Abstract
Heparin and its low molecular weight heparin derivatives, widely used as clinical anticoagulants, are acidic polysaccharide members of a family of biomacromolecules called glycosaminoglycans (GAGs). Heparin and the related heparan sulfate are biosynthesized in the Golgi apparatus of eukaryotic cells. Heparin is a polycomponent drug that currently is prepared for clinical use by extraction from animal tissues. A heparin pentasaccharide, fondaparinux, has also been prepared through chemical synthesis for use as a homogenous anticoagulant drug. Recent enabling technologies suggest that it may now be possible to synthesize heparin and its derivatives enzymatically. Moreover, new technologies including advances in synthetic carbohydrate synthesis, enzyme-based GAG synthesis, micro- and nano-display of GAGs, rapid on-line structural analysis, and microarray/microfluidic technologies might be applied to the enzymatic synthesis of heparins with defined structures and exhibiting selected activities. The advent of these new technologies also makes it possible to consider the construction of an artificial Golgi to increase our understanding of the cellular control of GAG biosyntheses in this organelle.
Keywords: Heparin, enzymes, biosynthesis, Golgi, microfluidics
Heparin, a widely used anticoagulant, was introduced clinically soon after its discovery in 1916.1 For the next 70 years, it was the agent of choice for clinical anticoagulation. Heparin is an acidic polysaccharide isolated by extraction from animal tissues including porcine intestine.2 The heparin polysaccharide is a member of the glycosaminoglycan (GAG) family and comprises a repeating disaccharide structure of 1→4-linked hexuronic acid and glucosamine saccharide residues (Fig. 1). It is highly sulfated and has an average molecular weight of 10 to 15 kd.3 Although heparin exerts its anticoagulant effect in several ways, its most well-understood activity involves binding to the serine protease inhibitor antithrombin III (AT), which undergoes a conformational change, making it a potent inhibitor of thrombin and other serine proteases (i.e., factor [F] Xa) important in the blood coagulation cascade.4
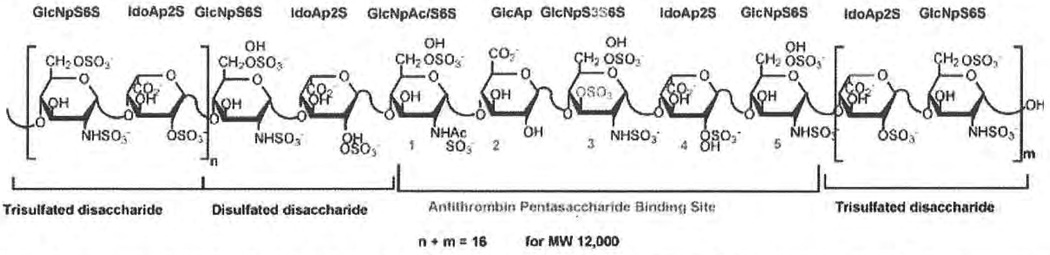
Figure 1.
Primary structure and structural variability of a heparin chain with high affinity for antithrombin III (AT). GlcNp, glucosamine; ldoAp, iduronic acid; GlcAp, glucuronic acid; S, sulfo; Ac, acetyl.
During the last 30 years, low molecular weight heparins (LMWHs) have been under extensive development as improved anticoagulant agents.5 Several LMWHs have been approved for clinical use and have begun to replace heparin as the clinical anticoagulant of choice.6 These LMWHs have a major advantage when compared with heparin: they are subcutaneously bio-available, resulting in their improved pharmacodynamics and allowing their application on an outpatient basis.5,6
CURRENT LIMITATIONS FOR DEVELOPMENT OF THE NEXT GENERATION OF AGENTS
LMWHs still retain several problems also associated with heparin, despite the major improvements associated with their introduction. Both heparin and LMWHs are prepared from animal sources, raising concerns about viral and prion-based contamination,2,6 and limiting the amount of drug that can be produced easily.2 Heparin and LMWHs are also polydisperse, polycomponent, polypharmacologic agents, making the analysis and control of these drugs difficult.1,5,6 Finally, both heparin and LMWHs can cause severe side effects such as hemorrhage and heparin-induced thrombocytopenia (HIT).7
Recognizing the limitation of these polycomponent drugs, a homogenous synthetic pentasaccharide, fondaparinux, was developed and introduced (Arixtra, GSK, Brentford, UK) that binds AT specifically and inhibits the serine protease inhibitor FXa.8 Fondaparinux, which has been approved for clinical use, has not been a commercial success. The chemical synthesis of fondaparinux is both complicated and costly,9 and the high cost of fondaparinux has not been justified by pharmacologic improvements compared with heparin and LMWHs.8,10 Furthermore, fondaparinux still retains the major side effect of hemorrhage and carries a slight risk of HIT.10 Other synthetic heparin analogs under development are also unlikely to be successful without major improvements in the field of synthetic carbohydrate chemistry.11
The need for improved heparin-based anticoagulants still remains. An ideal agent would (1) not be derived from animal sources; (2) be relatively inexpensive to prepare in amounts suitable for the growing market in anticoagulant drugs; (3) be a single component or a structurally defined multicomponent agent; (4) exhibit excellent bioavailability and pharmacodynamic/pharmacokinetic profiles; and (5) show reduced side effects, particularly with regard to hemorrhage and HIT. This article discusses recently developed enabling technologies that might pave the way for the development of new heparin-based anticoagulants beyond the LMWHs. These enabling technologies include major advances in (1) our understanding of GAG biosynthesis; (2) chemical carbohydrate synthesis; (3) enzyme-based GAG synthesis; (4) micro-and nano-display of GAGs; (5) online GAG analysis; and (6) microarray and microfluidic platforms.
The next generation of anticoagulants should show nearly ideal chemical, biochemical, and pharmacological properties to ensure their development as pharmaceuticals within the next few decades. Finally, a platform useful for preparing such carbohydrate-based anticoagulant agents would represent a form of artificial Golgi, and as such, would provide an improved understanding of the biosyntheses of heparin and the closely related GAG, heparan sulfate (HS), within the cell as well as the underlying controls of GAG biosynthesis.
GAG BIOSYNTHESIS
Lindahl et a112 have studied heparin/HS biosynthesis extensively. Heparin and HS are structurally similar in that all of the disaccharide units found in heparin are also observed in HS and all of the disaccharide units found in HS are also observed in the heparin.13 Moreover, several extended sequences, such as high/low-sulfated domains and the AT-binding site (Fig. 1), can be found in both heparin and HS.14 Heparin and HS differ primarily in the amount of each disaccharide that they contain.13 For example, in HS, d-glucuronic acid is the major uronic acid, more than half of the glucosamine residues are N-acetylated, and there is an average of one sulfo group per disaccharide. In contrast, heparin has higher concentrations of α-L-iduronic acid and N-sulfo-β-d-glu-cosamine residues, and an average of two to three sulfo groups per disaccharide.13 Heparin and HS contain at least 10 different sugar units,13,15 complicating efforts to sequence these polysaccharides.16–20
Similar biosynthetic pathways process heparin and HS.12,21–23 First, the core protein is synthesized and then a tetrasaccharide linker is attached to specific serine residues.24,25 From this linkage, a repeating 1→4 glycosidically linked copolymer of β-d-glucuronic acid and N-acetyl-β-d-glucosamine, called heparosan, is synthesized through the stepwise addition of uridine di-phosphate (UDP) -activated sugars.12 Heparosan can also be biosynthesized by bacteria, including Escherichia coli and Pasteurella multocida.26,27 During its synthesis in animal cells, the heparosan linear homocopolymer is modified sequentially through the action of N-deacetylase/N-sulfotransferase, C-5 epimerase, and 2-, 6-, and 3-O-sulfotransferases (OSTs). Complete or nearly complete modification of this nascent GAG chain results in a highly N- and O-sulfo—modified, α-L-iduronic acid—rich GAG called heparin.3 Partial modification of the same chain results in an N-acetyl-β-d-glucosamine, β-d-glucuronic acid—rich GAG called HS.25,29 All of the enzymes involved in the biosynthesis of heparin and HS have been cloned and expressed. Many of these biosynthetic enzymes (i.e., 3-OST, 6-OST) have multiple isoforms30–33 that show temporal- and tissue-specific expression.32,34 Despite these major advances in our understanding of GAG biosynthesis, it is still unclear how biosynthesis is controlled (i.e., extent, type of chain modification, and domain placement).21,29
Heparin is biosynthesized as a proteoglycan called serglycin,24 and pharmaceutical heparin generally is obtained from animal tissues rich in serglycin-containing mast cells,35 including porcine intestine or bovine lung. Pharmaceutical heparin, commercially prepared in metric ton quantities, ranges in molecular weight from 5000 to 40,000, with an average molecular weight of 12,000.1,2
CHEMICAL CARBOHYDRATE SYNTHESIS
Limitations in the Chemical Synthesis of Heparin
Despite recent advances,11,36 the total chemical synthesis of heparin, heparin oligosaccharides, and derivatives using current state-of-the-art techniques, has serious limitations. The multiple steps required by Sinaӱ9 and van Boecke137 to chemically synthesize an intricately substituted heparin oligosaccharide, although it displays elegant chemistry,9,36,37 results in a product that simply costs too much. Indeed, much synthetic work has focused on simplifying the target structure (i.e., replacing 2-amino-2-deoxy-d-glucose with d-glucose to prepare active analogs)11,37–39 rather than optimizing the synthesis of the natural product. Although recent advances in the chemical synthesis of heparin oligosaccharides by Thollas et a1,40 Jacquinet et a1,41 Prabhu et a1,42 Lohman et a1,43 Orgueira,44 and others45–47 have decreased the number of synthetic steps, increased stereocontrol, and enhanced yields, each new target still represents a major research commitment. Despite problems inherent to the chemical synthesis of heparin oligosaccharides, the synthetic AT-binding pentasaccharide, fondaparinux, was introduced as a clinical anticoagulant in both the United States and Europe.8 Although the approval of fondaparinux represents a drug development success, the commercial outlook for this expensive product is not bright.
Prospects for Synthetic UDP Sugars in Enzymatic Synthesis of GAGs
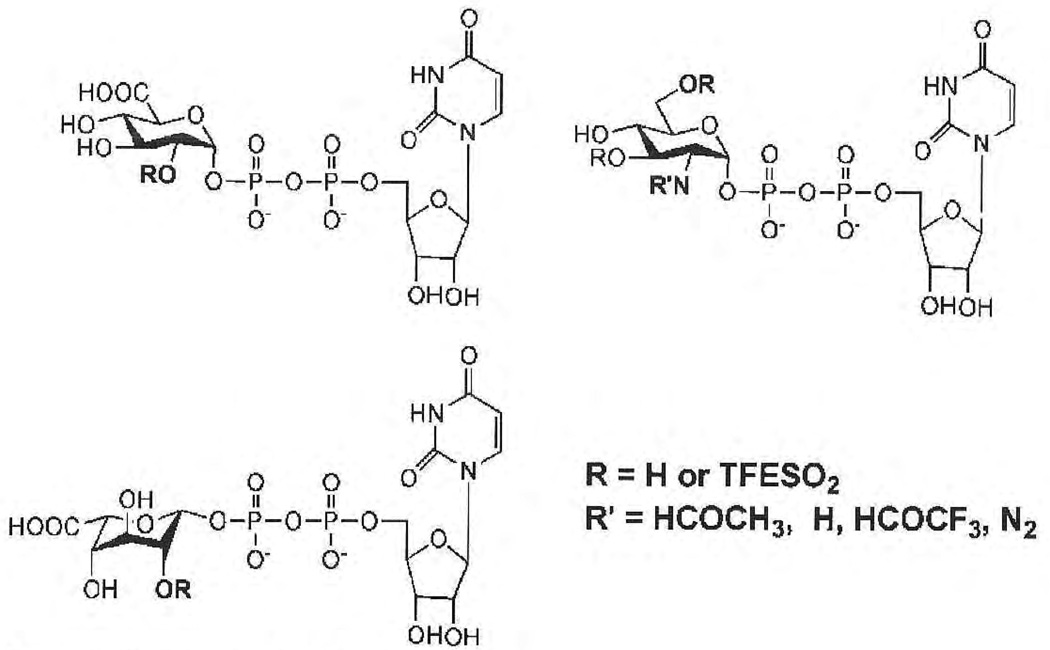
UDP sugar donors, UDP-N-acetylglucosamine and UDP-glucuronic acid, are routinely used in the laboratory for studying heparin/HS biosynthesis.23 Recently, unnatural UDP monosaccharide donors have been synthesized and used successfully to study the structure—activity relationship of an antibiotic glycosides, such as vancomycin.48 Other unnatural UDP monosaccharide donors, such as UDP-iduronic acid, UDP-2-azidoglucose, UDP-N-trifluoroacetylglucosamine, and donors containing trifluoroethyl (TFE) -protected sulfo groups,49 are currently under intensive investigation (Fig. 2). Although it is unclear if synthases/glycoltransferases will accept all of these unnatural donors in place of their natural UDP-N-acetylglucosamine and UDP-glucuronic acid substrates, preliminary studies demonstrate that UDP-N-trifluor-oacetylglucosamine is readily accepted in place of UDP—N-acetylglucosamine. Modeling of donors with neutral TFE-protected sulfo groups suggests that they can be accommodated by synthases/glycosyltransferases. These unnatural UDP donors can be synthesized chemically, by glycosylation of activated sugar with UDP, or enzymatically from the 1-phosphosugar. Given that these donors contain only a single glycosidic linkage, their synthesis in multiple-gram amounts is relatively uncomplicated, suggesting the potential application of these unnatural UDP donors in the preparation of chemo-enzymatically synthesized heparins.
Figure 2.
Natural and unnatural uridine diphosphate donors.
ENZYME-BASED GAG SYNTHESIS
Synthase-Catalyzed Preparation of Unsulfated GAGs
Polysaccharide synthases are the glycosyltransferase enzymes that catalyze the polymerization of activated sugars to form GAGs (Table 1).50–52 Synthase activity typically is associated with either the plasma membrane (in prokaryotes) or the Golgi apparatus membranes (in eukaryotes). Some species of pathogenic bacteria take advantage of the endogenous role of GAGs in cell—cell interaction and cellular communication in their animal hosts. The bacteria form surface coatings (polysaccharide capsules) that comprise GAGs identical or chemically similar to host molecules.53 Certain pathogenic strains of E. coli and P. multocida make capsules composed of polymers very similar to HS. These GAG-like polymers are not immunogenic and protect the bacteria from host defenses. Indeed, these microbes use their GAGs as molecular camouflage.
Table 1.
Acidic Glycosaminoglycan (GAG) Synthases/Glycosyltransferases and Substrates
| GAG | Species | GAG-Transferases | UDP Donors | Acceptors |
|---|---|---|---|---|
| Hyaluronan | Vertebrates | Class I HAS 1,2,3 | GlcNAc, GlcA | (GlcNAc-GlcA)n ≥ 1 |
| (GlcA-GlcNAc)n ≥ 1 | ||||
| Streptococcus | Class I spHAS | |||
| Chlorella | Class I cvHAS | |||
| Virus | ||||
| Pasteurella | Class II | pmHAS | GlcNAc, GlcA | |
| Chondroitin | Vertebrates | ChSy | GalNAc, GlcA | (GalNAc-GlcA)n ≥ 1 |
| (GlcA-GalNAc)n > 1 | ||||
| Pasteurella | pmCS, KfoC | GalNAc, GlcA | ||
| Escherichia | ||||
| Hyaluronan/chondroitin | Recombinant chimeric | pmHAS/pmCS | GalNAc | (HexNAc-GlcA)n ≥ 1 |
| GlcN, GlcNAc, GlcA | (GlcA-HexNAc)n ≥ 1 | |||
| Heparan | Vertebrates | EXT 1,2 | GlcNAc, GlcA | (GlcNAc-GlcA)n ≥ ? |
| (GlcA-GlcNAc)n ≥ 1 | ||||
| Heparosan | Pasteurella | pmHS 1,2 | GlcNAc, GlcA | (GlcNAc-GlcA)n ≥ 11 |
| (GlcA-GlcNAc)n > 1 | ||||
| Escherichia | KfiA + KfiC (2 proteins needed) | GlcNAc, GlcA |
UDP, uridine diphosphate; HAS, hyaluronan synthase; GlcNAC, N-acetylglucosamine; GlcA, glucuronic acid; ChSy, chondroitin synthase; CS, chrondroitin sulfate; Hs, heparan sulfate.
Membrane-bound synthase/glycosyltransferase proteins are difficult to handle, thus, few enzymes have been identified through biochemical purification. A larger number of synthases/glycosyltransferases have been sequenced at the nucleotide level, cloned, and expressed. Despite these advances, molecular details on their three-dimensional native structures, active sites, and catalytic mechanisms are very limited. The exploration of these enzymes for GAG synthesis is just beginning in our laboratories,54 but a few of these catalysts have been harnessed as practical chemoenzymatic tools.
Enzyme Catalyzed Synthesis of Sulfated GAGs
The HS biosynthetic enzymes have been expressed in E. coli as truncated (no transmembrane domain) fusion proteins using a bacterial cell line that coexpresses a chaperone.59–60 Chaperones help recombinant proteins fold correctly to obtain functional proteins. These recombinant proteins have comparable specific enzymatic activities and substrate specificity of those expressed in mammalian and insect cells.
Enzymatically Designing the Functions of Heparin/HS
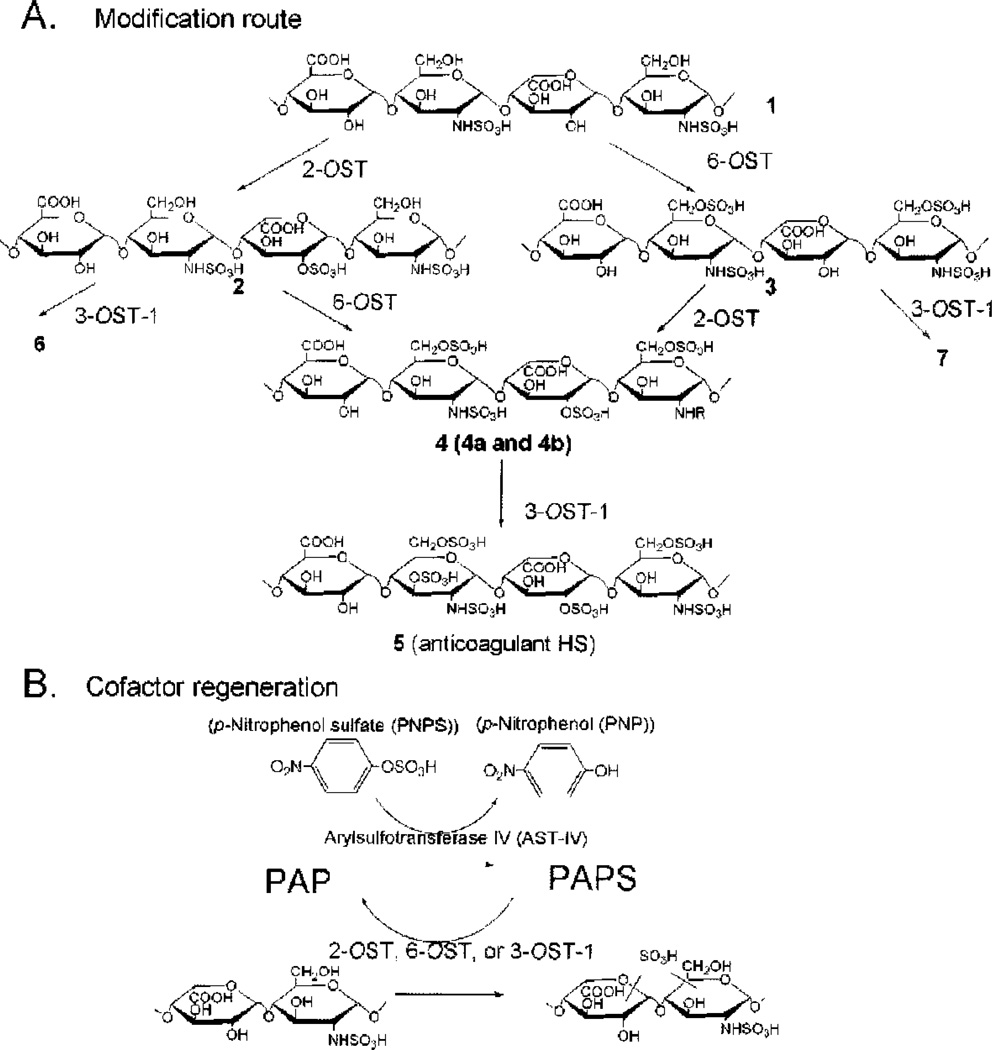
An enzymatic approach has been developed to synthesize heparin from completely desulfonated N-sulfonated heparin. Although this clearly is not a viable commercial approach, it serves to demonstrate that much of the technology is in hand to synthesize heparin chemo-enzymatically. Only three enzymatic steps are required for the synthesis of heparin with anticoagulant activity from desulfonated N-sulfonated heparin (Fig. 3), The current study demonstrates two major advances that may permit the large-scale synthesis of heparin. First, large amounts of all the required OST enzymes have been expressed successfully in E. coli (as described) and immobilized on solid supports packed into bioreactors. Second, a 3′-phosphoadenosine-5′-phosphosulfate (PAPS) regeneration system, developed by Burkart et al,61 has been coupled to the enzymatic synthesis reactions. This PAPS regeneration system alone reduces the cost of the synthesis by more than 1000-fold, making the large-scale enzymatic synthesis possible.55
Figure 3.
Schematic synthesis of sulfonated polysaccharides and 3′-phosphoadenosine-5′-phosphosultate (PAPS) regeneration system. (A) The stepwise enzymatic synthesis of sulfonated polysaccharides using heparan sulfate (HS) sulfotransferases. The description of intermediate polysaccharides is shown in the text. Compounds 4a and 4b were prepared by inverting the order of sulfonation steps. 4a was prepared by incubating compound 1 with 2-O-sullotransferase (2-OST) followed by 6-OST, whereas 4b was prepared by incubating compound 1 with 6-OST followed by 2-O5T. (B) The reaction catalyzed by arylsulfotransferase IV to generate PAPS.
PAP (3′,5′-phosphoadenosine) is formed when the sulfo group is transferred to an acceptor. A PAPS regeneration system converts PAP to PAPS through the action of recombinant arylsulfotransferase-IV, which catalyzes the transfer of sulfo group from p-nitrophenyl sulfate (PNPS) to PAP (Fig. 3B).61 This system is essential for the large-scale synthesis for the following reasons. First, PAP inhibits the HS OSTs with concentration that inhibits 50% values of ~100 µM, making milligram-scale synthesis difficult without continuously removing PAP.55 Second, a PAPS regeneration system permits the PNPS as the sulfo donor and catalytic amounts of PAP, consequently reducing the cost of synthesis.
The PAPS regeneration system performs very well with immobilized 2-OST, 6-OST, and 3-OST-1. Under the standard conditions, 2-OST, 6-OST, and 3-OST-1 afforded 98%, 97%, and 98% complete modification, respectively. The incorporation of 2-O-sulfo and 6-O-sulfo groups can be confirmed using disaccharide analysis of the structures of the newly synthesized poly-saccharide products 2, 3 and 4a and 4b. Approximately 1.5 mg of AT-binding HS (5, sec Fig. 1) was synthesized from 10 mg of completely desulfated N-sulfated (CDSNS) heparin (1) using this approach.55
Synthesized polysaccharide intermediates were digested with a mixture of heparin lyases, and the resulting disaccharides were analyzed using reversed-phase ion-pair partition high-performance liquid chromatography.55 Analysis of 1 showed a disaccharide component of the structure of ΔUAP-GlcNPS (ΔUA is 4-dcoxy-α-L-threo-hex-4-enopyranosyluronic acidp is pyranoside), residual unsulfated disaccharide, ΔUAP-GlcNpAc, and small amounts of sulfated disaccharides due to incomplete chemical desulfonation (Table 2). The modification by 2-OST elevated the level of ΔUAP2S-GlcNpS by ~ 5-fold (Table 3), confirming the structure of 2. Analysis of 3 afforded a 7-fold increase in the level of the disaccharide ΔUAP-GlcNpS6S compared with 1, consistent with 6-OST catalyzed modification (Table 2). The level of trisulfodisaccharide, ΔUAp2S-GlcNPS6S in 4a and 4b, was increased by ~ 10-fold compared with that of 1 (Table 2). These results clearly establish that the expected enzymatic modifications took place at each step. It is interesting to note that 6-O-sulfation occurs at N-sulfo-glucosamine, consistent with the substrate specificity of 6-OST in vitro.62 The 2-O-sulfation occurs predominantly at the uronic acid with an N-sulfoglucosamine residue at the reducing end. Interestingly, substantial amounts of ΔUAP-GlcNpS remain in 4a and 4b. This observation is not unexpected given that the HS from various tissues also contain ΔUAp-GlcPNS disaccharide unit, suggesting that the structures of 4a and 4b are similar to HS from natural sources. 1H-nucelar magnetic resonance analyses on these polysaccharides were found to be consistent with the disaccharide analysis.55
Table 2.
Summary of the Disaccharide Compositions of the Synthetic Polysaccharides*
| Compound | ΔUA-GlcNAc (nmole)† (%) |
ΔUA-GlcNS (nmole) (%) |
ΔUA2S-GlcNS (nmole) (%) |
ΔUA-GlcNS6S (nmole) (%) |
ΔUA2S-GlcNS6S (nmole) (%) |
Total Recovery (%)‡ |
|---|---|---|---|---|---|---|
| 1 | 12.5 (16.1%) | 50.0 (64.4%) | 6.3 (8.1%) | 6.3 (8.1%) | 2.5 (3.2%) | 54.0 |
| 2 | 12.5 (16.6%) | 27.5 (36.6%) | 30.0 (40.0%) | 2.5 (3.3%) | 2.5 (3.3%) | 49.2 |
| 3 | 10.0 (14.0%) | 27.5 (38.5%) | 5.0 (7.0%) | 25.0 (35.0%) | 3.8 (5.3%) | 51.0 |
| 4a | 12.5 (18.8%) | 17.5 (26.2%) | 7.5 (11.2%) | 10.0 (15.0%) | 20.0 (30.0%) | 50.0 |
| 4b | 17.5 (21.7%) | 15.0 (18.6%) | 8.0 (9.9%) | 15.0 (18.6%) | 25.0 (31.0%) | 48.0 |
Each synthesized polysaccharide (100 µg) was digested with a mixture of heparin lyases. The resultant disaccharides were purified by a BioGel P-2 column, and resolved by reversed-phase ion-pair partition high-performance liquid chromatograph.
The amount of each disaccharide was estimated by determining its peak area with a standard curve generated with known amount of the disaccharide standard.
A recovery yield was calculated by using 2-O-[35S]sulfo heparin as an internal standard.
ΔUA, 4-deoxy-α-L-threo-hex-4-enopyranosyluronic acid; GlcNAc, N-acetylglucosamine; GlcNS, N-sulfoglucosamine; ΔUA2S, 4-deoxy-α-L-threo-hex-4eno-2-sulfopyranosyluronic acid; GlcNS6S, N-sulfo-6-sulfoglucosamine.
Table 3.
Anti–Factor Xa and Antithrombin III (AT) Activities of Synthesized Polysaccharide Intermediates
| Sample | Factor Xa Inhibition* (IC50, ng/mL) |
Thrombin Inhibition* (IC50, ng/mL) |
Binding to AT (%)† |
|---|---|---|---|
| Heparin‡ | 20 | 10 | ND |
| HS§ | > 5000 | > 3000 | ND |
| Fondaparinux‖ | 58 | > 3000 | ND |
| 6 | > 2000 | > 3000 | 5% |
| 7 | 126 | 96 | 31% |
| 5 | 40 | 32 | 38% |
The procedures for measuring the activities of factor Xa and thrombin are described elsewhere.1
3-O-[35S]sulfo compounds were used to determine their bindings to AT as described elsewhere. 13H-labeled HS from CHO cells was used as a negative control for the AT binding. About 0.3% of 3H-labeled HS bound to AT.
From Sigma.
HS was isolated from bovine kidney.
Fondaparinux is the chemically synthesized antithrombin-binding pentasaccharide, which was obtained from local pharmacy.
IC50, concentration that inhibits 50%; HS, heparan sulfate; ND, not determined.
Characterization of the affinities of AT to heparin and enzymatically modified heparin derivatives (1–5) relied on surface plasmon resonance (SPR).55 A two-state reaction model was applied to the SPR study. None of the derivatives, with the exception of the 2,6,3-O-sulfo polysaccharide (5) and heparin, had high affinity to AT. The binding constant (KD) for the binding of 2,6,3-O-sulfo polysaccharide 5 to AT was 170 nM, comparable to that of pharmaceutical heparin (75 nM).
Because it is known that the introduction of the 3-O-sulfo group by 3-OST-1 is essential for the synthesis of anticoagulant heparin, the FXa and thrombin activities of polysaccharides 5, 6, and 7 were evaluated (Table 3). As expected, heparin was a potent activator for AT-mediated inhibition of FXa and thrombin, whereas fondaparinux specifically activates the AT-mediated inhibition of FXa.47 Polysaccharide 5 has very similar potency to heparin, inhibiting the activities of both FXa and thrombin, suggesting that an enzyme-based approach is indeed capable of synthesizing an anticoagulant heparin poly-saccharide. It has been reported that the presence of 2-O-sulfo groups is not essential for HS binding to AT and its resulting anticoagulant activity.63 Indeed, polysaccharide intermediate 7 lacks 2-O-sulfo groups but still exhibits anticoagulant activity, consistent with a previous report.63 In contrast, 6 lacks 6-O-sulfo groups, and thus has no anticoagulant activity, given that 6-O-sulfo groups are critical in AT binding.64 The AT binding affinities of the synthesized polysaccharides were also determined (Table 3), demonstrating that the anticoagulant activities of the compounds correlated to their binding affinity to AT. Taken together, these results demonstrate that the structure and anticoagulant activities of the enzymatically synthesized polysaccharide 5 were consistent with the known structure and activity of heparin (Fig. 1).
ON-LINE GAG ANALYSIS
Recent advances in mass spectrometry suggest that it is now possible to analyze intact heparin/HS chains. Electrospray ionization mass spectrometry (MS), useful as an on-line analytical tool, has been used effectively to analyze heparin oligosaccharides of molecular mass as high as 7 kd.65 MS-MS methods allow for the positioning of sulfo groups within the GAG chain.66 Electron capture dissociation—MS can be used to differentiate iduronic acid from glucuronic acid residues.67 These advances in MS analysis should be applicable to the real time analysis of polysaccharide chains synthesized in microfluidic devices. In addition, the recent use of matrix-assisted laser desorption ionization (MALDI)-MS for the direct analysis of uncomplexed sulfated oligosaccharides on a target surface68,69 suggests that it might be possible to use MALDI-MS for the analysis of heparin/HS on the surface of a microarray synthesizer (described in “Microarrays”).
MICRO- AND NANO-DISPLAY OF GAGs
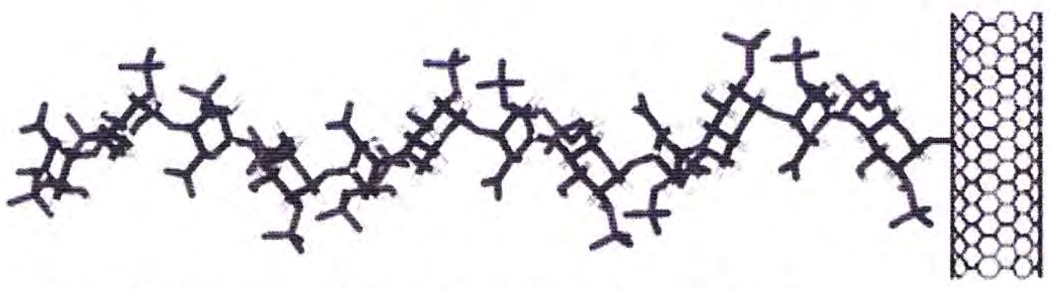
Solid-phase synthesis of biopolymers offers the same advantage as it does in peptide synthesis, maximizing yields and minimizing separation problems.70 Although heparin and heparin oligosaccharides have been coupled to macroscopic supports such as affinity resins71 for several years, such chemistry has only recently been applied to nano-based carriers.71,72 The size of such nanocarriers resembles the dimensions of the core protein of heparin/HS proteoglycans (Fig. 4). These nanoglycoconjugates are soluble72 in buffer solutions, allowing their movement in microfluidic systems. Furthermore, nanocarriers such as gold73 and quantum dots74 exhibit optical properties that facilitate their detection and analysis. These rapidly developing technologies suggest the use of nanocarriers as solid supports for the enzyme-based synthesis of heparin in microfluidic reactors. GAGs can be synthesized on a solid phase (plastic, glass, metal, etc.) using enzyme catalysts.75
Figure 4.
A schematic of a heparin glycosaminoglycan chain immobilized to a carbon nanotube.
MICROARRAY AND MICROFLUIDIC PLATFORMS
Microarrays
Microarrays have been used for the screening of protein binding to heparin and heparin oligosaccharides.76,77 The successful enzymatic modification of heparosan linked to a surface78 suggests that it might also be possible to synthesize libraries of hcparins in a micro-array format. A microarray of up to 5000 addressable spots can be printed on a glass microscope slide.79 Each spot can be designed to contain a polysaccharide for modification by C-5 epimerase and OSTs or an oligosaccharide for extension with glycosyltransferase and subsequent enzymatic modification. Such a large micro-array could then he screened and be probed with heparin-binding proteins. One might envision using microarrays to identify an enzymatically synthesized heparin able to bind AT and inhibit thrombin and FXa without binding platelet factor 4, thus preventing HIT.80 Glycoanalysis of each microarray spot would rely on MALDI-MS, and once characterized, a chain with desirable biochemical and pharmacological properties could be synthesized in large scale for exploitation as a novel pharmaceutical agent. Such devices might even be useful for introducing designer heparins, fine-tuned to an individual patient. Such a pharmacogenetic approach requires a high-throughput platform to be practicable.
Glycosylation Using Continuous-Flow Microfluidics
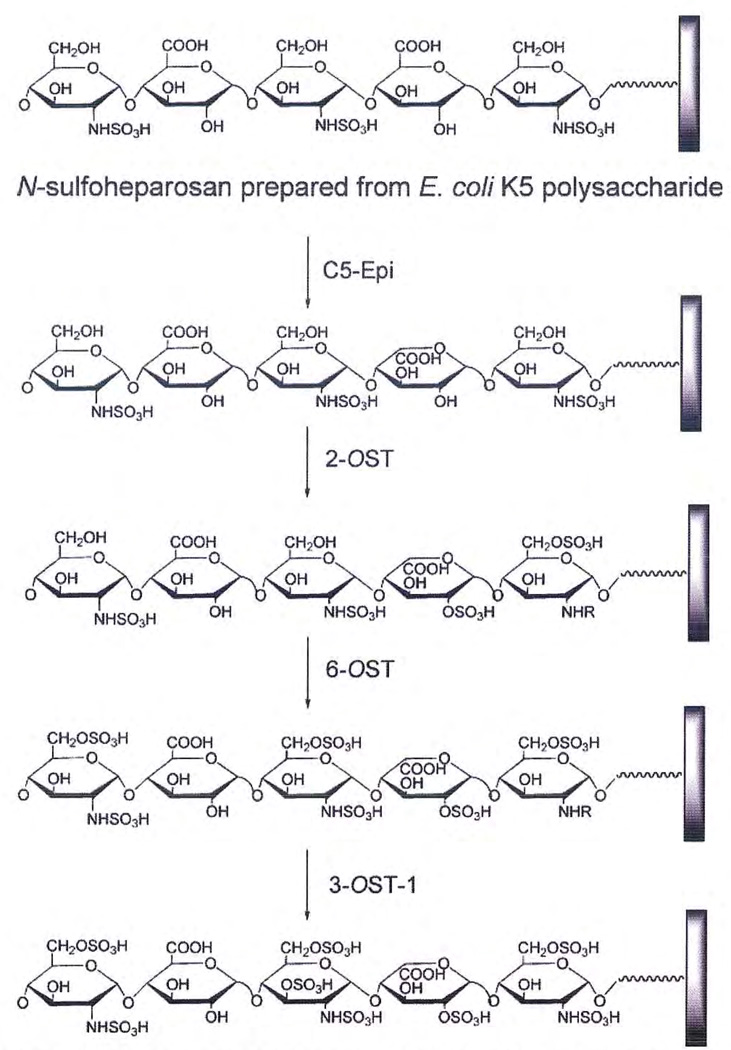
In early studies, Hernaiz et a178 demonstrated that 3-OST-1 could be used to convert inactive HS (incapable of binding AT) into active (AT binding) HS in a microfluidic system. On the basis of this initial success, four enzymes involved in heparin/HS biosynthesis were cloned and expressed in E. coli. These enzymes were then used in solution-phase chemistry to regenerate an active AT-binding site within a chemically modified heparin polysaccharide.55 This approach was next extended to modify heparosan, a bacterial capsular polysaccharide purified from E. coli K5, having the structure → 4)-α-d-N-acetylglucosamine (1→ 4)-β-d-glucuronic acid (1→.56 This polysaccharide was chemically de-N-acetylated and N-sulfonated to afford N-sulfoheparosan, having the structure → 4)-α-d-N-sulfoglucosamine (1→ 4)-β-d-glucuronic acid (1→. N-sulfoheparosan was immobilized onto a microfluid, flow cell-based, SPR sensor chip (Fig. 3). An interacting AT analyte was flowed over the chip and AT-polysaccharide (ligand) binding was measured using SPR.78 As expected, AT failed to bind to N-sulfoheparosan because this ligand lacks the requisite structure comprising the AT-binding site (Fig. 5).56,78 Biosynthetic enzymes and appropriate cofactors, which had been applied previously to solution-phase syrithesis,55 were used to modify the N-sulfohcparosan ligand immobilized to the surface of the microfluidic biosensor chip. When the correct enzymes and cofactors were applied in the correct sequence, an AT-binding site was introduced into the immobilized ligand. After this modification, the AT analyte bound to the ligand on the biosensor chip with the expected affinity, confirming that the biosynthetic enzymes were capable of acting in this micro-fluidic reactor. Unfortunately, because there was insufficient control within the reactor, only a small number of the resulting chains contained the desired AT binding site. Furthermore, the placement of this site at a precise position within the chain could not be accomplished.
Figure 5.
Solid-phase synthesis of heparin from N-sulfoheparosan. Epi, ???; OST, O-sulfotransferase; AT, antithrombin III.
Related preliminary experiments have also demonstrated that heparin polysaccharides can be immobilized covalently onto nanoparticle carriers and that enzymes are capable of acting on the nanoparticle-immobilized polysaccharide.72 Furthermore, the heparin/HS biosynthetic enzymes have been expressed as fission proteins, immobilized to solid supports through their fusion protein domains, and demonstrate high stability and retention of activity.55
FUTURE PROSPECTS
Glyosylation in the Golgi
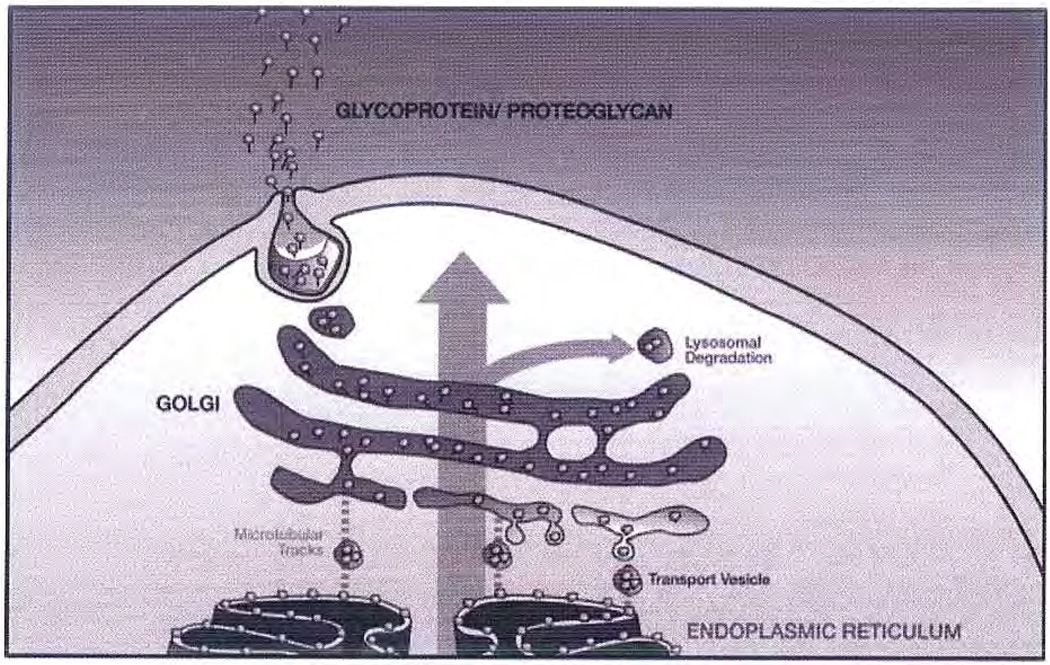
The Golgi is an intracellular stack of membrane-hounded vesicles, responsible for the modification and packaging of secreted proteins.81 This organelle is the subcellular entity controlling all glycosylation in eukaryotes (Fig. 6). With the successful determination of multiple genomes and the ongoing successes in proteomics, the comprehensive study of carbohydrate/glycan structure-function relationships, also known as glycomics, is considered the last frontier in the biomacromolecular sciences. A unique feature of glycomics is that there does not exist a straightforward correlation between the glycome and the proteome or genome. Unraveling the multifaceted relationships among the glycome, proteome, and genome is difficult because the extremely complex process of glycan synthesis within the Golgi is not well understood. The primary obstacles to understanding are (1) the intricate and fluid structural organization of this organelle; (2) the complex trafficking of molecules (enzymes, chaperones, cofactors, and substrates) within the Golgi and between the Golgi and the endoplasmic reticulum (the organelle responsible for protein bio-synthesis and the initial steps in glycosylation); (3) a limited understanding of the regulation and interplay between glycan biosynthetic enzymes; and (4) the difficulties associated with elucidating glycan structure and function.
Figure 6.
Simplified drawing of the Golgi. Core protein (gray ball) is biosynthesized in the endoplasmic reticulum (ER), glycosylation is initiated and core protein decorated with oligosaccharide acceptor is transferred in budded transport vesicle along microtubule tracks to the Golgi (some retrieval from the Golgi by the ER is also possible). As the glycoprotein moves through the Golgi stacks (cis to medial to trans), the glycan chain (black line) is elongated and elaborated. Mature glycoprotein exits the trans Golgi in vesicles for transport to the cell surface (some removal of excess or damaged glycoprotein takes place through lysosomal degradation).
Among the most important glycoconjugates biosynthesized in the Golgi are proteoglycans. An improved understanding of hcparan/HS biosynthesis will lead inevitably to an improved understanding of normal and pathophysiological processes. This under-standing will contribute to the solution of both historic and emerging health problems in the 21st century.
Controlled artificial synthesis of heparin/HS is extremely challenging because they are the most structurally complex biopolymcrs in nature. Their biosynthesis in the Golgi involves the condensation of nucleotide sugars on specific serine residues within a core protein and the subsequent modification (re-modeling) of the growing polysaccharide chain through the action of membrane-bound enzymes. This is the most complex example of glycan biosynthesis, involving as many as 20 different enzymes found in multiple isoforms, and these enzymes are differentially expressed in a spatial and temporal fashion within all animals. Moreover, the underlying coordination of these biosynthetic enzymes within the Golgi (i.e., as complexes, in specific compartments, on templates, with chaperones, etc.,) is not well understood. An improved understanding of the parameters that control glycan synthesis is needed, including controlled access of enzymes to protein-core—containing oligosaccharide acceptors, assembly of multienzymc complexes, and access to cofactors and substrates.
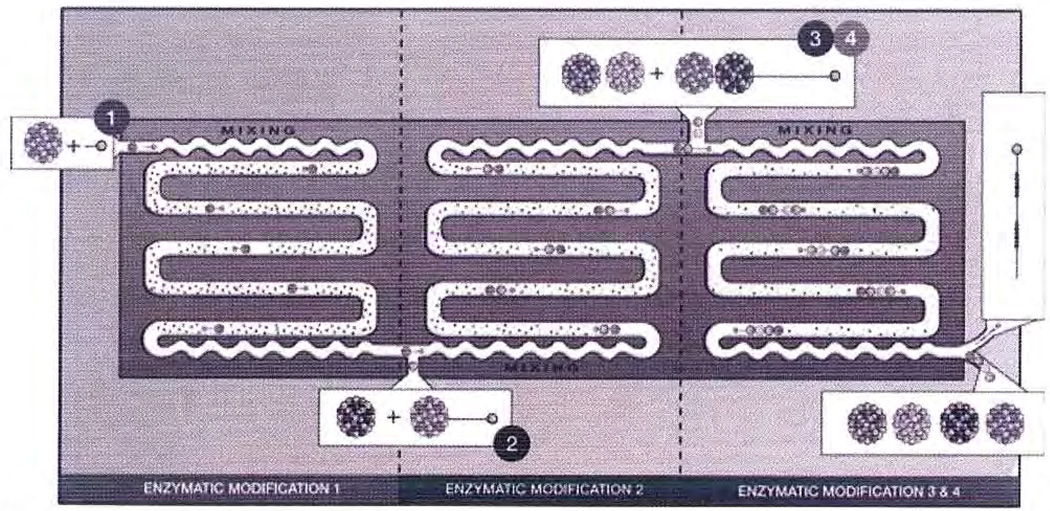
Research currently is underway to construct an artificial Golgi using microfluids for the controlled synthesis of heparin on nanosupports and using on-line product analysis (Fig. 7). Such a system will require additional development and integration of all of the enabling technologies discussed in this article. The resulting device will serve as both a test device for the understanding of Golgi function and regulation as well as a reactor for the generation of structurally defined heparins with discrete biological activities.
Figure 7.
Schematic representation of an artificial Golgi.
REFERENCES
- 1.Linhardt RJ. Heparin: an important drug enters its seventh decade. Chem Ind. 1991;2:45–50. [Google Scholar]
- 2.Linhardt RJ, Gunay NS. Production and chemical processing of low molecular weight heparins. Semin Thromb Hemost. 1999;25:5–14. [PubMed] [Google Scholar]
- 3.Casu B. Structure and biological activity of heparin. Adv Carbohydr Chem Biochem. 1985;43:51–134. doi: 10.1016/s0065-2318(08)60067-0. [DOI] [PubMed] [Google Scholar]
- 4.Munoz EM, Linhardt RJ. Heparin binding domains in vascular biology. Arterioscler Thromb Vascular Biol. 2004;24:1549–1557. doi: 10.1161/01.ATV.0000137189.22999.3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fareed J, Jeske W, Hoppensteadt D, Clarizio R, Walgenga JM. Low molecular weight heparins: pharmacologic profile and product differentiation. Am J Cardiol. 1998;82:3L–10L. doi: 10.1016/s0002-9149(98)00105-2. [DOI] [PubMed] [Google Scholar]
- 6.Linhardt RJ, Claude S. Hudson Award address in carbohydrate chemistry. Heparin: structure and activity. J Med Chem. 2003;46:2551–2564. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 7.Warkentin TE, Cook DJ. Heparin, low molecular weight heparin, and heparin-induced thrombocyropenia in the ICU. Crit Care Clin. 2005;21:513–529. doi: 10.1016/j.ccc.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Vuillemenot A, Schiele 1, Meneveau N, et al. Efficacy of a synthetic pentasaccharide, a pure factor Xa inhibitor, as an antithrombotic agent—a pilot study in the setting of coronary angioplasty. Thromb Haemost. 1999;81:214–220. [PubMed] [Google Scholar]
- 9.Sinaÿ P, Jacquinet J-C. Total synthesis of a heparin pentasaccharide fragment having high affinity for antithrombin III. Carbohydr Res. 1984;132:C5–C9. [Google Scholar]
- 10.Efird LE, Kockler DR. Fondaparinux for thromboembolic treatment and prophylaxis of heparin-induced thrombocytopenia. Ann Pharmacother. 2006;40:1383–1387. doi: 10.1345/aph.1G738. [DOI] [PubMed] [Google Scholar]
- 11.Avci FY, Karst NA, Linhardt RJ. Synthetic oligosaccharides as heparin-mimetics displaying anticoagulant properties. Curr Pharm Des. 2003;9:2323–2335. doi: 10.2174/1381612033453929. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl U, Feingold DS, Roden L. Biosynthesis of heparin. Trends Biochem Sci. 1986;11:221–225. [Google Scholar]
- 13.Griffin CC, Linhardt RJ, VanGorp CL, et al. Isolation and characterization of heparan sulfate from crude porcine intestinal mucosa peptidoglycan heparin. Carbohydr Res. 1995;276:183–197. doi: 10.1016/0008-6215(95)00166-q. [DOI] [PubMed] [Google Scholar]
- 14.Marcum JA, Rosenberg RD. Anticoagulantly active heparan sulfate proteoglycan and the vascular endothelium. Semin Thromb Hemost. 1987;13:464–474. doi: 10.1055/s-2007-1003523. [DOI] [PubMed] [Google Scholar]
- 15.Linhardt RJ, Liu J, Han X-J. Mapping and sequencing of oligosaccharides by electrophoresis. Trends Glycosci Glycotechnol. 1993;5:181–192. [Google Scholar]
- 16.Merry CL, Lyon M, Deakin JA, Hopwood JJ, Gallagher JT. Highly sensitive sequencing of the sulfated domains of heparan sulfate. J Biol Chem. 1999;274:18455–18462. doi: 10.1074/jbc.274.26.18455. [DOI] [PubMed] [Google Scholar]
- 17.Vives RR, Pye DA, Salmivirta M, Hopwood JJ, Lindahl U, Gallagher JT. Sequence analysis of heparan sulphate and heparin oligosaccharides. Biochem J. 1999;339:767–773. [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbull JE, Hopwood JJ, Gallagher JT. A strategy for rapid sequencing of heparan sulfate and heparin saccharides. Proc Natl Acad Sci USA. 1999;96:2698–2703. doi: 10.1073/pnas.96.6.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Sequencing complex polysaccharides. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Desai UR, Han X, Linhardt RJ. Sequencing of heparin. Glycobiology. 1995;5:765–774. doi: 10.1093/glycob/5.8.765. [DOI] [PubMed] [Google Scholar]
- 21.Gallagher JT. The extended family of proteoglycans: social residents of the pericellular zone. Curr Opin Cell Biol. 1989;1:1201–1218. doi: 10.1016/s0955-0674(89)80072-9. [DOI] [PubMed] [Google Scholar]
- 22.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 23.Lidholt K. Biosynthesis of glycosaminoglycans in mammalian cells and in bacteria. Biochem Soc Trans. 1997;25:866–870. doi: 10.1042/bst0250866. [DOI] [PubMed] [Google Scholar]
- 24.Robinson HC, Horner AA, Höök M, Ogren S, Lindahl UA. Proteoglycan form of heparin and its degradation to single-chain molecules. J Biol Chem. 1978;253:6687–6693. [PubMed] [Google Scholar]
- 25.Fransson L-A. Heparan sulfate proteoglycans: structure and properties. In: Lane DA, Lindahl U, editors. Heparin Chemical and Biological Properties, Clinical Applications. Boca Raton, FL: E. Arnold; 1989. pp. 115–133. [Google Scholar]
- 26.DeAngelis PL, Gunay NS, Toida T, Mao W-J, Linhardt RJ. Identification of the capsular polysaccharides of type D and F Pasteurella multocida as unmodified heparin and chondroitin, respectively. Carbohydr Res. 2002;337:1547–1552. doi: 10.1016/s0008-6215(02)00219-7. [DOI] [PubMed] [Google Scholar]
- 27.DeAngelis PL. Microbial glycosaminoglycan glycosyltransferases. Glycobiology. 2002;12:9R–16R. doi: 10.1093/glycob/12.1.9r. [DOI] [PubMed] [Google Scholar]
- 28.Gallagher JT, Walker A. Molecular distinctions between heparan sulphate and heparin. Biochem J. 1985;230:665–674. doi: 10.1042/bj2300665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallagher JT, Turnbull JE, Lyon M. Patterns of sulphation in heparan sulphate: polymorphism based on a common structural theme. Int J Biochem. 1992;24:553–560. doi: 10.1016/0020-711x(92)90326-v. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Shworak NW, Sinaÿ P, et al. Expression of heparan sulfate D-glucosaminyi 3-O-sulfotransferase isoforms reveals novel substrate specificities. J Biol Chem. 1999;274:5185–5192. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi M, Sugumaran G, Liu J, Shworak NW, Silbert JE, Rosenberg RD. Molecular cloning and characterization of a human uronyl 2-sulfotransferase that sulfates iduronyl and glucuronyl residues in dermatan/chondroitin sulfate. J Biol Chem. 1999;274:10474–10480. doi: 10.1074/jbc.274.15.10474. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi M, Habuchi H, Habuchi O, Saito M, Kimata K. Purification and characterization of heparan sulfate 2-sulfotransferase from cultured Chinese hamster ovary cells. J Biol Chem. 1996;271:7645–7653. doi: 10.1074/jbc.271.13.7645. [DOI] [PubMed] [Google Scholar]
- 33.Forsberg E, Pejler G, Ringvall M, et al. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400:773–776. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- 34.Stringer SE, Mayer-Proschel M, Kalyani A, Rao M, Gallagher JT. Heparin is a unique marker of progenitors in the glial cell lineage. J Biol Chem. 1999;274:25455–25460. doi: 10.1074/jbc.274.36.25455. [DOI] [PubMed] [Google Scholar]
- 35.Humphries DE, Wong GW, Friend DS, et al. Heparin is essential for the storage of specific granule protcases in mast cells. Nature. 1999;400:769–772. doi: 10.1038/23481. [DOI] [PubMed] [Google Scholar]
- 36.Karst NA, Linhardt RI. Recent chemical and enzymatic approaches to the synthesis of glycosaminoglycan oligosaccharides. Curr Med Chem. 2003;10:1993–2031. doi: 10.2174/0929867033456891. [DOI] [PubMed] [Google Scholar]
- 37.Van Boeckel CAA, Petitou M. The unique antithrombin III binding domain of heparin: a lead to new synthetic antithrombotics. Angew Chem Int Ed Engl. 1993;32:1671–1690. [Google Scholar]
- 38.Ichikawa Y, Monden R, Kuzuhara H. Synthesis of methyl glycoside derivatives of tri- and penta-saccharides related to the antithrombin III-binding sequence of heparin employing cellobiose as a key starting-material. Carbohydr Res. 1988;172:37–64. doi: 10.1016/s0008-6215(00)90842-5. [DOI] [PubMed] [Google Scholar]
- 39.Lucas H, Basten JEM, Konradsson P, Van Boeckel CAA. A short synthetic route towards a biologically active heparin-like pentasaccharide with a pseudo-alternating sequence. Angew Chem Int Ed Engl. 1993;32:434–436. [Google Scholar]
- 40.Thollas B, Jacquinet J-C. Synthesis of various sulfoforms of the trisaccharide β-D-GLcpA (1→3)-β-D-Galp-(1→3)-β-D-Galp-(1→OMP) as probes for the study of the biosynthesis and sorting of proteoglycans. Org Biomol Chem. 2004;2:434–442. doi: 10.1039/b314244b. [DOI] [PubMed] [Google Scholar]
- 41.Jacquinet J-C. An expeditious preparation of various sulfoforms of the disaccharide β-D Galp-(1→3)-D-Galp, a partial structure of the linkage region of proteoglycans, as their 4-methoxyphenyl β-D-glycosides. Carbohydr Res. 2004;339:349–359. doi: 10.1016/j.carres.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Prabhu A, Venot A, Boons G-J. New set of orthogonal protecting groups for the modular synthesis of heparan sulfate fragments. Org Lett. 2003;5:4975–4978. doi: 10.1021/ol0359261. [DOI] [PubMed] [Google Scholar]
- 43.Lohman GJS, Seeberger PH. A stercochemical surprise at the late stage of the synthesis of fully N-differentiated heparin oligosaccharides containing amino, acetamido, and N-sulfonatc groups. J Org Chem. 2004;69:4081–4093. doi: 10.1021/jo035732z. [DOI] [PubMed] [Google Scholar]
- 44.Orgueira HA, Bartolozzi A, Schell P, Litjens EJN, Palmacci ER, Seeberger PH. Modular synthesis of heparin oligosaccharides. Chemistry. 2003;9:140–169. doi: 10.1002/chem.200390009. [DOI] [PubMed] [Google Scholar]
- 45.Koshida S, Suda Y, Sobel M, Ormsby J, Kusumoto S. Synthesis of heparin partial structures and their binding activities to platelets. Bioorg Med Chem Lett. 1999;9:3127–3132. doi: 10.1016/s0960-894x(99)00550-8. [DOI] [PubMed] [Google Scholar]
- 46.Tabeur C, Mallet J-M, Bono F, Herbert J-M, Petitou M, Sinaÿ P. Oligosaccharides corresponding to the regular sequence of heparin: chemical synthesis and interaction with FGF-2. Bioorg Med Chem. 1999;7:2003–2012. doi: 10.1016/s0968-0896(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 47.Petitou M, van Boeckel CAA. A synthetic antithrombin ITT binding pentasaccharide is now a drug! What comes next? Angew Chem Int Ed Engl. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 48.long SD, Oberthur M, Losey HC, et al. The structural basis for induction of VanB resistance. J Am Chem Soc. 2002;124:9064–9065. doi: 10.1021/ja026342h. [DOI] [PubMed] [Google Scholar]
- 49.Karst N, Islam IF, Linhardt RJ. Sulfo-protected hexosamine monosaccharides: potentially versatile building blocks for glycosaminoglycan synthesis. Org Lett. 2003;5:4839–4842. doi: 10.1021/ol035882w. [DOI] [PubMed] [Google Scholar]
- 50.Jin W, DeAngelis PL. Analysis of the two active sites of the hyaluronan synthase and the chondroitin synthase of Pasteurella multocida. Glycobiology. 2003;13:661–671. doi: 10.1093/glycob/cwg085. [DOI] [PubMed] [Google Scholar]
- 51.DeAngelis PL, Oatrnan LC, Gay DF. Rapid chemoenzymatic synthesis of monodisperse hyaluronan oligosaccharides with immobilized enzyme reactors. J Biol Chem. 2003;278:35199–35203. doi: 10.1074/jbc.M306431200. [DOI] [PubMed] [Google Scholar]
- 52.Pummill PE, DeAngelis PL. Alteration of polysaccharide size distribution of a vertebrate hyaluronan synthase by mutation. J Biol Chem. 2003;278:19808–19814. doi: 10.1074/jbc.M301097200. [DOI] [PubMed] [Google Scholar]
- 53.DeAngelis PL. Microbial glycosaminoglycan glycosvltransferases. Glycobiology. 2002;12:9R–16R. doi: 10.1093/glycob/12.1.9r. [DOI] [PubMed] [Google Scholar]
- 54.DeAngelis FL, Gunay NS, Toida T, Mao W-J, Linhardt RJ. Identification of the capsular polysaccharides of type D and F Pasteurella multocida as unmodified heparin and chondroitin, respectively. Carbohydr Res. 2002;337:1547–1552. doi: 10.1016/s0008-6215(02)00219-7. [DOI] [PubMed] [Google Scholar]
- 55.Chen J, Avci FY, Muñoz EM, et al. Enzymatically redesigning of biologically active heparan sulfate. J Biol Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muñoz E, Xu D, Avci F, Kemp M, Liu J, Linhardt RJ. Enzymatic synthesis of heparin related polysaccharides on sensor chips: rapid screening of heparin-protein interactions. Biochem Biophys Res Commun. 2006;339:597–602. doi: 10.1016/j.bbrc.2005.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duncan MB, Liu M, Fox C, Liu J. Characterization of the N-deacetylase domain from the heparan sulfate N-deacetylase/N-sulfotransferase 2. Biochem Biophys Res Commun. 2006;339:1232–1237. doi: 10.1016/j.bbrc.2005.11.142. [DOI] [PubMed] [Google Scholar]
- 58.Kakuta Y, Sueyoshi T, Negishi M, Pedersen LC. Crystal structure of the sulfotransferase domain of human heparan sulfate N-deacetylase/N-sulfotransferase 1. J Biol Chem. 1999;274:10673–10676. doi: 10.1074/jbc.274.16.10673. [DOI] [PubMed] [Google Scholar]
- 59.Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J Biol Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- 60.Moon AF, Edavettal SC, Krahn JM, et al. Structural analysis of the sulfotransferase (3-OST-3) involved in the biosynthesis of an entry receptor of herpes simplex virus 1. J Biol Chem. 2004;279:45185–45193. doi: 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burkart MD, Izumi M, Chapman E, Lin C-H, Wong C-H. Regeneration of PAPS for the enzymatic synthesis of sulfated oligosaccharides. J Org Chem. 2000;65:5565–5574. doi: 10.1021/jo000266o. [DOI] [PubMed] [Google Scholar]
- 62.Smeds E, Habuchi H, Do A-T, et al. Substrate specificities of mouse heparan sulphate glucosaminyl 6-O-sulfotransferases. Biochem J. 2003;372:371–380. doi: 10.1042/BJ20021666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, Lawrence R, Schwartz JJ, et al. The effect of precursor structures on the action of glucosaminyl 3-O-sulfotransferase-1 and the biosynthesis of anticoagulant heparan sulfate. J Biol Chem. 2001;276:28806–28813. doi: 10.1074/jbc.M100204200. [DOI] [PubMed] [Google Scholar]
- 64.Atha DH, Lormeau J-C, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 65.Thanawiroon C, Rice KG, Toida T, Linhardt RJ. LC/MS sequencing of highly sulfated heparin-derived oligosaccharides. J Biol Chem. 2004;279:2608–2615. doi: 10.1074/jbc.M304772200. [DOI] [PubMed] [Google Scholar]
- 66.Chi L, Amster J, Linhardt RJ. Mass spectrometry for the analysis of highly charged sulfated carbohydrates. Curt Anal Chem. 2005;1:223–240. doi: 10.2174/157341105774573929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolff JJ, Chi L, Linhardt RJ, Amster IJ. Distinguishing glucuronic from iduronic acid in glycosaminoglycan tetrasaccharides by using electron detachment dissociation. Anal Chem. 2007;79:2015–2022. doi: 10.1021/ac061636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laremore TN, Murugesan S, Park T-1, Avci FY, Zagorevski DV, Linhardt RJ. Matrix assisted laser desorption/ionization mass spectrometric analysis of uncomplexed highly sulfated oligosaccharides using ionic liquid matrices. Anal Chem. 2006;78:1774–1779. doi: 10.1021/ac051121q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laremore TN, Zhang F, Linhardt RJ. New ionic liquid matrix for direct UV-MALDT TOE-MS analysis of dermatan sulfate and chondroitin sulfate oligosaccharides. Anal Chem. 2007;79:1604–1610. doi: 10.1021/ac061688m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seeberger PH, Werz DB. Automated synthesis of oligosaccharides as a basis for drug discovery. Nat Res Drug Discov. 2005;4:751–763. doi: 10.1038/nrd1823. [DOI] [PubMed] [Google Scholar]
- 71.Murugesan S, Xie J, Linhardt RJ. Immobilization of heparin-approaches and applications. Curr Top Med Chem. 2006 doi: 10.2174/156802608783378891. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murugesan S, Park T, Yang H, Mouse S, Linhardt RJ. Nano-based neoproteoglycans blood compatible carbon nanotubes. Langmuir. 2006;22:3461–3463. doi: 10.1021/la0534468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar A, Murugesan S, Pushparaj VL, et al. Conducting organic-inorganic submicron rods based on ionic liquids. Small. 2006;3:429–433. doi: 10.1002/smll.200600442. [DOI] [PubMed] [Google Scholar]
- 74.Psaltis D, Quake SR, Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature. 2006;442:381–386. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 75.DeAngelis PL. Polymer grafting by polysaccharide syntheses. 6,444,447. U.S. Patent. 1999
- 76.Zhi ZL, Powell AK, Turnbull JE. Fabrication of carbohydrate microarrays on gold surfaces: Direct attachment of nonderivatized oligosaccharides to hydrazide modified self-assembled monolayers. Anal Chem. 2006;78:4786–4793. doi: 10.1021/ac060084f. [DOI] [PubMed] [Google Scholar]
- 77.de Paz JL, Noti C, Seeberger PH. Microarrays of synthetic heparin oligosaccharides. J Am Chem Soc. 2006;128:2766–2767. doi: 10.1021/ja057584v. [DOI] [PubMed] [Google Scholar]
- 78.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparan sulfate on a biochip promotes its interaction with antithrombin III. Biochem Biophys Res Common. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 79.Bertone P, Snyder M. Advances in functional protein microarray technology. FEBS J. 2005;272:5400–5411. doi: 10.1111/j.1742-4658.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- 80.Mikhailov D, Young HC, Linhardt RJ, Mayo KH. Heparin dodecasaccharide binding to platelet fector-4 and growth-related protein-α: Introduction of a partially folded state and implications for heparin induced thrombocytopenia. J Biol Chem. 1999;274:25317–25329. doi: 10.1074/jbc.274.36.25317. [DOI] [PubMed] [Google Scholar]
- 81.De Graffenried CL, Bertozzi CR. The roles of enzyme localization and complex formation in glyean assembly within the Golgi apparatus. Curt Opin Cell Biol. 2004;16:356–363. doi: 10.1016/j.ceb.2004.06.007. [DOI] [PubMed] [Google Scholar]