Abstract
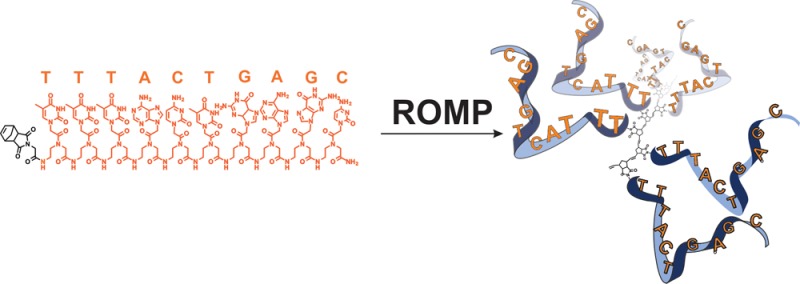
Here we report the preparation of poly(oligonucleotide) brush polymers and amphiphilic brush copolymers from nucleic acid monomers via graft-through polymerization. We describe the polymerization of PNA-norbornyl monomers to yield poly-PNA (poly(peptide nucleic acid)) via ring-opening metathesis polymerization (ROMP) with the initiator, (IMesH2)(C5H5N)2(Cl)2RuCHPh.1 In addition, we present the preparation of poly-PNA nanoparticles from amphiphilic block copolymers and describe their hybridization to a complementary single-stranded DNA (ssDNA) oligonucleotide.
The display of chemical functionality in a multivalent fashion on surfaces and particles and as brushes on polymer backbones is a common theme in nature as well as for synthetic systems.2−4 Such systems take advantage of the unique properties that arise when monomeric species are incorporated into a densely packed three-dimensional (3D) architecture. Here we describe the preparation of polymeric nucleic acids wherein single-stranded sequences of peptide nucleic acids (PNAs)102 are incorporated as polymer brushes via graft-through polymerization using the ROMP initiator (IMesH2)(C5H5N)2(Cl)2RuCHPh (Figure 1). Nucleic acids, both natural and synthetic, standout as the quintessential carriers of chemical information stored as specific sequences of bases positioned along a backbone.4−7 As such, synthetic oligonucleotides and nucleic acid bioconjugates are powerful tools in a range of fields including in biotechnology (e.g., PCR),8,9 in materials science as programmable structural synthons10−16 and as aptamers selected by in vitro evolution.17−21 In each application the nucleic acid functions to enrich a chemical system with information, facilitating predictable interactions with complementary sequences,22 or with other molecules including enzymes, proteins, and small molecules.18,23−25 We reasoned that the graft-through polymerization of an oligonucleotide sequence would provide a powerful new approach for the multivalent display of chemical information on a synthetic template.
Figure 1.

Synthesis and characterization of a poly(oligonucleotide). (A) PNA-norbornyl monomer (PNA-Nb) polymerized using ROMP initiator (IMesH2)(C5H5N)2(Cl)2Ru=CHPh (“Ru”) to form poly-PNA homopolymer, I, and poly-PNA block copolymer, II. (B) Representative percent conversion for I determined by the disappearance of the olefin signal associated with PNA-Nb in 1H NMR. (C) Representative SEC-MALS for II. Mn = 28,270 indicating a degree of polymerization of 5 for the PNA block.
There have been extensive efforts to prepare nucleic acid inspired synthetic polymers involving the direct polymerization of appropriately modified monomers, generating synthetic polymers with single nucleobases as side-chains.26−31 Although this approach allows the integration of purine and pyrimidine bases onto a synthetic backbone, it does not allow the incorporation of sequences containing multiple bases and thus does not result in informational polymeric systems. In addition to strategies involving directly polymerized nucleobases, there are an increasing number of examples of oligonucleotide-polymer bioconjugates in the literature reliant upon post-polymerization conjugation reactions.32−34 These approaches, shown in Scheme 1, seek to fix recognition elements natural to DNA and RNA along a synthetic polymer or polymeric nanoparticle template and have found use in an array of arenas including the programmed assembly of nanoparticles,2,35−37 in delivery vehicles,5,38−40 and as effective DNA-probes.34,41,42 Furthermore, the function of these materials is intrinsically governed by the information within the nucleic acid sequence itself as well as the dense and multivalent 3D array induced by the polymer scaffold. Indeed, function dictated by 3D biomolecular display is not unique to nucleic acids, rather this concept extends to all classes of biomolecules, most effectively demonstrated in the past with peptides and proteins.3,43−47 Strategies for the polymerization of (graft-through) and polymerization from (graft-from) proteins and peptides have been used to build macromolecules through sequential addition of monomers to a growing chain, taking advantage of catalyst proficiency and avoiding kinetically unfavorable conjugations (graft-to) between multiple large macromolecules.48−54 However, unlike for other biomolecules (saccharides,55,57 peptides,58 and proteins59,60), there are no examples of graft-through polymerization of nucleic acids and very few examples of graft-from polymerization off of nucleic acids.100,101 Therefore, despite their promise, polymer bioconjugates of true nucleic acid sequences have been limited to those prepared via post-polymerization modification and hence are limited in terms of maximum achievable DNA density, are difficult to reproduce, and suffer from incomplete incorporation of the nucleic acid at each position of the polymer (Scheme 1A).
Scheme 1. Known Methods for the Incorporation of Multiple Nucleic Acids or Nucleobases into Polymers.

(A) Post-polymerization modification of a polymer with a nucleic acid.51−53 (B) Polymerization of a pyrimidine base as a modified monomer.44−50
Figure 2.
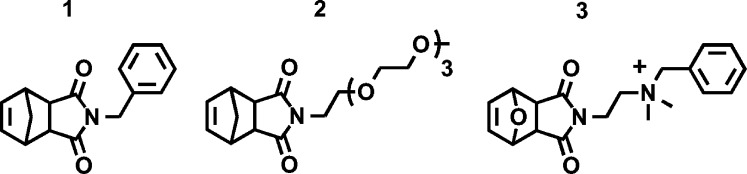
Structures of monomers used for block copolymer preparation.
In order to avoid shortcomings associated with post-polymerization modification reactions, a nucleic acid monomer capable of undergoing direct graft-through polymerization was synthesized. Initial studies attempting direct-polymerization of DNA-based monomers via ROMP were met with limited success. Therefore, a PNA-based monomer was chosen as an ideal target for this study for three key reasons: (1) PNA can be prepared in significant quantities via standard peptide bond-forming reactions on solid support; (2) PNA is soluble in DMF making it readily compatible with (IMesH2)(C5H5N)2(Cl)2RuCHPh (Figure 1 for structure); and (3) we hypothesized that the neutral N-(2-amino-ethyl)-glycine backbone would be more compatible with the ruthenium-based catalyst than the polyanionic phosphate backbone of DNA.
A 10-base PNA sequence (Figure 1) was designed to discourage self-hybridization while providing a sufficient number of bases for efficient hybridization with a complementary sequence of DNA at room temperature. Moreover, this 10-base sequence represents a sequence encoded with each of the four letters of the genetic alphabet and enough information to communicate specifically with other nucleic acids and proteins, a function not possible via the polymerization of a single purine or pyrimidine monomer (as in Scheme 1B). After the complete PNA sequence was prepared, N-(glycine)-cis-5-norbornene-exo-dicarboximide was coupled to the N-terminus while on solid support. The PNA norbornene monomer (PNA-Nb) was then cleaved and deprotected from the solid support using TFA:cresol (80:20), purified by HPLC, and the mass confirmed by ESI-MS (see Supporting Information (SI)). Following HPLC purification, PNA-Nb was lyophilized to afford a white powder.
For polymerization studies, PNA-Nb was resuspended in dry, degassed N,N-dimethylformamide-d7 in a J. Young NMR tube in a glovebox. An appropriate amount of ruthenium initiator was added to the solution, and the disappearance of the norbornene olefin resonance was then monitored by 1H NMR. Complete disappearance of the monomeric olefin peak indicated complete polymerization of PNA-Nb into poly-PNA. A series of experiments were carried out to determine reproducibility of polymerization reactions with respect to both the preparation of homopolymers as well as block copolymers (Table 1). For PNA homopolymers, degrees of polymerization of 10 (polymer I) and 5 (polymer III) were targeted with complete consumption of PNA-Nb confirmed by 1H NMR for both reactions. In preparation of a PNA-containing block copolymer, a phenyl-functionalized norbornene (1) was polymerized as the first block followed by PNA-Nb incorporated as the second block (as shown in polymer II (Figure 1, Table 1). To achieve this, PNA-Nb was added to the living phenyl polymer chain, and the disappearance of the norbornene olefin of PNA-Nb was monitored by 1H NMR. The resulting polymeric species were then analyzed by SEC-MALS (see SI). Having established that PNA-Nb could be successfully polymerized into block copolymers, we sought to assess the reproducibility of these reactions by attempting to synthesize block copolymers of identical composition. Using a live ruthenium catalyst on a phenyl homopolymer with a degree of polymerization of 30, three separate but identical reactions were set up in which the attempted degree of polymerization of the PNA monomer was 7.5 (Table 1, polymers IV–VI). The degree of polymerization of the PNA block ranged from 5 to 6 (60–80% completion), indicating a good degree of reproducibility and predictability for these reactions. In addition, higher degrees of polymerization could be achieved for this type of block copolymer as illustrated by the preparation of polymers VII and VIII (Table 1). To examine the compatibility of PNA-Nb polymerization with other block copolymer systems, an oligoethylene glycol functionalized norbornene (2) and a quaternary amine-functionalized norbornene (3) were synthesized as monomers for incorporation into block copolymer scaffolds as the initial block. The resulting block polymers (IX and X in Table 1) showed percent conversions of PNA-Nb comparable to the phenyl-based block copolymers, with the amine-functionalized system demonstrating the lowest percent conversion. Given the slight variation in PNA-Nb percent conversion between these three different block copolymer systems (VII, IX and X in Table 1), the identity of the non-PNA block may dictate PNA conversion efficiency and should be taken into consideration for future studies.
Table 1. Polymers and Copolymers of PNA with Monomers Shown in Figure 2.
| polymer | mon1a | mon2c | md | nd | % con.e |
|---|---|---|---|---|---|
| I | PNA-Nb (10:1)b | – | 10 | – | 99 |
| II | 1 (35:1) | PNA-Nb (5:1) | 35 | 5 | 97 |
| III | PNA-Nb (5:1) | – | 5 | – | 97 |
| IV | 1 (30:1) | PNA-Nb (7.5:1) | 30 | 5 | 65 |
| V | 1 (30:1) | PNA-Nb (7.5:1) | 30 | 5 | 65 |
| VI | 1 (30:1) | PNA-Nb (7.5:1) | 30 | 6 | 79 |
| VII | 1 (36:1) | PNA-Nb (9:1) | 35 | 8 | 88 |
| VIII | 1 (36:1) | PNA-Nb (18:1) | 35 | 16 | 87 |
| IX | 2 (36:1) | PNA-Nb (9:1) | 33 | 7 | 74 |
| X | 3 (36:1) | PNA-Nb (9:1) | 41 | 5 | 56 |
Indicates indentity of monomer polymerized first (degree of polymerization, DP = m).
Ratios shown indicate monomer to initiator ratio or intended DP.
Indicates identity of monomer polymerized second (DP = n).
Observed degree of polymerization of mon1 (m) or mon2 (n).
Percent conversion of PNA-Nb determined by 1H NMR.
To assess the DNA-binding capability of these systems, block copolymer II was chosen. The assembly of II to generate spherical nanoparticles was achieved by dissolving II in DMSO and then dialyzing into aqueous solution (see SI for details).61 The resulting nanoparticle (PNA-NP) was characterized by DLS and TEM (Figure 3). DLS data support the formation of an aggregated species in solution. TEM reveals the existence of nanoparticles on the order of 20 nm in diameter. The melting temperature (Tm) of PNA-NP hybridized with its complementary DNA sequence was determined to be 58.1 °C, an ∼8 °C increase over an identical, nonparticulate, unpolymerized PNA sequence. These melting data suggest cooperative binding and accessible PNA forming the shell of the nanoparticles. In support of this model, we conducted a molecular dynamics simulation of PNA-NP(62−64) assembled from 60 amphiphiles giving a structure that equilibrated into a spherical particle ∼21 nm in diameter. Polynorbornyl chains packed well to form a compact hydrophobic core largely protected from contact with water. The simulations show the hydrophilic PNA chains solvated in water forming the shell of the micellar nanoparticles.
Figure 3.

Poly-PNA amphiphile II was dialyzed from DMSO into H2O to generate nanoparticles. (A) DLS data indicating a hydrodynamic radius of 25 nm. (B) Tm of PNA-NP with a complementary DNA sequence was found to be 58.1 °C. (C) Negative-stained TEM of PNA-NP provided evidence of spherical 20 nm diameter nanoparticles. Atomistic models of (D) II and (E) PNA-NP. II is shown in a conformation present within PNA-NP.
In summary, we have shown that one can prepare nucleic acid brush polymers and amphiphilic brush copolymers by direct, graft-through polymerization of an oligonucleotide. To our knowledge, this is the first example of a polymer-nucleic acid bioconjugate generated via direct polymerization of an oligonucleotide monomer. In addition, these materials show cooperative hybridization to complementary DNA oligonucleotides. We believe this type of approach provides an efficient synthetic strategy for the incorporation of nucleic acids into particle and polymer-based materials. The interest in doing so is driven by potential applications including the facile preparation of materials for affinity purification of DNA,65,66 gene and nucleic acid delivery to cells,5,38−40,67−70 and in the development of materials capable of programmed self-assembly.12−16,71−75
Acknowledgments
We acknowledge support for this work from the AFOSR via a PECASE (FA9550-11-1-0105) and from the ARO (W911NF-11-1-0264 and W911NF-14-1-0169). We acknowledge the UCSD Cryo-Electron Microscopy Facility, supported by NIH grants to T.S.B. and the Agouron Institute. N.C.G acknowledges the Alfred P. Sloan Foundation. Computational work was supported by an ACS PRF grant no. 53062-ND6 to P.K. This research used the Extreme Science and Engineering Discovery Environment (XSEDE): NSF OCI-1053575 to P.K.
Supporting Information Available
Experimental details and data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Love J. A.; Sanford M. S.; Day M. W.; Grubbs R. H. J. Am. Chem. Soc. 2003, 125, 10103. [DOI] [PubMed] [Google Scholar]
- Lytton-Jean A. K. R.; Gibbs-Davis J. M.; Long H.; Schatz G. C.; Mirkin C. A.; Nguyen S. T. Adv. Mater. 2009, 21, 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard H. D.; Okada S. Y.; Grubbs R. H. Macromolecules 2000, 33, 6239. [Google Scholar]
- Mammen M.; Choi S. K.; Whitesides G. M. Angew. Chem., Int. Ed. 1998, 37, 2755. [DOI] [PubMed] [Google Scholar]
- Nielsen P.; Egholm M.; Berg R.; Buchardt O. Science 1991, 254, 1497. [DOI] [PubMed] [Google Scholar]
- Rosi N. L.; Giljohann D. A.; Thaxton C. S.; Lytton-Jean A. K. R.; Han M. S.; Mirkin C. A. Science 2006, 312, 1027. [DOI] [PubMed] [Google Scholar]
- Lockhart D. J.; Dong H. L.; Byrne M. C.; Follettie M. T.; Gallo M. V.; Chee M. S.; Mittmann M.; Wang C. W.; Kobayashi M.; Horton H.; Brown E. L. Nat. Biotechnol. 1996, 14, 1675. [DOI] [PubMed] [Google Scholar]
- Lipshutz R. J.; Fodor S. P. A.; Gingeras T. R.; Lockhart D. J. Nat. Genet. 1999, 21, 20. [DOI] [PubMed] [Google Scholar]
- Saiki R. K.; Scharf S.; Faloona F.; Mullis K. B.; Horn G. T.; Erlich H. A.; Arnheim N. Science 1985, 230, 1350. [DOI] [PubMed] [Google Scholar]
- Orum H.; Nielsen P. E.; Egholm M.; Berg R. H.; Buchardt O.; Stanley C. Nucleic Acids Res. 1993, 21, 5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish R. D.; Schulman R.; Rothemund P. W. K.; Winfree E. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H.; Chao J.; Xiao S.-J.; Seeman N. C. Nat. Nanotechnol. 2009, 4, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin C. A.; Letsinger R. L.; Mucic R. C.; Storhoff J. J. Nature 1996, 382, 607. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. Nature 2006, 440, 297–302. [DOI] [PubMed] [Google Scholar]
- Dietz H.; Douglas S. M.; Shih W. M. Science 2009, 325, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao-Ping C.; Rush A. M.; Thompson M. P.; Gianneschi N. C. Angew. Chem., Int. Ed. 2010, 49, 5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaye F. A.; Palmer A. L.; Sleiman H. F. Science 2008, 321, 1795. [DOI] [PubMed] [Google Scholar]
- Joyce G. F. Curr. Opin. Struct. Biol. 1994, 4, 331. [DOI] [PubMed] [Google Scholar]
- Patil S. D.; Rhodes D. G.; Burgess D. J. AAPS J. 2005, 7, E61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpashchikov D. M.; Stojanovic M. N. J. Am. Chem. Soc. 2005, 127, 11348. [DOI] [PubMed] [Google Scholar]
- Breaker R. R. Chem. Rev. 1997, 97, 371. [DOI] [PubMed] [Google Scholar]
- Hermann T.; Patel D. J. Science 2000, 287, 820. [DOI] [PubMed] [Google Scholar]
- Tyagi S.; Kramer F. R. Nat. Biotechnol. 1996, 14, 303. [DOI] [PubMed] [Google Scholar]
- Breaker R. R.; Joyce G. F. J. Mol. Evol. 1995, 40, 551. [DOI] [PubMed] [Google Scholar]
- Liou H.-C.; Baltimore D. Curr. Opin. Cell Biol. 1993, 5, 477. [DOI] [PubMed] [Google Scholar]
- Wilson G. G.; Murray N. E. Annu. Rev. Genet. 1991, 25, 585. [DOI] [PubMed] [Google Scholar]
- Davies R. G.; Gibson V. C.; Hursthouse M. B.; Light M. E.; Marshall E. L.; North M.; Robson D. A.; Thompson I.; White A. J. P.; Williams D. J.; Williams P. J. J. Chem. Soc., Perkin Trans. 1 2001, 26, 3365. [Google Scholar]
- Gibson V. C.; Marshall E. L.; North M.; Robson D. A.; Williams P. J. Chem. Commun. 1997, 12, 1095. [Google Scholar]
- Lutz J. F.; Thunemann A. F.; Nehring R. J. Polym. Sci., Part A: Polym. Chem. 2005, 43, 4805. [Google Scholar]
- Monnard P. A.; Kanavarioti A.; Deamer D. W. J. Am. Chem. Soc. 2003, 125, 13734. [DOI] [PubMed] [Google Scholar]
- Spijker H. J.; van Delft F. L.; van Hest J. C. M. Macromolecules 2007, 40, 12. [Google Scholar]
- McHale R.; O’Reilly R. K. Macromolecules 2012, 45, 7665. [Google Scholar]
- Alemdaroglu F. E.; Herrmann A. Org. Biomol. Chem. 2007, 5, 1311. [DOI] [PubMed] [Google Scholar]
- Kwak M.; Herrmann A. Angew. Chem., Int. Ed. 2010, 49, 8574. [DOI] [PubMed] [Google Scholar]
- Gibbs J. M.; Park S. J.; Anderson D. R.; Watson K. J.; Mirkin C. A.; Nguyen S. T. J. Am. Chem. Soc. 2005, 127, 1170. [DOI] [PubMed] [Google Scholar]
- Farokhzad O. C.; Jon S. Y.; Khademhosseini A.; Tran T. N. T.; LaVan D. A.; Langer R. Cancer Res. 2004, 64, 7668. [DOI] [PubMed] [Google Scholar]
- Park S. Y.; Lytton-Jean A. K. R.; Lee B.; Weigand S.; Schatz G. C.; Mirkin C. A. Nature 2008, 451, 553. [DOI] [PubMed] [Google Scholar]
- Storhoff J. J.; Mirkin C. A. Chem. Rev. 1999, 99, 1849. [DOI] [PubMed] [Google Scholar]
- Zilong Z.; Hongmin M.; Nannan W.; Donovan M. J.; Ting F.; Mingxu Y.; Zhuo C.; Xiaobing Z.; Weihong T. Angew. Chem., Int. Ed. 2013, 52, 7487. [Google Scholar]
- Jeong J. H.; Park T. G. Bioconjugate Chem. 2001, 12, 917. [DOI] [PubMed] [Google Scholar]
- Rush A. M.; Nelles D. A.; Blum A. P.; Barnhill S. A.; Tatro E. T.; Yeo G. W.; Gianneschi N. C. J. Am. Chem. Soc. 2014, 136, 7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaylord B. S.; Heeger A. J.; Bazan G. C. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret B.; Calame M.; Libchaber A. J. Nat. Biotechnol. 2001, 19, 365. [DOI] [PubMed] [Google Scholar]
- Biagini S. C. G.; Parry A. L. J. Polym. Sci., Part A: Polym. Chem. 2007, 45, 3178. [Google Scholar]
- Fraley A. W.; Pons B.; Dalkara D.; Nullans G.; Behr J.-P.; Zuber G. J. Am. Chem. Soc. 2006, 128, 10763. [DOI] [PubMed] [Google Scholar]
- Hahn M. E.; Randolph L. M.; Adamiak L.; Thompson M. P.; Gianneschi N. C. Chem. Commun. 2013, 49, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliceti P.; Veronese F. M. Adv. Drug Delivery Rev. 2003, 55, 1261. [DOI] [PubMed] [Google Scholar]
- Liu F. T.; Zinnecker M.; Hamaoka T.; Katz D. H. Biochemistry 1979, 18, 690. [DOI] [PubMed] [Google Scholar]
- Broyer R. M.; Quaker G. M.; Maynard H. D. J. Am. Chem. Soc. 2008, 130, 1041. [DOI] [PubMed] [Google Scholar]
- Becker M. L.; Liu J. Q.; Wooley K. L. Biomacromolecules 2005, 6, 220. [DOI] [PubMed] [Google Scholar]
- Lele B. S.; Murata H.; Matyjaszewski K.; Russell A. J. Biomacromolecules 2005, 6, 3380. [DOI] [PubMed] [Google Scholar]
- De P.; Li M.; Gondi S. R.; Sumerlin B. S. J. Am. Chem. Soc. 2008, 130, 11288. [DOI] [PubMed] [Google Scholar]
- Mihov G.; Grebel-Koehler D.; Lubbert A.; Vandermeulen G. W. M.; Herrmann A.; Klok H. A.; Mullen K. Bioconjugate Chem. 2005, 16, 283. [DOI] [PubMed] [Google Scholar]
- Becker M. L.; Liu J. Q.; Wooley K. L. Chem. Commun. 2003, 2, 802. [DOI] [PubMed] [Google Scholar]
- Averick S. E.; Bazewicz C. G.; Woodman B. F.; Simakova A.; Mehl R. A.; Matyjaszewski K. Eur. Polym. J. 2013, 49, 2919. [Google Scholar]
- Pohl N. L.; Kiessling L. L. Synthesis 1999, SI, 1515. [Google Scholar]
- Nomura K.; Schrock R. R. Macromolecules 1996, 29, 540. [Google Scholar]
- Lee Y.; Sampson N. S. ChemBioChem 2009, 10, 929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammeyer J. K.; Blum A. P.; Adamiak L.; Hahn M. E.; Gianneschi N. C. Polym. Chem. 2013, 4, 3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumerlin B. S. ACS Macro Lett. 2012, 1, 141. [DOI] [PubMed] [Google Scholar]
- Heredia K. L.; Bontempo D.; Ly T.; Byers J. T.; Halstenberg S.; Maynard H. D. J. Am. Chem. Soc. 2005, 127, 16955. [DOI] [PubMed] [Google Scholar]
- He P.; He L. Biomacromolecules 2009, 10, 1804–1809. [DOI] [PubMed] [Google Scholar]
- Averick S. E.; Dey S. K.; Grahacharya D.; Matyjaszewski K.; Das S. R. Angew. Chem. Int. Ed. 2014, 53, 2739–2744. [DOI] [PubMed] [Google Scholar]
- Mai Y.; Eisenberg A. Chem. Soc. Rev. 2012, 41, 5969. [DOI] [PubMed] [Google Scholar]
- Bae J. W.; Pearson R. M.; Patra N.; Sunoqrot S.; Vukovic L.; Kral P.; Hong S. Chem. Commun. 2011, 47, 10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukovic L.; Khatib F. A.; Drake S. P.; Madriaga A.; Brandenburg K. S.; Kral P.; Onyuksel H. J. Am. Chem. Soc. 2011, 133, 13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukovic L.; Madriaga A.; Kuzmis A.; Banerjee A.; Tang A.; Tao K.; Shah N.; Kral P.; Onyuksel H. Langmuir 2013, 29, 15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann R.; Arnold L.; Ruth J. DNA 1984, 3, 122. [Google Scholar]
- Lesignoli F.; Germini A.; Corradini R.; Sforza S.; Galaverna G.; Dossena A.; Marchelli R. J. Chromatogr. A 2001, 922, 177. [DOI] [PubMed] [Google Scholar]
- Cutler J. I.; Auyeung E.; Mirkin C. A. J. Am. Chem. Soc. 2012, 134, 1376. [DOI] [PubMed] [Google Scholar]
- Koppelhus U.; Nielsen P. E. Adv. Drug Delivery Rev. 2003, 55, 267. [DOI] [PubMed] [Google Scholar]
- Pooga M.; Soomets U.; Hallbrink M.; Valkna A.; Saar K.; Rezaei K.; Kahl U.; Hao J. X.; Xu X. J.; Wiesenfeld-Hallin Z.; Hokfelt T.; Bartfai T.; Langel U. Nat. Biotechnol. 1998, 16, 857. [DOI] [PubMed] [Google Scholar]
- Davis M. E. Curr. Opin. Biotechnol. 2002, 13, 128. [DOI] [PubMed] [Google Scholar]
- Kershner R. J.; Bozano L. D.; Micheel C. M.; Hung A. M.; Fornof A. R.; Cha J. N.; Rettner C. T.; Bersani M.; Frommer J.; Rothemund P. W. K.; Wallraff G. M. Nat. Nanotechnol. 2009, 4, 557. [DOI] [PubMed] [Google Scholar]
- Gu H.; Chao J.; Xiao S.-J.; Seeman N. C. Nature 2010, 465, 202–U86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C. Nano Lett. 2010, 10, 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaoxiang D.; Seung-Hyun L.; Chengde M. J. Nanosci. Nanotechnol. 2005, 5, 1954.16430130 [Google Scholar]
- Pianowski Z. L.; Winssinger N. Chem. Soc. Rev. 2008, 37, 1330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.