Abstract
The Fragile X-related disorders are X-linked disorders resulting from the inheritance of FMR1 alleles with >54 CGG/CCG repeats in their 5′ UTR. The repeats expand both somatically and on intergenerational transmission and increased repeat numbers are associated with increased risk of disease and increased risk of further expansion. The mechanism responsible for expansion is unknown. Here, we show in a knockin mouse model of these disorders that somatic expansion is much less common in females than in males. We show that this is due in large part to the fact that expansions occur only when the repeat is on the active X chromosome. However, even when this is taken into account, expansions in females are still less common than expected. This additional gender effect is not due to a protective effect of estrogen, a deleterious effect of testosterone or to differences in the expression of the Fmr1 gene or a variety of X-linked and autosomal DNA repair genes. However, our data do suggest that a higher level of expression of genes that protect against oxidative damage in females may contribute to their lower levels of expansion. Whatever the basis, our data suggest that the risk for somatic expansion may be lower in women than it is in men. This could help explain the reduced penetrance of some aspects of disease pathology in women. The fact that expansion only occurs when the Fmr1 allele is on the active X chromosome has important implications for the mechanism of repeat expansion.
INTRODUCTION
The Fragile X-related disorders (FXDs) result from the inheritance of FMR1 alleles that have a CGG/CCG-repeat tract in the 5′ UTR that has >54 repeats (reviewed in 1). This repeat tract is expansion-prone with the risk of expansion increasing with increasing repeat number. Disease pathology is related to repeat number with premutation (PM) alleles, those alleles having 55–200 repeats, being associated with an increased risk of a neurodegenerative condition, Fragile X-associated tremor and ataxia syndrome (FXTAS; MIM #300623) (2). Women with the PM are also at risk of a form of ovarian dysfunction known as Fragile X-associated primary ovarian insufficiency (FXPOI) (3,4). In addition, female PM carriers are at risk of transmitting much larger, so-called full mutation (FM) alleles with >200 repeats, to their children. FM alleles cause Fragile X syndrome (FXS; MIM #300624), a moderate to severe learning disability (reviewed in 1).
The FXDs belong to a larger group of human diseases known as the repeat expansion diseases (5). These diseases, which include progressive myoclonus epilepsy (EPM1; MIM #254800), myotonic dystrophy type 1 (DM1; MIM #160900) and 2 (DM2; MIM #602668), and Huntington disease (HD; MIM #143100), involve a variety of different repeats that are predominantly G+C-rich and have repeat units that range in size from triplets to dodecamers. These repeats all share the ability to form unusual intrastrand and interstrand structures of one sort or another (6), but whether they share a common expansion mechanism is still an open question.
We have previously shown in a knockin mouse model of the FX PM that expansion occurs both in the germ line and in somatic cells and that some cell types are particularly expansion prone including some that are post-mitotic like neurons and oocytes (7). This would be consistent with the idea that replication is not absolutely required for expansion. We have also demonstrated that oxidative damage increases expansion risk (8) and that the mismatch repair protein MSH2 is absolutely required for expansion in this mouse model (9). Furthermore, we have identified a second genetic factor that contributes to repeat instability, CSB, a protein essential for transcription coupled repair (TCR). The loss of CSB reduces the intergenerational transmission of expanded alleles in older female mice and the extent of somatic expansion seen in some organs in males (10). However, very little else is known about the genetic or environmental factors that affect expansion in the FX PM mice. In this article, we show that somatic expansions are much more extensive in males than in females. We show that this is due in part to the fact that expansions are only seen when the PM is on the active X chromosome. However, our data also suggest that there are additional factors that contribute to the increased risk of expansion in males. Our results have implications for the mechanism of repeat expansion. They also raise the possibility that the lower risk of somatic expansion contributes to the lower penetrance of FXTAS seen in women with the PM.
RESULTS
Somatic expansion is less extensive in females than in males
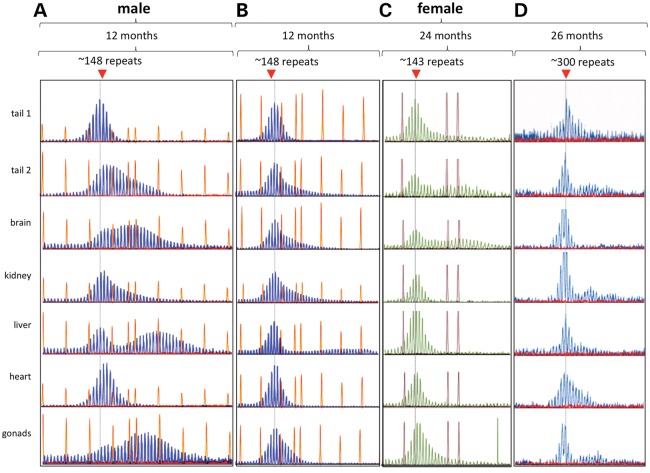
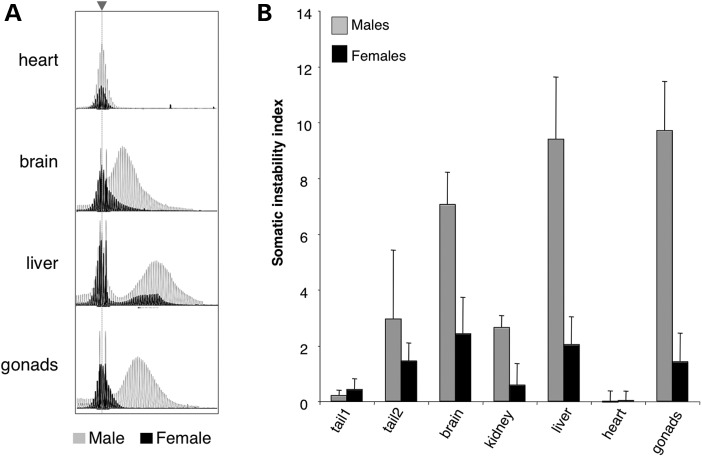
We have previously shown that there is no gender difference in the number of expanded alleles seen in the progeny of in FX PM mice (11). However, somatic expansion in adult FX PM males is always more extensive than it is in females (Fig. 1), although the tissues that are at most risk for expansion remain the same. For example, the most expansion-prone organs in both males and females are the liver, brain and gonads, while expansions are rarely seen in the heart. Despite the fact that expansion increases with age in males (7,10), very little expansion is seen even in females that are almost twice as old (24 versus 12 months) or that have more than twice as many repeats (300 repeats versus 148 repeats; Fig. 1, right-hand panels). These differences can be clearly seen when the GeneMapper traces from males and females are superimposed as shown in Figure 2A. As can be seen from the average somatic instability index (SII), a quantitative measure of the extent of somatic instability (12), from eight males and eight females, this gender difference ranges from 2-fold in tail DNA to ∼4.7-fold in the kidney and liver (Fig. 2B).
Figure 1.
Somatic instability is more extensive in male FX PM mice than in females. Comparison of the GeneMapper profiles for different organs of a 12-month-old male mouse (A) with the profiles for different organs from an age-matched female with a similar repeat number (B), a 24-month-old female with a similar repeat number (C) and a 26-month-old female with ∼300 repeats (D). The dotted gray line indicates the position of the original inherited allele. Either 500 ROX™ or the 1200 LIZ® molecular weight markers were used. The choice of marker did not affect the GeneMapper profile. Tail 1 refers to the tail DNA at weaning, while tail 2 is the tail DNA sample taken at the time of euthanasia.
Figure 2.
Comparison of somatic instability and SII for male and female mice 12 months of age. (A) Overlay of representative GeneMapper profiles from the heart, brain, liver and gonads of a 12-month-old male (gray) and female (black) mouse with ∼146 repeats. The dotted gray line and arrow head indicates the position of the original inherited allele. (B) The SII in different organs of 12-month-old males (n = 8) and females (n = 8) with ∼145 repeats. Tail 1 refers to the tail DNA at weaning, while tail 2 is the tail DNA sample taken at 12 months of age.
Expansion only occurs when the PM allele is on the active X chromosome
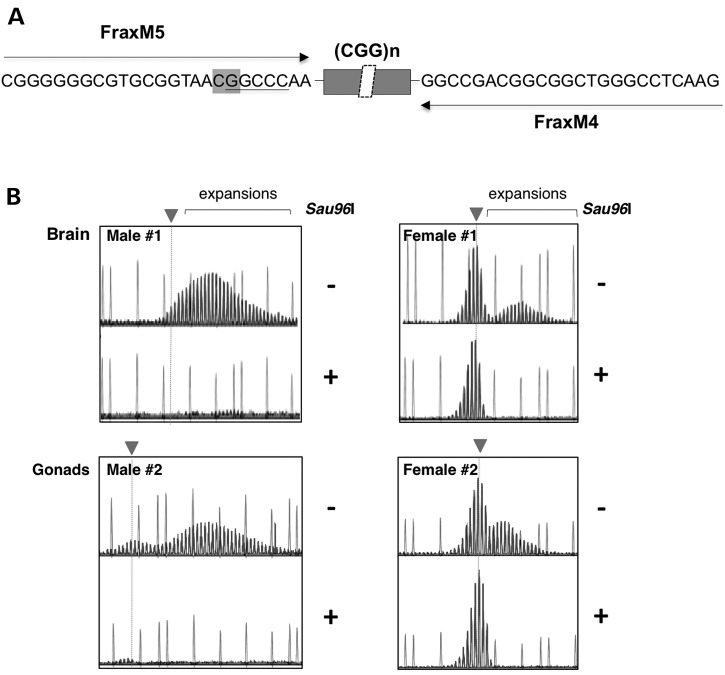
Since the Fmr1 gene is on the X chromosome, it follows that in females heterozygous for the PM, the PM allele will be on the inactive X chromosome in ∼50% of cells. To ascertain whether there are differences between the expansion profiles of the PM on the active and inactive X chromosome, we examined the GeneMapper profile generated by PCR of the repeat from female DNAs that had either been mock digested or digested with Sau96I, a methylation-sensitive restriction enzyme that has a recognition site in the binding site for one of the PM-specific primers used for the PCR. The GeneMapper profile in the mock digested sample would be a composite of the repeats present on both the active (unmethylated) and inactive (methylated) X chromosomes, while the GeneMapper profile generated after Sau96I digestion would reflect only the repeats present on the inactive X chromosomes. As can be seen in the left-hand panels of Figure 3, digestion with Sau96I abolishes all trace of the PM allele in male mice, consistent with the fact that in males no X inactivation and thus no methylation of the PM allele occurs. In contrast, the PCR profile for all females tested showed significant amounts of Sau96I-resistant DNA as would be expected if the PM allele was sometimes located on the inactive X chromosome. However, the GeneMapper profiles differed for the mock digested and Sau96I-digested material. Specifically, while the mock digested sample contained PCR products that were larger than the original allele and thus diagnostic of expansions, these products were missing from the Sau96I-digested sample. Similar results were seen when a different methylation-sensitive enzyme that has a recognition site that overlaps one of the primers was used (EaeI, data not shown). Thus, whatever somatic expansion occurs in the female PM mouse occurs on the active X chromosome.
Figure 3.
Relationship between expansion and presence of the PM on the active X chromosome. (A) Schematic showing the region of the FX PM mouse Fmr1 gene amplified with FraxM4 and FraxM5, illustrating the principle behind using Sau96I-predigestion to examine expansion on the inactive X chromosome. FraxM4 and FraxM5 amplify the PM allele but not the WT allele since the 3′ ends of each primer correspond to the bases added to the PM allele when the mouse line was first generated. The sequence corresponding to the primer in the case of FraxM5 and the primer binding site for FraxM4 is shown. The position of a CpG residue that overlaps with a Sau96I cleavage site in the FraxM5 sequence is indicated in the boxed region. Since Sau96I cleavage is blocked by overlapping methylation, the FraxM5 region will be cut by Sau96I only when the PM allele is on the active X chromosome. As a result when the DNA is digested with Sau96I prior to PCR, the FraxM5 primer will only be able to amplify the PM on the inactive X chromosome. Note that a Sau96I cleavage site is also present in the region corresponding to the FraxM4 sequence. However, this site does not overlap a CpG residue and thus is cleaved whether the allele is methylated or not but since the cleavage site leaves 18 bases adjacent to the repeat for the primer to bind, this primer amplifies the repeat in Sau96I treated material whether the allele is methylated or not. Since males only possess an active X chromosome, Sau96I digestion eliminates all traces of the PM allele. In females, only the PM allele on the inactive X chromosome remains to be amplified after digestion by Sau96I. (B) GeneMapper profiles of two males and two females without (−) and with (+) pretreatment with Sau96I along with the 1200 LIZ® molecular weight markers. The dotted gray line and arrowhead indicates the position of the original inherited allele.
X inactivation does not account completely for the difference in the extent of expansion
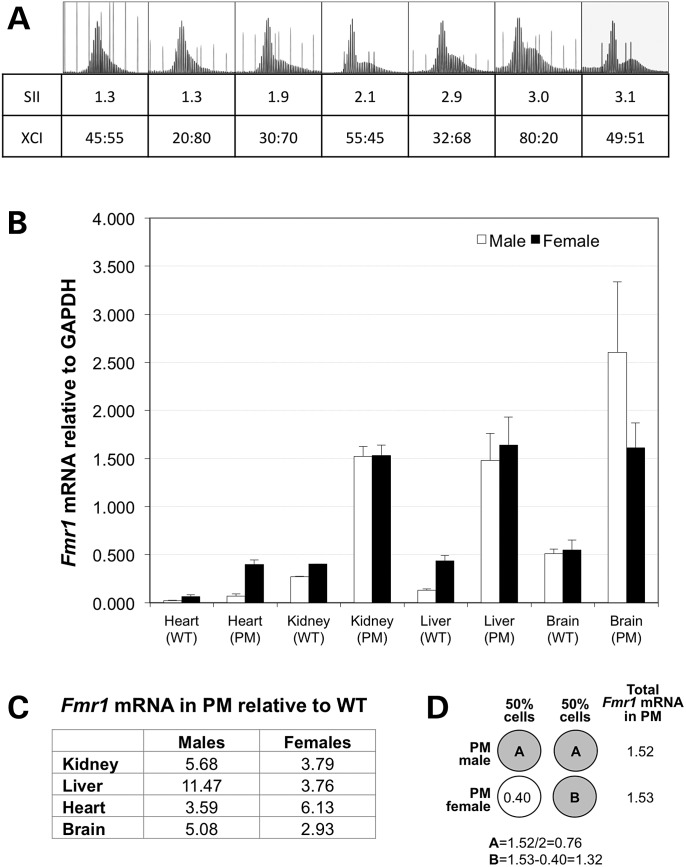
However, the greater than 2-fold differences in the SII cannot be completely explained by this fact unless there was significant skewing of X chromosome inactivation (XCI). Since amplification of the repeat is very sensitive to repeat length and DNA concentration (Lokanga, Zhao and Usdin, unpublished observations), we designed a different assay to monitor XCI that utilized the cleavage site for the methylation-sensitive restriction enzyme EaeI that is located in the neomycin marker in intron 1 that was part of the construct used to generate the FX PM mouse line (13). Details of this assay are described in Materials and Methods section. Using this assay, we found no evidence of skewing of XCI (data not shown). Rather, a range of XCIs were obtained for individual animals that were consistent with the observation that at the time of X inactivation only a small number of cells are present in the embryo (14) with the PM allele being on the active X chromosome on average 50% of the time (data not shown). If the SII was simply due to the proportion of cells in which the PM allele was on the active X chromosome, the extent of expansion in females would be on average ∼50% that seen in males, whereas in some tissue the reduction in SII in females was much lower than this. For example, the average SII in male kidney was ∼4.7 times that in females. Furthermore, there was no clear relationship between whether the PM allele was on the active allele and the SII (Fig. 4). For example, one of the mice with the lowest SII (1.3) and the mouse with the highest SII (3.1) had very similar XCIs with the PM being on the inactive X ∼50% of the time. The fact that expansion in organs like kidney and liver is ∼4.7-fold more extensive in males suggest that X inactivation does not fully account for the gender difference in expansion in all tissue.
Figure 4.
SII, XCI ratios and Fmr1 transcription. (A) The GeneMapper profiles, SII and XCI for the brains of seven age-matched females with similar repeat sizes run with either the 500 ROX™ or 1200 LIZ® molecular weight markers. The XCI ratio is shown as the fraction of the PM allele on the active X versus the fraction of the PM on the inactive X. (B) Comparison of Fmr1 mRNA expression in various organs of male and female mice with and without the PM allele. The Fmr1 mRNA levels were determined by quantitative real-time PCR and are expressed relative to GAPDH. The standard deviation for each group is shown. (C) The Fmr1 mRNA levels in PM mice relative to the levels of Fmr1 mRNA in WT mice. The Fmr1 levels in different organs of male and female mice were normalized to GAPDH and then expressed as the ratio of the levels in PM to the levels in WT mice. (D) Illustration of how the amount of mRNA produced from the PM allele was deduced in female kidney by using the level of mRNA seen in WT mice. The open circle represents the WT allele and the grey circles the PM alleles.
Gender differences in transcription of the Fmr1 gene do not account for differences in expansion
To assess whether gender differences in transcription of the Fmr1 gene could contribute to the gender difference in somatic expansion, we compared the levels of Fmr1 mRNA in different organs (Fig. 4B). For both males and females, the levels of transcription were much higher in PM mice than in WT mice consistent with the increase levels of transcription initiation seen in human PM carriers (15). In males, the Fmr1 transcript levels ranged from 3.6-fold higher in PM kidney to 11.5-fold higher in the PM liver (Fig. 4C). In general, the difference in mRNA levels between PM and WT mice was smaller in females than it was in males. This likely reflects, at least in part, the fact that in PM males the more transcriptionally active PM allele is expressed in all their cells, while in females heterozygous for the PM, the total level of Fmr1 transcript reflects contributions from both the hyper-expressed PM allele and the WT allele that is expressed at much lower levels. For this reason, when evaluating potential gender differences in transcription neither the absolute levels of transcript in the PM mice or the levels relative to WT can be directly compared. Thus while the total amount of Fmr1 transcript in males and females are comparable for organs like kidney and liver, the level of transcript that can be attributed to the PM allele in females must be higher, since the hyper-expressed PM allele is only expressed in 50% of their cells as illustrated in Figure 4D. Thus, if anything, females make more Fmr1 transcript from the PM allele in those organs not less. Furthermore, the total amount of transcript in the heart of females is approximately six times higher than in males. Yet the levels of somatic expansion in that tissue are not higher than they are in males. This is consistent with our earlier observation that Fmr1 transcription in different organs of males does not account for organ-specific differences in the propensity to undergo somatic expansion (10). Thus, while chromatin that is capable of being transcribed is required, the absolute level of transcription does not determine the extent of somatic expansion.
The remaining gender difference cannot be explained by the presence of estrogen or testosterone
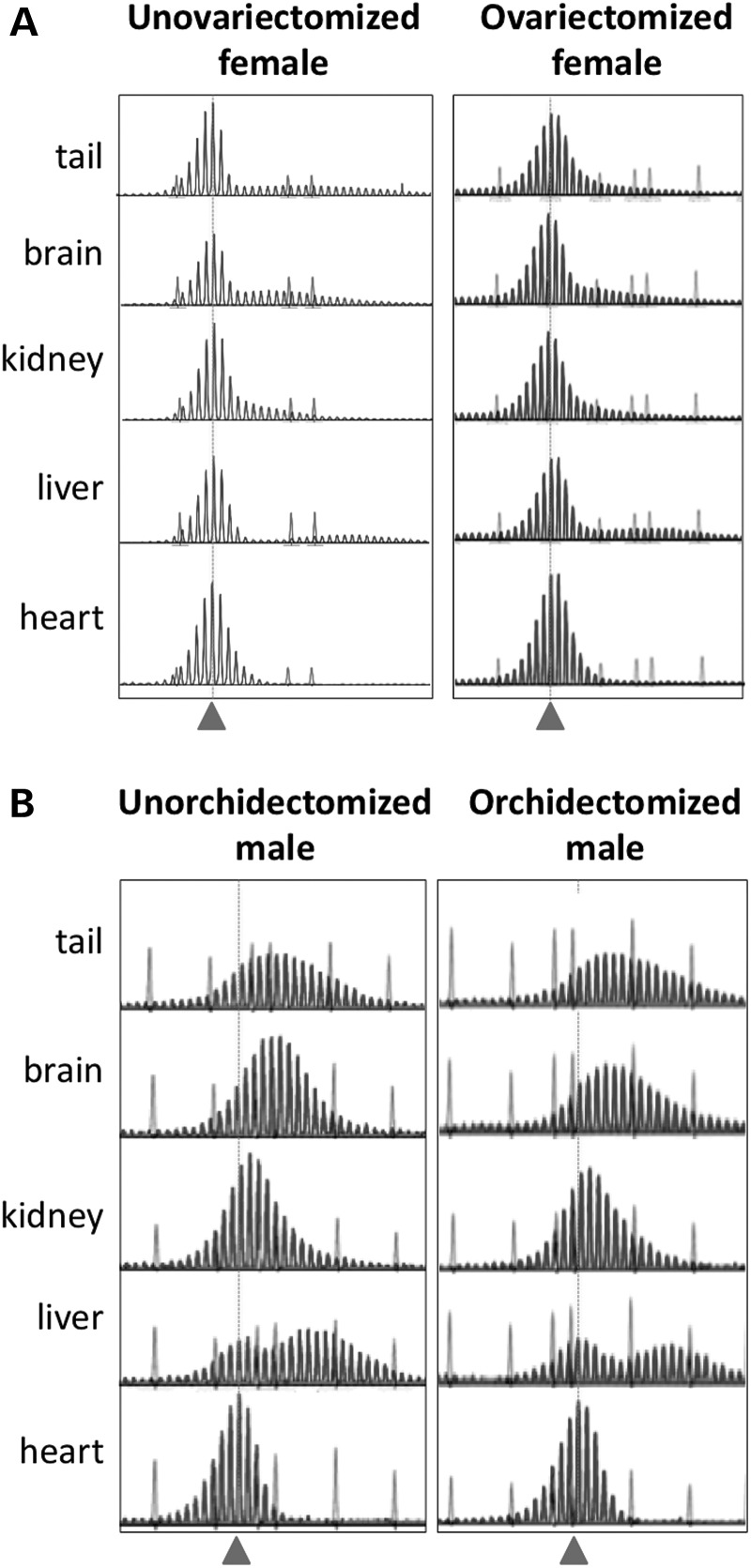
To test whether a protective effect of estrogen could contribute to the gender difference in expansion, we ovariectomized 16-day-old females and examined the extent of expansion in various organs of those animals at 12 months of age. No difference was seen between ovariectomized and non-ovariectomized mice in the extent of expansion (Fig. 5A). To test whether testosterone exacerbated expansion, we examined the extent of expansion in castrated males. No difference was seen between these males and uncastrated males in the extent of expansion either (Fig. 5B). Thus, expression of archetypal male and female hormones does not explain the gender difference in the extent of expansion.
Figure 5.
Effect of removal of female and male gonads on the extent of somatic expansion. The GeneMapper profiles for 12-month-old male and female mice with repeat numbers of ∼145 who were either untreated or had their gonads removed at 7 weeks of age. Samples were run with either a 500 ROX™ (unovariectomized female) or 1200 LIZ® molecular weight marker. The dotted gray line and arrowhead indicate the position of the original inherited allele.
Gender differences in the expression of a number of candidate DNA repair proteins also do not account for the remaining gender differences
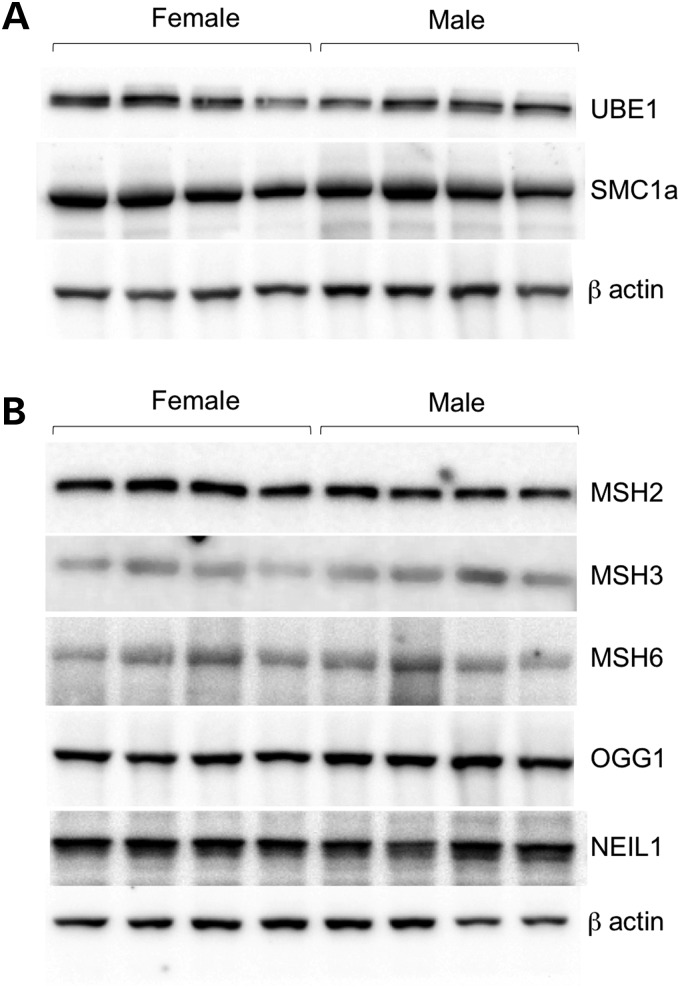
A number of genes involved in DNA repair are located on the X chromosome. These genes, which include the Ubiquitin-activating enzyme 1 (Ube1; Uba1; Smax), the Structural maintenance of chromosomes protein 1A (Smc1a) and the alpha thalassemia/mental retardation syndrome X-linked (Atrx) genes, are potentially good candidates for the source of gender difference. However, neither UBE1 nor SMC1a showed significant differences in protein expression in the livers of male and female mice by western blotting (Fig. 6A). ATRX levels were below the limit of detection in liver. However, we also found no gender difference in the expression of this protein in the brain (data not shown). We also tested the levels of proteins thought to be important in the expansion process. Specifically, we have previously shown that the mismatch repair protein MSH2 is essential for expansion in the FX PM mouse and that the extent of somatic expansion in different organs in males correlated with the levels of the mismatch repair proteins MSH2 and its binding partners MSH3 and MSH6 (9). While their roles in CGG-repeat expansion have not been tested, the DNA glycosylases OGG1 and NEIL1 have been implicated in expansion in mouse models of a repeat expansion disease resulting from CAG/CTG-repeat expansion (16,17). The levels of these proteins were thus also tested. However, none of these proteins showed any gender differences that could explain the difference in expansion either (Fig. 6B).
Figure 6.
Western blots showing the levels of various DNA repair proteins in male and female FX PM mice. Protein isolated from the livers of 12-month-old male and female FX PM mice was subjected to electrophoresis, transferred to membrane and challenged with antibodies to the indicated proteins as described in Materials and Methods. (A) Levels of X-linked DNA repair genes. (B) Levels of DNA repair genes implicated in the repeat expansion diseases.
Gender differences exist in the expression of genes involved in the response to oxidative damage that may contribute to the differences in expansion
We have previously shown that oxidative damage exacerbates germline expansion in both males and females (8). Some studies of gender differences in the extent of oxidative DNA damage in C57BL/6 mice show more oxidative damage in females (18), while others show more oxidative damage in males (19). Furthermore, the PM allele itself has been reported to be responsible for increased oxidative damage (20). It may be that either inherent gender differences in the relative expression of different oxidative damage response proteins or gender differences in the effects of the PM on these genes could account for the gender difference in the extent of somatic expansion. We therefore determined the levels of expression of 84 different genes known to be important in the response to oxidative damage in the liver of male and female PM mice. Of these 84 genes, 29 (34.5%) showed a >1.5-fold difference between genders (P < 0.05) and 14 (16.6%) a >2-fold change (P < 0.05). Table 1 lists the genes with the most significant gender differences in expression in the liver of PM animals. Glutathione S-transferase Pi 1 (Gstp1) expression was 5.3-fold higher in males than in females (P = 0.000546) consistent with previous reports (21). Conversely, Flavin containing monooxygenase 2 (Fmo2) mRNA levels were ∼11 times higher in females than in males [again consistent with previous reports (21)]. NAD(P)H dehydrogenase, Quinone 1 (Nqo1) mRNA levels were also elevated in females consistent with previous reports (22). However, while expression of Superoxide dismutase 1 (Sod1) and glutathione peroxidase 1 (Gpx1) is lower in female WT C57BL/6 mice than in males (18), expression of both genes is significantly higher in female C57BL/6 mice carrying the PM than in males with the PM. This may reflect a specific response of female mice to the presence of the PM. While transcript levels do not always reflect the protein level and/or activity, evidence suggests that many genes involved in the response to oxidative stress are regulated at the level of transcription (23,24). While some of the genes whose expression is upregulated in PM females can be induced by oxidative stress and thus may reflect exposure to oxidative damage, many other genes that are also upregulated by oxidative stress are not elevated. Furthermore, the expression of many of the genes expressed more highly in females are upregulated in males in response to caloric restriction (25), a treatment believed to reduce oxidative stress (reviewed in 26). These data lend support to the idea that the elevated levels of many of these genes in female PM mice may represent a higher basal level of protection against oxidative damage.
Table 1.
Gene expression differences in the liver between male and female PM mice
| Gene | Fold changea | P-value |
|---|---|---|
| Gstp1 | 5.33 | 0.0005 |
| Nox4 | 4.64 | 0.0012 |
| Aox1 | 2.35 | 0.0001 |
| Sod1 | −1.51 | 0.0021 |
| Slc38a1 | −1.53 | 0.0420 |
| Ctsb | −1.53 | 0.0154 |
| Xpa | −1.53 | 0.0058 |
| Ercc2 | −1.56 | 0.0031 |
| Hmox1 | −1.62 | 0.0150 |
| Cygb | −1.70 | 0.0059 |
| Noxo1 | −1.71 | 0.0484 |
| Vim | −1.76 | 0.0097 |
| Ccs | −1.83 | 0.0000 |
| Gpx3 | −1.83 | 0.0022 |
| Cyba | −1.86 | 0.0211 |
| Sod3 | −1.89 | 0.0038 |
| Ncf2 | −1.98 | 0.0171 |
| Noxa1 | −1.98 | 0.0263 |
| Ngb | −2.12 | 0.0299 |
| Ptgs1 | −2.24 | 0.0041 |
| Fancc | −2.36 | 0.0004 |
| Ucp2 | −2.38 | 0.0021 |
| Gpx6 | −2.46 | 0.0314 |
| Gpx7 | −2.51 | 0.0193 |
| Ncf1 | −2.54 | 0.0238 |
| Nqo1 | −2.59 | 0.0477 |
| Nos2 | −2.86 | 0.0457 |
| Ccl5 | −2.87 | 0.0060 |
| Fmo2 | −11.36 | 0.0028 |
aA positive value indicates a higher transcript level in males relative to females and a negative value a higher level of transcript in females.
DISCUSSION
We show here that while the same organs are prone to somatic expansion in male and female FX PM mice, females show much less expansion in those organs than males of the same age and with the same repeat number (compare panels A and B in Fig. 1). While expansions continue to increase with age even females that are twice as old do not show the same extent of expansion as younger males (Fig. 1 A and C). Furthermore, while the extent of somatic expansion is dependent on repeat number, even females with much larger repeat numbers show very little expansion (Fig. 1D).
This gender difference is quite different from the gender effects reported in a mouse model for HD, a CAG/CTG-repeat expansion disease. Specifically, in that mouse model, no significant effect of gender is seen in the extent of somatic expansion in adult HD mice, although male embryos show more expansions than female embryos from the same father (17).
We have shown that a significant fraction of the gender difference in the FX PM model can be attributed to the fact that the Fmr1 gene is located on the X chromosome; in female, FX PM mice somatic expansion occurs exclusively on PM alleles that are on the active X chromosome (Fig. 3), and presumably an open chromatin configuration and/or transcriptional activity of the gene is required for expansion. This is consistent with the observation that the repeat is stable in human carriers of methylated FM alleles (27). Since both X chromosomes are active in oocytes, this may explain why the frequency of germ-line expansions on maternal transmission of PM alleles is comparable to those seen on paternal transmission.
A role for transcription/open chromatin in the CTG/CAG-repeat expansion diseases has been previously suggested from work using various bacterial/yeast/tissue culture/fly models (reviewed in 28). Recently, the effect of unidirectional and bidirectional transcription was examined in a human fibroblast model system using an integrated construct expressing 800 repeats under the control of CMV and ROSA26 promoters situated on either side of the repeat (29). In this assay system constructs in which the repeat was transcribed showed higher numbers of both expansions and contractions than cell lines in which the repeat was not transcribed and this effect was magnified when transcription of the repeat occurred bidirectionally. In transgenic mouse models of DM1 and HD, a role for transcription/open chromatin was inferred from the fact that in various mouse lines more extensive somatic instability in a particular tissue was generally correlated with higher levels of transcription of the transgene (30,31). However, in both the fibroblast system and these mouse lines, the chromosomal context of the constructs used was different in each case. Since chromosomal context can affect expansion in a variety of ways, it is difficult to definitively attribute the differences observed solely to the transcriptional activity of the locus particularly since there is not always a good correlation between transcription rates in specific tissue and the propensity to expand in that tissue (29–31). Since in the FX PM mouse, we are able to compare the expansion of the identical PM allele in the same chromosome context on active and inactive versions of the X chromosome in the same animal, our data firmly establish a role for transcription/open chromatin in expansion.
The molecular basis for this dependence on transcription/open chromatin is not yet clear. Since heterochromatin is less vulnerable to certain forms of DNA damage and also less accessible to some repair enzymes (32), it may be that heterochromatin formation itself accounts for the stability of the repeat on the inactive X. Alternatively, it may be transcription that triggers expansion. It has been suggested from work in models of the CAG/CTG-repeat expansion diseases (33–38) that expansion proceeds via TCR, a form of DNA repair that occurs only on the template strand of actively transcribed genes (39). However, we have previously have shown that CSB, a protein essential for TCR, plays no role in expansion in male or young female FX PM mice (10).
There are a number of other transcription-dependent processes that have the potential to generate expansions (reviewed in 28,40). For example, co-transcriptionally formed R-loops that form in G-rich regions have the potential to generate DNA strand breaks by causing replication fork stalling that could ultimately lead to expansion (41,42). However, since expansion occurs in oocytes and neurons (7,9), cells that are not replicating, it suggests that replication fork stalling is not required for expansion. The single-stranded region generated on the non-template strand during transcription may also be vulnerable to strand breaks and other forms of DNA damage. Transcription through this region may also favor expansion since it would increase the levels of H3K36me3, a histone modification that recruits MSH2-containing DNA repair complexes (43) that we have shown to be essential for expansion (9). However, the simplest model that would account for the role of transcription in repeat expansion is that transcription creates an opportunity for the non-template strand of the repeat to form the hairpins that are thought to be the substrate for expansion. Once formed, the hairpins would then be recognized by the MSH2-dependent process that generates expansions (9). The recent demonstration that the related repeats (CAG)n and (CTG)n can trigger the activation of the MutLα endonuclease to generate an incision suggests one way that the MMR machinery could process the hairpins on templates outside of DNA replication (44).
However, while transcription/open chromatin is required for expansion, it is not sufficient; expansion is not seen in all tissue and there is not a simple relationship between Fmr1 transcription and extent of expansion in these tissues. Furthermore, there are no significant gender differences in the transcription of the Fmr1 gene that could account for the gender difference in somatic expansions that we observed (Fig. 4). We have also previously shown that there is not a straightforward relationship between the amount of transcription of the Fmr1 gene in males and the extent of expansion in different organs either (7,9,10). Thus it may be that some other factor in the expansion process is rate-limiting and that these factors may show gender differences.
The other factors contributing to the gender difference in the risk of expansion remain to be ascertained. It does not seem to be related to levels of either estrogen or testosterone or to be related to the expression of any of the known DNA repair genes on the X chromosome or DNA repair genes on autosomes that have been implicated in expansions. Given our demonstration that oxidative stress can increase expansion risk, the gender differences in the expression of a number of other genes involved in the response to oxidative damage in FX PM mice may be relevant. Females showed significantly higher levels of expression of a number of genes able to remove potentially damaging reactive oxygen species (ROS) (Table 1). Of the 29 genes that showed a >1.5-fold difference in expression between males and females, 26 are more highly expressed in females and most of these are involved with protection from oxidative stress rather than the generation of ROS. The list of genes that are more highly expressed in FX PM females includes genes belonging to the Sod and Gpx families that are among the most important genes involved in protection against oxidative damage. The higher level of expression of enzymes like this that reduce ROS levels may result in less oxidative damage to DNA, which in turn results in fewer expansion events in females. A number of genes involved in DNA damage repair are also more highly expressed in female PM mice, including Fancc, Ercc2 and Xpa. Thus it could also be that the elevated level of expression of these genes also allows the error-free repair of any DNA damage that may otherwise give rise to expansions. Given that oxidative stress is elevated in human PM carriers (20), it may be that the elevated level of expression of genes that protect against this stress may contribute to the lower penetrance of FXTAS in human female PM carriers.
Whatever the basis of the gender difference in the extent of somatic expansion may be, the fact that somatic expansions occur less frequently in female PM mice may be relevant clinically for women carrying a PM allele. A lower frequency of somatic expansions would mean that there would be greater concordance between repeat numbers measured in blood and repeat numbers in the brain. This would mean that FXTAS risk assessment may be simpler for females than for males, at least for those with high repeat numbers who would otherwise be at risk of somatic expansion.
MATERIALS AND METHODS
Mouse maintenance and surgery
The generation of the FX PM mice was described previously (13). Mice were maintained in accordance with the guidelines of the NIDDK Animal Care and Use Committee and with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85-23, revised 1996). Ovariectomy was performed via a dorsal midline incision. Briefly, 16-day-old females were anesthetized by intra-peritoneal injection of 0.5 ml of Avertin. After the onset of anesthesia, the animals were placed under infrared lamp to prevent heat loss, and then the fur was shaved bilaterally over the lumbar spine to expose the skin. After swabbing the shaved skin with 70% (v/v) ethanol followed by sterile PBS, a 5–8 mm incision was made in the midline of the lower back, in the subcutaneous tissue and peritoneum. Ovaries were removed by gently severing the oviduct, using sterile, small scissors. The uterus and the remaining part of the oviduct are replaced back into the abdominal cavity and the muscle layer sutured using 5-0 vicryl absorbable suture. The testes of 5–7-week-old male mice were removed via a ventral midline incision. Briefly, the mice were anesthetized by IP Avertin injection, the ventral surface area shaved and cleaned with Betadine surgical scrub followed by a 70% alcohol wipe, to remove loose hair. A 5–8 mm incision was made in the ventral midline at the point level with the top of legs, followed by a 5–8 mm incision in the subcutaneous tissue and peritoneum. The vas deferens and testis were exposed, the vas deferens cauterized and the testes excised. The fat pads were pushed back into the cavity and the muscular layer closed with 5-0 vicryl absorbable suture. The skin incision was closed with sterile wound clips, which are removed after 7–10 days.
Genotyping and analysis of repeat number
Genomic DNA was extracted using a Maxwell®16 Mouse tail DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. The presence of the PM allele and its repeat number was determined using a fluorescent PCR assay and FraxM4 and FraxM5 primer pair as described previously (7). The SII was calculated from the GeneMapper profiles of DNA from different organs as previously described and used to evaluate the extent of somatic expansion in adult mice.
A modification of the fluorescent PCR assay used to determine the repeat number was used to examine the relationship between expansion and the location of the PM allele on the active or inactive X chromosome. The FraxM5 primer binding site has a recognition site for the methylation-sensitive enzyme Sau96I that contains a CpG residue. Digestion with Sau96I thus prevents PCR amplification of the PM when it is on the active X chromosome. Briefly, 1 µg of genomic DNA was digested in New England Biolabs buffer 4 (New England BioLabs, Ipswich, MA, USA) with 10U of Sau96I in a total volume of 50 µl. For each sample, a mock digested control was prepared the same way except for the omission of enzyme. The extent of digestion was monitored by the addition of pUC19, a plasmid that has six Sau96I restriction sites. The digests were then monitored by agarose gel electrophoresis to ensure that the pUC19 digestion was complete.
Determination of the X inactivation ratio
Genomic DNA was mock digested or digested with the methylation-sensitive enzyme EaeI (New England Biolabs) prior to real-time qPCR. A methylation-sensitive amplicon was amplified with primers XCI-TF (5′-TGGACGAAGAGCATCAGGGG-3′) and XCI-TR (5′-GCGATACCGTAAAGCACGAG-3′). This amplicon corresponds to a region of the neomycin resistance marker from the targeting vector used to generate the mouse line. It contains two EaeI sites with CpG residues that will be digested only if the template is on the active X chromosome. A PCR control for EaeI digestion was included. This PCR reaction was carried out using primers XCI-CF (5′-CGGTTCTTTTTGTCAAGACCGA-3′) and XCI-CR (5′-CAGGAGCAAGGTGAGATGACA-3′) that amplifies a 198 bp region just upstream of the first amplicon. This region that contains an EaeI site that contains no CpG and is thus digested whether the region is methylated/on the inactive X or not. The ratio of the yield of the EaeI digested methylation-sensitive amplicon to the yield of the same amount of mock digested material was used to determine the X inactivation ratio.
Evaluation of potential gender differences in protein expression by western blotting
Protein extracts were prepared from flash frozen tissues of 12-month-old females (four) and males (four) that had ∼140 repeats. Tissues were homogenized using a tissue homogenizer (Precellys® 24,Bertin Technologies) with T-PER protein extraction reagent (Pierce Biotechnology, Inc, Rockford, IL, USA) supplemented with complete, Mini, EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA) and phosphatase inhibitor cocktail-3 (Sigma-Aldrich) according to the supplier's instructions. The protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). Proteins were heated for 10 min at 70°C in LDS-Sample Buffer (Life Technologies) supplemented with 50 mm DTT, resolved by electrophoresis on a 3–8% NuPAGE Novex Tris-Acetate gels (Life Technologies) and electro-blotted onto nitrocellulose membranes using NuPAGE Transfer Buffer (Life Technologies) supplemented with 10% methanol. Transfer was carried out at 100 V at room temperature for 1 h. Membranes were blocked for 1 h at room temperature in 5% ECL Prime blocking agent (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) in TBST pH 7.5 (10 mm Tris–HCl, 0.15 mm NaCl and 0.01% Tween 20), and then incubated overnight at 4°C with antibodies to the following proteins ATRX (Ab97508), MSH2 (ab70270) and MSH3 (ab74607), NEIL1 (ab21337) and SMC1 (Ab52324) from Abcam (Cambridge, MA, USA), MSH6 (3995S) and Ube1a/b (#4891) from Cell Signaling (Boston, MA, USA) and OGG1 (15125-1-AP) from Proteintech Group (Chicago, IL, USA). After addition of the ECL Prime detection reagent (GE Healthcare Bio-Sciences), the blot was imaged using a Fluorchem™ M imaging system (Proteinsimple, Santa Clara, CA, USA). The protein levels were then normalized to the levels of β-actin (ab8227; Abcam).
Evaluation of gender differences in the expression of genes involved in the response to oxidative stress
The livers of 12-month-old males (four) and females (four) with PM alleles that had ∼145 repeats were homogenized using a Precellys® lysing kit (Bertin Technologies) according to the manufacturer’s instructions. Total RNA was isolated from the homogenate using Maxwell®16 LEV simply RNA purification Kits (Promega). The RNA concentration and quality was determined using an Agilent Bioanalyzer (Agilent, Santa Clara, CA, USA). The RNA was converted into cDNA using the RT2 first strand kit (Qiagen, Valencia, CA, USA) and analyzed using the Mouse Oxidative Stress RT2 Profiler™ PCR array (PAMM-065ZE-4, Qiagen) according to the manufacturer's instructions. The real-time PCR array data were analyzed using a web-base version of RT2 profiler PCR array data analysis version 3.5 (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php).
FUNDING
The work described in this article was funded by a grant from the Intramural Program of the NIDDK to K.U. (DK057808-07).
ACKNOWLEDGEMENTS
The Usdin lab would like to acknowledge all the hard work by the people that take care of the mice used in this study. Without their help, this work would not have been possible.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Chonchaiya W., Schneider A., Hagerman R.J. Fragile X: a family of disorders. Adv. Pediatr. 2009;56:165–186. doi: 10.1016/j.yapd.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagerman P. Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol. 2013;126:1–19. doi: 10.1007/s00401-013-1138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman S.L. Premature ovarian failure in the fragile X syndrome. Am. J. Med. Genet. 2000;97:189–194. doi: 10.1002/1096-8628(200023)97:3<189::AID-AJMG1036>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 4.Murray A., Webb J., Grimley S., Conway G., Jacobs P. Studies of FRAXA and FRAXE in women with premature ovarian failure. J. Med. Genet. 1998;35:637–640. doi: 10.1136/jmg.35.8.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry M., Usdin K. Human Nucleotide Expansion Disorders. Springer, Heidelberg; 2006. [Google Scholar]
- 6.Usdin K. Bending the rules: unusual nucleic acid structures and human disease. In: Wells R.D., Ashizawa T., editors. Genetic Instabilities and Hereditary Neurological Diseases. Elsevier-Academic Press; 2006. [Google Scholar]
- 7.Lokanga R.A., Entezam A., Kumari D., Yudkin D., Qin M., Smith C.B., Usdin K. Somatic expansion in mouse and human carriers of Fragile X premutation alleles. Hum. Mutat. 2013;34:157–166. doi: 10.1002/humu.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Entezam A., Lokanga A.R., Le W., Hoffman G., Usdin K. Potassium bromate, a potent DNA oxidizing agent, exacerbates germline repeat expansion in a fragile X premutation mouse model. Hum. Mutat. 2010;31:611–616. doi: 10.1002/humu.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lokanga R.A., Zhao X.-N., Usdin K. The mismatch repair protein, MSH2, is rate-limiting for repeat expansion in a Fragile X premutation mouse model. Hum. Mutat. 2014;35:129–136. doi: 10.1002/humu.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X.-N., Usdin K. Gender and cell-type specific effects of the transcription coupled repair protein, ERCC6/CSB, on repeat expansion in a mouse model of the Fragile X-related disorders. Hum. Mutat. 2014;35:341–349. doi: 10.1002/humu.22495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Entezam A., Usdin K. ATR protects the genome against CGG.CCG-repeat expansion in Fragile X premutation mice. Nucleic Acids Res. 2008;36:1050–1056. doi: 10.1093/nar/gkm1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.M., Zhang J., Su A.I., Walker J.R., Wiltshire T., Kang K., Dragileva E., Gillis T., Lopez E.T., Boily M.J., et al. A novel approach to investigate tissue-specific trinucleotide repeat instability. BMC Syst. Biol. 2010;4:29. doi: 10.1186/1752-0509-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Entezam A., Biacsi R., Orrison B., Saha T., Hoffman G.E., Grabczyk E., Nussbaum R.L., Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amos-Landgraf J.M., Cottle A., Plenge R.M., Friez M., Schwartz C.E., Longshore J., Willard H.F. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am. J. Hum. Genet. 2006;79:493–499. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tassone F., Beilina A., Carosi C., Albertosi S., Bagni C., Li L., Glover K., Bentley D., Hagerman P.J. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. 2007;13:555–562. doi: 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovtun I.V., Liu Y., Bjoras M., Klungland A., Wilson S.H., McMurray C.T. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mollersen L., Rowe A.D., Illuzzi J.L., Hildrestrand G.A., Gerhold K.J., Tveteras L., Bjolgerud A., Wilson D.M., 3rd, Bjoras M., Klungland A. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum. Mol. Genet. 2012;21:4939–4947. doi: 10.1093/hmg/dds337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali S.S., Xiong C., Lucero J., Behrens M.M., Dugan L.L., Quick K.L. Gender differences in free radical homeostasis during aging: shorter-lived female C57BL6 mice have increased oxidative stress. Aging Cell. 2006;5:565–574. doi: 10.1111/j.1474-9726.2006.00252.x. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton M.L., Van Remmen H., Drake J.A., Yang H., Guo Z.M., Kewitt K., Walter C.A., Richardson A. Does oxidative damage to DNA increase with age? Proc. Natl Acad. Sci. USA. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross-Inta C., Omanska-Klusek A., Wong S., Barrow C., Garcia-Arocena D., Iwahashi C., Berry-Kravis E., Hagerman R.J., Hagerman P.J., Giulivi C. Evidence of mitochondrial dysfunction in fragile X-associated tremor/ataxia syndrome. Biochem. J. 2010;429:545–552. doi: 10.1042/BJ20091960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Z.D., Csanaky I.L., Klaassen C.D. Effects of aging on mRNA profiles for drug-metabolizing enzymes and transporters in livers of male and female mice. Drug Metab. Dispos. 2012;40:1216–1225. doi: 10.1124/dmd.111.044461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y.Q., Zhang D., Jin T., Cai D.J., Wu Q., Lu Y., Liu J., Klaassen C.D. Diurnal variation of hepatic antioxidant gene expression in mice. PLoS ONE. 2012;7:e44237. doi: 10.1371/journal.pone.0044237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreger H., Westphal K., Wilck N., Baumann G., Stangl V., Stangl K., Meiners S. Protection of vascular cells from oxidative stress by proteasome inhibition depends on Nrf2. Cardiovasc. Res. 2010;85:395–403. doi: 10.1093/cvr/cvp279. [DOI] [PubMed] [Google Scholar]
- 24.Kwak M.K., Itoh K., Yamamoto M., Sutter T.R., Kensler T.W. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol. Med. 2001;7:135–145. [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Z.D., Klaassen C.D. Short-term calorie restriction feminizes the mRNA profiles of drug metabolizing enzymes and transporters in livers of mice. Toxicol. Appl. Pharmacol. 2013;274:137–146. doi: 10.1016/j.taap.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pamplona R., Barja G. Mitochondrial oxidative stress, aging and caloric restriction: the protein and methionine connection. Biochim. Biophys. Acta. 2006;1757:496–508. doi: 10.1016/j.bbabio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Glaser D., Wohrle D., Salat U., Vogel W., Steinbach P. Mitotic behavior of expanded CGG repeats studied on cultured cells: further evidence for methylation-mediated triplet repeat stability in fragile X syndrome. Am. J. Med. Genet. 1999;84:226–228. [PubMed] [Google Scholar]
- 28.Lin Y., Hubert L., Jr, Wilson J.H. Transcription destabilizes triplet repeats. Mol. Carcinog. 2009;48:350–361. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamori M., Pearson C.E., Thornton C.A. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats. Hum. Mol. Genet. 2011;20:580–588. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lia A.S., Seznec H., Hofmann-Radvanyi H., Radvanyi F., Duros C., Saquet C., Blanche M., Junien C., Gourdon G. Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum. Mol. Genet. 1998;7:1285–1291. doi: 10.1093/hmg/7.8.1285. [DOI] [PubMed] [Google Scholar]
- 31.Goula A.V., Stys A., Chan J.P., Trottier Y., Festenstein R., Merienne K. Transcription elongation and tissue-specific somatic CAG instability. PLoS Genet. 2012;8:e1003051. doi: 10.1371/journal.pgen.1003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falk M., Lukasova E., Kozubek S. Higher-order chromatin structure in DSB induction, repair and misrepair. Mutat. Res. 2010;704:88–100. doi: 10.1016/j.mrrev.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Salinas-Rios V., Belotserkovskii B.P., Hanawalt P.C. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011;39:7444–7454. doi: 10.1093/nar/gkr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung J., Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 35.Hubert L., Jr, Lin Y., Dion V., Wilson J.H. Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum. Mol. Genet. 2011;20:4822–4830. doi: 10.1093/hmg/ddr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y., Wilson J.H. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y., Wilson J.H. Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair. 2009;8:878–885. doi: 10.1016/j.dnarep.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Y., Dion V., Wilson J.H. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 39.Hanawalt P.C. Preferential DNA repair in expressed genes. Environ. Health Perspect. 1987;76:9–14. doi: 10.1289/ehp.87769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khobta A., Epe B. Interactions between DNA damage, repair, and transcription. Mutat. Res. 2012;736:5–14. doi: 10.1016/j.mrfmmm.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 41.Aguilera A., Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 42.McIvor E.I., Polak U., Napierala M. New insights into repeat instability: role of RNA*DNA hybrids. RNA Biol. 2010;7:551–558. doi: 10.4161/rna.7.5.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F., Mao G., Tong D., Huang J., Gu L., Yang W., Li G.M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pluciennik A., Burdett V., Baitinger C., Iyer R.R., Shi K., Modrich P. Extrahelical (CAG)/(CTG) triplet repeat elements support proliferating cell nuclear antigen loading and MutLalpha endonuclease activation. Proc. Natl Acad. Sci. USA. 2013;110:12277–12282. doi: 10.1073/pnas.1311325110. [DOI] [PMC free article] [PubMed] [Google Scholar]