Abstract

Oxidation of 5-methylcytosine in DNA by ten-eleven translocation (Tet) family of enzymes has been demonstrated to play a significant role in epigenetic regulation in mammals. We found that Tet enzymes also possess the activity of catalyzing the formation of 5-hydroxymethylcytidine (5-hmrC) in RNA in vitro. In addition, the catalytic domains of all three Tet enzymes as well as full-length Tet3 could induce the formation of 5-hmrC in human cells. Moreover, 5-hmrC was present at appreciable levels (∼1 per 5000 5-methylcytidine) in RNA of mammalian cells and tissues. Our results suggest the involvement of this oxidation in RNA biology.
It is known that RNA carries more than 100 distinct types of modifications, and these modifications modulate the structure and functions of RNA. (1) In this vein, it was found that methylation at the N6 of adenine and oxidative demethylation of the resulting N6-methyladenine by two members of the ALKBH family dioxygenases, i.e., FTO and ALKBH5, may be relevant in the epigenetic control of gene regulation.2−5 Aside from N6-methyladenosine, 5-methylcytidine (5-mrC) has long been known to be present in RNA.6 Recent sequencing studies revealed the widespread presence of 5-mrC in both coding and noncoding RNA,7,8 with more than 8000 candidate 5-mrC sites being identified in mRNA, implicating this RNA methylation in gene regulation.7−9
Recent studies showed that the ten-eleven translocation (Tet) family of Fe(II)- and 2-oxoglutarate-dependent dioxygenases in mammals could induce the sequential oxidation of 5-methyl-2′-deoxycytidine (5-mdC) to yield 5-hydroxymethyl-2′-deoxycytidine (5-hmdC), 5-formyl-2′-deoxycytidine (5-fodC), and 5-carboxyl-2′-deoxycytidine (5-cadC).10−15 In this context, it is worth noting that 5-hmdC, instead of dC, is incorporated into genomic DNA of T-eleven bacteriophage from the 5-hmdC triphosphate, and the 5-hmdC in DNA is further glucosylated, which serves as an important mechanism for the bacteriophages to protect their DNA from degradations by host and phage factors.16 In addition, a recent study revealed that cytosine 5-methyltransferases were capable of adding formaldehyde to the C5 position of cytosine to yield 5-hydroxymethylcytosine in DNA.17 In mammalian cells, the oxidized derivatives of 5-mdC may constitute alternative epigenetic marks as they could be recognized by unique cellular proteins.13,18−20 In addition, 5-formylcytosine and 5-carboxylcytosine are readily recognized by thymine DNA glycosylase, and the subsequent action by the base excision repair machinery converts an initially methylated cytosine to its unmethylated counterpart,13,21 which may contribute to active cytosine demethylation in mammals. Aberrant Tet-mediated oxidation of 5-mdC in DNA is known to be associated with human diseases including cancer.22−24 In addition, the genome of Drosophila melanogaster lacks a homologue of the mammalian DNA methyltransferases Dnmt1, Dnmt3a, or Dnmt3b, but it encodes the RNA methyltransferase Dnmt2 and a conserved Tet homologue.25 Interestingly, ALKBH family enzymes, which are another family of Fe(II)- and 2-oxoglutarate-dependent enzymes, can oxidize the N-alkylated nucleobases in both DNA and RNA.26−31 These findings, along with the structural similarity between human Tet2 and ALKBH-family enzymes,32 prompted us to hypothesize that the Tet family enzymes may also be capable of oxidizing the methyl group of 5-mrC in RNA (Figure 1a).
Figure 1.
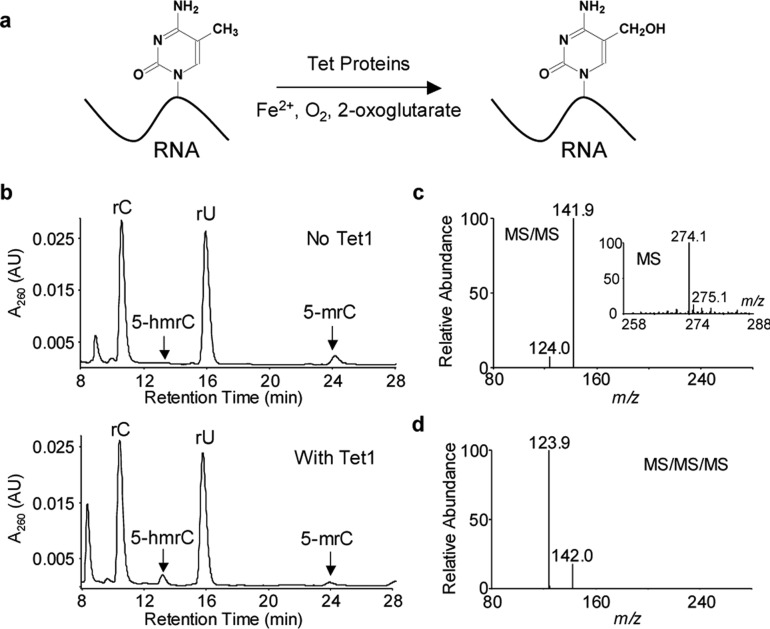
Catalytic domain of Tet1 can catalyze the formation of 5-hmrC from 5-mrC in RNA in vitro. (a) Tet-catalyzed formation of 5-hmrC. (b) HPLC traces for the separation of the nucleoside mixtures of single-stranded RNA, UUUCAGCUC(5-mrC)GGUCACGCUC, without Tet1 treatment and the same RNA after the Tet1-mediated oxidation. The peaks corresponding to 5-hmrC, 5-mrC, and canonical nucleosides are labeled. (c,d) MS/MS and MS/MS/MS characterizations of 5-hmrC, which monitor the fragmentation of the [M + H]+ ion of the 5-hmrC (c) and the further fragmentation of the protonated nucleobase (d), respectively. Displayed in the inset of (c) is the positive ion electrospray ionization mass spectrum for 5-hmrC.
To explore this possibility, we first assessed the capability of recombinant catalytic domain of mouse Tet1 protein in inducing the oxidation of 5-mrC in RNA by conducting an in vitro reaction with the use of a single-stranded RNA carrying a single 5-mrC. HPLC analysis of the nucleoside mixture from the enzymatic digestion of the RNA isolated from the reaction mixture revealed the formation of 5-hydroxymethylcytidine (5-hmrC), which is accompanied by a decrease in the level of 5-mrC (Figure 1b). The identities of the two nucleosides (i.e., 5-hmrC and 5-mrC) were confirmed by mass spectrometric analyses (Figures 1c,d and S1 and Scheme S1). Thus, this biochemical assay demonstrated that Tet1 is able to oxidize 5-mC in single-stranded RNA in vitro.
We next investigated the relative efficiencies of the catalytic domain of Tet1 in oxidizing 5-mrC in RNA and 5-mdC in DNA. To this end, we conducted another in vitro reaction by using a 11-mer RNA sequence, AGCUC(5-mrC)GGUCA, or a duplex DNA with a single 5-mdC situated in the same sequence context. We then subjected the reaction mixtures directly to LC-MS and MS/MS analyses (Figures 2 and S2 and S3). Quantification results based on LC-MS data revealed 5-hmrC as the major product formed when single-stranded RNA was employed as the substrate, though we were able to detect very low level of 5-forC at 40 min (Figures 2 and 3a). It is of note that omitting Fe2+ in the reaction buffer led to a decrease in the formation of 5-hmrC by ∼4-fold, whereas exclusion of 2-oxoglutarate in the reaction buffer nearly abolished the Tet1-catalyzed formation of 5-hmrC (Figure S4), supporting that 5-hmrC arises from the Fe2+- and 2-oxoglutarate-dependent dioxygenase activity of Tet1. For the duplex DNA substrate, we, however, observed a rapid formation of 5-hmdC and then 5-fodC, which is accompanied by the nearly complete loss of 5-mdC. Furthermore, 5-hmdC and 5-fodC were almost completely converted to 5-cadC at later time points (Figures 2 and 3b). This finding is consistent with Tet enzyme’s capability in the sequential oxidation of 5-mdC to 5-hmdC, 5-fodC, and 5-cadC.33,34 These results, therefore, supported that the Tet-mediated oxidation of 5-mrC in RNA is much less efficient than the corresponding oxidation of 5-mdC in duplex DNA. We also found that Tet1 displayed a higher activity toward single-stranded DNA than single-stranded RNA in the same sequence context (Figure S5). Comparison of the extents of oxidation of 5-mdC in single- vs double-stranded DNA showed that the oxidation of 5-mdC is more facile in the latter substrate, which could be attributed to the preferential binding of Tet1 to duplex DNA. Thus, the less efficient oxidation of 5-mrC to 5-hmrC in RNA than the corresponding oxidation of 5-mdC in duplex DNA could be partly due to the less favorable binding of Tet1 to single-stranded RNA. Future structural determination of the Tet protein-RNA complex, along with the known structure of Tet2-DNA complex,32 may provide additional mechanistic insights into the difference in Tet-mediated oxidation of 5-mC in DNA and RNA.
Figure 2.
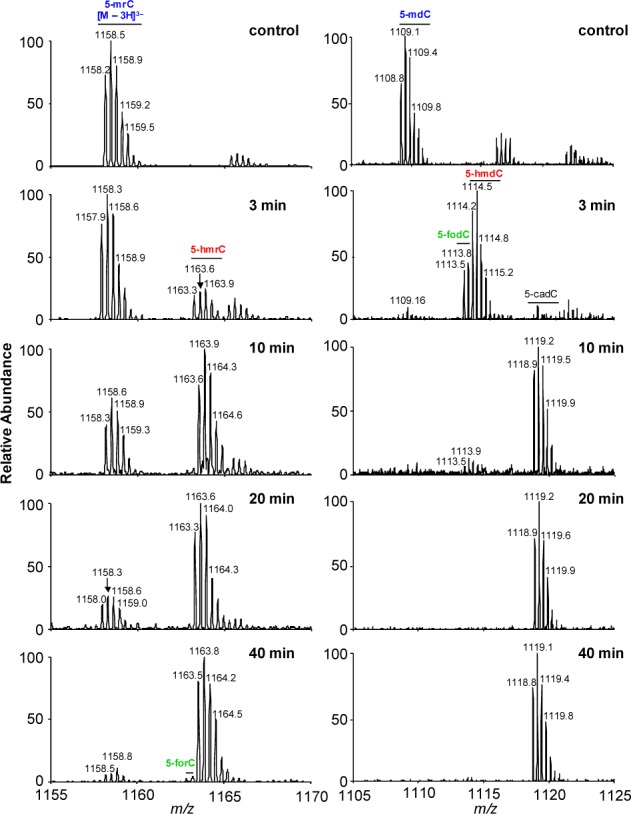
LC-MS for monitoring the Tet1-mediated oxidation of 5-mrC in a single-stranded RNA, AGCUC(5-mrC)GGUCA (left) and a duplex DNA, d(AGCTC(5-mdC)GGTCA) /d(TGACCGGAGCT) (right). Shown are the higher resolution “ultra-zoom-scan” MS results for monitoring the [M – 3H]3– ions of the initial 5-mC-bearing 11mer RNA (left) or DNA (right), together with their oxidation products, where the 5-mC is oxidized to 5-hmC, 5-foC, and 5-caC. The peaks at around m/z 1166 and 1117 for the control samples in the left and right panels are attributed to the Na+ ion adduct, i.e., the [M + Na+ – 4H]3– ions, of the 5-mrC-containing RNA and 5-mdC-bearing DNA strand, respectively.
Figure 3.
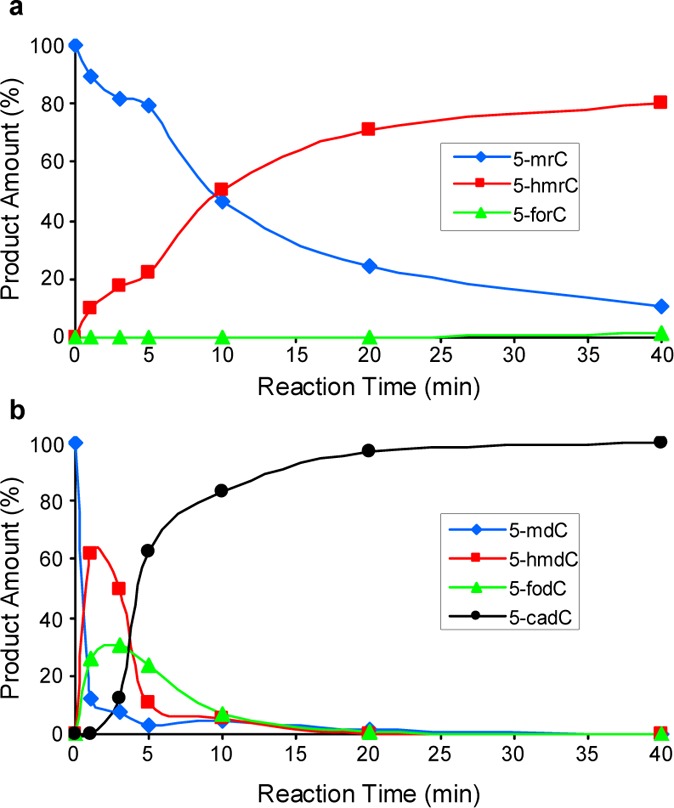
Time-dependent formation of oxidation products of 5-mrC in single-stranded RNA, AGCUC(5-mrC)GGUCA (a) and of 5-mdC in duplex DNA, d(AGCTC(5-mdC)GGTCA) /d(GTGACCGGAGCTG) (b). The products were quantified from LC-MS analyses (Figure 2).
To further assess the function of Tet1 in this oxidation, we overexpressed the catalytic domain of Tet1 (Tet1-CD) or its inactive mutant (Tet1-m) in HEK293T cells,35 isolated total RNA from the cells, digested it to mononucleosides, and quantified the levels of 5-hmrC in the resulting nucleoside mixture by using LC-MS/MS/MS with the isotope dilution method (Figures S6–S8). The coelution of the analyte with the stable isotope-labeled standard at 10.1–10.2 min, together with the similar fragment ions for the analyte and internal standard, allowed for the unambiguous identification of 5-hmrC (Figure S7). Similar as what we described previously for the quantification of 5-hmdC in DNA,35 we monitored the fragmentation of the protonated ion of modified nucleobase (i.e., the ion of m/z 142, which is the major fragment ion found in the MS/MS of the protonated ion of 5-hmrC, Figure 1c) in MS/MS/MS, which displayed the facile loss of a H2O molecule (i.e., the ion of m/z 124, Figure S7a, inset, and Scheme S1). The corresponding fragment ion was found for the isotope-labeled standard, with the exception of a 2 Da mass shift introduced by 15N-labeling to the nucleobase portion (Figure S7b, inset, and Scheme S1). Our LC-MS/MS/MS quantification results revealed that the catalytic activity of Tet1 conferred a marked elevation in the level of 5-hmrC, as the RNA samples isolated from HEK293T cells transfected with wild-type Tet1 carried significantly higher levels of 5-hmrC (11.9 modifications per 106 ribonucleosides) than those isolated from cells transfected with the mutant form of Tet1 or a control pGEM-T vector (at 2.0 and 1.9 modifications per 106 ribonucleosides, respectively, Figure S9 and Table S1). Likewise, overexpression of the catalytic domains of Tet2 and Tet3 also led to significant elevations in the levels of 5-hmrC in HEK293T cells (Figure S9 and Table S1).
Considering that other domains of Tet proteins may also be involved in regulating their substrate accessibility, we next assessed the levels of 5-hmrC in cells overexpressing individually the three full-length Tet proteins. Indeed our results demonstrated that the overexpression of full-length Tet3, but not Tet1 or Tet2, could result in substantially elevated level of 5-hmrC in RNA, where the levels of 5-hmrC were 4.1 and 1.8 modifications per 106 nucleosides in HEK293T cells overexpressing the full-length Tet3 and its catalytically inactive mutant, respectively (Figure 4a and Table S1). In this regard, it is important to note that all three full-length Tet proteins are functional, as manifested by marked increases in the levels of 5-hmdC in genomic DNA isolated from cells overexpressing any of the three full-length Tet proteins (Figure 4b and Table S2). Along this line, it is worth noting that Tet1 and Tet2 are localized in the nucleus, whereas Tet3 is localized in both the cytosol and the nucleus.36
Figure 4.
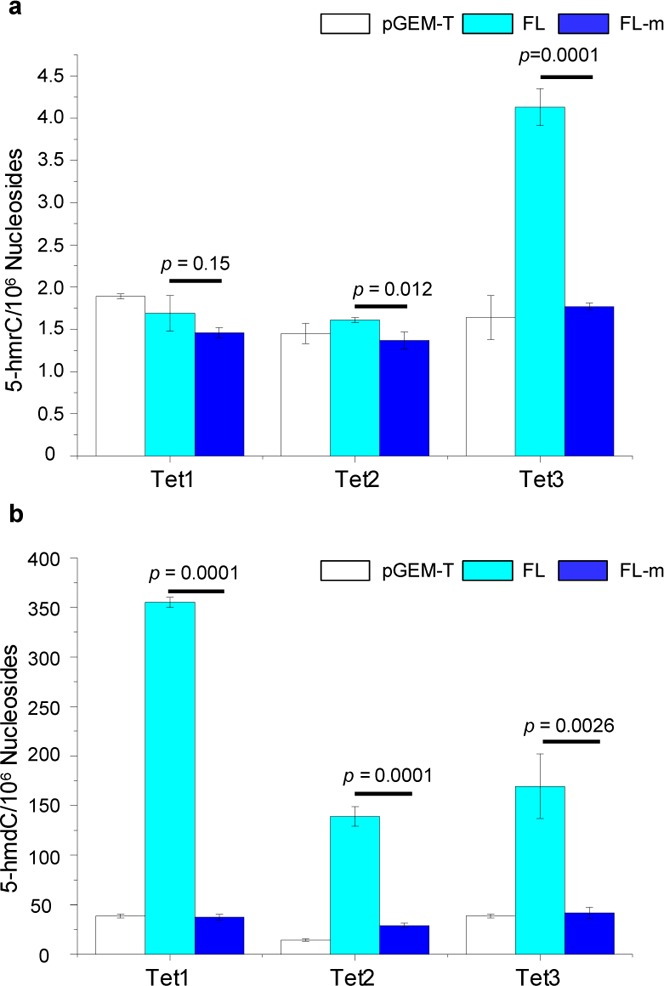
Levels of 5-hmrC and 5-hmdC in HEK293T cells overexpressing individually the full-length (FL) Tet proteins, or their catalytically inactive mutants (FL-m). “pGEM-T” refers to DNA samples from HEK293T cells transfected with the control pGEM-T Easy plasmid. The data represent the means and standard deviations of three independent transfection and measurement results. The p values were calculated using unpaired two-tailed Student’s t-test.
To further exploit the roles of Tet enzymes in inducing 5-hmrC in vivo, we measured the levels of 5-hmrC in total RNA isolated from wild-type mouse embryonic stem (ES) cells and Tet-null ES cells where Tet1, Tet2, and Tet3 were genetically deleted (Tet–/–). Our results demonstrated that removal of all three Tet activities led to a significant decline in the level of 5-hmrC in total RNA (from 1.4 to 0.82 modifications per 106 ribonucleosides, Figure S10a and Table S3), whereas knockout of the thymine DNA glycosylase gene (Tdg–/–) did not lead to apparent change in 5-hmrC level (Figure S10a and Table S3). The relatively small difference in the levels of 5-hmrC in the wild-type and Tet–/– ES cells is in line with the relatively low level of expression of Tet3 in ES cells.12 In addition, the presence of appreciable levels of 5-hmrC in Tet–/– ES cells suggests that other enzyme(s) might also be involved in oxidizing 5-mC to 5-hmrC in mammalian cells, though we cannot formally exclude the possibility that some 5-hmrC may also be induced by cellular reactive oxygen species. Thus, the above results support that Tet enzymes contribute to the oxidation of 5-mrC in RNA to 5-hmrC in vivo.
Having demonstrated the enzymatic activity of Tet1 toward 5-mC in RNA, we next assessed the occurrence of 5-hmrC in RNA isolated from various mouse and human tissues by using LC-MS/MS/MS (Figure S10 and Table S3). In this vein, it is of note that 5-hmrC was previously detected in rRNA isolated from wheat seedlings.37 Our results showed that 5-hmrC could be readily detected in RNA samples isolated from all the tissue types we tested, including brain, heart, pancreas, and spleen, with the level being the highest in the heart (3.9 modifications per 106 ribonucleosides, Figure S10c and Table S3). In addition, 5-hmrC could be detected in human brain RNA at a frequency of 1.4 per 106 ribonucleosides (Figure S10c and Table S3). 5-hmrC could also be found in cultured human cancer cells, including the HeLa cervical cancer cells (at 0.68 modifications per 106 ribonucleosides) and WM-266–4 melanoma cells (1.6 modifications per 106 ribonucleosides, Figure S10b and Table S3). For comparison, we also quantified 5-mrC in these RNA samples using HPLC analysis (Figure S6). The 5-mrC levels also varied among the different tissues (0.5–2.2% of rC) and cancer cells (0.5–0.7%), and the relative levels of 5-mrC parallel the relative levels of 5-hmrC in these tissue and cell samples (Figure S10b,c, Figure S11, and Table S3). Considering the levels of 5-mrC and 5-hmrC, we conclude that ∼0.02% of 5-mrC is modified to 5-hmrC in these tissue and cellular RNA samples.
Taken together, we demonstrate that Tet enzymes can catalyze the formation of 5-hmrC from 5-mrC both in vitro and in vivo. We also determined, for the first time, the levels of 5-hmrC in tissue and cellular RNA by using a sensitive and accurate LC-MS/MS/MS with the isotope dilution method. Our results revealed that the level of this modification occurs at a frequency of approximately one 5-hmrC per 5000 5-mrC. Recent bisulfite sequencing data showed the widespread presence of 5-mrC in both coding and noncoding RNA.7,8 The presence of appreciable level of 5-hmrC in cellular RNA and the involvement of Tet-family enzymes in inducing this modification suggest that the function of Tet enzymes is not restricted to the epigenetic regulation at the DNA level, but perhaps can also be extended to RNA. In addition, 5-hmrC may also participate in the epigenetic regulation of gene expression. The present work sets the stage for future studies in defining the distribution and site-specific localization of 5-hmrC in different RNA species (i.e., rRNA, mRNA and tRNA), and the function of this 5-mrC oxidation in RNA biology. The relative levels of 5-hmrC in RNA are lower than those of 5-hmdC in DNA. It will be of particular importance to determine whether the 5-hmrC is a stable oxidation product or occurs transiently, possibly as an intermediate step in a pathway leading toward 5-mC decay in RNA, or is perhaps a signal that mediates RNA degradation. Both scenarios could explain the relatively low level of this modification at steady state in vivo.
Acknowledgments
This work was supported by the National Institutes of Health (CA160965 to G.P.P., CA101864 to Y.W., and AG043376 to L.J.N. and Y.W.). The authors would like to thank Prof. Stephen Baylin for kindly providing the Tet2 expression plasmids.
Supporting Information Available
Detailed experimental conditions, HPLC, mass spectrometry and quantification results. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Cantara W. A.; Crain P. F.; Rozenski J.; McCloskey J. A.; Harris K. A.; Zhang X.; Vendeix F. A.; Fabris D.; Agris P. F. Nucleic Acids Res. 2011, 39, D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Fu Y.; Zhao X.; Dai Q.; Zheng G.; Yang Y.; Yi C.; Lindahl T.; Pan T.; Yang Y. G.; He C. Nat. Chem. Biol. 2011, 7, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.; Dahl J. A.; Niu Y.; Fedorcsak P.; Huang C. M.; Li C. J.; Vagbo C. B.; Shi Y.; Wang W. L.; Song S. H.; Lu Z.; Bosmans R. P.; Dai Q.; Hao Y. J.; Yang X.; Zhao W. M.; Tong W. M.; Wang X. J.; Bogdan F.; Furu K.; Fu Y.; Jia G.; Zhao X.; Liu J.; Krokan H. E.; Klungland A.; Yang Y. G.; He C. Mol. Cell 2013, 49, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Fu Y.; He C. Trends Genet. 2013, 29, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.; Agarwala S. D.; Mumbach M. R.; Jovanovic M.; Mertins P.; Shishkin A.; Tabach Y.; Mikkelsen T. S.; Satija R.; Ruvkun G.; Carr S. A.; Lander E. S.; Fink G. R.; Regev A. Cell 2013, 155, 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y.; Lyko F.; Helm M. Nucleic Acids Res. 2010, 38, 1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires J. E.; Patel H. R.; Nousch M.; Sibbritt T.; Humphreys D. T.; Parker B. J.; Suter C. M.; Preiss T. Nucleic Acids Res. 2012, 40, 5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amort T.; Souliere M. F.; Wille A.; Jia X. Y.; Fiegl H.; Worle H.; Micura R.; Lusser A. RNA Biol. 2013, 10, 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S.; Sajini A. A.; Blanco S.; Dietmann S.; Lombard P.; Sugimoto Y.; Paramor M.; Gleeson J. G.; Odom D. T.; Ule J.; Frye M. Cell Rep. 2013, 4, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T.; Hackner B.; Truss M.; Munzel M.; Muller M.; Deiml C. A.; Hagemeier C.; Carell T. Angew. Chem., Int. Ed. Engl. 2011, 50, 7008. [DOI] [PubMed] [Google Scholar]
- Tahiliani M.; Koh K. P.; Shen Y.; Pastor W. A.; Bandukwala H.; Brudno Y.; Agarwal S.; Iyer L. M.; Liu D. R.; Aravind L.; Rao A. Science 2009, 324, 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S.; D’Alessio A. C.; Taranova O. V.; Hong K.; Sowers L. C.; Zhang Y. Nature 2010, 466, 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. F.; Li B. Z.; Li Z.; Liu P.; Wang Y.; Tang Q.; Ding J.; Jia Y.; Chen Z.; Li L.; Sun Y.; Li X.; Dai Q.; Song C. X.; Zhang K.; He C.; Xu G. L. Science 2011, 333, 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S.; Shen L.; Dai Q.; Wu S. C.; Collins L. B.; Swenberg J. A.; He C.; Zhang Y. Science 2011, 333, 1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S.; Heintz N. Science 2009, 324, 929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. A. Annu. Rev. Microbiol. 1980, 34, 137. [DOI] [PubMed] [Google Scholar]
- Liutkeviciute Z.; Lukinavicius G.; Masevicius V.; Daujotyte D.; Klimasauskas S. Nat. Chem. Biol. 2009, 5, 400. [DOI] [PubMed] [Google Scholar]
- Yildirim O.; Li R.; Hung J. H.; Chen P. B.; Dong X.; Ee L. S.; Weng Z.; Rando O. J.; Fazzio T. G. Cell 2011, 147, 1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauer C.; Hoffmann T.; Bultmann S.; Casa V.; Cardoso M. C.; Antes I.; Leonhardt H. PLoS One 2011, 6, e21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt C. G.; Gnerlich F.; Smits A. H.; Pfaffeneder T.; Jansen P. W.; Bauer C.; Munzel M.; Wagner M.; Muller M.; Khan F.; Eberl H. C.; Mensinga A.; Brinkman A. B.; Lephikov K.; Muller U.; Walter J.; Boelens R.; van Ingen H.; Leonhardt H.; Carell T.; Vermeulen M. Cell 2013, 152, 1146. [DOI] [PubMed] [Google Scholar]
- Maiti A.; Drohat A. C. J. Biol. Chem. 2011, 286, 35334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M.; Huang Y.; Jankowska A. M.; Pape U. J.; Tahiliani M.; Bandukwala H. S.; An J.; Lamperti E. D.; Koh K. P.; Ganetzky R.; Liu X. S.; Aravind L.; Agarwal S.; Maciejewski J. P.; Rao A. Nature 2010, 468, 839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. G.; Jiang Y.; Qiu R.; Rauch T. A.; Wang Y.; Schackert G.; Krex D.; Lu Q.; Pfeifer G. P. Cancer Res. 2011, 71, 7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. L.; Shen F.; Huang W.; Qi J. H.; Wang Y.; Feng Y. Q.; Liu S. M.; Yuan B. F. Clin. Chem. 2013, 59, 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell T. L.; McGuffin L. J.; Dunwell J. M.; Pfeifer G. P. Cell Cycle 2013, 12, 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas P. A.; Otterlei M.; Falnes P. O.; Vagbo C. B.; Skorpen F.; Akbari M.; Sundheim O.; Bjoras M.; Slupphaug G.; Seeberg E.; Krokan H. E. Nature 2003, 421, 859. [DOI] [PubMed] [Google Scholar]
- Falnes P. O.; Bjoras M.; Aas P. A.; Sundheim O.; Seeberg E. Nucleic Acids Res. 2004, 32, 3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. H.; Jin S. G.; Cai S.; Chen Y.; Pfeifer G. P.; O’Connor T. R. J. Biol. Chem. 2005, 280, 39448. [DOI] [PubMed] [Google Scholar]
- Westbye M. P.; Feyzi E.; Aas P. A.; Vagbo C. B.; Talstad V. A.; Kavli B.; Hagen L.; Sundheim O.; Akbari M.; Liabakk N. B.; Slupphaug G.; Otterlei M.; Krokan H. E. J. Biol. Chem. 2008, 283, 25046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G.; Yang C. G.; Yang S.; Jian X.; Yi C.; Zhou Z.; He C. FEBS Lett. 2008, 582, 3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C.; Yang C. G.; He C. Acc. Chem. Res. 2009, 42, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.; Li Z.; Cheng J.; Rao Q.; Gong W.; Liu M.; Shi Y. G.; Zhu J.; Wang P.; Xu Y. Cell 2014, 155, 1545. [DOI] [PubMed] [Google Scholar]
- Hashimoto H.; Pais J. E.; Zhang X.; Saleh L.; Fu Z. Q.; Dai N.; Correa I. R. Jr.; Zheng Y.; Cheng X. Nature 2013, 506, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizaki S.; Sugiyama H. Org. Biomol. Chem. 2014, 12, 104. [DOI] [PubMed] [Google Scholar]
- Liu S.; Wang J.; Su Y.; Guerrero C.; Zeng Y.; Mitra D.; Brooks P. J.; Fisher D. E.; Song H.; Wang Y. Nucleic Acids Res. 2013, 41, 6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioka Y.; Watanabe A.; Saito K.; Yamada Y. PLoS One 2012, 7, e45031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz I.; Kiraly I.; Lasztily D. Planta 1978, 142, 263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.