA World (Gilbert 1986; Joyce, 1991). Most likely, modern coenzymes emerged as chemical helpers of ribozymes long before proteins began to take over the majority of enzyme functions. Although protein enzymes have largely displaced ribozymes, they still make use of the original RNA World coenzymes to carry out their catalytic tasks. An interesting aside, most vitamins are precursors of these coenzymes, and ubiquity of these ancient compounds along with their catalytic functions (required in small amounts) are the characteristics one would expect for organisms such as humans to safely rely on diet (and probably also our own microbiomes) as their sources.
If this evolutionary history is true, then perhaps some of the common riboswitches we see today may have their origins in the RNA World (Breaker, 2009; 2011). Instead of controlling the transcription progression of modern RNA polymerases, or acting as the gatekeeper for ribosomes associating with mRNAs, some riboswitch aptamers originally could have served as allosteric regulatory domains for ribozymes. Some ancient aptamers could have been positioned just as they are in modern bacteria and could have regulated ancient transcription to control the production of ribozymes and other noncoding RNAs, rather than mRNAs.
The fact that coenzymes are ancient, ubiquitous, and important can be used to rationalize why some coenzyme-sensing riboswitches are so common. However, there is a glaring hole in this hypothesis. Where are the riboswitches for some of the fundamental coenzymes such as nicotinamide adenine dinucleotide (NAD) and coenzyme A (CoA)? These two coenzymes so intimately involved in redox and acyl group transformations, respectively, so far appear to be ignored by the known collection of riboswitches. Perhaps there is a need to sense a diversity of acyl-CoA derivatives, and so there may not be a single common class for the parent CoA moiety but rather a variety of rare RNA aptamers that each selectively recognize a specific acyl-CoA compound. However, NAD and its phosphorylated derivative NADP are ubiquitous and obvious riboswitch ligand candidates, yet they lack any partner riboswitches. It seems reasonable to speculate that one or more riboswitches will eventually be identified that sense nicotinamide-based coenzymes. Alternatively, cells may exclusively use other mechanisms involving protein factors to serve this purpose.
The arguments above can be greatly expanded to take account of other very common and ancient metabolites, including amino acids as discussed below. For example, where are the riboswitches for all other coenzymes, for key nucleoside derivatives, for various sugars and lipids? Not all compounds, particularly metabolic intermediates, need to have their concentrations constantly monitored by genetic factors. However, metabolites that are at key nodes in interconnected biochemical pathways seem like ideal compounds to be sensed by riboswitches. Most commonly, the known riboswitch classes respond to critical end products of metabolic pathways, and such compounds should comprise the short list of metabolites for testing with orphan riboswitch candidates.
How Many Riboswitches Sense Amino Acids?
Lysine and glycine represent the 6th and 7th most common riboswitch classes, while glutamine-sensing riboswitches (Ames and Breaker, 2011) are far less common. Glutamine is a key compound in the metabolism of nitrogen, and therefore may have been important for controlling metabolic processing of nitrogen in RNA World organisms. Also, a complete set of the 20 amino acids would have needed to be present in the last common ancestor of modern organisms for protein synthesis. Thus, there has been plenty of time for riboswitches to emerge that sense the complete set of amino acids.
The need to respond genetically to amino acid concentrations is made very evident by the fact that many amino acid biosynthesis and transport enzyme classes are controlled at one time or another by T box RNAs (Grundy and Henkin, 1993; Green et al., 2010) as described earlier. T box regulators are usually ‘ON’ switches that activate genes that code for proteins to import or manufacture more of the needed amino acid. They are so common and diverse in Gram-positive organisms that there seems to be little need for a large diversity of riboswitches that directly bind to each amino acid. However, T box RNAs are absent in most Gram-negative lineages of bacteria, and so it remains unclear whether these organisms rely on riboswitches or perhaps protein-based genetic factors to maintain amino acid homeostasis. If direct RNA-amino acid interactions are exploited, then there are as many as 17 more amino acids in need of a corresponding riboswitch discovery.
Riboswitch Selectivity and Sensitivity: Some Counterintuitive Characteristics
Riboswitches must accurately control gene expression in response to the appropriate concentrations of the metabolite they target. In some instances, the riboswitch aptamers may have evolved to exclude all natural analogs of the target ligand to avoid misregulation. Again, we can look to the many SAM riboswitch classes for examples. SAM-I riboswitches are strikingly proficient at excluding close natural analogs of SAM, such as SAH and the two precursors of SAM (methionine and ATP) (Winkler et al., 2003). This riboswitch discriminates against SAH by more than two orders of magnitude, which should be more than sufficient to preclude genes such as those that encode SAM synthase from being repressed by the metabolic byproduct of SAM use. If unwanted SAH repression of SAM synthase production were to occur, then the cell would be denied the production of this key coenzyme just when it is being used at a rate that causes a build-up of SAH.
Conceivably, riboswitch aptamers would not always need to be perfected to most tightly bind their target ligand. Although SAM-I appears to appropriately exploit a strong discriminatory capability against SAH, another riboswitch class that apparently controls SAM synthase expression actually has a better affinity for SAH than for SAM (Weinberg et al., 2010). We speculate that this more rare SAM/SAH class (Figure 2) most likely responds to changing SAM concentrations in cells, and never has the opportunity to bind SAH. This makes sense because SAH, which is toxic to cells, most likely is never permitted to rise to a level that approaches the concentration of SAM.
Another initially puzzling observation is that some riboswitch aptamers exhibit extraordinarily high affinities for their target ligands. The champion so far is a c-di-GMP aptamer from Vibrio cholera (Sudarsan et al., 2008), which has an apparent dissociation constant of no poorer than 10 pM (Smith et al., 2009). By comparison, a single molecule present in a volume encompassed by a typical bacterial cell would be ~1000 times higher in concentration! Furthermore, the residency for c-di-GMP on this aptamers is measured on a timescale of months, which is far greater than the minutes timescale for a rapidly-dividing cell.
To address this type of paradox, we (Wickiser et al., 2005a; 2005b) and others (Gilbert et al., 2006; Garst and Batey, 2009; Haller et al., 2011) have collected data indicating that some riboswitches do not reach thermodynamic equilibrium with their target ligands, but rather make use of the rate constant for ligand association relative to the speed of RNA transcription to control gene expression using concentrations of ligands that are biologically relevant. In other words, some riboswitches are kinetically rather than thermodynamically driven. For riboswitches that directly control transcription termination (Figure 1B), the faster RNA polymerase proceeds up to (and eventually past) the terminator stem, the higher the concentration of ligand needs to be to successfully influence gene expression. This has been demonstrated in vitro for or an FMN-responsive “OFF” riboswitch, whereby removing natural transcriptional pause sites or increasing the speed of RNA polymerase by optimizing reaction conditions results in a need for higher FMN concentrations to trigger transcription termination (Wickiser et al., 2005a).
Spoofing Riboswitch Ligands by Nature and by Design
After the first riboswitch validation studies, it quickly became apparent that these RNAs should be attractive targets for drug development (Breaker, 2009; Blount and Breaker, 2006; Deigan and Ferré-D’Amaré, 2011). First, many riboswitches repress the expression of genes whose protein products are involved in the transport or biosynthesis of essential metabolites. Therefore, compounds that trick riboswitches by mimicking the natural ligand might inhibit bacterial growth by starving the cells for that essential metabolite. Second, medicinal chemists already have a “hit” compound (the natural ligand) for each validated riboswitch class that they can begin to chemically alter to create new antibiotics. In this regard, riboswitches are almost unique among noncoding RNAs classes because they have evolved pockets to purposefully bind a small molecule, and therefore should be more easily drugged.
One of the first demonstrations that riboswitches can be broadly drugged came with the recognition that bacterial resistance to the toxic lysine analog called aminoethycysteine (AEC) could be attained by acquiring mutations in lysine riboswitches (Sudarsan et al., 2003a). Similarly, resistance to the inhibitory effects of the thiamin analog called pyrithiamine is acquired by mutations in TPP riboswitches of fungal and bacterial species (Kubodera et al., 2003; Sudarsan et al., 2005). Since compounds such as AEC and pyrithiamine are so similar to lysine and thiamin, respectively, these compounds likely do more than just bind their corresponding riboswitches to cause growth inhibition. For example, AEC also is recognized by aminoacyl-tRNA synthases, and therefore may cause further problems for cells by becoming incorporated into proteins (Ataide et al., 2007). Thus, riboswitch mutations either prevent the analog from suppressing metabolite biosynthesis, disregulate metabolite production to yield an excess that swamps the concentration of the analog, or both.
Chemists since the 1940s and 1950s have been producing compounds like pyrithiamine or AEC that have antimicrobial activity and that are now known to trigger riboswitch action. However, nature has been using ligand analogs to target riboswitches most likely for billions of years. The natural product roseoflavin (a close analog of riboflavin) was isolated from a species of Streptomyces and reported in 1974 to have antimicrobial activity (Otani et al., 1974). It is now known that roseoflavin is bound by FMN riboswitches (Serganov et al., 2009; Lee et al., 2009; Ott et al., 2009; Mansjö and Johansson, 2011) and that resistance to this compound (or more likely its phosphorylated derivative) is acquired by mutating the riboswitch aptamers to preclude ligand binding (Lee et al., 2009). Again, given the chemical similarity between roseoflavin phosphate and FMN, it seems possible that the compound could suppress FMN production as well as interfere with enzymes that use this coenzyme.
The full spectrum of modern drug development strategies can now be brought to bear on the development of new compounds that target riboswitches. For example, atomic resolution structures are available for most validated riboswitch classes (Edwards et al., 2007; Montange and Batey, 2008; Serganov, 2010), which guides chemists in their efforts to make rational changes to compounds that are bound by riboswitch aptamers. This approach has been used most extensively for guanine riboswitches, whose aptamer structures were used to guide the design of numerous purine analogs that are retain binding activity (Kim et al., 2009; Mulhbacher et al., 2010; Daldrop et al., 2011). Some of these guanine analogs inhibit bacterial growth (Kim et al., 2009; Mulhbacher et al., 2010) presumably by suppressing the production of proteins involved in the recycling and de novo biosynthesis of purines. Also, other drug discovery strategies such as high-throughput screening have been demonstrated with riboswitches (Blount et al., 2006; Mayer and Famulok, 2006), and such approaches should lead to the identification of new chemical scaffolds that trigger riboswitch-mediated gene regulation.
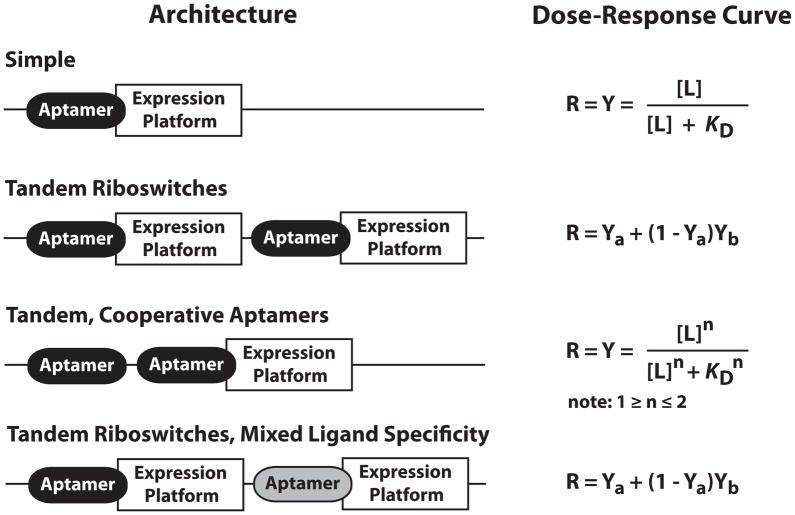
Tandem Riboswitches Increase Functional Complexity
Simple one-to-one interactions between a metabolite and a riboswitch, it takes an 81-fold change in ligand concentration to exploit 80% of the gene expression dynamic range (Figure 5). However, some organisms break through this functional barrier by stacking riboswitches or their substructures in tandem to yield gene control systems that are far more sophisticated. The most common way that cells create systems that require smaller changes in ligand concentration to trigger gene expression (or more “digital” function) is to stack two homologous riboswitches in tandem (Welz and Breaker, 2007). This allows cells to exploit 80% of their expression dynamic range in response to only a 40-fold change in ligand concentration. A more sophisticated and more effective way to achieve this goal is to integrate two homologous aptamers in tandem with only one expression platform, such that the aptamers bind two identical ligands cooperatively. This type of cooperative riboswitch architecture is commonly used by glycine riboswitches (Mandal et al., 2003), wherein the binding of one amino acid improved folding and affinity of the adjacent aptamer (Huang et al., 2010; Butler et al., 2011; Sudarsan et al., 2006). These arrangements can approach the theoretical performance wherein only a 9-fold change in ligand concentration would be needed to exploit 80% of their expression dynamic range.
Figure 5. Architectures of riboswitches and their effects on ligand-mediated gene control.
The expression platforms can be conventional systems as listed in Figure 1, or can be self-processing ribozymes. R designates the fraction of gene regulation, Y designates the fraction of aptamer bound by ligand, [L] designates ligand concentration, KD designates apparent dissociation constant. For all riboswitch architectures other than cooperative systems, values for Y are determined by using the equation for a simple riboswitch.
A variety of tandem riboswitches with mixed ligand specificities have also been observed. One example (Sudarsan et al., 2006) functions as a two-input Boolean NOR gate, wherein either the coenzymes AdoCbl or SAM will suppress the expression of the downstream gene. A larger variety of complex riboswitch arrangements are theoretically possible (Stoddard and Batey, 2006) and at least some of these are likely to be exploited by cells to create other forms of Boolean logic gates or more digital switches. Another intriguing possibility is that the use of tandem riboswitches that control gene expression at two different levels (e.g. transcription termination and translation initiation) could function as a natural “band-pass” switch, wherein activation demands a certain window of ligand concentration. Such band-pass RNA switches have been successfully engineered to activate expression at a median ligand concentration, while suppressing expression when concentrations become either too low or too high (Muranaka and Yokobayashi, 2010).
A series of tandem SAM-II and SAM-IV riboswitches have recently been found (Poiata et al., 2009) that appear to control transcription and translation, respectively. This arrangement is like that expected for natural band-pass function. However, it is likely that these tandem arrangements, found in slow-growing bacteria that thrive in ocean water, have evolved to regulate SAM coenzyme synthesis with extreme efficiency. Like many single riboswitch arrangements, the SAM-II riboswitch simply controls transcription to prevent unwanted SAM synthase mRNA production when SAM levels are adequate. If full-length mRNAs are produced, the SAM-V riboswitch then suppresses translation when SAM levels are sufficient, rather than suppressing translation by rapid mRNA turnover. This type of tandem arrangement could substantially reduce the amount of energy wasted by simpler riboswitch architectures.
Integrating Riboswitches with Ribozymes
The vast majority of riboswitches control gene expression by controlling transcription termination or translation initiation. However, fusing riboswitches with ribozymes seems like a logical way to directly regulate the chemical reactions catalyzed by RNA. Engineered examples of “allosteric” ribozymes can routinely be created in the laboratory (Soukup and Breaker, 1999; Breaker, 2002), and the relative ease of their assembly suggests that cells could similarly generate ligand-controlled ribozymes through natural processes.
Two classes of ribozymes that are activated by the presence of specific metabolites have been known for some time. The self-splicing reactions of group I ribozymes commonly are triggered by the selective binding of guanosine or one of its 5′ phosphorylated derivatives such as GTP (Nielsen and Johansen, 2009). Guanosine actually serves as a substrate and becomes covalently attached to the intron RNA during the first step of splicing. In contrast, self-cleaving glmS ribozymes are activated by the selective binding of glucosamine-6-phosphate (GlcN6P) (Winkler et al., 2004), which functions as a coenzyme to promote phosphoester transfer (McCarthy et al., 2005). However, these ligand-triggered ribozymes do not qualify as true allosteric enzymes wherein ligand binding at one site triggers chemical transformation at a distal active site.
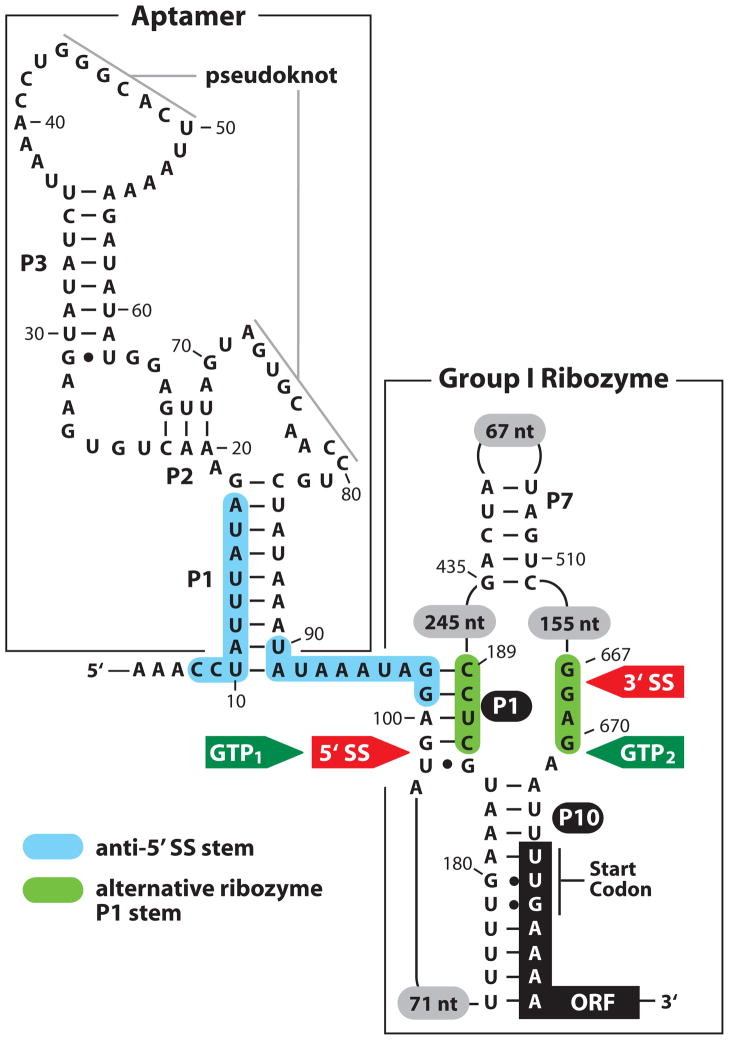
Recently, a series of closely-related group I ribozymes in various strains of Clostridium difficile bacteria have been demonstrated to function as allosteric ribozymes that are activated by the second messenger c-di-GMP (Lee et al., 2010). In each instance, a class II c-di-GMP riboswitch is located several nucleotides upstream of the 5′ splice site for the ribozyme (Figure 6). When c-di-GMP is bound by the aptamer, the resulting structure presents the splice site for processing by the ribozyme. In contrast, low concentration of second messenger causes the 5′ splice site to be masked by an alternative structure, and therefore proper splicing of the precursor mRNA and subsequent protein translation cannot be completed. Thus, both c-di-GMP and GTP molecules are required to trigger gene expression, meaning this tandem riboswitch-ribozyme arrangement functions as a Boolean AND gate. Intriguingly, since each c-di-GMP molecule is made from two GTP molecules, it seems possible that the cell makes use of this allosteric ribozyme to express the adjoining ORF as c-di-GMP accumulates, unless GTP levels have been substantially decreased by biosynthesis of this second messenger.
Figure 6. Allosteric control mechanism of a group I self-splicing ribozyme by a c-di-GMP riboswitch aptamer.
Sequence and secondary structure model for the allosteric ribozyme from the bacterium C. difficile (Baker et al., 2010). A class II c-di-GMP riboswitch aptamer, formed by three conventional base-paired regions (P1 through P3) and one pseudoknot, resides only six nucleotides upstream of the initial structured portion (shaded P1) of a group I ribozyme. The 5′ splice site (red “5′ SS” between nucleotides 100 and 101) is a target for GTP attack (“GTP1”) when c-di-GMP is bound by the aptamer. Following this first step of splicing, the second step promotes attack by G101 of the 5′ exon at the 3′ splice site (red “3′ SS” between nucleotides 667 and 668) to yield spliced exons. In the absence of c-di-GMP binding, alternative base pairing (blue “anti-5′ SS stem) occurs between aptamer and ribozyme nucleotides. This anti-5′ SS stem displaces the ribozyme P1 stem, which precludes normal splicing. In contrast, an “alternative ribozyme P1 stem” (green) forms and promotes GTP attack (GTP2) after nucleotide 670. The products of normal splicing, promoted by c-di-GMP, are efficiently translated because they carry a typical ribosome binding site located the proper distance upstream of the unusual UUG start codon. The products of alternative splicing, which occurs when c-di-GMP is low in concentration, lack a ribosome binding site and therefore are not translated.
This natural fusion between a riboswitch and a ribozyme is exceptionally rare and complex, and therefore simpler arrangements between other ribozymes classes and riboswitches should be more likely to occur. In this regard, it is notable that large numbers of self-cleaving ribozymes have been reported recently (e.g. see Webb et al., 2009; Seehafer et al., 2011; Perreault et al., 2011), and the precise roles in the organisms that carry them remain to be elucidated for the vast majority of these examples. It seems reasonable that at least a few of these ribozymes will be found to serve as the “catalytic platform” through which some riboswitch aptamers control RNA processing.
Where are the Eukaryotic Riboswitches?
It is not surprising that TPP riboswitches, which are the most numerous of all riboswitch classes, are present in organisms from all three domains of life (Sudarsan et al., 2003b). However, it seems very strange that this class is the only one so far that has been convincingly demonstrated to function in eukaryotes. To understand why there is a dearth of de novo eukaryotic riboswitch discovery, it is important to consider some hard facts. Compared to bacterial genomes, eukaryotic genomes can be several orders of magnitude larger, fewer of such genomes are sequenced, gene annotations are typically less comprehensive, and genetics studies can be more challenging. Regardless, bioinformatics can easily reveal homologs of validated bacterial riboswitches that are hidden in eukaryotic genomes. Since these searches have been done, it seems clear that, of the 24 riboswitch classes known, only TPP riboswitches frequently occur in eukaryotic genomes (largely in fungi and plants).
When TPP riboswitches occur in eukaryotes, they usually control alternative splicing of precursor mRNAs (Kubodera et al., 2003; Cheah et al., 2007; Wachter et al., 2007; Bocobza et al., 2007; Croft et al., 2007). In some fungal and plant TPP riboswitches, a 5′ splice site is blocked by base pairing with nucleotides of the aptamer (Cheah et al., 2007; Wachter et al., 2007). The spliceosome can only gain access to this 5′ splice site if TPP is bound by the aptamer and precludes these nucleotides from base pairing with splice site nucleotides. Alternative splicing of the Neurospora crassa NMT1 pre-mRNA controlled by a TPP riboswitch causes retention of short open reading frames located upstream of the main ORF when TPP concentration is high. This causes ribosomes to translate these short “uORFs” rather than express the protein encoded by the main ORF.
The substantial conformational changes that eukaryotic riboswitches undergo in response to ligand binding could be harnessed to control many different processes that influence gene expression. However, the control of alternative splicing seems like an ideal way for metabolites to regulate RNA processing and protein expression. Introns also can be quite long, and therefore could easily encode metabolite-binding aptamer and expression platform domains that are ultimately removed from mature mRNAs. If more classes of eukaryotic riboswitches exist, a rich hunting ground could be the extensive amount of intronic sequences present in many higher organisms.
Regardless of the potential for eukaryotic riboswitch discovery, it still seems strange that only one of the 24 riboswitch classes that serve bacterial so well is commonly found in eukaryotes. Perhaps eukaryotic cells rely more heavily on other regulatory strategies such as uORFs (Wethmar et al., 2010). Alternatively, most eukaryotic cells are far better than bacteria at avoiding nutrient restrictions, and therefore are less dependent on genetic systems that precisely monitor fundamental metabolites and shut down various processes in difficult times. Perhaps, eukaryotic cells may have a greater need to sense and respond to signaling compounds that are not present in bacteria. If true, then it may be more likely to find eukaryotic riboswitches for second messengers and other signaling compounds rather than common metabolites.
Conclusions
Many bacterial species, including well-studied organisms such as Bacillus subtilis and Escherichia coli, have a variety of riboswitch classes controlling key metabolic pathways. Since these bacteria grow rapidly and have the potential to evolve rapidly, the presence of so many riboswitches suggests that these RNA domains are not primitive and ineffectual genetic elements, but rather are highly refined sensors and switches that are competitive with protein factors through evolution. Even simple riboswitch discoveries can yield knowledge of previously obscured signaling networks (e.g. c-di-GMP), or help reveal the biosynthetic and transport proteins for fundamental metabolites (e.g. preQ1, MoCo). Moreover, given the diversity of known riboswitches and the striking level of complexity of some examples, it seems reasonable to conclude that far more biochemical and functional diversity will be uncovered as new riboswitch discoveries are made.
Key challenges for researchers in this area will involve the development of efficient ways to identify new riboswitch candidates, linking these orphan riboswitches to their natural ligands, and validating the possible exotic functions of these RNA-metabolite complexes. Bioinformatics methods are likely to play a major role in revealing conserved riboswitches and in establishing how widespread these classes are among organisms from the three domains of life. Of particular concern is that rare riboswitch candidates may respond to compounds that are not commercially available or that are not known to science. Methods to culture organisms carrying these riboswitches and their ligands may need to be developed to match each new riboswitch class with its natural ligand.
The current collection of riboswitches has increased our knowledge of RNA structure and function, and some discoveries have resolved long-standing mysteries on how cells sense and respond to various stresses and signals. Intriguingly, natural riboswitches can be examined to teach us what types of mechanisms for RNA-based gene control are preferred, and these are already are being reverse engineered to help guide the designs of synthetic riboswitches (e.g. see Muranaka et al., 2009; Wieland et al., 2009). In some instances, tricking riboswitches with ligand analogs may yield novel antibacterial agents (Blount and Breaker, 2006). Therefore, each new riboswitch class discovery offers new insights on the diverse functions of RNA and offers opportunities for technical advances.
Acknowledgments
The author thanks past and present members of the Breaker laboratory for many contributions to riboswitch research. Current research on riboswitch discovery is supported by an NIH grant (GM022778), and by the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ames TD, Breaker RR. Bacterial riboswitch discovery and analysis. In: Meyer G, editor. The Chemical Biology of Nucleic Acids. John Wiley and Sons, Ltd; 2010. pp. 433–452. [Google Scholar]
- Ames TD, Breaker RR. Bacterial aptamers that selectively bind glutamine. RNA Biol. 2011;8:82–89. doi: 10.4161/rna.8.1.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide SF, Wilson SN, Dand S, Rogers TE, Roy B, Banerjee R, Henkin TM, Ibba M. Mechanisms of resistance to an amino acid antibiotic that targets translation. ACS Chem Biol. 2007;2:819–827. doi: 10.1021/cb7002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial gene control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, Brown TS, Nakano S, Yajima R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- Block KF, Hammond MC, Breaker RR. Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol. 2010;192:3983–3989. doi: 10.1128/JB.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Blount K, Puskarz I, Penchovsky R, Breaker R. Development and application of a high-throughput assay for glmS riboswitch activators. RNA Biol. 2006;3:77–81. doi: 10.4161/rna.3.2.3102. [DOI] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Engineered allosteric ribozymes as biosensor components. Curr Opin Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol. 2009;4:771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches and the RNA World. In: Atkins JF, Gesteland RF, Cech TR, editors. RNA Worlds. CSH Press; 2011. pp. 63–77. [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daldrop P, Reyes FE, Robinson DA, Hammond CM, Lilley DM, Batey RT, Brenk R. Novel ligands for a purine riboswitch discovered by RNA-ligand docking. Chem Biol. 2011;18:324–335. doi: 10.1016/j.chembiol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambach MD, Winkler WC. Expanding roles for metabolite-sensing regulatory RNAs. Curr Opin Microbiol. 2009;12:161–169. doi: 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Ferré-D’Amaré AR. Riboswitches: Discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011 doi: 10.1021/ar200039b. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Klein DJ, Ferré-D’Amare AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Mironov AS, Nudler E. The riboswitch control of sulfur metabolism in bacteria. Proc Natl Acad Sci USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim Biophys Acta. 2009;1789:584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamers domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Gilbert W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- Gold L, Brown D, He Y, Shtatland T, Singer BS, Wu Y. From oligonucleotide shapes to genomic SELEX: Novel biological regulatory loops. Proc Natl Acad Sci USA. 1997a;94:59–64. doi: 10.1073/pnas.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L, Singer B, He Y, Brody E. SELEX and the evolution of genomes. Curr Opin Genet Dev. 1997b;7:848–851. doi: 10.1016/s0959-437x(97)80050-0. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Lehman SC, Henkin TM. The L box regulons: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci USA. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- Haller A, Soulière MF, Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res. 2011 doi: 10.1021/ar200035g. (in press) [DOI] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Joyce GF. The rise and fall of the RNA world. New Biologist. 1991;3:399–407. [PubMed] [Google Scholar]
- Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci USA. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Breaker RR. Purine sensing by riboswitches. Biol Cell. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert B, Narberhaus F. Microbial thermosensors. Cell Mol Life Sci. 2009;66:2661–2676. doi: 10.1007/s00018-009-0041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004a;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004b;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mansjö M, Johansson J. The riboflavin analog roseoflavin targets an FMN-riboswitch and blocks Listeria monocytogenes growth, but also stimulates virulence gene-expression and infection. RNA Biol. 2011 doi: 10.4161/rna.8.4.15586. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer G, Famulok M. High-throughput-compatible assay for glmS riboswitch metabolite dependence. Chem Bio Chem. 2006;7:602–604. doi: 10.1002/cbic.200500490. [DOI] [PubMed] [Google Scholar]
- McCarthy TJ, Plog MA, Floy SA, Jansen JA, Soukup JK, Soukup GA. Ligand requirements for glmS ribozymes self-cleavage. Chem Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA. 2008;14:685–695. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. Challenges of ligand identification for riboswitch candidates. RNA Biol. 2011;8:5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Rios J, Navarro M, Soberón M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Riboswitches: emerging theses in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010;6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranaka N, Yokobayashi Y. A synthetic riboswitch with chemical band-pass response. Chem Commun. 2010;46:6825–6827. doi: 10.1039/c0cc01438a. [DOI] [PubMed] [Google Scholar]
- Muranaka N, Abe K, Yokobayashi Y. Mechanism-guided library design and dual genetic selection of synthetic OFF riboswitches. Chem Bio Chem. 2009;10:2375–2381. doi: 10.1002/cbic.200900313. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Johansen SD. Group I introns: Moving in new directions. RNA Biol. 2009;6:375–383. doi: 10.4161/rna.6.4.9334. [DOI] [PubMed] [Google Scholar]
- Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Katagai T, Tomura D, Shoun H, Hoshino T, Furukawa T. Analysis of the transcriptional activity of the hut promoter in Bacillus subtilis and identification of a cis-acting regulatory region associated with catabolite repression downstream from the site of transcription. Mol Microbiol. 1992;6:2573–2582. doi: 10.1111/j.1365-2958.1992.tb01434.x. [DOI] [PubMed] [Google Scholar]
- Otani S, Takatsu M, Nakano M, Kasai S, Miura R. Roseoflavin, a new antimicrobial pigment from Streptomyces. J Antibiot (Tokyo) 1974;27:86–87. [PubMed] [Google Scholar]
- Ott E, Stolz J, Lehmann M, Mack M. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 2009;6:276–280. doi: 10.4161/rna.6.3.8342. [DOI] [PubMed] [Google Scholar]
- Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Febeyre G, Breaker RR. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol. 2011;7:e1002031. doi: 10.1371/journal.pcbi.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA. 2009;15:2046–2056. doi: 10.1261/rna.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Welz R, Iwata-Reuyl D, Breaker RR. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehafer C, Kalweit A, Steger G, Gräf S, Hammann C. From Alpaca to zebrafish: Hammerhead ribozymes wherever you look. RNA. 2011;17:21–26. doi: 10.1261/rna.2429911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A. The long and short of riboswitches. Curr Opin Struct Biol. 2009;15:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A. Determination of riboswitch structures: light at the end of the tunnel? RNA Biol. 2010;7:98–103. doi: 10.4161/rna.7.1.10756. [DOI] [PubMed] [Google Scholar]
- Shimotsu H, Kuroda MI, Yanofsky C, Henner DJ. Novel form of transcription attenuation regulates expression of the Bacillus subtilis tryptophan operon. J Bacteriol. 1986;166:461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. Nucleic acid molecular switches. Trends Biotechnol. 1999;17:469–476. doi: 10.1016/s0167-7799(99)01383-9. [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Batey RT. Mix-and-match riboswitches. ACS Chem Biol. 2006;1:751–754. doi: 10.1021/cb600458w. [DOI] [PubMed] [Google Scholar]
- Stormo GD, Ji Y. Do mRNAs act as direct sensors of small molecules to control their expression? Proc Natl Acad Sci USA. 2001;98:9465–9467. doi: 10.1073/pnas.181334498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure that controls gene expression by binding lysine. Genes Dev. 2003a;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003b;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacterial sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Breaker RR. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol. 2008;86:157–168. doi: 10.1139/O08-008. [DOI] [PubMed] [Google Scholar]
- Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell. 2008;29:691–702. doi: 10.1016/j.molcel.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CH, Riccetelli NJ, Ruminski DJ, Lupták A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA. 2008;14:822–828. doi: 10.1261/rna.988608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wethmar K, Smink JJ, Leutz A. Upstream open reading frames: Molecular switches in (patho)physiology. Bio Essays. 2010;32:885–893. doi: 10.1002/bies.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HB., III Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005a;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005b;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew Chem Int Ed Engl. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002a;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002b;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozymes. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]