Abstract
Editing of the pre-mRNA for the serotonin receptor 2C (5-HT2CR) by site-specific adenosine deamination (A-to-I pre-mRNA editing) substantially increases the functional plasticity of this key neurotransmitter receptor and is thought to contribute to homeostatic mechanisms in neurons. 5-HT2CR mRNA editing generates up to 24 different receptor isoforms. The extent of editing correlates with 5-HT2CR functional activity: more highly edited isoforms exhibit the least function. Altered 5-HT2CR editing has been reported in postmortem brains of suicide victims. We report a comparative analysis of the connections among 5-HT2CR editing, genome-wide gene expression and DNA methylation in suicide victims, individuals with major depressive disorder and non-psychiatric controls. The results confirm previous findings of an overrepresentation of highly edited mRNA variants (which encode hypoactive 5-HT2CR receptors) in the brains of suicide victims. A large set of genes for which the expression level is associated with editing was detected. This signature set of editing-associated genes is significantly enriched for genes that are involved in synaptic transmission, genes that are preferentially expressed in neurons, and genes whose expression is correlated with the level of DNA methylation. Notably, we report that the link between 5-HT2CR editing and gene expression is disrupted in suicide victims. The results suggest that the postulated homeostatic function of 5-HT2CR editing is dysregulated in individuals who committed suicide.
INTRODUCTION
The serotonin receptor 2C (5-HT2CR) is abundantly expressed in neurons of the central nervous system (CNS), where it modulates diverse processes that affect sleep, sexual activity, feeding behavior, anxiety and depressive state (reviewed in 1,2). The 5-HT2CR belongs to the G-protein-coupled receptor (GPCR) superfamily and is involved in multiple signal transduction pathways through coupling to different G proteins, which allows 5-HT2CR to mediate intracellular responses via various effector molecules (reviewed in 3,4). Several of these second messengers have been reported to influence the activity of ion channels, and thus affect the excitability of the 5-HT2CR-expressing neurons (5).
The 5-HT2CR shows remarkable functional variability that derives from editing of its pre-mRNA by site-specific adenosine deamination (A-to-I pre-mRNA editing) (4), which is catalyzed by RNA-specific adenosine deaminases (ADAR1 and ADAR2) (reviewed in 6,7). The A-to-I editing most frequently occurs in non-coding Alu repetitive elements (8–10). Although the biological relevance of this editing remains uncertain, its overall level has been reported to be dramatically higher in primates than in other mammals, and moreover, in humans compared with non-human primates, thus suggesting a possible contribution of editing to the development of higher-order cognitive processes (8,11,12). Site-specific mRNA editing resulting in re-coding has been reliably confirmed in only a few transcripts, most of which encode proteins that are expressed in the CNS (reviewed in 13,14). Notably, this type of editing occurs at functionally critical positions in proteins involved in synaptic transmission, including 5-HT2CR, many non-NMDA (AMPA and kainate) glutamate receptor ion channel subunits, α3 subunit of GABAA receptor ion channel, and voltage-gated potassium (Kv1.1.) and calcium (Cav1.3) channels. The protein isoforms resulting from mRNA editing profoundly influence neuronal excitability and signal transduction. Hence, it has been hypothesized that RNA editing provides efficient means for fine-tuning neurophysiological properties in response to environmental stimuli (15). In line with this hypothesis, recent work in cultured neurons and hippocampal slices has demonstrated that pharmacologically induced changes in neuronal activity can impact the extent of the ADAR-dependent editing of specific target mRNAs (16,17).
The 5-HT2CR can be edited at up to five closely-spaced (within a 15 nucleotide sequence) adenine positions that have been denoted A, B, E (also known as C′), C, and D sites. Because inosine is read as guanosine during translation (6), editing can alter codons for three amino acids in the second intracellular loop of the receptor (18,19), a region involved in the coupling with G-proteins (20). Combinatorial editing at the five positions can generate up to 32 mRNA variants encoding 24 different receptor isoforms (sites A and B as well as sites E and C are in the same codons). The extent of editing is inversely correlated with the 5-HT2CR function such that the more highly edited isoforms are less active than less extensively edited ones (reviewed in 4). The unedited Ile156-Asn158-Ile160 (INI) isoform possesses considerable constitutive and agonist-stimulated activity. In contrast, when the mRNA is edited, the coupling of 5-HT2CR to G-proteins and its affinity for serotonin are drastically reduced. Specifically, experiments in heterologous expression systems have shown that, compared with the INI isoform, the VSV (Val156-Ser158-Val160) isoform, which is the most common isoform in the human prefrontal cortex (21–23) and can be encoded by two different mRNA variants (ACD or ABCD), shows 4–5-fold decreased coupling to signaling pathways and 4-fold reduced constitutive activity (18,24).
Studies in animals have revealed variations in 5-HT2CR editing patterns among different strains of laboratory mice, indicating that editing can be genetically regulated (25). Experiments in different animal models have also suggested that 5-HT2CR editing is influenced by exposure to acute stress and chronic treatment with antidepressants (26) as well as by other pharmacological treatments (reviewed in 4, although see 27). A recent study of spinal cord injury (SCI) in a rat model has established a link between an editing-mediated increase in the spinal cord 5-HT2CR activity and the recovery of the spinal motoneuron excitability after SCI (28). Taken together, the diverse nature of 5-HT2CR signaling, its involvement in the regulation of complex behaviors, and its remarkable capacity to vary/adjust its activity via editing based on genetic status or in response to environmental impact suggest that 5-HT2CR editing is a homeostatic mechanism that allows 5-HT2CR-expressing neurons to stably maintain their functional characteristics.
Postmortem studies in several laboratories have reported an association between 5-HT2CR editing and suicide (22,23,29–31). Specifically, our previous findings indicate that in the three major psychiatric diseases, major depressive disorder (MDD), bipolar disorder (BPD) and schizophrenia (SZ), suicide is consistently associated with higher level of 5-HT2CR editing (and accordingly lower receptor activity) in the prefrontal cortex (PFC) (22,23,29–31). Suicide is linked to numerous, diverse risk factors. Among the strongest associations is the presence of psychopathology, most notably MDD, BPD, SZ or substance use disorder (32). Genetic epidemiological data provide strong evidence for a heritable component of suicidal behavior which is in part independent from the familial clustering of mental disorders (33). Nevertheless, environmental variables explain a substantial fraction of the total variance in suicidal behavior (34). Recent studies have increasingly emphasized the role of interactions between genetic vulnerability and environmental impact in suicide, suggesting that environmental stressors could influence subsequent behavior by altering gene function through epigenetic re-programming (35,36). Epigenetic processes link environmental factors to gene function via chromatin modifications (i.e. DNA methylation and histone modifications) that regulate gene expression. Recent reports describe suicide-associated alterations of DNA methylation in the promoter regions of several genes and the effects of these alterations on gene expression (reviewed in 37). These findings imply that epigenetic modifications form an interface through which suicide-associated environmental impacts affect gene expression. Little is known, however, about the biological mechanisms which translate gene-environmental interactions into modulation of neuronal functions that ultimately govern complex behaviors, e.g. suicide. The A-to-I editing in general, and 5-HT2CR editing in particular, is a potentially important mechanism of this type as it could be involved in homeostatic regulation of neurons by linking environmental stimuli to signal transduction and neuronal excitability.
Here we investigated (i) whether 5-HT2CR editing is linked to pathways that are important for neuronal activity and (ii) whether this link is altered in the brain of people that committed suicide. We first quantified and compared the levels of 5-HT2CR editing in the PFC between individuals who died of suicide or non-suicide causes using autopsy brain specimens obtained from two independent cohorts. The results confirm the overrepresentation of highly edited mRNA variants in suicide brains that was previously observed in different studies and populations. We then investigated the relationship between 5-HT2CR editing and gene expression in both study cohorts and detected numerous genes whose expression level was associated with editing. The set of these editing-associated signature genes was significantly enriched for genes that are involved in synaptic transmission, genes that are preferentially expressed in neurons, and genes whose expression is associated with the level of DNA methylation. Finally, we found that the link between 5-HT2CR editing and gene expression was (nearly) eliminated in suicide victims.
RESULTS
Overall analysis of RNA editing
We used massive parallel sequencing (MPS) to investigate 5-HT2CR editing in the brain samples obtained from two cohorts described in Materials and Methods, Supplementary Methods and Supplementary Material, Tables S1 and S2. Cohort 1 consisted of subjects who died of suicide (N = 22) or non-suicide-related causes (control group, NC; N = 29). Cohort 2 consisted of patients with a history of MDD who died of suicide (MDDSU, N = 10) or of non-suicide causes (MDDNon-SU; N = 24) and control individuals without any psychiatric diagnoses (DX) who died of non-suicide causes (NC; N = 31). The samples were dissected from sub-regions of the PFC–orbital frontal cortex (OFC; Brodmann area 11) for Cohort 1 and the subgenual anterior cingulate cortex (ACC; Brodmann area 25) for Cohort 2.
On average, 331 924 ± 3494 and 452 450 ± 44 881 (mean ± SE) MPS reads were obtained for each subject in Cohorts 1 and 2, respectively. The high sequencing depth ensured highly reliable detection of all of the 32 5-HT2CR mRNA editing variants, including those with abundance of <0.001% (38).
We detected only small (<1.3%), although significant for some variants, differences in the variant frequencies between the cohorts (Fig. 1, Supplementary Material, Table S3 and Fig. S1). Accordingly, the patterns of 5-HT2CR variant expression were highly similar between the cohorts and were also consistent with the data obtained in our previous studies that had been performed in yet another PFC sub-region, dorsolateral PFC (DLPFC) (22). In particular, only 3 of the 32 mRNA variants (editing combinations ABCD, ABD and NONE) were observed at ∼10% or greater frequency; ABCD was the most highly expressed variant in all three areas of the PFC.
Figure 1.
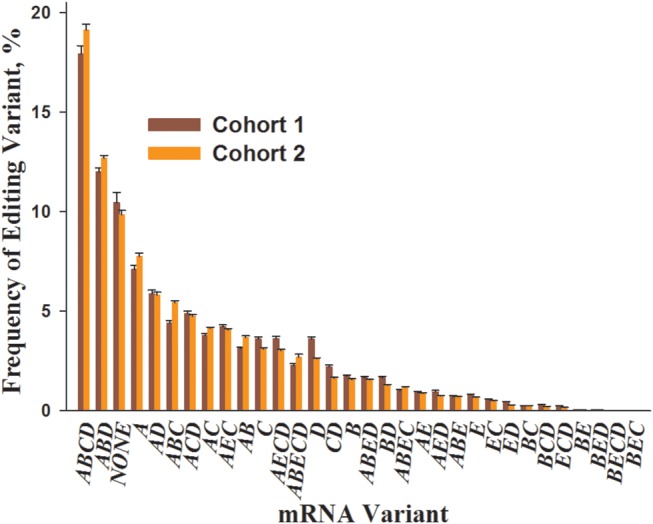
Comparison of the 5-HT2CR mRNA editing patterns between Cohort 1 and Cohort 2. Shown are mean ± SEM of variant frequencies in the postmortem OFC (Cohort 1) and ACC (Cohort 2) samples of the clinically unremarkable non-suicide individuals (N = 29 and N = 31 for Cohorts 1 and 2, respectively).
In the principal component analysis (PCA) of all 5-HT2CR editing variants, the ABCD (edited at sites A, B, C, D) and NONE (non-edited variant) aligned along the first principal component which by itself explained 62.7 and 62.6% of the total data variance in Cohorts 1 and 2, respectively (Supplementary Material, Fig. S2). Therefore, in all further analyses, the level of editing in each individual was represented by the frequency of the ABCD variant (hereinafter ‘ABCD’).
ABCD is increased in the brains of suicide patients
In our previous studies in the DLPFC of BPD and SZ patients, we detected more ABCD in suicides than in non-suicides with the same diagnosis but detected no differences between the non-suicides with BPD or SZ and psychiatrically normal controls (22). In our subsequent study of MDD patients, we found significantly less editing in MDDNon-SU than in MDDSU, whereas the MDDSU did not significantly differ from psychiatrically normal controls (39). Thus, both studies have shown that, compared with non-suicides with the same diagnosis, suicides express higher levels of the ABCD variant that encodes a hypoactive 5-HT2CR protein isoform, suggesting that 5-HT2CR editing (and the receptor function) is altered in the PFC of suicide victims regardless of their underlying psychiatric illness.
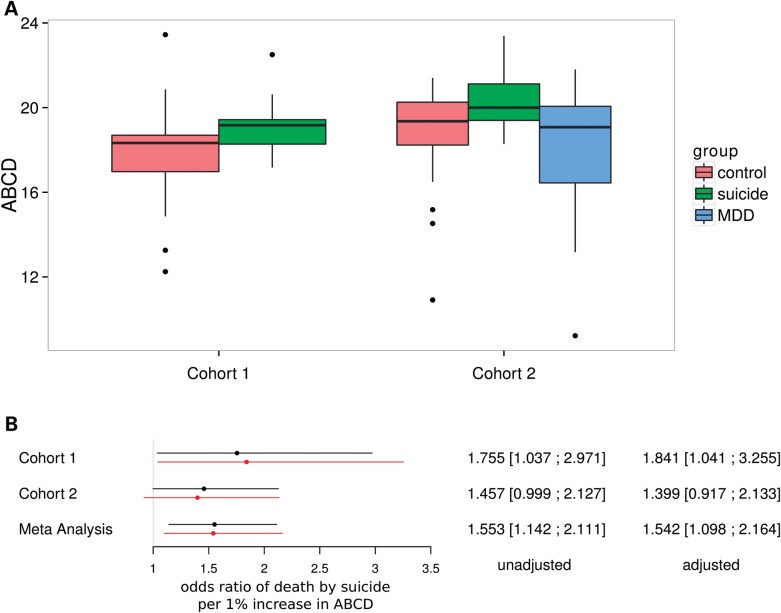
Here we first asked if our previous findings could be validated in different cohorts and/or different areas of the PFC. In Cohort 1, we detected a higher ABCD frequency in the suicide compared with the control group (P = 0.0217 by Wilcoxon test) (Fig. 2A). In Cohort 2, we detected a higher ABCD frequency in the MDDSU patients compared with MDDNon-SU individuals [P = 0.0216 by Tukey's honest significant difference (HSD) post hoc test]; there was also more ABCD in the MDDSU compared with the NC group but this comparison was not statistically significant (P = 0.3326 by Tukey's HSD post hoc test).
Figure 2.
Association between 5-HT2CR editing and suicide. (A) Comparison of the ABCD frequencies among different sub-groups in Cohorts 1 and 2; Cohort 1: suicides, N = 22; NCs, N = 29. Cohort 2: MDDSU, N = 10; MDDNon-SU, N = 24; NCs, N = 31. (B) Odds ratios of death by suicide per 1% increase in ABCD estimated using a logistic regression model; black line: univariate model. Red line: multivariate model (adjusting for age, sex, pH, PMI and DX in Cohort 2).
Using logistic regression analysis, we then compared ABCD frequencies between suicides and non-suicides, adjusting for covariates [i.e. age, sex, pH, postmortem interval (PMI) as well as DX in Cohort 2]. We detected higher ABCD in suicides for both cohorts (P values < 0.07) (Fig. 2B, Supplementary Material, Table S4). The meta-analysis of both cohorts showed P values of 0.005 and 0.012 for unadjusted and adjusted models, respectively. Importantly, among all regressors, ABCD was the only variable that was significantly associated with suicide in the meta-analysis (Supplementary Material, Table S4).
In Cohort 1, the presence of alcohol in blood or urine (Supplementary Material, Table S1) showed no association with ABCD (P = 0.34 by Wilcoxon test). In Cohort 2, ABCD frequencies did not vary among MDD subjects with different severity of depression (Supplementary Material, Table S2) [P = 0.96 by one-way analysis of variance (ANOVA)]. In addition, within each group (all MDD subjects, MDDNon-SU, or MDDSU), there were no associations between ABCD and antidepressant medications (all P values > 0.48 by Wilcoxon test). The latter were determined by positive toxicology for at least one antidepressant at the time of death (Supplementary Material, Table S2). Collectively, these results confirm our previous findings of increased 5-HT2CR editing in the PFC of suicide victims irrespective of the influence of the underlying psychiatric illness (22,39).
5-HT2CR editing shows a robust gene expression signature
We then performed an analysis of association between genome-wide gene expression and ABCD in Cohorts 1 and 2, controlling for age, sex, PMI, brain pH as well as DX in Cohort 2. We first identified genes whose expression correlated with ABCD using a liberal criterion of significance (unadjusted, one-tailed P-value <0.05). Among 10 140 genes that were present in the expression data sets of both Cohorts 1 and 2 (Supplementary Material, Table S5), the expression of 684 genes was positively associated with ABCD in both cohorts [odds ratio (OR) = 2.54, Fisher's exact test overlap P-value <1E−15], and the expression of 346 genes was negatively associated with ABCD in both cohorts (OR = 4.43, Fisher's exact test overlap P-value <1E−15) (Fig. 3, Supplementary Material, Table S6). Collectively 1030 genes were significantly associated with ABCD in both cohorts. We denoted this set of genes as ‘ABCD expression signature’ (or simply ‘ABCD signature’) genes (Supplementary Material, Table S7).
Figure 3.
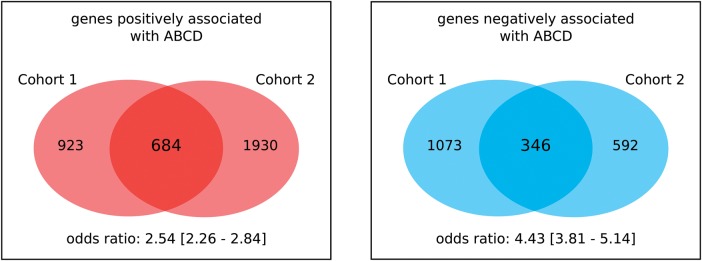
Identification of the ABCD expression signature. Shown are the number of genes significantly associated with ABCD by study cohort and direction of association. The overlap between the results from the two cohorts is highly significant for both signs of association (odds ratios >2.5; Fisher's exact test P-values <1E−15).
Signature genes are involved in neurotransmission and are linked to mental disorders
We then performed a functional annotation analysis of the ABCD signature genes using MetaCore (www.genego.com), and detected highly significant enrichment for neuron- and nervous system-related terms in all examined ontology databases (Supplementary Material, Table S8). Specifically, the ABCD signature was significantly enriched for genes involved in synaptic transmission and neurotransmitter secretion as well as genes localized within synapse and synaptic vehicle membranes, and genes associated with serious mental illnesses (such as SZ, MDD and BPD). The enrichment for neural-related categories was detected mostly among genes that were positively associated with ABCD, whereas among genes that were negatively associated with ABCD, a number of inflammation and immunity related categories were enriched (Supplementary Material, Table S9).
We detected a strong enrichment of ABCD signature genes for genes involved in both GABA-ergic and glutamatergic synaptic transmission (Supplementary Material, Tables S8 and S9). In particular, the expression of GAD1 (the major GABA-synthesizing enzyme, glutamic acid decarboxylase), SLC32A1 (a vesicular inhibitory acid transporter that is involved in GABA and glycine uptake in synaptic vesicles), GABRA1, GABRA5, GABRB2, GABRB3, GABRD, GABRG2 and GABARAPL1 (multiple subunits of GABAA receptors and GABAA receptor-associated protein-like 1) as well as GRIA4, GRIK2, GRINA and GRM7 (different subunits of ionotropic glutamate receptors and the metabotropic receptor 7) was positively associated with ABCD.
In addition to factors related to specific neurotransmitter systems, synaptic transmission is prominently regulated by presynaptic factors controlling the vesicle-mediated release of neurotransmitters (40). In line with these observations, the ABCD signature included numerous genes that encode various components of synaptic vesicles such as N-ethylmaleimide sensitive fusion protein (NSF), NSF attachment receptor (SNARE) proteins and other vesicle proteins, including synaptosomal-associated protein 25 kDa (SNAP25), synaptobrevins (VAMP1, VAMP2, VAMP5, VAMP8), synapsin (SYN2), syntaxins (STX1B, STX6, STX12), syntaxin binding (or MUNC18) proteins (STXBP1, STXBP2), synaptogyrins (SYNGR1, SYNGR3), synaptotagmins (SYT1, SYT11, SYT13), clathrin proteins (CLTB, CLTC), the Rab family of small GTPases (RAB2A, RAB11A), and double C2-like domain-containing protein alpha (DOC2A).
As mentioned above, 5-HT2CR is coupled to several G proteins that stimulate intracellular responses via effector molecules which influence the activity of ion channels (5). In line with these data, the ABCD signature included genes that encode several G proteins (GNAO1, GNAI1, GNA12, GNB5, GNG3, GNG5, GNG11) as well as multiple (mostly voltage-gated) potassium channels (KCNC1, KCNC2, KCNC4, KCNA1, KCNS3, KCNQ5, KCNF1, KCNK4, KCNJ12, KCTD8, HCN1) and a voltage-gated potassium channel-interacting protein (KCNIP4).
The ABCD signature also included many genes that encode proteins involved in Ca2+ signaling, such as calmodulin (CALM2, CALM3) and Ca2+/calmodulin-dependent protein kinases (CAMKK1, CAMK2D, CAMK2G, MKNK2). Notably, calmodulin has been reported to bind to the proximal region of the 5-HT2CR C-terminus upon receptor activation by 5-HT, which is critical for G protein-independent, arrestin-dependent ERK1,2 signaling mediated by 5-HT2CR (41).
Although in most cases, expression of genes with neuronal functions was found to be positively associated with ABCD, a variety of neuronal genes within the signature showed a negative association (e.g. GNA12, GNG5, GNG11, PLCD1, MKNK2, VAMP5), implying a complex relationship between editing and neuronal functional pathways. Last but not least, the ABCD signature included ADARB1, which encodes ADAR2, but did not include ADAR, which encodes ADAR1 or ADARB2, which encodes ADAR3. ADAR3 is a brain-specific protein whose sequence is closely related to ADAR2, but whose activity has not yet been demonstrated (42). Expression of ADARB1 was positively associated with ABCD.
A recent report identified 41 blood mRNA biomarkers of suicidality using a combination of different designs and approaches (43). This set of blood biomarker genes was significantly enriched for genes related to inflammatory disease and inflammatory response categories. Interestingly, 4 of these 41 genes were present in the ABCD signature (GBP1, ATP6V0E1, EPHX1, TNFSF10) and showed a negative association with ABCD. This is in line with our finding that the set of genes that was negatively associated with ABCD is highly enriched for inflammation and immunity related ontologies.
Distinct cell-type specificity of ABCD signature genes
In our recent study, we compared DNA methylation patterns between Fluorescence-Activated Cell Sorting (FACS)-sorted neuronal and non-neuronal (glial) cells using sister-aliquots of the OFC specimens obtained from six non-suicide subjects from Cohort 1 (44). Classification of the differentially methylated CpG sites into those undermethylated in neurons (neuronal type) and those undermethylated in non-neuronal cells (glial type), combined with findings of others that methylation within control elements typically negatively correlates with gene expression, yielded large sets of predicted neuron-specific and glia-specific genes (5098 and 6821 genes, respectively). These sets of genes were in excellent agreement with the available direct measurements of gene expression in human and mouse. In order to explore cell-specificity of the ABCD signature genes, we investigated the overlap between the signature genes and these DNA methylation data. We detected a highly significant overlap between those signature genes that were positively associated with ABCD (684 genes) and the neuron-specific gene set (OR = 1.988, P-value < 1E−15 by Fisher's exact test), whereas there was no significant overlap between these signature genes and the glial-specific genes. In contrast, the group of the signature genes that were negatively associated with ABCD (346 genes) showed a non-random overlap with the glial-specific gene set of (OR = 3.312, P-value < 1E−15) but not the neuronal-specific set (Supplementary Material, Table S10).
We next compared the ABCD signature genes with the cell type-specific gene-expression patterns derived from the analysis of the human transcriptome. We applied an in silico tissue dissection statistical approach that is based on ‘weighted gene co-expression network analysis’ (WGCNA) (45). The WGCNA identifies biologically relevant patterns in expression datasets by grouping genes into modules with strongly covarying patterns across the samples. Consequently, WGCNA can distinguish gene-expression networks associated with specific cell types (e.g. neurons or glia) that are present in a heterogeneous sample (e.g. whole human cortex) thanks to the distinct transcriptional profiles of these cell types and variation in the relative proportions of neurons and glial cells across samples (46). In particular, WGCNA co-expression modules can be examined for cell-type specificity using cell type-enriched gene sets identified in previous studies (47–53). We have previously applied WGCNA to the postnatal sub-cohort (N = 220) of the gene expression microarray dataset from the BrainCloud study which analyzed PFC samples obtained from a large collection of autopsy brain specimens (Gene Expression Omnibus; accession GSE30272) (54). Consistent with previous studies in the brain tissues (46,51), the WGCNA yielded well-defined gene co-expression modules (N = 77) with specific anatomical distributions frequently related to specific cell types (44). We detected numerous modules consisting of genes with enhanced expression in neurons or in different types of glia (microglia, astrocytes and oligodendrocytes) (44). A comparison of the coexpression modules with the ABCD geneexpression signature showed that the signature genes positively associated with ABCD (N = 684) were mostly over-represented in the modules related to glutamatergic or GABAegric (e.g. parvalbumin-expressing) neurons (adjusted P-values <0.01), whereas those signature genes that were negatively associated with ABCD (346 genes) were over-represented in microglia and astrocyte modules (all adjusted P-values <0.05) (Supplementary Material, Table S11). We did not detect an enrichment of the negatively associated signature genes in modules related to oligodendrocytes. Instead and in line with our gene ontology findings which linked the negatively associated signature genes with categories related to inflammation, 5 of the 8 modules that were enriched for the negatively associated signature genes were related to microglia.
Signature genes are over-represented among genes which show association between expression and DNA methylation among different individuals
Animal studies indicate that 5-HT2CR editing (and therefore, function) can be regulated by environmental factors (4). Because environment can influence gene expression via epigenetic processes, we asked if the ABCD signature was enriched for genes whose expression is linked to epigenetic modifications. Although there are many epigenetic marks that potentially could be studied, DNA methylation is the major mark that has already been demonstrated to be stable postmortem (55). Thus, we investigated the relationship between DNA methylation and expression of the ABCD signature genes.
The present results indicate that the majority of the ABCD signature genes are expressed in neurons. In addition, a dramatic difference in the DNA methylation landscapes between neuronal and glia cells has been recently reported (44,56). Therefore, we assessed DNA methylation within the PFC neuronal population. Neuronal nuclei were isolated from the sister aliquots of the OFC specimens from Cohort 1 (N = 51), which were used in our 5-HT2CR editing and RNA-Seq studies described above. We employed Illumina HM450K array technology, which examines the methylation status of ∼485 000 individual cytosine positions (mostly CpG sites). Although the array contains only ∼2% of the total CpG sites present in the genome, it spans the entire genome covering 99% of the RefSeq protein-coding genes with multiple probes per gene, 96% of the CpG islands from the UCSC database, high and low CpG density promoters and predicted enhancers.
We first identified those CpG sites in the HM450K array that belonged to the 10 140 genes whose expression were analyzed in both cohorts and were used to delineate the ABCD signature (see Supplementary Material, Supplementary Methods for CpG site-to-gene annotation). Next, regression analysis was applied to determine which of these 10 140 genes had at least one CpG site whose methylation level correlated with the expression of the respective gene in Cohort 1. To avoid spurious associations, we restricted the analysis to those sites which showed at least 10% range of variation in the beta values (that is, the sites for which the difference between the maximum and minimum beta value was at least 0.1). We detected a set of 551 such genes (hereinafter, methylation-linked genes). Among the methylation-linked genes, 63 were present in the ABCD signature, and 56 of these 63 genes were positively associated with ABCD (Supplementary Material, Table S12). Thus, although there was no significant overlap between the signature as a whole (1080 genes) and the set of the methylation-linked genes (OR = 1.150, P = 0.173), there was a significant overlap between the 684 positively associated signature genes and this set (OR = 1.644, P = 8.584E−4). No significant overlap was detected with negatively associated signature genes (OR = 0.340, P = 0.999). Because DNA methylation was measured in neurons, this observation is compatible with our finding that the positively associated signature genes are preferentially expressed in neurons. Thus, the ABCD signature (or at least its neuronal component) is enriched for genes whose expression is linked to DNA methylation.
We also tested the overlap between the ABCD signature and a list of genes involved in epigenetic regulation (57–60), whose expression was detected in both cohorts. Of the 69 such genes, only 3 were present in the signature: HDAC1, SMYD2 and SMYD3 (Supplementary Material, Table S13). This overlap, however, was not significant (OR = 0.40, Fisher's exact test P-value = 0.98).
Association between editing and gene expression is altered in suicide
We then asked whether or not the relationship between editing and gene expression differs in people who died of suicide compared with individuals who died of non-suicide causes. We first developed an overall ABCD expression signature score for each individual. The score was computed as the difference between the average standardized expression of the signature genes positively associated with ABCD and the average standardized expression of the signature genes negatively associated with ABCD (see Materials and Methods). Accordingly, for each individual, the signature score summarized the expression pattern of all signature genes, taking into account the sign of their association with ABCD. A high signature score for a specific individual indicated high expression of the signature genes positively associated with ABCD and low expression of the signature genes negatively associated with ABCD. Conversely, a low signature score indicated low expression of the genes positively associated with ABCD and high expression of the genes negatively associated with ABCD.
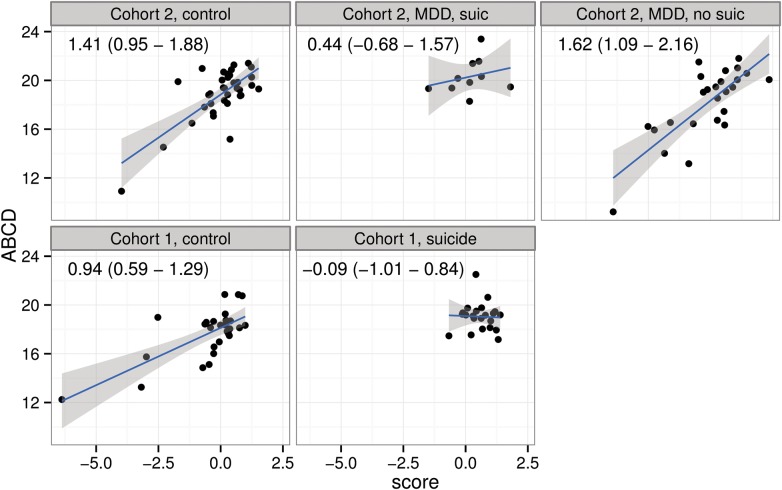
We then compared the signature scores between the suicides and non-suicides in each cohort. As expected, we observed significant association between the signature score and ABCD in non-suicide groups in both cohorts (all P-values < 1E−4) (Fig. 4). In a striking contrast, among the suicide victims from both cohorts, there was no association between the score and ABCD (P-values > 0.462). In other words, the range of variation for both ABCD frequencies and signature scores was dramatically lower in the suicide compared with the non-suicide groups.
Figure 4.
Association between ABCD frequencies and gene expression signature score in different clinical sub-populations of Cohorts 1 and 2. Least squares regression lines (blue lines) and 95% error intervals (gray shade) are superimposed to each scatterplot. Estimated slopes and their 95% confidence intervals are reported as text within each panel.
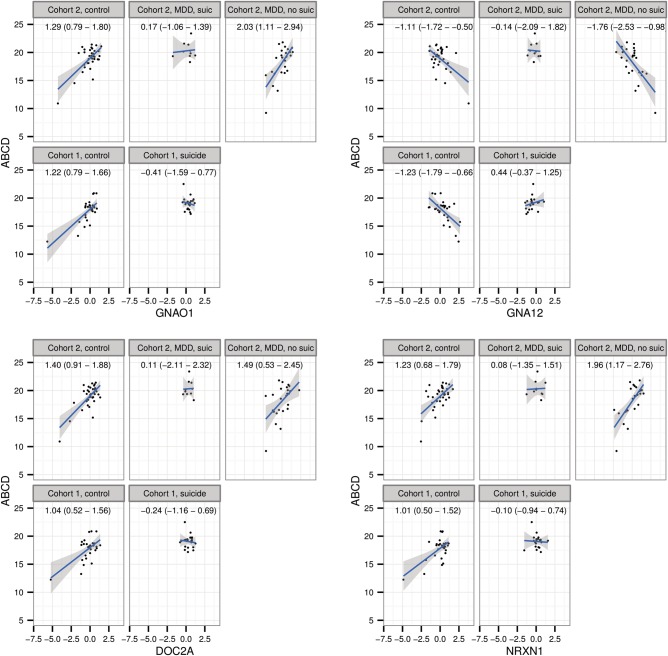
We then attempted to identify the individual genes that were associated with ABCD in non-suicide but not in suicide subjects. To this end, we performed an association analysis between ABCD and gene expression in both cohorts introducing ‘Suicide’ as an interaction term. This analysis yielded no significant overlap between the cohorts, probably because much larger samples are needed to detect interactions (61). However, because the signature score captures the expression patterns for all ABCD signature genes, the differences in the patterns of association between the suicide and non-suicide groups for the majority of the genes in the signature were very similar to each other and to the overall pattern (Fig. 5 and Supplementary Material, Fig. S3).
Figure 5.
Association between expression of several signature genes and ABCD frequencies in different clinical sub-populations of Cohorts 1 and 2 (see Fig. 4 for details). Similar to the data obtained for the overall signature score (Fig. 4), we detected significant association between expression of the signature genes and ABCD in non-suicide but not in suicide groups from both cohorts. More examples are shown in Supplementary Material, Figure S3.
DISCUSSION
RNA editing is one of the major mechanisms by which environmental signals overwrite genetically encoded information to modify gene function and regulation, particularly in the brain, thus providing a possible interface between environmental inputs and behavior. Higher overall editing levels have been reported for the human brain compared with the brains of other mammals including non-human primates (8,11), suggesting that RNA editing might underpin higher-order brain function and complex behaviors, and could be involved in pathophysiology of psychiatric and neurological diseases.
Previous studies have shown that 5-HT2CR editing is altered in the PFC of suicide victims (22,23,29–31), and we confirmed these findings in the present work. Hence, we speculate that A-to-I editing in general, and 5-HT2CR editing in particular, could act as a homeostatic mechanism that dampens the effect of a wide range of environmental and genetic factors that impair neuronal function. This hypothesis implies that 5-HT2CR editing is linked to the signaling pathways that are important for neuronal activity and consequently to the expression of genes whose products are involved in these pathways. Similar hypotheses have been recently proposed on the basis of a comparative study of A-to-I editing in autistic and non-autistic cerebella (62). We further speculated that the relationship between 5-HT2CR editing and gene expression would differ between individuals who died of suicide versus those who died of non-suicide-related causes.
The present study aimed to test these hypotheses in two cohorts of individuals who died of suicide or other causes. Although the cohorts were, out of necessity, relatively small, the results show that 5-HT2CR editing is associated with a robust gene expression signature, i.e. a large set of genes whose expression was significantly correlated with editing in both cohorts. This signature set was significantly enriched for genes involved in synaptic function and genes associated with serious mental illnesses, such as SZ, MDD and BPD, as well as suicide (Supplementary Material, Tables S8 and S9). Psychological autopsy studies show that >90% of all individuals who die by suicide meet criteria for at least one psychiatric disorder, most commonly MDD, BPD, SZ or substance use disorder (32).
Our analysis also demonstrated that the signature set was enriched for genes that are preferentially expressed in neurons and for those neuronal genes whose expression is negatively correlated with their DNA methylation. In the brain, DNA methylation within promoters and distal regulatory regions is associated with repression of gene expression (44,56). In addition, although DNA methylation marks in postmitotic cells may persist throughout the cellular lifetime, the methylation status is a flexible substrate for epigenetic modification that can be altered by cellular response to environmental stimuli (63). Thus, our findings imply a link between environmental impact and 5-HT2CR editing through chromatin remodeling and the associated gene expression changes.
The key finding of this study is that the association between editing and gene expression is disrupted in suicide, suggesting a dysregulation of the putative homeostatic function of 5-HT2CR editing in the PFC of suicide victims. Specifically, compared with the non-psychiatric controls and especially to the MDD without suicide group, the suicide group included no individuals with low levels of editing and low ABCD signature scores (Figs 4 and 5). We speculate that in the brain of suicide victims, a mechanism that enhances the function of 5-HT2CR through lowering the level of editing and hence prevents neuron malfunction is impaired. Elucidation of the nature of this putative mechanism(s) is a goal for subsequent work.
The editing-associated signature included ADARB1, which encodes ADAR2, but not ADAR which encodes ADAR1. A recent study has reported that the dysfunctional isoform of ADAR2 is over-represented in postmortem cerebella from individuals with autism (62). Thus, the pathways related to the regulation of ADAR2 might be important for gene-expression mediated crosstalk between neuronal function and 5-HT2CR editing. These, still virtually uncharacterized ADAR2-related pathways (64) could be compromised in suicide. Indeed, as shown here, the apparent dysregulation of editing in the PFC of suicide victims is characterized by a notable increase in the frequency of the ABCD variant the formation of which requires editing by both ADAR1 and ADAR2 because site D is primarily edited by ADAR2 (65).
The most obvious shortcoming of the present work appears to be that the majority of the MDD subjects in Cohort 2 were receiving antidepressant medications. Therefore it was difficult to ascertain whether medication, rather than an innate pathophysiology, accounted for the differences in editing between MDDNon-SU and MDDSU. Multiple lines of evidence, however, argue against the possibility that suicide-related differences in editing observed in our study could be caused by medication: (1) Similar to the results obtained in earlier animal studies (reviewed in 4), a recent report in mice that employed MPS (and therefore, achieved high precision in assessing 5-HT2CR editing levels) showed that chronic treatments with antidepressant drugs resulted in an increase in 5-HT2CR mRNA editing (66). Specifically, in that study increased editing at the A and B sites was observed in the striatum and hippocampus following fluoxetine and only in the hippocampus following amitriptyline treatment. Neither drug influenced editing in the neocortex; in addition, no editing alterations were observed in any brain region following treatment with antipsychotic medications (olanzapine or clozapine); (2) In the present study, within each group in Cohort 2 (all MDD subjects, MDDNon-SU, or MDDSU), there was no association between ABCD and antidepressant medications. Moreover, the percentage of subjects that showed positive toxicology for at least one antidepressant medication at the time of death was higher in the MDDNoSuic (75%) than in the MDDSuic (60%) group; however, more editing was detected in the MDDSuic than in the MDDNoSuic; (3) In our previous work on 5-HT2CR editing in SZ and BPD patients with and without suicide, the history of antidepressant treatment among suicide victims had no discernible impact on editing (22). Also, no differences were observed between non-psychiatric controls and SZ or BPD patients that did not commit suicide, although more than half of the non-suicide BPD patients received antidepressant medication, and all non-suicide SZ patients received antipsychotic medications; (4) In the present study, we observed difference in editing between suicide victims and subjects without any antemortem DX in Cohort 1. Although 6 of the 22 suicide subjects in this cohort had a history of depression, only one suicide subject showed positive toxicology for medications (olanzapine and clozapine). Based on this collective evidence, we believe that the editing alterations in suicide subjects that we identified in this work are unlikely to be related to their treatment with antidepressant or antipsychotic medications.
To summarize, the results of this work imply that the postulated homeostatic function of 5-HT2CR editing is dysregulated in the brains of suicide victims. The resulting increased editing (or more precisely, relative paucity of unedited variants) could be a synergistic effect of suicide-associated mutations and environmental stimuli. The relatively small number of samples analyzed in this report does not allow us to examine this possibility. In studies of suicide, which is a rare and unpredictable event, obtaining postmortem samples is an obvious challenge. We attempted to partially overcome this problem by studying the link between editing, gene expression and DNA methylation. Further, more detailed and accurate analysis of the multiple connections between genetic and molecular phenotypic signatures of suicide seems to have potential to unravel the molecular mechanisms underpinning this complex behavior and ultimately develop effective interventions.
MATERIALS AND METHODS
Subjects and brain regions
Human postmortem brain specimens from the PFC of Caucasian individuals were provided by the Brain Collection of Dr Yasmin Hurd (Icahn School of Medicine at Mount Sinai, New York, NY) (Cohort 1) (67,68) and by the University of Pittsburgh Brain Tissue Donation Program (Pittsburgh, PA) (Cohort 2) (69,70). Detailed description of the cohorts and brain regions is provided in Supplementary Methods and Supplementary Material, Tables S1 and S2. All brains were analyzed for adequate brain pH [6.8 ± 0.2 and 6.6 ± 0.3, mean ± standard deviation (SD) for Cohorts 1 and 2, respectively].
In Cohort 1, 6 of the 22 suicide subjects had a history of depression. The NC subjects did not have any antemortem DX. At the time of death, all subjects showed negative toxicology for opiates and other illicit drugs. Three control subjects and five suicide subjects were positive for alcohol in blood or urine, and therapeutic agents were detected in one control and one suicide subject.
In Cohort 2, control subjects showed negative toxicology for illicit drugs and antidepressant drugs, while 18 MDDNon-SU and 6 MDDSU individuals showed positive toxicology for at least one antidepressant medication at the time of death.
RNA extraction, quality control and analysis of 5-HT2CR RNA editing
Total RNA was extracted using Invitrogen ToTally RNA Kit (Cohort 1) or TRIzol reagent (Cohort 2) (both reagents from Life Technologies). The quality of RNA was assessed using Agilent Bioanalyzer. RNA integrity number (RIN) for all specimens was >6.4 (7.6 ± 0.6 and 8.1 ± 0.6, mean ± SD for Cohorts 1 and 2, respectively). Complimentary DNA (cDNA) was synthesized from equal quantities of RNA from each subject (1 μg of RNA per 10 μl of reaction) using High Capacity cDNA Reverse Transcription kit (Life Technologies).
5-HT2CR RNA editing was assessed using MPS of PCR-amplified fragments in the region of editing as described in refs (39,71) and detailed in Supplementary Methods. Because inosine is read as guanine during reverse transcription (6), the sequencing enables to assess the pattern and the level of editing in each experimental sample. Specifically, MPS provides the number of reads for each of the 32 5-HT2CR mRNA editing variants in each subject. The relative frequency of each variant in each subject was calculated as the ratio of the number of reads detected for a particular variant to the total number of reads for all variants in the subject.
Gene-expression analysis
Transcriptional profiling of Cohort 1 was performed using mRNA-Seq. For each subject, the same RNA preparations were used for both RNA editing and RNA-Seq assays. Libraries were prepared from 1 μg of total RNA using Illumina's TruSeq RNA library preparation kit V2, and sequencing was performed using an Illumina HiSeq2000. After de-multiplexing of each library pool according to its barcode sequence, from 8 to 20 million (M) (mean 15.1 M) reads per sample were available for analysis. Alignment, assembly and quantification of the data were performed using the TopHat 2.0.8b and Cufflinks 2.1.1 (72). Only transcripts with the maximum value of the fragments per kilobase of transcript per million mapped reads (FPKM) >2 among all subjects were analyzed in the study. Gene expression profiles of Cohort 2 specimens have been previously assessed using Illumina microarray chip Human v3, which contains 48 803 RefSeq probes (73).
For each gene that was represented by several transcripts (Cohort 1) or probes (Cohort 2), only one transcript/probe that resulted in a signal with the highest SD was retained. After this filtering, the data for 13 101 and 24 022 unique gene symbols were retained for Cohorts 1 and 2, respectively. Among these, there were 10 140 gene symbols that were present in both datasets (cohorts), and those genes were used in subsequent association analyses with 5-HT2CR editing and DNA methylation data (see below). The list of genes that were present in both datasets (cohorts) with associated P-values is reported in Supplementary Material, Table S5.
Analysis of association between editing and gene expression
The association between gene expression and editing was assessed for Cohorts 1 and 2 by fitting linear regression models of gene expression level versus editing level. The editing level was represented by the frequency of the ABCD mRNA variant in each individual. The model also included other factors which could affect gene expression as covariates, such as age, sex, PMI, brain pH and DX (suicide or NC in Cohort 1; MDDSU, MDDNon-SU or NC in Cohort 2). The categorical variables (DX and sex) were expanded into k− 1 indicator variables, k being the number of levels observed for that variable.
Calculation of ABCD gene expression signature score
The ABCD gene expression signature score was derived in order to summarize the expression pattern of the ABCD signature genes in each individual. The score allowed us to analyze the relationship between the ABCD signature and the clinical covariates (DX and suicide status). To calculate the score, the expression values for signature genes were first standardized to a mean of ‘0’ and a SD of ‘1’. In Cohort 1, the FPKM expression values (incremented by 1) were log-2 transformed prior to the standardization. The score was then computed for each individual as the difference between the average standardized expression of the signature genes that were positively associated with ABCD and the average standardized expression of the signature genes that were negatively associated with ABCD.
DNA methylation analysis of neuronal cells
The DNA methylation of neuronal cells was measured in Cohort 1 only. Neuronal nuclei were separated from non-neuronal (glial) nuclei using FACS with DNA-binding dye 7-AAD and anti-NeuN neuron-specific antibodies mostly as described in ref. (44) and as detailed in Supplementary Methods.
Genomic scale single-site resolution DNA methylation profiling of neuronal cells was performed using Illumina's Infinium Human Methylation450K array (HM450K) (Illumina Inc.) as described in refs (74,75) and detailed in Supplementary Methods. Methylation level at each individual CpG site in each specimen was measured as ‘beta value’ (β), which is defined as the ratio of the methylated probe intensity to the sum of the methylated and unmethylated probe intensities. The resulting β can range from 0 (no methylation) to 1 (complete methylation). The CpG site-to-gene annotation was performed as described in Supplementary Methods.
Analysis of association between DNA methylation and gene expression
The association between gene expression and DNA methylation was analyzed in Cohort 1 (N = 51) using a linear regression model. Only those 10 140 genes that were detected in both cohorts were analyzed. Age, sex, PMI and brain pH were included in the statistical model. The analysis was performed for cis associations only. Specifically, expression of each gene was correlated with the methylation level at the CpG sites that were annotated for the given gene.
Analysis of difference in editing among disease groups
The differences in the average level of the ABCD mRNA variant among disease groups were tested using the two-sided Wilcoxon test in Cohort 1 and the one-way ANOVA followed by the Tukey's HSD post hoc pairwise comparisons in Cohort 2. Prior to each test, within each Cohort–disease group combination, observations below the 25th percentile minus 1.5 times the interquartile range, and above the 75th percentile plus 1.5 times the interquartile range were identified as outliers (resulting in four outliers in Cohort 1 and four outliers in Cohort 2) and were removed from the analysis.
Suicide propensity modeling
Univariate (one regressor at a time) and multivariate (all regressors: ABCD, age, sex, PMI as well as DX for Cohort 2) logistic regression models were fitted for the probability of suicide against the available regressors in Cohorts 1 and 2.
Software
Linear and logistic modeling, and PCA as well as visualization were performed using R, version 3.0.0 (http://www.R-project.org/). The PCA of the frequencies of 5-HT2CR mRNA editing variants was performed using the ‘prcomp’ function in R. Linear modeling of gene expression data was performed using Limma, version 3.12.1.
SUPPLEMENTARY MATERIAL
FUNDING
This work was funded by VISN3 Mental Illness Research and Education Clinical Center (MIRECC), VA Merit Review Award (BX001829 to S.D.); National Institute of Health (R21MH090352 and R21DA031557 to S.D.; and R01DA015446 to Y.H.); and Hope for Depression Research Foundation grant to S.D. E.V.K. is supported by intramural funds of the US Department of Health and Human Services to National Library of Medicine.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Alisa Timashpolsky for superb technical assistance. The work was supported with resources and the use of facilities at the James J Peters VA Medical Center, Bronx, New York.
Conflict of Interest statement. Authors have no competing financial interests in relation to the work described.
REFERENCES
- 1.Giorgetti M., Tecott L.H. Contributions of 5-HT(2C) receptors to multiple actions of central serotonin systems. Eur. J Pharmacol. 2004;488:1–9. doi: 10.1016/j.ejphar.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 2.Millan M.J. Serotonin 5-HT2C receptors as a target for the treatment of depressive and anxious states: focus on novel therapeutic strategies. Therapie. 2005;60:441–460. doi: 10.2515/therapie:2005065. [DOI] [PubMed] [Google Scholar]
- 3.Berg K.A., Clarke W.P., Cunningham K.A., Spampinato U. Fine-tuning serotonin2c receptor function in the brain: molecular and functional implications. Neuropharmacology. 2008;55:969–976. doi: 10.1016/j.neuropharm.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werry T.D., Loiacono R., Sexton P.M., Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol. Ther. 2008;119:7–23. doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Harvey P.J., Li X., Li Y., Bennett D.J. Endogenous monoamine receptor activation is essential for enabling persistent sodium currents and repetitive firing in rat spinal motoneurons. J. Neurophysiol. 2006;96:1171–1186. doi: 10.1152/jn.00341.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gott J.M., Emeson R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 7.Valente L., Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 8.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blow M., Futreal P.A., Wooster R., Stratton M.R. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 11.Paz-Yaacov N., Levanon E.Y., Nevo E., Kinar Y., Harmelin A., Jacob-Hirsch J., Amariglio N., Eisenberg E., Rechavi G. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenberg E., Nemzer S., Kinar Y., Sorek R., Rechavi G., Levanon E.Y. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Penn A.C., Balik A., Greger I.H. Reciprocal regulation of A-to-I RNA editing and the vertebrate nervous system. Front. Neurosci. 2013;7:61. doi: 10.3389/fnins.2013.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenthal J.J., Seeburg P.H. A-to-I RNA editing: effects on proteins key to neural excitability. Neuron. 2012;74:432–439. doi: 10.1016/j.neuron.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattick J.S., Mehler M.F. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31:227–233. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Balik A., Penn A.C., Nemoda Z., Greger I.H. Activity-regulated RNA editing in select neuronal subfields in hippocampus. Nucleic Acids Res. 2013;41:1124–1134. doi: 10.1093/nar/gks1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanjana N.E., Levanon E.Y., Hueske E.A., Ambrose J.M., Li J.B. Activity-dependent A-to-I RNA editing in rat cortical neurons. Genetics. 2012;192:281–287. doi: 10.1534/genetics.112.141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., Sanders-Bush E., Emeson R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q., O'Brien P.J., Chen C.X., Cho D.S., Murray J.M., Nishikura K. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J. Neurochem. 2000;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- 20.Moro O., Lameh J., Hogger P., Sadee W. Hydrophobic amino acid in the i2 loop plays a key role in receptor-G protein coupling. J. Biol. Chem. 1993;268:22273–22276. [PubMed] [Google Scholar]
- 21.Dracheva S., Elhakem S.L., Marcus S.M., Siever L.J., McGurk S.R., Haroutunian V. RNA editing and alternative splicing of human serotonin 2C receptor in schizophrenia. J. Neurochem. 2003;87:1402–1412. doi: 10.1046/j.1471-4159.2003.02115.x. [DOI] [PubMed] [Google Scholar]
- 22.Dracheva S., Patel N., Woo D.A., Marcus S.M., Siever L.J., Haroutunian V. Increased serotonin 2C receptor mRNA editing: a possible risk factor for suicide. Mol. Psychiatry. 2008;13:1001–1010. doi: 10.1038/sj.mp.4002081. [DOI] [PubMed] [Google Scholar]
- 23.Lyddon R., Dwork A.J., Keddache M., Siever L.J., Dracheva S. Serotonin 2c receptor RNA editing in major depression and suicide. World J. Biol. Psychiatry. 2013;14:590–601. doi: 10.3109/15622975.2011.630406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niswender C.M., Copeland S.C., Herrick-Davis K., Emeson R.B., Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem. 1999;274:9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- 25.Du Y., Davisson M.T., Kafadar K., Gardiner K. A-to-I pre-mRNA editing of the serotonin 2C receptor: comparisons among inbred mouse strains. Gene. 2006;382:39–46. doi: 10.1016/j.gene.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Englander M.T., Dulawa S.C., Bhansali P., Schmauss C. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J. Neurosci. 2005;25:648–651. doi: 10.1523/JNEUROSCI.3895-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H., Urban D.J., Blashka J., McPheeters M.T., Kroeze W.K., Mieczkowski P., Overholser J.C., Jurjus G.J., Dieter L., Mahajan G.J., et al. Quantitative analysis of focused a-to-I RNA editing sites by ultra-high-throughput sequencing in psychiatric disorders. PLoS One. 2012;7:e43227. doi: 10.1371/journal.pone.0043227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray K.C., Nakae A., Stephens M.J., Rank M., D'Amico J., Harvey P.J., Li X., Harris R.L., Ballou E.W., Anelli R., et al. Recovery of motoneuron and locomotor function after spinal cord injury depends on constitutive activity in 5-HT2C receptors. Nat. Med. 2010;16:694–700. doi: 10.1038/nm.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niswender C.M., Herrick-Davis K., Dilley G.E., Meltzer H.Y., Overholser J.C., Stockmeier C.A., Emeson R.B., Sanders-Bush E. RNA editing of the human serotonin 5-HT2C receptor. Alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24:478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 30.Gurevich I., Tamir H., Arango V., Dwork A.J., Mann J.J., Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 31.Iwamoto K., Kato T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci. Lett. 2003;346:169–172. doi: 10.1016/s0304-3940(03)00608-6. [DOI] [PubMed] [Google Scholar]
- 32.Nock M.K., Borges G., Bromet E.J., Cha C.B., Kessler R.C., Lee S. Suicide and suicidal behavior. Epidemiol. Rev. 2008;30:133–154. doi: 10.1093/epirev/mxn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brent D.A., Mann J.J. Family genetic studies, suicide, and suicidal behavior. Am. J. Med. Genet. C Semin. Med. Genet., 2005;133C:13–24. doi: 10.1002/ajmg.c.30042. [DOI] [PubMed] [Google Scholar]
- 34.Qin P., Agerbo E., Mortensen P.B. Suicide risk in relation to socioeconomic, demographic, psychiatric, and familial factors: a national register-based study of all suicides in Denmark, 1981–1997. Am. J Psychiatry. 2003;160:765–772. doi: 10.1176/appi.ajp.160.4.765. [DOI] [PubMed] [Google Scholar]
- 35.Tsai S.J., Hong C.J., Liou Y.J. Recent molecular genetic studies and methodological issues in suicide research. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2011;35:809–817. doi: 10.1016/j.pnpbp.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Labonte B., Turecki G. Epigenetic effects of childhood adversity in the brain and suicide risk. In: Dwivedi Y., editor. The Neurobiological Basis of Suicide. Boca Raton, FL, USA: CRC Press. Chapter 13; 2012. [PubMed] [Google Scholar]
- 37.Labonte B., Turecki G. The epigenetics of suicide: explaining the biological effects of early life environmental adversity. Arch. Suicide Res. 2010;14:291–310. doi: 10.1080/13811118.2010.524025. [DOI] [PubMed] [Google Scholar]
- 38.Morabito M.V., Ulbricht R.J., O'Neil R.T., Airey D.C., Lu P., Zhang B., Wang L., Emeson R.B. High-throughput multiplexed transcript analysis yields enhanced resolution of 5-hydroxytryptamine 2C receptor mRNA editing profiles. Mol. Pharmacol. 2010;77:895–902. doi: 10.1124/mol.109.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyddon R., Dwork A.J., Keddache M., Siever L.J., Dracheva S. Serotonin 2c receptor RNA editing in major depression and suicide. World J. Biol. Psychiatry. 2013;14:590–601. doi: 10.3109/15622975.2011.630406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L.G., Hamid E., Shin W., Chiang H.C. Exocytosis and endocytosis: modes, function, and coupling mechanisms. Annu. Rev. Physiol. 2014;76:301–331. doi: 10.1146/annurev-physiol-021113-170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labasque M., Reiter E., Becamel C., Bockaert J., Marin P. Physical interaction of calmodulin with the 5-hydroxytryptamine2C receptor C-terminus is essential for G protein-independent, arrestin-dependent receptor signaling. Mol. Biol. Cell. 2008;19:4640–4650. doi: 10.1091/mbc.E08-04-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C.X., Cho D.S., Wang Q., Lai F., Carter K.C., Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains 1. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le-Niculescu H., Levey D.F., Ayalew M., Palmer L., Gavrin L.M., Jain N., Winiger E., Bhosrekar S., Shankar G., Radel M., et al. Discovery and validation of blood biomarkers for suicidality. Mol. Psychiatry. 2013;18:1249–1264. doi: 10.1038/mp.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozlenkov A., Roussos P., Timashpolsky A., Barbu M., Rudchenko S., Bibikova M., Klotzle B., Byne W., Lyddon R., Di Narzo A.F., et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 2014;42:109–127. doi: 10.1093/nar/gkt838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 46.Oldham M.C., Konopka G., Iwamoto K., Langfelder P., Kato T., Horvath S., Geschwind D.H. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugino K., Hempel C.M., Miller M.N., Hattox A.M., Shapiro P., Wu C., Huang Z.J., Nelson S.B. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 48.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 49.Winden K.D., Oldham M.C., Mirnics K., Ebert P.J., Swan C.H., Levitt P., Rubenstein J.L., Horvath S., Geschwind D.H. The organization of the transcriptional network in specific neuronal classes. Mol. Syst. Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller J.A., Horvath S., Geschwind D.H. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colantuoni C., Lipska B.K., Ye T., Hyde T.M., Tao R., Leek J.T., Colantuoni E.A., Elkahloun A.G., Herman M.M., Weinberger D.R., Kleinman J.E. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ernst C., McGowan P.O., Deleva V., Meaney M.J., Szyf M., Turecki G. The effects of pH on DNA methylation state: in vitro and post-mortem brain studies. J. Neurosci. Methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 56.Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D., et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fournier A., Sasai N., Nakao M., Defossez P.A. The role of methyl-binding proteins in chromatin organization and epigenome maintenance. Brief. Funct. Genomics. 2012;11:251–264. doi: 10.1093/bfgp/elr040. [DOI] [PubMed] [Google Scholar]
- 58.Kooistra S.M., Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 59.Gavin D.P., Chase K.A., Sharma R.P. Active DNA demethylation in post-mitotic neurons: a reason for optimism. Neuropharmacology. 2013;75:233–245. doi: 10.1016/j.neuropharm.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huynh J.L., Casaccia P. Epigenetic mechanisms in multiple sclerosis: implications for pathogenesis and treatment. Lancet Neurol. 2013;12:195–206. doi: 10.1016/S1474-4422(12)70309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith P.G., Day N.E. The design of case-control studies: the influence of confounding and interaction effects. Int. J. Epidemiol. 1984;13:356–365. doi: 10.1093/ije/13.3.356. [DOI] [PubMed] [Google Scholar]
- 62.Eran A., Li J.B., Vatalaro K., McCarthy J., Rahimov F., Collins C., Markianos K., Margulies D.M., Brown E.N., Calvo S.E., et al. Comparative RNA editing in autistic and neurotypical cerebella. Mol. Psychiatry. 2013;18:1041–1048. doi: 10.1038/mp.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Labonte B., Suderman M., Maussion G., Navaro L., Yerko V., Mahar I., Bureau A., Mechawar N., Szyf M., Meaney M.J., Turecki G. Genome-wide epigenetic regulation by early-life trauma. Arch. Gen. Psychiatry. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orlandi C., Barbon A., Barlati S. Activity regulation of adenosine deaminases acting on RNA (ADARs) Mol. Neurobiol. 2012;45:61–75. doi: 10.1007/s12035-011-8220-2. [DOI] [PubMed] [Google Scholar]
- 65.Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 66.Abbas A.I., Urban D.J., Jensen N.H., Farrell M.S., Kroeze W.K., Mieczkowski P., Wang Z., Roth B.L. Assessing serotonin receptor mRNA editing frequency by a novel ultra high-throughput sequencing method. Nucleic Acids Res. 2010;38:e118. doi: 10.1093/nar/gkq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nikoshkov A., Drakenberg K., Wang X., Horvath M.C., Keller E., Hurd Y.L. Opioid neuropeptide genotypes in relation to heroin abuse: dopamine tone contributes to reversed mesolimbic proenkephalin expression. Proc. Natl. Acad. Sci. U. S. A. 2008;105:786–791. doi: 10.1073/pnas.0710902105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drakenberg K., Nikoshkov A., Horvath M.C., Fagergren P., Gharibyan A., Saarelainen K., Rahman S., Nylander I., Bakalkin G., Rajs J., et al. Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7883–7888. doi: 10.1073/pnas.0600871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guilloux J.P., Douillard-Guilloux G., Kota R., Wang X., Gardier A.M., Martinowich K., Tseng G.C., Lewis D.A., Sibille E. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Mol. Psychiatry. 2012;17:1130–1142. doi: 10.1038/mp.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tripp A., Oh H., Guilloux J.P., Martinowich K., Lewis D.A., Sibille E. Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. Am. J. Psychiatry. 2012;169:1194–1202. doi: 10.1176/appi.ajp.2012.12020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Navarrett S., Collier L., Cardozo C., Dracheva S. Alterations of serotonin 2C and 2A receptors in response to T10 spinal cord transection in rats. Neurosci. Lett. 2012;506:74–78. doi: 10.1016/j.neulet.2011.10.052. [DOI] [PubMed] [Google Scholar]
- 72.Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X., Lin Y., Song C., Sibille E., Tseng G.C. Detecting disease-associated genes with confounding variable adjustment and the impact on genomic meta-analysis: with application to major depressive disorder. BMC Bioinformatics. 2012;13:52. doi: 10.1186/1471-2105-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bibikova M., Le J., Barnes B., Saedinia-Melnyk S., Zhou L., Shen R., Gunderson K.L. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 75.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L., et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.