Abstract
The biological roles of heparin (HP) and heparan sulfate (HS) are mediated mainly through their interaction with proteins. In the present work, we provide a rapid method for screening HP/HS–protein interactions providing structural data on the key sulfo groups that participate in the binding. A library of polysaccharides structurally related to HP was prepared by immobilizing the biotinylated N-sulfated K5 polysaccharide (N-sulfoheparosan) on sensor chips followed by selective modification of this polysaccharide with enzymes that participate in HP/HS biosynthesis. The polysaccharides synthesized on the surface of the sensor chips differ in the number and position of sulfo groups present both on uronic acid and glucosamine residues. Surface plasmon resonance was used to measure the interaction of each member of this polysaccharide library with antithrombin III (ATIII), to afford structural information on sulfo groups required for this HP/HS–protein interaction. This method is viewed as widely applicable for the study of the structure–activity relationship (SAR) of HP/HS–protein interactions.
Keywords: Heparin, Surface plasmon resonance, Sulfo group, Enzymatic synthesis antithrombin
Heparin (HP) and heparan sulfate (HS) are structurally related glycosaminoglycans (GAGs) that participate in numerous important biological processes such as blood coagulation, viral and bacterial entry and infection, angiogenesis, and cancer development [1–6]. They are linear polysaccharides with a repeating disaccharide unit of 1,4-linked uronic acid (D-glucuronic or L-iduronic acid) and D-glucosamine residues. Both uronic acid and glucosamine can contain sulfo groups at different positions of the pyranose ring including 2-O-sulfo substitution of the uronic acid residue, and 2-N-, 3-O-, and 6-O-sulfo substitution in the glucosamine residue. All these structural variations make HP/HS a very heterogeneous highly charged polysaccharide [7]. HP/HS carry out their biological functions primarily by their interaction with proteins in which the sulfo groups of the GAG electrostatically interact or hydrogen bond with basic amino acids of the target protein [8]. Frequently, heparin-binding proteins interact with HP/HS specifically and with high affinity, as dictated by the number and relative position of the sulfo groups in the interacting oligosaccharide sequence [9,10]. The most widely studied example is the highly specific binding of HP/HS with the serine protease inhibitor, ATIII. The effective interaction between ATIII and HP/HS requires the presence of a sulfo group in position 3 of the central glucosamine of a pentasaccharide of sequence GlcNS6S-GlcA-GlcNS3S ± 6S-IdoA2S-GlcNS6S [11,12]. In addition to ATIII, there are a large and increasing number of HP/HS-binding proteins involved in important biological processes [8]. Structural information has been reported in some of the HP/HS–protein complexes revealing the relative importance of certain sulfo groups in specific positions of the uronic acid and/or glucosamine residues [10,13]. Unfortunately, in most cases there is a lack of general approaches for identifying key sulfo groups for protein-binding specificities.
Here we present a method for a rapid screening of HP/HS–protein interactions. A small library of polysaccharides structurally related to HP/HS was synthesized on a sensor chip using bacterially expressed C5-epimerase and sulfotransferases, and the ability of each polysaccharide to bind ATIII was analyzed using surface plasmon resonance (SPR). This method measures the interaction between an immobilized ligand on a sensor chip and an analyte that is passed over the surface of the sensor chip. The mass of analyte that binds to the immobilized ligand is monitored as a function of time (sensorgram). Thus, binding is determined in real time and detailed analysis of the resulting sensorgrams provides quantitative kinetic and binding-affinity data [14]. In the present work, heparosan, bacterial capsular polysaccharide from Escherichia coli K5 strain, was chemically modified through de-N-acetylation and N-sulfonation to afford N-sulfoheparosan. After immobilization of N-sulfoheparosan, it was enzymatically modified with C5-epimerase and various O-sulfotransferases involved in HP/HS biosynthesis. Selective O-sulfonation of N-sulfoheparosan afforded sensor chips containing a variety of polysaccharides structurally related to HP/HS. These polysaccharides are missing sulfo groups at defined positions in their uronic acid and/or glucosamine residues. The screening of this library relied on the well-studied HP–ATIII interaction to validate the utility of this approach for understanding the SAR of HP/HS–protein interactions.
Materials and methods
Materials
Heparosan was purified from E. coli K5 strain (from American Type Culture Collection) as described in a previous report [15]. Porcine intestinal heparin (sodium salt) was from Sigma (St. Louis, MO). DEAE Sephadex ion exchange resin (50 g) and S200 HR Sephacryl GPC resin (250 ml) were from Sigma (St. Louis, MO). Sulfo-N-hydroxy-succinimide-long chain-biotin (sulfo-NHS-LC-biotin) was from Pierce (Rockford, IL). Streptavidin (SA) sensor chips and HBS–EP buffer (0.01M Hepes, pH 7.4, 0.15M NaCl, 3 mM EDTA, and 0.005% surfactant P20) were from BIAcore (Biacore AB, Uppsala, Sweden). Double distilled water and HPLC grade water were used. All buffers were filtered (0.22 μm) and degassed prior to using. SPR measurements were performed on a BIAcore 3000 instrument with BIAcore 3000 control and BIA evaluation software (ver. 4.0.1).
Expression and purification of HS biosynthesis enzymes
The catalytic domain of human C5-epimerase (E53-N609) was cloned into pMAL-c2X vector (New England Biolabs) using the BamH1 and HindIII sites to generate a maltose-binding protein (MBP)-epimerase fusion protein. The full length cDNA of C5-epimerase was a gift from Dr. Rosenberg (Massachusetts Institute of Technology). Expression of C5-epimerase was achieved in Origami-B cells (Novagen) carrying pGro7 plasmid (Takara, Japan) which expresses chaperonin proteins GroEL and GroES. The bacteria were grown in LB medium supplemented with 2 mg/ml glucose, 12.5 μg/ml tetracycline, 15 μg/ml kanamycin, 35 μg/ml chloramphenicol, and 50 μg/ml carbenicillin at 37 °C. When the OD 600 nm reached 0.6–0.8, IPTG and L-arabinose were added to a final concentration of 0.1 mM and 1 mg/ml, respectively. The protein was purified following the protocol provided by the manufacturer. The purified proteins migrated at 100 kDa on SDS–PAGE with the purity greater than 60%.
The expression and purification of mouse 6-OST-1 and Chinese hamster 2-OST were described in our previous publication [16]. The expression and purification of mouse 3-OST-1 were described elsewhere [17].
Determination of the activities of HS biosynthetic enzymes
The activity of C5-epimerase was determined by coupling the reactions of epimerization and 2-O-sulfation using N-sulfoheparosan as a substrate. Briefly, purified MBP-epimerase (~6 μg) was incubated with N-sulfoheparosan (5 μg) in 100 μl of a buffer containing 50 mM Mes, 10 mM MnCl2, 5 mM MgCl2, and 1% Triton X-100 (pH 7) at 37 °C for 2 h. MBP-2-OST (4 μg) was then added into the reaction mixture along with [35S]PAPS (1 × 106 cpm). The mixture was incubated at 37 °C for another 1 h, and the mixture was subjected to a 200 μl DEAE–Sepharose column to purify [35S]HS. The [35S]HS was degraded to disaccharides by nitrous acid (pH 1.5). The identities (either GlcA2S-AnMan or IdoA2S-AnMan) of the resultant 35S-labeled disaccharides were determined by coeluting with appropriate 3H-labeled disaccharides on reverse-phase ion-pairing HPLC [18]. After the treatment of N-sulfoheparosan by epimerase followed by 2-OST, greater than 80% of the 35S-labeled disaccharide mixture was IdoA2S-AnMan, whereas, less than 5% of the 35S-labeled disaccharides was IdoA2S-AnMan in 2-OST-treated N-sulfoheparosan. The results suggest that the expressed protein carries the anticipated C5-epimerase activity.
The activities of 2-OST and 6-OST-1 were determined following the published procedures using completely desulfated N-sulfonated heparin as a substrate [19,20]. The activity of 3-OST-1 was determined following the procedure described elsewhere [21].
Preparation and purification of heparosan
Heparosan was prepared from E. coli K5 strain as explained previously [15]. In addition to following the published procedure, the isolated polysaccharide (20 mg) was further purified by S200-HR size exclusion column (4.8 × 75 cm) eluted with 200 mM sodium chloride at a flow rate of 1 ml/min. The fractions containing heparosan were detected by carbazole assay [22] and then these fractions were dialyzed against distilled water (MWCO 1000 Da) and lyophilized to obtain the purified product (7.5 mg) for further modifications.
Preparation of N-deacetylated heparosan
Heparosan (7.0 mg) was dissolved in sodium hydroxide (2.0 ml, 2 N), incubated for 24 h at 60 °C, and cooled to room temperature, and was adjusted to pH 7 with concentrated hydrochloric acid. The reaction product was dialyzed against distilled water (MWCO 1000 Da) and lyophilized to obtain completely N-deacetylated heparosan intermediate (5.0 mg).
Partial biotinylation of N-deacetylated heparosan and heparin
The free amino groups of the unsubstituted glucosamine residues in heparin [23] were biotinylated following a procedure previously described by our group [24]. N-Deacetylated heparosan (4.0 mg, in 2 ml of 50 mM sodium bicarbonate) was treated with sulfo-NHS-LC-biotin (1.6 mg, in 20 μL dimethylformamide ~4 equiv. biotin/polysaccharide chain) to react with the free amino groups of glucosamine residues of the polysaccharide, following the general procedure for heparin biotinylation. The reaction product was dialyzed (MWCO 1000 Da) and lyophilized to obtain pure partially biotinylated, N-deacetylated heparosan (4.0 mg).
N-Sulfonation of biotinylated N-deacetylated heparosan
The partially biotinylated N-deacetylated heparosan intermediate (4.0 mg in 1 ml distilled water) was treated with sodium bicarbonate (10 mg) and trimethylamine–sulfurtrioxide complex (10 mg) in a single step and incubated at 50 °C for 12 h. Equal portion of sodium bicarbonate and trimethylamine–sulfur trioxide was added two more times at 12 h intervals. The solution was then brought to room temperature and dialyzed overnight against distilled water (MWCO 1000 Da). The dialyzate was lyophilized to obtain pure, completely N-substituted, N-sulfoheparosan with less than 4 biotin tags per polysaccharide chain.
Immobilization of biotinylated, N-sulfoheparosan to the SA sensor chips
The SA sensor chip was pre-treated with three consecutive injections (5 μl) of 50 mM NaOH in 1M NaCl at 5 μl/min, to remove any non-specifically bound contaminants. Biotinylated, N-sulfoheparosan (0.1 mg/ml in HBS–EP running buffer) was flowed at 37 °C (5 μl/min, manual injection) over one or two flow cells simultaneously followed by a 5μl injection of 2M NaCl. The binding of biotinylated, N-sulfoheparosan and heparin–biotin was confirmed by the observation of a 570–813 RU response.
Enzymatic modification of N-sulfoheparosan on sensor chip surfaces
Each enzyme was diluted in buffer containing 50 mM Mes, 0.1% Triton X-100, 5 mM MgCl2, 10 mM MnCl2, and 0.1 mg/ml bovine serum albumin. C5-epimerase was diluted to a final concentration of 0.5 μM; 2-O-, 6-O-, and 3-O-sulfotransferases were combined with PAPS (0.08 mM) prior to injection and diluted to a final concentration of 0.6 μM. Every modification occurred by injecting 15 μl of the enzymatic solution three times for epimerase, and four times for the sulfotransferases, consecutively at a flow rate of 1 μl/min and temperature of 37 °C. After every injection, the sensor chip was cleaned with 5 μl of 2 M NaCl.
Binding studies of the interaction between ATIII and modified N-sulfoheparosan polysaccharides
ATIII (500 nM in HBS–EP running buffer) was injected at a flow rate of 5 μl/min over the four flow cells of every sensor chip at 25 °C. The protein was injected for 3 min following surface regeneration by injection of 5 μl of 2M NaCl, followed by 5 μl NaOH (50 mM) containing 1M NaCl. The response was monitored as a function of time (sensorgram).
Affinity and kinetic study of the interaction of ATIII with fully (C5-epi,2,6,3-OST) modified N-sulfoheparosan and HP
Different concentrations of ATIII (10, 25, 50, 75, and 100 nM in HBS–EP running buffer) were injected at a flow rate of 30 μl/min over a chip with fully modified N-sulfoheparosan or HP immobilized to one of the flow cells at 25 °C. A kinetic injection mode was used, leading the protein to flow for 3 min and to be dissociated for the next 3 min. The surface was regenerated by injection of 30 μl of 2M NaCl, followed by 30 μl NaOH (50 mM) containing 1 M NaCl. The response was monitored as a function of time (sensorgram).
Results
Enzymatic modification of N-sulfoheparosan polysaccharide on sensor chip surfaces
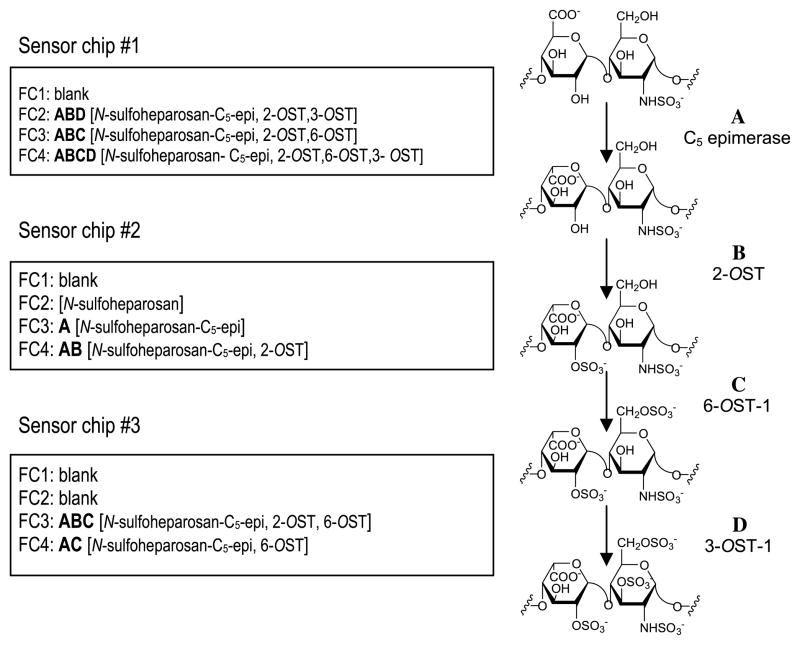
In the biosynthesis of HP/HS, a copolymer of 1,4-linked GlcA and N-acetylated glucosamine (GlcNAc) is modified by several enzymes including N-deacetylase/N-sulfotransferase, C5-epimerase, and 2-O-, 6-O-, and 3-O-sulfotransferases (ST) [25,26]. The starting polysaccharide (heparosan) used in the present study is structurally related to HP/HS. Heparosan, a copolymer of 1,4-linked GlcA and GlcNAc, was obtained as a capsular K5 polysaccharide from E. coli, and chemically N-deacetylated, and N-sulfonated to afford N-sulfoheparosan. Biotinylated, N-sulfoheparosan was immobilized in three streptavidin (SA) sensor chips through a strong, non-covalent, biotin–streptavidin interaction. Four different enzymes were next used to generate a small polysaccharide library. C5-epimerase converts glucuronic into iduronic acid and sulfotransferases 2-OST, 6-OST-1, and 3-OST-1 transfer sulfo groups from PAPS to the positions 2-O- of uronic acid, and 6-O- and 3-O- of the glucosamine. Each modification of the N-sulfoheparosan on the sensor surface was carried out by injecting the enzyme for 15 min at least three times (in the presence of PAPS for the O-ST enzymes) under experimental conditions designed to maximize the efficiency of the enzymatic reaction. Consecutive injections of C5-epimerase, 2-OST, 6-OST-1, and/or 3-OST-1 in each flow cell mimicked the biosynthetic pathway in which heparosan is converted to HP/HS in the Golgi [26,27]. From our previous solution-phase studies on these enzymes, we estimate the percent conversion at each step was ~10% GlcA to IdoA, ~80% IdoA to IdoA2S, ~30% GlcNS to GlcNS6S, and ~10% GlcNS ± 6S to GlcNS ± 6S3S [16]. In one flow cell on the sensor chip N-sulfoheparosan was fully modified, affording a heparin-like polysaccharide. This fully modified polysaccharide, as well as intermediate polysaccharides, N-sulfoheparosan (control), N-sulfoheparosan modified with C5-epi, N-sulfoheparosan modified with C5-epi and 2-OST, N-sulfoheparosan modified with C5-epi, 2-OST and 6-OST, N-sulfoheparosan modified with C5-epi, 2-OST, and 3-OST, and N-sulfoheparosan modified with C5-epi and 6-OST, lacking sulfo groups at specific positions of the polysaccharide chain were prepared as a small polysaccharide library for evaluation by SPR (Fig. 1).
Fig. 1.
Schematic representation of the enzymatic synthesis of polysaccharides structurally related with HP on sensor chip surfaces. Three sensor chips containing immobilized polysaccharides were prepared.
Binding of ATIII to the N-sulfoheparosan derived polysaccharide library
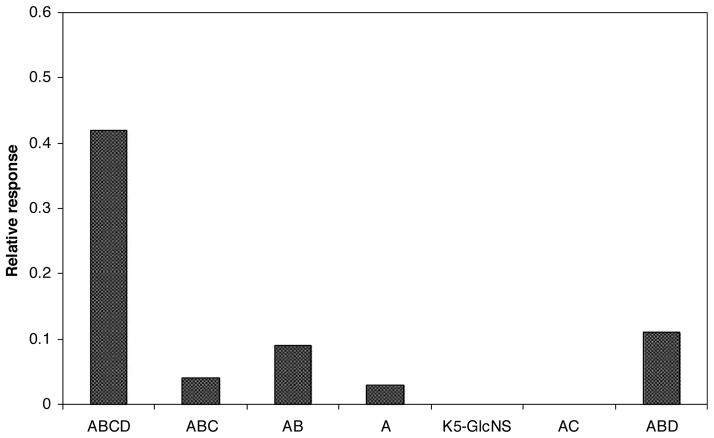
Interaction between ATIII and HP/HS has been studied in great detail [28]. ATIII binds specifically to the HP pentasaccharide GlcNS6S-GlcA-GlcNS3S ± 6S-IdoA2S-GlcNS6S. The sulfo group in position 3 of the central glucosamine residue (bolded) is required for the binding. Sulfo groups in positions 2-O- of uronic acid and N-positions of the glucosamine residue are known to play a less critical role in binding to ATIII while 6-O-sulfonation of glucosamine residue appears to be essential [29–32]. These structural characteristics make the ATIII-HP/HS interaction a good model for the evaluation of our small polysaccharide library. A solution of ATIII (500 nM in HBS–EP buffer) was injected for 3 min over the three sensor chips at a flow rate of 5 μl/min. The ability of the modified N-sulfoheparosan polysaccharides to bind ATIII was observed, monitoring the response as a function of time (sensorgram). No detectable binding was observed between ATIII and N-sulfoheparosan or N-sulfoheparosan modified with C5-epi and 6-OST and a weak response was monitored when ATIII was injected over N-sulfoheparosan modified with C5-epi, N-sulfoheparosan modified with C5-epi and 2-OST, N-sulfoheparosan modified with C5-epi, 2-OST, and 6-OST, and N-sulfoheparosan modified with C5-epi, 2-OST, and 3-OST. A much higher response was obtained when ATIII was flowed over the flow cell containing N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST, resulting in a fully modified, heparin-like polysaccharide (Fig. 2).
Fig. 2.
Diagram of the response obtained in the sensorgrams of the binding of ATIII (500 nM) with modified heparosan after 3 min injection. Enzymatic modifications: A (N-sulfoheparosan modified with C5-epi), AB (N-sulfoheparosan modified with C5-epi and 2-OST), ABC (N-sulfoheparosan modified with C5-epi, 2-OST, and 6-OST), ABCD (N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST and 3-OST), ABD (N-sulfoheparosan modified with C5-epi, 2-OST, and 3-OST), AC (N-sulfoheparosan modified with C5-epi, and 6-OST). For a better comparison a relative response (RU of the binding/RU of the polysaccharide immobilization) has been used.
Affinity and kinetic study of the interaction of ATIII with N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST (heparin-like polysaccharide)
N-Sulfoheparosan modified with C5-epi, 2-OST, 6-OST and 3-OST corresponds to a fully modified, heparin-like polysaccharide. Thus, its interaction with ATIII should present binding and kinetic behavior comparable to the HP/HS–ATIII interaction. Different concentrations of ATIII (10, 25, 50, 75, and 100 nM in HBS–EP) were injected over the sensor chip containing the heparin-like polysaccharide for 3 min and the dissociation was monitored for another 3 min. The sensorgrams were globally fitted to a conformational change model (Fig. 3A) and a dissociation constant of 61.7 nM was calculated (Table 1). This value is in the same range as the value previously published of the interaction between ATIII and HP (KD = 50 nM) [12,33,34] or HS (KD = 118 nM) [24]. Additionally we monitored the interaction of ATIII with HP under the same experimental conditions using SPR technology obtaining comparable affinity and kinetic data as the ATIII interaction with N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST (Table 1).
Fig. 3.
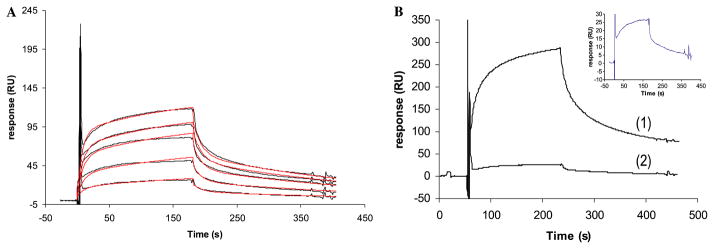
(A) Sensorgrams of the interaction of ATIII (100, 75, 50, 25, and 10 nM) with N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST. The theoretical curves obtained after the global fitting are in red. (B) Sensorgrams of the interaction of ATIII (10 nM) with HP (1) and N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST and 3-OST (2). The sensorgram of the interaction between ATIII and N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST has been expanded. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
Table 1.
Kinetic and affinity data of the interaction of ATIII with N-sulfoheparosan-C5-epi, 2-OST, 6-OST, 3-OST, and HP
| Kona (×106 M−1 s−1) | Koffa (×10−2 s−1) | K+1b (×10−3 s−1) | K−1b (×10−3 s−1) | KD (nM) | χ2 | |
|---|---|---|---|---|---|---|
| ATIII with N-sulfoheparosan-C5-epi, 2-OST, 6-OST, 3-OST | 0.67 ± 0.01 | 0.62 ± 0.02 | 7.10 ± 0.11 | 3.61 ± 0.04 | 61.7 | 2.67 |
| ATIII with HP | 21.6 ± 0.03 | 3.63 ± 0.06 | 5.81 ± 0.10 | 2.85 ± 0.05 | 8.5 | 29.70 |
Kon,Koff: on and off rate constants of the binding step.
K+1,K−1: on and off rate constants of the conformational change.
Discussion
HP and HS carry out their biological functions primarily by their interaction with proteins [8]. The affinity and specificity of the binding depends on the sulfation pattern of the oligosaccharide sequence [9,10]. Consequently, to establish the SAR of HP/HS protein interaction, it is important to obtain structural information of the sulfo groups that are relevant for the protein binding. In the present study, we provide a rapid method for screening HP/HS–protein interactions to get structural data of key sulfo groups that participate in the binding event. A library of polysaccharides structurally related to HP was prepared by immobilizing biotinylated N-sulfoheparosan to sensor chips and subsequent selective modification with enzymes that participate in the biosynthesis of HP/HS. The solution-phase, enzymatic synthesis of a heparin-like polysaccharide starting from both completely de-sulfated heparin and heparosan has been reported [16,35] and the enzymatic modification of HS by 3-OST-1 on a sensor chip surface has been previously described by our group [24]. These previous successes suggested it might be possible to synthesize a small polysaccharide library on sensor chip surface. SPR experiments of the interaction of ATIII with this small polysaccharide library showed that ATIII bound with high affinity only to the heparin-like polysaccharide, N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST (Fig. 2). The absence of the sulfo group in position 3 of the glucosamine residues, N-sulfoheparosan modified with C5-epi, 2-OST, and 6-OST, decreased the SPR response by 10-fold, confirming the essential nature of this sulfo group for ATIII binding. When 3-O-sulfo groups are present but 6-O-sulfo groups are absent, N-sulfoheparosan modified with C5-epi, 2-OST, and 3-OST, a reduced sensor response was also observed. Thus, confirming that the 6-O-sulfo group in the glucosamine residue also makes a significant contribution to ATIII binding, in agreement with published data [29]. Furthermore, only the sensorgrams obtained for the interaction of ATIII and the heparin-like polysaccharide, N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST, resembled the sensorgrams obtained for HP/HS with ATIII (Fig. 3B). The affinity and kinetic data of the interaction of ATIII with N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST and HP were obtained by global fitting of the resulting sensorgrams and corresponded to a conformational change model. The excellent fit obtained is in agreement with its well-known conformational change in ATIII that occurs on ATIII binding to HP [12]. For a given ATIII concentration, the intensity of the sensorgrams obtained for heparin-like polysaccharide, N-sulfoheparosan modified with C5-epi, 2-OST, 6-OST, and 3-OST, was lower than that observed for ATIII–HP binding (Fig. 3B), nevertheless, comparable affinity and kinetic data were obtained (Table 1). These results clearly demonstrate that modification of N-sulfoheparosan with C5-epimerase, and 2-O-, 6-O-, and 3-OST-1 afforded a functional ATIII-binding site. Thus, all the enzymatic modifications must have taken place, supporting the formation of the correct structures for all the polysaccharide intermediates comprising this small library.
In conclusion, we have developed a method for the chip-based synthesis of a polysaccharide library as well as a rapid screening method for HP/HS–protein interactions. This method can be used to identify critical sulfo groups involved in binding. This method has been validated using with the well-studied ATIII–HP/HS interaction. Future studies will utilize this smaller chip-based library to screen other less studied heparin–protein interactions.
References
- 1.Folkman J, Weisz PB, Joullie MM, Li WW, Ewing WR. Control of angiogenesis with synthetic heparin substitutes. Science. 1989;243:1490–1493. doi: 10.1126/science.2467380. [DOI] [PubMed] [Google Scholar]
- 2.Lin YL, Lei HY, Lin YS, Yeh TM, Chen SH, Liu HS. Heparin inhibits dengue-2 virus infection of five human liver cell lines. Antiviral Res. 2002;56:93–96. doi: 10.1016/s0166-3542(02)00095-5. [DOI] [PubMed] [Google Scholar]
- 3.Marino M, Banerjee M, Jonquieres R, Cossart P, Ghosh P. GW domains of the Listeria monocytogenes invasion protein InlB are SH3-like and mediate binding to host ligands. EMBO J. 2002;21:5623–5634. doi: 10.1093/emboj/cdf558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 5.Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1. J Gen Virol. 2004;85:805–809. doi: 10.1099/vir.0.19641-0. [DOI] [PubMed] [Google Scholar]
- 6.Tyrrell DJ, Kilfeather S, Page CP. Therapeutic uses of heparin beyond its traditional role as an anticoagulant. Trends in Pharmacol Sci. 1995;16:198–204. doi: 10.1016/s0165-6147(00)89022-7. [DOI] [PubMed] [Google Scholar]
- 7.Comper WD. Heparin and Related Polysaccharides. Gordon Breach; NY: 1981. [Google Scholar]
- 8.Capila I, Linhardt RJ. Heparin–protein interactions. Angew Chem Int Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 9.Muñoz EM, Linhardt Robert J. Heparin-binding domains in vascular biology. Arterioscler Thromb Vascular Biol. 2004;24:1549–1557. doi: 10.1161/01.ATV.0000137189.22999.3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raman R, Sasisekharan V, Sasisekharan R. Structural insights into biological roles of protein–glycosaminoglycan interactions. Chem Biol. 2005;12:267–277. doi: 10.1016/j.chembiol.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Casu B, Oreste P, Torri G, Zoppetti G, Choay J, Lormeau JC, Petitou M, Sinaÿ P. The structure of heparin oligosaccharide fragments with high anti-(factor Xa) activity containing the minimal antithrombin III-binding sequence. Chemical and carbon-13 nuclear-magnetic-resonance studies. Biochem J. 1981;197:599–609. doi: 10.1042/bj1970599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. The anticoagulant activation of antithrombin by heparin. Proc Natl Acad Sci USA. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noti C, Seeberger PH. Chemical approaches to define the structure–activity relationship of heparin-like glycosaminoglycans. Chem Biol (Cambridge, MA, United States) 2005;12:731–756. doi: 10.1016/j.chembiol.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Karlsson R. SPR for molecular interaction analysis: a review of emerging application areas. J Mol Recogn. 2004;17:151–161. doi: 10.1002/jmr.660. [DOI] [PubMed] [Google Scholar]
- 15.Vann WF, Schmidt MA, Jann B, Jann K. The structure of the capsular polysaccharide (K5 antigen) of urinary-tract-infective Escherichia coli 010:K5:H4. A polymer similar to desulfo-heparin. Eur J Biochem. 1981;116:359–364. doi: 10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Avci FY, Munoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. Enzymatic redesigning of biologically active heparan sulfate. J Biol Chem. 2005 doi: 10.1074/jbc.M504338200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J Biol Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Duncan MB, Carrick K, Pope RM, Liu J. Biosynthesis of 3-O-sulfated heparan sulfate: unique substrate specificity of heparan sulfate 3-O-sulfotransferase isoform 5. Glycobiology. 2003;13:785–794. doi: 10.1093/glycob/cwg101. [DOI] [PubMed] [Google Scholar]
- 19.Habuchi H, Habuchi O, Kimata K. Purification and characterization of heparan sulfate 6-sulfotransferase from the culture medium of Chinese hamster ovary cells. J Biol Chem. 1995;270:4172–4179. doi: 10.1074/jbc.270.8.4172. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi M, Habuchi H, Habuchi O, Saito M, Kimata K. Purification and characterization of heparan sulfate 2-sulfotransferase from cultured Chinese hamster ovary cells. J Biol Chem. 1996;271:7645–7653. doi: 10.1074/jbc.271.13.7645. [DOI] [PubMed] [Google Scholar]
- 21.Xia G, Chen J, Tiwari V, Ju W, Li JP, Malmström A, Shukla D, Liu J. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J Biol Chem. 2002;277:37912–37919. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 22.Bitter T, Muir HM. A modified uronic acid carbazole reaction. Anal Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 23.Toida T, Yoshida H, Toyoda H, Koshiishi I, Imanari T, Hileman RE, Fromm JR, Linhardt RJ. Structural differences and the presence of unsubstituted amino groups in heparan sulphates from different tissues and species. Biochem J. 1997;322(Pt. 2):499–506. doi: 10.1042/bj3220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparan sulfate on a biochip promotes its interaction with antithrombin III. Biochem Biophys Res Commun. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 25.Sasisekharan R, Venkataraman G. Heparin and heparan sulfate: biosynthesis, structure and function. Curr Opin Chem Biol. 2000;4:626–631. doi: 10.1016/s1367-5931(00)00145-9. [DOI] [PubMed] [Google Scholar]
- 26.Sugahara K, Kitagawa H. Heparin and heparan sulfate biosynthesis. IUBMB Life. 2002;54:163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- 27.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 28.Olson ST, Bjork I, Bock SC. Identification of critical molecular interactions mediating heparin activation of antithrombin implications for the design of improved heparin anticoagulants. Trends Cardiovasc Med. 2002;12:198–205. doi: 10.1016/s1050-1738(02)00160-3. [DOI] [PubMed] [Google Scholar]
- 29.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 30.Beetz T, Van Boeckel CAA. Synthesis of antithrombin binding heparin-like pentasaccharide lacking 6-O sulfate at its reducing end. Tetrahedron Lett. 1986;27:5889–5892. [Google Scholar]
- 31.Lindahl U, Bäckström G, Thunberg L. The antithrombin-binding sequence in heparin. Identification of an essential 6-O-sulfate group. J Biol Chem. 1983;258:9826–9830. [PubMed] [Google Scholar]
- 32.Riesenfeld J, Thunberg L, Höök M, Lindahl U. The antithrombin-binding sequence of heparin. Location of essential N-sulfate groups. J Biol Chem. 1981;256:2389–2394. [PubMed] [Google Scholar]
- 33.Desai UR, Petitou M, Bjork I, Olson ST. Mechanism of heparin activation of antithrombin, role of individual residues of the pentasaccharide activating sequence in the recognition of native and activated states of antithrombin. J Biol Chem. 1998;273:7478–7487. doi: 10.1074/jbc.273.13.7478. [DOI] [PubMed] [Google Scholar]
- 34.Olson ST, Srinivasan KR, Björk I, Shore JD. Binding of high affinity heparin to antithrombin III. Stopped flow kinetic studies of the binding interaction. J Biol Chem. 1981;256:11073–11079. [PubMed] [Google Scholar]
- 35.Rusnati M, Oreste P, Zoppetti G, Presta M. Biotechnological engineering of heparin/heparan sulphate: a novel area of multi-target drug discovery. Curr Pharmac Des. 2005;11:2489–2499. doi: 10.2174/1381612054367553. [DOI] [PubMed] [Google Scholar]