Abstract
Small ruminant lentivirus (SRLV), also called ovine progressive pneumonia virus or maedi-visna, is present in 24% of US sheep. Like human immunodeficiency virus, SRLV is a macrophage-tropic lentivirus that causes lifelong infection. The production impacts from SRLV are due to a range of disease symptoms, including pneumonia, arthritis, mastitis, body condition wasting and encephalitis. There is no cure and no effective vaccine for preventing SRLV infection. However, breed differences in prevalence and proviral concentration indicate a genetic basis for susceptibility to SRLV. Animals with high blood proviral concentration show increased tissue lesion severity, so proviral concentration represents a live animal test for control post-infection in terms of proviral replication and disease severity. Recently, it was found that sheep with two copies of TMEM154 haplotype 1 (encoding lysine at position 35) had lower odds of SRLV infection. In this study, we examined the relationship between SRLV control post-infection and variants in two genes, TMEM154 and CCR5, in four flocks containing 1403 SRLV-positive sheep. We found two copies of TMEM154 haplotype 1 were associated with lower SRLV proviral concentration in one flock (P < 0.02). This identified the same favorable diplotype for SRLV control post-infection as for odds of infection. However, frequencies of haplotypes 2 and 3 were too low in the other three flocks to test. The CCR5 promoter deletion did not have consistent association with SRLV proviral concentration. Future work in flocks with more balanced allele frequencies is needed to confirm or refute TMEM154 association with control of SRLV post-infection.
Keywords: chemokine (C-C motif) receptor 5, disease susceptibility, domestic sheep, maedi–visna virus, ovine lentivirus, ovine progressive pneumonia virus, transmembrane protein 154, viremia
Introduction
Small ruminant lentivirus (SRLV), also known as maedi–visna virus or ovine progressive pneumonia virus, is a common pathogen in the US, where an estimated 24% of sheep are seropositive (Cutlip et al. 1992). Transmission of SRLV is thought to occur via the respiratory route with one major source being colostrum/milk from infected ewes (Reina et al. 2009), and the virus can be transmitted throughout an animal's lifespan (De Boer et al. 1979). In addition to inducing lifelong infection, SRLV causes varying clinical manifestations of pneumonia, arthritis, mastitis, cachexia, dyspnea and/or encephalitis (Blacklaws 2012). Small ruminant lentivirus disease symptoms generally become more evident with advancing age, and infected ewes are culled approximately one year earlier than uninfected ewes (Peterhans et al. 2004), which can be a large proportion of a ewe's productive lifetime (Annett et al. 2011; Byun et al. 2012). Additional adverse sheep production impacts from SRLV include reduced birth rates, birth weights and lamb growth as well as import restrictions (Keen et al. 1997; Arsenault et al. 2003; Reina et al. 2009). Methods of controlling or preventing SRLV infection, such as (i) repeated serological testing of adults with culling seropositive sheep or (ii) artificial rearing of lambs deprived of colostrum from infected dams, are expensive and have limited applicability in large production flocks (Houwers et al. 1983, 1984, 1989).
Breed differences in both SRLV seroprevalence and proviral concentration have suggested that genetics may play an important role in susceptibility (Gates et al. 1978; Cutlip et al. 1986; Houwers et al. 1989; Herrmann-Hoesing et al. 2008). Because SRLV induces lifelong infection, serological status has high concordance with direct viral measures of infection and can be used to measure odds of infection (Herrmann-Hoesing et al. 2007). Certain breeds have been associated consistently with higher or lower odds of infection. For example, Rambouillet has lower odds of infection than do other breeds (Gates et al. 1978; Cutlip et al. 1986; Houwers et al. 1989; Herrmann-Hoesing et al. 2008), and Columbia sheep have higher odds of infection (Herrmann-Hoesing et al. 2008). Proviral concentration significantly associates with severity of disease pathology, with high proviral concentration corresponding to high lesion score (Herrmann-Hoesing et al. 2009). Breed differences in level of proviral concentration are consistent with differences in odds of infection (Herrmann-Hoesing et al. 2008). Identifying genetic markers in specific breeds that associate with high proviral concentrations and removal or separation of these animals could result in reducing transmission and associated pathology of SRLV.
Recently, it has been discovered that variants in the transmembrane protein 154 (TMEM154) gene were associated with odds of SRLV infection, and the association has been validated in multiple large animal sets (Heaton et al. 2012). Little is known about the function of TMEM154 beyond transmembrane domain prediction, but its association with asthma severity in humans suggests a possible conserved role in airway immunity (Slager et al. 2011). Marker validation is important to reduce the probability of false-positive results and to improve reliability of predictive use (Hirschhorn et al. 2002; Li & Meyre 2013; White & Knowles 2013). TMEM154 haplotypes with strong supporting data include haplotype 1 [containing lysine (K) at position 35] and haplotypes 2 and 3 [containing glutamic acid (E) at position 35]. Of these, sheep with two copies of haplotype 1 were less susceptible to SRLV infection, but sheep with at least one copy of either haplotypes 2 or 3 were more susceptible (Heaton et al. 2012). A standardized genotyping method has been developed and commercialized for sheep producers to take advantage of marker-assisted selection to reduce susceptibility (Heaton et al. 2013). In addition, it is possible that sheep homozygous for TMEM154 haplotype 1 may also have lower proviral concentrations and lesion severity among infected sheep. However, sheep with these TMEM154 haplotypes have not been examined for control of SRLV post-infection.
Additional opportunities exist for developing genetic markers connected with control of SRLV post-infection based on the ovine chemokine (C-C motif) receptor 5 (CCR5) gene. Both SRLV and HIV are macrophage-tropic lentiviruses (Gendelman et al. 1986; Gorrell et al. 1992; Alkhatib & Berger 2007). Individual human beings with the delta-32 frame-shift deletion in CCR5 show high natural resistance against HIV, and that resistance has been associated with lack of functional CCR5 protein on the cell surface (Kaslow et al. 2005; Alkhatib & Berger 2007). A more subtle relationship between SRLV and CCR5 has been identified in sheep. Specifically, a four base-pair promoter deletion in ovine CCR5 has been associated with lower SRLV proviral concentration; this promoter deletion was also associated with lower expression of CCR5 (White et al. 2009). However, this association has been identified in only one flock, and it needs to be validated with additional flocks.
In the current study, we hypothesized that the previously defined low-risk TMEM154 diplotype (two copies of haplotype 1) would be associated with lower SRLV proviral concentration in multiple flocks of sheep. Also, we hypothesized that a four base-pair deletion in the promoter of ovine CCR5 would confirm an association with lower SRLV proviral concentration in multiple flocks of sheep. If either of these hypotheses were confirmed, then these data would provide evidence supporting one or more genetic markers for marker-assisted selection to achieve lower proviral concentration of SRLV.
Materials and methods
Phenotype and populations
Blood was collected by jugular venipuncture from four different flocks from three US states totaling 2236 ewes. Genomic DNA was extracted from peripheral blood leukocytes (PBLs) using previously described protocols (Herrmann-Hoesing et al. 2008; Heaton et al. 2012; White et al. 2012). In 2004, 353 ewes with a mean age of 4.35 years were sampled from an Idaho flock and included approximately equal numbers of Rambouillet, Polypay and Columbia breeds (Herrmann-Hoesing et al. 2008). A 2008 cohort from the same Idaho flock containing 947 ewes of Rambouillet, Polypay and Columbia breeds with a mean age of 2.42 years were also sampled (White et al. 2012). The average generation interval in the Idaho flock was approximately one every two to three years, and none of the animals were duplicated between the 2004 and 2008 Idaho sheep flocks. In 2009, 340 Polypay ewes with a mean age of 3.16 years were sampled from Iowa (Heaton et al. 2012), and 596 Rambouillet–Columbia crossbred ewes with a mean age of 3.18 years were sampled from Montana.
Proviral concentration of SRLV was determined by a validated qPCR method with over 95% concordance with serological methods (Herrmann-Hoesing et al. 2007). Briefly, PBLs were isolated from sheep blood, and DNA was extracted following the manufacturer's directions for Puregene (Genra System, Inc.). Real-time qPCR was performed using amplification primers for SRLV of a forward primer from the transmembrane gene, tm (5′-TCATAGTGCTTGCTATCATGGCTA-3′), and a reverse primer from the envelope glycoprotein gene, env (5′CCGTCCTTGTGTAGGATTGCT-3′). The SRLV tm-specific TaqMan probe, 5′-5′ hexachlorofluorescein-AGCAACACCGAGACCAGCTCCTGC-3′ Black Hole Quencher-1 (Integrated DNA Technologies) was used to quantify SRLV copy number (Herrmann-Hoesing et al. 2007). Up to 1 μg of DNA, 300 nm (final volume) of amplification primers, 250 nm of probe and TaqMan master mix diluted according to manufacturer's instructions (Applied Biosystems) were used in 50-μl reactions. Cycling conditions were 95 °C for 10 min followed by 60 cycles of 95 °C for 60 s and 55 °C for 60 s and 4 °C indefinitely for triplicate reactions (Herrmann-Hoesing et al. 2007). Known amounts (100–107 copies) of SRLV-containing plasmid were used in similar, triplicate reactions to generate standard curves. The SRLV copy numbers of unknown samples were determined using the mean threshold cycle value and the equation of the line generated in the standard curve (Herrmann-Hoesing et al. 2007). Proviral concentrations of zero were treated as SRLV negative for prevalence calculations.
Sequence variants and genotyping
TMEM154 genotypes were generated by Sanger sequencing of PCR fragments amplified from genomic DNA for 947 samples from Idaho 2008, 340 samples from Iowa, 353 samples from Idaho 2004 and 352 samples from Montana, as previously described (Heaton et al. 2012). Partway through the study, a faster and cheaper genotyping method became commercially available with greater than 98.5% concordance to the earlier PCR/sequencing method (Heaton et al. 2013). Therefore, another 244 samples from Montana were genotyped commercially by GeneSeek, Inc., using the new mass-spectrometric genotyping method (Heaton et al. 2013).
A deletion (g.52945778_52945781delATTC relative to accession NC_019476.1) in the promoter region of ovine CCR5 was genotyped as previously described (White et al. 2009). Briefly, fluorescent TaqMan genotyping was performed per manufacturer's protocol (Applied Biosystems) as previously described (White et al. 2009). An earlier restriction fragment length polymorphism assay (White et al. 2009) was used as a supplementary genotyping approach in some samples either as a primary genotyping method for the oldest samples or for verification purposes on some later samples (Idaho 2004 and Iowa 2009).
Statistical and bioinformatic analyses
phase 2.1 (Stephens et al. 2001; Stephens & Donnelly 2003) was used to determine TMEM154 diplotypes for 352 ewes from the Montana flock that were genotyped by sequencing. The confounded genotypes from these sequenced animals were processed using phase-known settings with diplotypes from commercial genotyping of additional animals from the same flock (Stephens et al. 2001; Stephens & Donnelly 2003). Compliance with Hardy–Weinberg proportions was determined for all genotypes using chi-square tests prior to further statistical analysis.
Mean proviral concentrations shown in Fig. 1 were calculated by simple average of log10-transformed proviral concentrations that were then reverse-transformed to copies/μg DNA scale. The scale transformation was performed to reduce the influence of outlier individuals (extremely high proviral concentration) in calculating mean proviral concentration. The mixed model procedure of sas v9.2 (SAS Institute) was used to examine association of genotypic variants with log10-transformed proviral concentration among positive animals. Proviral concentration was the dependent variable, and breed, age and genotype were included as fixed effects in the association models. Genotypes of interest were defined as previously reported for the CCR5 insertion/deletion (White et al. 2009). For TMEM154, only diplotypes 1,1, 1,2, 1,3, 2,2, 2,3 and 3,3 were included in analyses. Simultaneous testing was performed to analyze information content of TMEM154 haplotypes and CCR5 promoter deletion in one test, and the rest of the association model was as stated above. Furthermore, joint analysis of genotypes from all flocks was performed using the mixed model procedure of sas 9.2. Models were similar to those described above, but they also included random terms for location and year. All reported P-values were nominal and were not adjusted for multiple testing. The ggplot2 graphics package (Wickham 2009) in r v3.0.1 (Team 2011) was used for figure construction.
Figure 1.
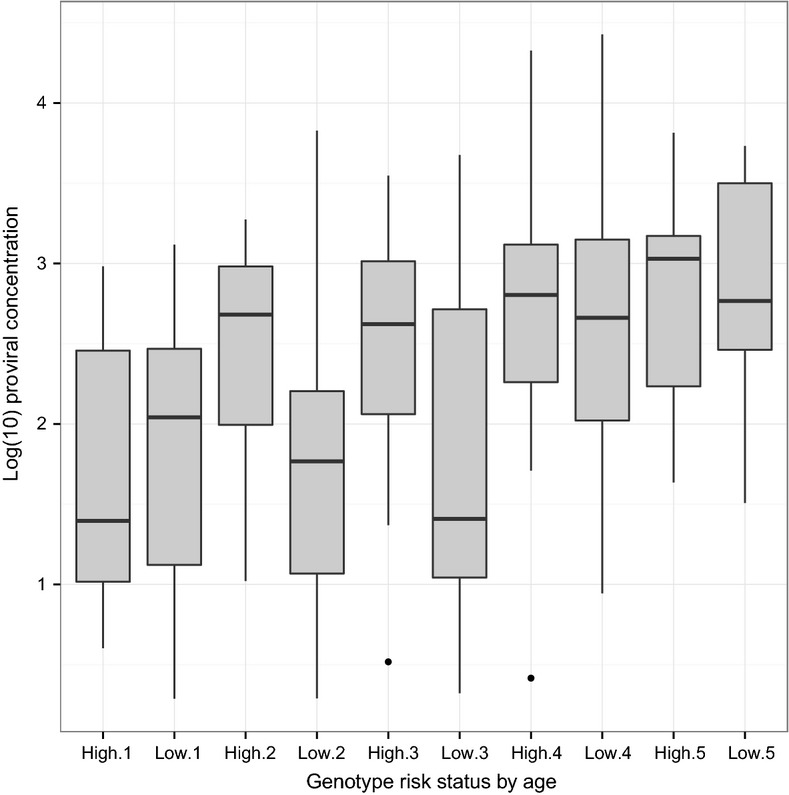
Boxplot of log10-transformed proviral concentration by age and TMEM154 diplotype risk status in the Iowa flock.
Results
Only the 1403 SRLV-positive sheep from Idaho 2004, Idaho 2008, Iowa and Montana were included in association analysis for proviral concentration and TMEM154 mutations or CCR5 promoter deletion. Adjusted mean proviral concentrations and other summary data by flock are shown in Table 1. A significant association was identified between TMEM154 and lower proviral concentration in the Iowa flock (P = 0.017; Table 2). Specifically, sheep with two copies of haplotype 1 had half the adjusted mean proviral concentration compared to sheep with one or more copies of haplotypes 2 or 3 (Table 2). The other three flocks had no identified association between TMEM154 and proviral concentration (P > 0.05; Table 2); however, these flocks had very low haplotype frequencies for haplotypes 2 and 3 (Table 3). Specifically, one ewe in the Montana flock, 19 ewes in Idaho 2004 and 32 ewes in Idaho 2008 had haplotype 2 or 3 (Table 3). The nonsignificant results were: Idaho 2004 (P = 0.064) with adjusted mean proviral concentrations of 304 and 774 for diplotypes 1,1 and those containing haplotypes 2 or 3 respectively, Idaho 2008 (P = 0.63) with adjusted means of 123 and 102 respectively, and Montana was inestimable on an individual flock basis with mean proviral concentration of 1329 for 1,1 diplotypes. The joint analysis of all flocks was significant (P = 0.013; Table 2).
Table 1.
Average proviral concentration from different flocks of sheep
| Animal set | Montana | Idaho 2004 | Idaho 2008 | Iowa |
|---|---|---|---|---|
| Total animals genotyped | 596 | 353 | 947 | 340 |
| Year of sampling | 2009 | 2004 | 2008 | 2009 |
| Breeds included | Rambouillet–Columbia crossbred | Rambouillet, Polypay and Columbia | Rambouillet, Polypay and Columbia | Polypay |
| Proviral concentration | 4770 | 1530 | 1250 | 1210 |
| qPCR-positive sheep | 607/620 = 97.9% | 226/377 = 60.0% | 368/947 = 38.9% | 202/340 = 59.4% |
Table 2.
Association between TMEM154 diplotypes and small ruminant lentivirus proviral concentration, including adjusted mean proviral concentration1 by diplotype
| Diplotype | |||
|---|---|---|---|
| Flock | P-value | 1,1 | Contain 2 or 3 |
| Iowa | 0.017 | 131 | 270 |
| Idaho 2004 | NS2 | ||
| Idaho 2008 | NS2 | ||
| Montana | NS2 | ||
| All flocks | 0.013 | 311 | 508 |
Adjusted means were reverse-transformed to the copies/μg DNA scale.
Not significant (P > 0.05)
Table 3.
Number of small ruminant lentivirus infected individuals bearing different TMEM154 diplotypes by flock
| Flock | Montana | Idaho 2004 | Idaho 2008 | Iowa | Total |
|---|---|---|---|---|---|
| Diplotype 1,1 | 595 | 171 | 333 | 88 | 1187 |
| Diplotypes containing 2 or 3 | 1 | 19 | 32 | 94 | 146 |
| Total animals | 596 | 190 | 365 | 182 | 1333 |
A significant association was identified between the CCR5 promoter insertion and proviral concentration in the Iowa flock (P < 0.05; Table 4). The adjusted mean log10 proviral concentration was higher in the deletion homozygotes than in the insertion homozygotes and heterozygotes (Table 4). The Montana and Idaho 2008 sheep flocks showed no significant association between CCR5 promoter deletion and proviral concentration (P > 0.05; Table 4). Specifically, for Montana, the association significance was P = 0.55 with adjusted means 1202, 1479 and 1288 for insertion homozygote, heterozygote and deletion homozygote respectively. For Idaho 2008, the association was significance was P = 0.96 with adjusted means of 120, 115 and 107 respectively. Numbers of animals by CCR5 genotype are shown in Table S1. The joint analysis of CCR5 including all flocks was significant (P = 0.028; Table 4). Simultaneous testing of markers in both genes was significant in the joint all-flocks analysis: TMEM154 (P = 0.016) and CCR5 (P = 0.023). Individual flock analyses provided similar results to the individual gene, single flock analyses: TMEM154 (P = 0.023) and CCR5 (P = 0.011) in the Iowa flock, TMEM154 (P > 0.05) and CCR5 (P = 0.010) for Idaho 2004 and (P > 0.05) for both genes in the other flocks. The breed, proviral concentration, TMEM154 diplotypes, age and CCR5 promoter variant genotypes for all animals in this study are shown in Table S2.
Table 4.
Association between CCR5 promoter deletion and small ruminant lentivirus proviral concentration, including adjusted mean proviral concentration1 by genotype
| Genotype2 | ||||
|---|---|---|---|---|
| Flock | P-value | II | ID | DD |
| Iowa | 0.041 | 126 | 288 | 371 |
| Idaho 20043 | 0.00773 | 3123 | 4943 | 1113 |
| Idaho 2008 | NS4 | |||
| Montana | NS4 | |||
| All flocks | 0.028 | 280 | 377 | 270 |
Adjusted means were reverse-transformed to the copies/μg DNA scale.
DD is homozygous deletion, II is homozygous insertion, and ID is heterozygous insertion/deletion.
Data from White et al. (2009).
Not significant (P > 0.05).
Discussion
This study examined the association between specific variants in two genes and SRLV proviral concentration using multiple flocks of sheep. Markers in the first gene, TMEM154, had already been validated for SRLV odds of infection but had never been examined for any measure of control post-infection. A marker in the second gene, CCR5, was associated with proviral concentration in one flock. If either variant was consistently associated with proviral concentration in multiple flocks of sheep, it would support potential use of one or more validated genetic markers for post-infection control of SRLV in sheep.
Sheep with two copies of TMEM154 haplotype 1 had half the proviral concentration compared with those with at least one copy of haplotype 2 or 3 in the Iowa flock (Table 2). Thus, the same diplotype previously identified with lower risk of initial SRLV infection (Heaton et al. 2012) also had improved control of SRLV post-infection in the Iowa flock. Boxplots by age and TMEM154 risk status are shown in Fig. 1. Only ages one to five years were included due to very low numbers of sheep age six to eight years. Almost all ages showed higher median proviral concentration in high-risk compared with low-risk diplotypes; the only exception was one year of age, when the high-risk diplotypes had lower median proviral concentration. Ages two and three years show the largest differences in median proviral concentration. At more advanced ages, smaller numbers of ewes with TMEM154 high-risk haplotype 2 or haplotype 3 could be due to culling or premature death (Fig. 1). Further, haplotype 1 appears to have a recessive mode of inheritance because two copies of haplotype 1 were required to show lower proviral concentration, which was also true for lower odds of infection (Heaton et al. 2012).
Although the joint analysis of all flocks showed association between TMEM154 diplotypes and SRLV proviral concentration (P = 0.013; Table 2), this analysis was dominated by the Iowa flock. The Idaho 2004, Idaho 2008 and Montana sheep flocks did not show significant association between TMEM154 haplotypes and proviral concentration in individual flock analysis (Table 2). However, the lack of association may be due to low frequencies of haplotypes 2 and 3 in these three sheep flocks (Table 3). None of these flocks had more than 32 individuals with haplotypes 2 or 3, compared with the Iowa flock, which had 94 ewes with haplotype 2 and/or 3. Other flocks with similar breed composition have been found to have higher allele frequencies of haplotype 2 and 3 (Heaton et al. 2013), and the low frequencies here are consistent with a hypothesis of historical selection against haplotypes 2 and 3 in these flocks that had a high occurrence of SRLV infection. It is also possible that the different allele frequencies between flocks could be due to genetic drift, as from founder effects. Without access to historical samples, it is not possible at present to distinguish selection from other potential explanations. In order to validate the association between TMEM154 haplotypes and proviral concentration in the Iowa flock, additional sheep flocks with higher frequencies of haplotype 2 and/or haplotype 3 among SRLV infected sheep will need to be identified and tested. If this genetic marker is validated for SRLV proviral concentration, it can be used for marker-assisted selection to not only reduce susceptibility but also to lower proviral concentration and severity of disease.
Previously, the CCR5 promoter deletion homozygotes were associated with lower proviral concentration in one flock (Idaho 2004) (White et al. 2009). Here, the Iowa animal set showed significant association with genotypes at this locus, but the direction of the association was opposite compared with previous findings (Table 4). Insertion homozygotes had lower proviral concentration than either deletion homozygotes or insertion/deletion heterozygotes. Although the joint analysis of all flocks was significant (P = 0.028; Table 4), the genotypic adjusted means showed higher proviral concentration in heterozygotes than in either homozygote. This can be explained by the opposite directions of association between the Idaho 2004 and Iowa flocks. In each case, one homozygote and the heterozygote showed high proviral concentration. When considered in an overall joint analysis, only the heterozygote was consistently associated with high proviral concentration. Further, current data showed no significant association in the other sheep flocks, even with testable frequencies for all genotypes (Table S1). Clearly, there was no consistent association between the CCR5 deletion and control of SRLV post-infection (Table 4).
There are multiple possible explanations for the inconsistent association of the CCR5 promoter deletion and SRLV proviral concentration. One possible reason is that the CCR5 promoter variant might occur in differing degrees of linkage disequilibrium with one or more additional underlying functional variants that are important for control of SRLV in different sheep flocks (Thormar 2005; Li & Meyre 2013), despite gene expression differences noted previously (White et al. 2009). Because CCR5 functions in signaling pathways that promote chemotaxis, lower expression of CCR5 may reduce chemotaxis of macrophages and other target leukocytes toward infected cells and slow cellular spread (Locati et al. 2002; Ma et al. 2005). Alternatively, a second explanation for the differing directions of association might occur because of one or more virus subtypes. Retroviruses mutate rapidly, and it is possible that some subtypes adapted to the insertion allele instead of the deletion allele. A third possibility could be unidentified co-infections with other, as yet unknown, immunomodulatory pathogens (Walson & John-Stewart 2007). Co-infection with such pathogens could change immune responses and SRLV disease severity in some sheep populations but not others where such co-infections were absent and, thus, change the direction of genetic variant association. Therefore, additional studies are necessary to clarify the role of the CCR5 promoter deletion in SRLV infection of sheep and in the immune system more generally.
Summary
In conclusion, testing for association between control of SRLV post-infection and genotypes for TMEM154 and CCR5 variants suggests different approaches to further work in each case. This is the first report of an association between TMEM154 diplotypes and SRLV proviral concentration, and the same desirable TMEM154 haplotypes associated with decreased proviral concentration in this flock were also associated with decreased SRLV odds of infection in a previous report (Heaton et al. 2012). Future work with additional sheep flocks possessing more balanced diplotype frequencies is needed to confirm or refute the association of TMEM154 with SRLV proviral concentration. For CCR5, a promoter deletion was not consistently associated with proviral concentration in multiple flocks of sheep. Further work with additional variants in the genomic region, SRLV strain data and/or co-infection data concerning other pathogens may help explain the complicated patterns of association observed here.
Acknowledgments
The authors wish to thank Codie Durfee, Caylee Birge, Salim Alhajri, Nic Durfee, Ralph Horn and James Allison for technical assistance; Mark Williams and the USSES technical staff for the Idaho sample collections; and Tom Kellom for USSES data collection. The genotyping of TMEM154 diplotypes for Iowa and Idaho sheep was performed at USMARC. The authors thank Lucia Sider and Michael Clawson for provision of reagent designs. S.N. White, L.M. Herrmann-Hoesing, M.R. Mousel and G.S. Lewis were co-authors on the paper describing the association of TMEM154 with odds of SRLV infection, but there is no intellectual property restricting its use. Otherwise, the authors have no conflicts of interest to declare. This research was funded by USDA-ARS grant 5348-32000-031-00D. Fahad Alshanbari's graduate training was funded by the Saudi Ministry of Higher Education.
Supporting information
Additional supporting information may be found in the online version of this article.
Table S1. Numbers of positive individuals by CCR5 genotype from four flocks.
Table S2. All animals included in the study with breed, mean qPCR, age, CCR5 genotype and TMEM154 diplotype.
References
- Alkhatib G, Berger EA. HIV coreceptors: from discovery and designation to new paradigms and promise. European Journal of Medical Research. 2007;12:375–84. [PubMed] [Google Scholar]
- Annett RW, Carson AF, Dawson LE, Irwin D, Gordon AW, Kilpatrick DJ. Comparison of the longevity and lifetime performance of Scottish Blackface ewes and their crosses within hill sheep flocks. Animal: An International Journal of Animal Bioscience. 2011;5:347–55. doi: 10.1017/S1751731110002107. [DOI] [PubMed] [Google Scholar]
- Arsenault J, Dubreuil P, Girard C, Simard C, Belanger D. Maedi-visna impact on productivity in Quebec sheep flocks (Canada) Preventive Veterinary Medicine. 2003;59:125–37. doi: 10.1016/s0167-5877(03)00086-2. [DOI] [PubMed] [Google Scholar]
- Blacklaws BA. Small ruminant lentiviruses: immunopathogenesis of visna-maedi and caprine arthritis and encephalitis virus. Comparative Immunology, Microbiology, and Infectious Diseases. 2012;35:259–69. doi: 10.1016/j.cimid.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Byun SO, Forrest RH, Frampton CM, Zhou H, Hickford JG. An association between lifespan and variation in insulin-like growth factor I receptor in sheep. Journal of Animal Science. 2012;90:2484–7. doi: 10.2527/jas.2011-4148. [DOI] [PubMed] [Google Scholar]
- Cutlip RC, Lehmkuhl HD, Brogden KA, Sacks JM. Breed susceptibility to ovine progressive pneumonia (maedi/visna) virus. Veterinary Microbiology. 1986;12:283–8. doi: 10.1016/0378-1135(86)90057-x. [DOI] [PubMed] [Google Scholar]
- Cutlip RC, Lehmkuhl HD, Sacks JM, Weaver AL. Seroprevalence of ovine progressive pneumonia virus in sheep in the United States as assessed by analyses of voluntarily submitted samples. American Journal of Veterinary Research. 1992;53:976–9. [PubMed] [Google Scholar]
- De Boer GF, Terpstra C, Houwers DJ, Hendriks J. Studies in epidemiology of maedi/visna in sheep. Research in Veterinary Science. 1979;26:202–8. [PubMed] [Google Scholar]
- Gates NL, Winward LD, Gorham JR, Shen DT. Serologic survey of prevalence of ovine progressive pneumonia in Idaho range sheep. Journal of the American Veterinary Medical Association. 1978;173:1575–7. [PubMed] [Google Scholar]
- Gendelman HE, Narayan O, Kennedy-Stoskopf S, Kennedy PG, Ghotbi Z, Clements JE, Stanley J, Pezeshkpour G. Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation of monocytes to macrophages. Journal of Virology. 1986;58:67–74. doi: 10.1128/jvi.58.1.67-74.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrell MD, Brandon MR, Sheffer D, Adams RJ, Narayan O. Ovine lentivirus is macrophagetropic and does not replicate productively in T lymphocytes. Journal of Virology. 1992;66:2679–88. doi: 10.1128/jvi.66.5.2679-2688.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton MP, Clawson ML, Chitko-McKown CG, et al. Reduced lentivirus susceptibility in sheep with TMEM154 mutations. PLoS Genetics. 2012;8:e1002467. doi: 10.1371/journal.pgen.1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton MP, Kalbfleisch TS, Petrik DT, Simpson B, Kijas JW, Clawson ML, Chitko-McKown CG, Harhay GP, Leymaster KA. Genetic testing for TMEM154 mutations associated with lentivirus susceptibility in sheep. PLoS ONE. 2013;8:e55490. doi: 10.1371/journal.pone.0055490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann-Hoesing LM, White SN, Lewis GS, Mousel MR, Knowles DP. Development and validation of an ovine progressive pneumonia virus quantitative PCR. Clinical and Vaccine Immunology. 2007;14:1274–8. doi: 10.1128/CVI.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann-Hoesing LM, White SN, Mousel MR, Lewis GS, Knowles DP. Ovine progressive pneumonia provirus levels associate with breed and Ovar-DRB1. Immunogenetics. 2008;60:749–58. doi: 10.1007/s00251-008-0328-9. [DOI] [PubMed] [Google Scholar]
- Herrmann-Hoesing LM, Noh SM, White SN, Snekvik KR, Truscott T, Knowles DP. Peripheral ovine progressive pneumonia provirus levels correlate with and predict histological tissue lesion severity in naturally infected sheep. Clinical and Vaccine Immunology. 2009;16:551–7. doi: 10.1128/CVI.00459-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genetics in Medicine. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Houwers DJ, Konig CD, de Boer GF, Schaake J., Jr Maedi-visna control in sheep. I. Artificial rearing of colostrum-deprived lambs. Veterinary Microbiology. 1983;8:179–85. doi: 10.1016/0378-1135(83)90064-0. [DOI] [PubMed] [Google Scholar]
- Houwers DJ, Schaake J, Jr, de Boer GF. Maedi-visna control in sheep II. Half-yearly serological testing with culling of positive ewes and progeny. Veterinary Microbiology. 1984;9:445–51. doi: 10.1016/0378-1135(84)90065-8. [DOI] [PubMed] [Google Scholar]
- Houwers DJ, Visscher AH, Defize PR. Importance of ewe/lamb relationship and breed in the epidemiology of maedi-visna virus infections. Research in Veterinary Science. 1989;46:5–8. [PubMed] [Google Scholar]
- Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. Journal of Infectious Diseases. 2005;191(Suppl 1):S68–77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- Keen JE, Hungerford LL, Littledike ET, Wittum TE, Kwang J. Effect of ewe ovine lentivirus infection on ewe and lamb productivity. Preventive Veterinary Medicine. 1997;30:155–69. doi: 10.1016/s0167-5877(96)01101-4. [DOI] [PubMed] [Google Scholar]
- Li A, Meyre D. Challenges in reproducibility of genetic association studies: lessons learned from the obesity field. International Journal of Obesity. 2013;37:559–67. doi: 10.1038/ijo.2012.82. [DOI] [PubMed] [Google Scholar]
- Locati M, Otero K, Schioppa T, Signorelli P, Perrier P, Baviera S, Sozzani S, Mantovani A. The chemokine system: tuning and shaping by regulation of receptor expression and coupling in polarized responses. Allergy. 2002;57:972–82. doi: 10.1034/j.1398-9995.2002.02166.x. [DOI] [PubMed] [Google Scholar]
- Ma B, Kang MJ, Lee CG, et al. Role of CCR5 in IFN-gamma-induced and cigarette smoke-induced emphysema. Journal of Clinical Investigation. 2005;115:3460–72. doi: 10.1172/JCI24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhans E, Greenland T, Badiola J, et al. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Veterinary Research. 2004;35:257–74. doi: 10.1051/vetres:2004014. [DOI] [PubMed] [Google Scholar]
- Reina R, Berriatua E, Lujan L, Juste R, Sanchez A, de Andres D, Amorena B. Prevention strategies against small ruminant lentiviruses: an update. Veterinary Journal. 2009;182:31–7. doi: 10.1016/j.tvjl.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Slager RE, Li X, Meyers DA, Bleecker ER. Recent developments in the genetics of asthma susceptibility and severity. In: Wenzel SE, editor; Chung KF, Bel EH, editors. Difficult-to-Treat Severe Asthma. Plymouth, UK: European Respiratory Society; 2011. pp. 82–96. [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. American Journal of Human Genetics. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RDC. r: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- Thormar H. Maedi-visna virus and its relationship to human immunodeficiency virus. AIDS Reviews. 2005;7:233–45. [PubMed] [Google Scholar]
- Walson JL, John-Stewart G. Treatment of helminth co-infection in individuals with HIV-1: a systematic review of the literature. PLoS Neglected Tropical Diseases. 2007;1:e102. doi: 10.1371/journal.pntd.0000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SN, Knowles DP. Expanding possibilities for intervention against small ruminant lentiviruses through genetic marker-assisted selective breeding. Viruses. 2013;5:1466–99. doi: 10.3390/v5061466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SN, Mousel MR, Reynolds JO, Lewis GS, Herrmann-Hoesing LM. Common promoter deletion is associated with 3.9-fold differential transcription of ovine CCR5 and reduced proviral level of ovine progressive pneumonia virus. Animal Genetics. 2009;40:583–9. doi: 10.1111/j.1365-2052.2009.01882.x. [DOI] [PubMed] [Google Scholar]
- White SN, Mousel MR, Herrmann-Hoesing LM, Reynolds JO, Leymaster KA, Neibergs HL, Lewis GS, Knowles DP. Genome-wide association identifies multiple genomic regions associated with susceptibility to and control of ovine lentivirus. PLoS ONE. 2012;7:e47829. doi: 10.1371/journal.pone.0047829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Numbers of positive individuals by CCR5 genotype from four flocks.
Table S2. All animals included in the study with breed, mean qPCR, age, CCR5 genotype and TMEM154 diplotype.


