Abstract
Heparan sulfate carries a wide range of biological activities, regulating blood coagulation, cell differentiation, and inflammatory responses. The sulfation patterns of the polysaccharide are essential for the biological activities. In this study, we report an enzymatic method for the sulfation of multimilligram amounts of heparan sulfate with specific functions using immobilized sulfotransferases combined with a 3′-phosphoadenosine 5′-phosphosulfate regeneration system. By selecting appropriate enzymatic modification steps, an inactive precursor has been converted to the heparan sulfate having three distinct biological activities, associated with binding to antithrombin, fibroblast growth factor-2, and herpes simplex virus envelope glycoprotein D. Because the recombinant sulfotransferases are expressed in bacteria, and the method uses a low cost sulfo donor, it can be readily utilized to synthesize large quantities of anticoagulant heparin drug or other biologically active heparan sulfates.
Heparan sulfate (HS)3 is a ubiquitous component of the cell surface and extracellular matrix. It regulates a wide range of physiologic and pathophysiologic functions, including embryonic development and blood coagulation and can facilitate viral infection (1, 2). HS exerts its biological effects by interacting with the specific proteins involved in a given process (3). HS is a highly charged polysaccharide consisting of 1←4-linked glucosamine and glucuronic/iduronic acid units that contain both N- and O-sulfo groups. Unique saccharide sequences within HS determine the specificity of the binding of HS to its target proteins (4). Heparin, a specialized form of HS, is a commonly used anticoagulant drug. Thus, new methods for the synthesis of heparin and HS attract considerable interest for those developing anticoagulant drugs having improved pharmacological effects.
Chemical synthesis has been the major route to obtain structurally defined heparin and HS oligosaccharides (5). The most important example involves the structure antithrombin-binding pentasaccharide. Asynthetic pentasaccharide, based on this structure, has been marketed in the United States under the trade name Arixtra. Arixtra is a specific factor Xa inhibitor that is used clinically to prevent venous thromboembolic incidents during surgery. Unfortunately, the total synthesis of heparin and HS oligosaccharides, larger than pentasaccharides, is extremely difficult. HS analogues with 14 saccharide units inhibit the activity of thrombin, but these synthetic analogues are simplified hybrid molecules of HS oligosaccharides and highly sulfated glucose units (6, 7) and are not the naturally occurring structures. Although our laboratory and others continue to pursue the synthesis of heparin and HS oligosac-charides (8), it has become clear that chemical synthesis alone is currently incapable of generating most larger oligosaccharide structures. The application of HS biosynthetic enzymes for generating large heparin and HS oligosaccharides with desired biological activities offers a promising alternative approach.
Six classes of enzymes are involved in the HS biosynthesis. HS is initially synthesized as a copolymer of D-glucuronic acid and GlcNAc through the action of D-glucuronyl and N-acetyl-D-glucosaminyltransferase (9). Next, a series of modifications take place, including N-deacetylation andN-sulfation (carried out byN-deacetylase/N-sulfotransferase) of the glucosamine residue to form N-sulfoglucosamine (GlcNS), C5 epimerization of glucuronic acid (carried out by epimerase) to form L-iduronic acid (IdoUA), 2-O-sulfation of IdoUA (carried out by 2-O-sulfotransferase (2-OST)), 6-O-sulfation of glucosamine (carried out by 6-O-sulfotransferase (6-OST)), and 3-O-sulfation of glucosamine (carried out by 3-O-sulfotransferase (3-OST)) (10). The reactions catalyzed by 2-OST, 6-OST, and 3-OST are shown in Fig. 1A. The enzymes responsible for HS biosynthesis have been cloned and expressed and were employed in the synthesis of HS polysaccharides. Kuberan and Rosenberg (11–13) utilized these enzymes to synthesize the HS containing antithrombin binding sites with anticoagulant activity. Although this approach demonstrated for the first time the feasibility of enzymatic synthesis of HS, approximately 1 μg of product was generated, making extensive structural characterization and biological studies impossible. Recently, Lindahl and colleagues reported an alternative chemoenzymatic approach for the synthesis of anticoagulant heparin from heparosan, the E. coli K5 capsular polysaccharide (14). This method utilized the C5 epimerase to convert D-glucuronic acid to IdoUA, followed by the chemical persulfation and finally selective desulfation. Although this approach afforded approximately 5 g of a heparin-like polysaccharide with anticoagulant activity, unnatural saccharide units, such as 3-O-sulfo- D-glucuronic acid, were present in their product, suggesting a limitation in the selectivity of chemical sulfation/desulfation in HS synthesis. Thus, developing an effective and highly selective approach for O-sulfation of polysaccharides remains a challenge for the large scale synthesis of HS.
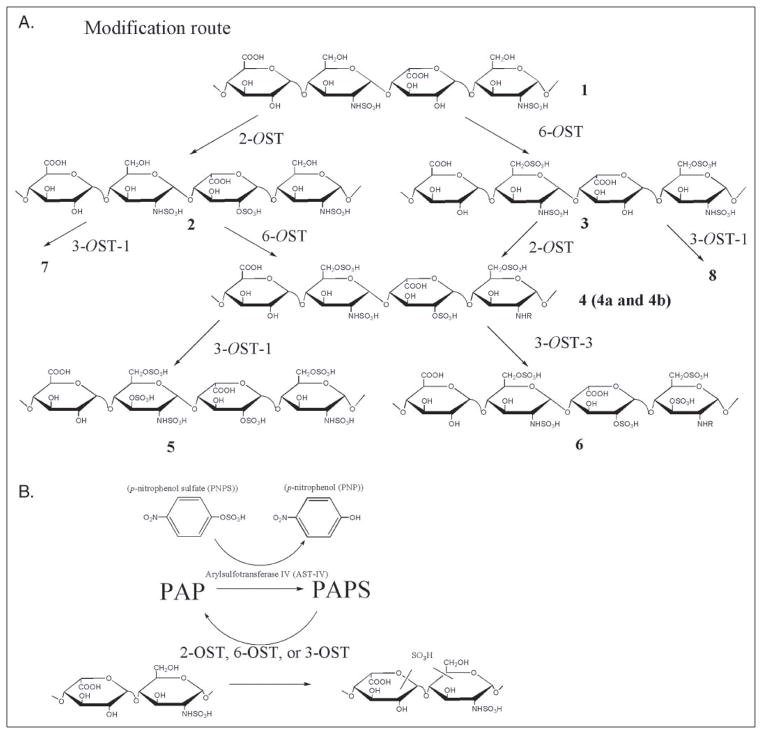
FIGURE 1. Schematic synthesis of sulfated polysaccharides and PAPS regeneration system.
A, the stepwise enzymatic synthesis of sulfated polysaccharides using HS sulfotransferases. The description of intermediate polysaccharides is given under “Results.” Compounds 4a and 4b were prepared by inverting the order of sulfation steps. 4a was prepared by incubating compound 1 with 2-OST followed by 6-OST, whereas 4b was prepared by incubating compound 1 with 6-OST followed by 2-OST. B, the reaction catalyzed by arylsulfotransferase IV (AST-IV) to generate PAPS. R represents -H or -SO3.
In this work, we describe an enzymatic approach to synthesize HS polysaccharides. Only two or three enzymatic steps are required for the synthesis of HS having three biological activities (Fig. 1). Two major advances have permitted the large scale synthesis of HS. First, large amounts of all the required HS sulfotransferases have been successfully expressed in Escherichia coli. Second, a PAPS regeneration system, developed by Burkhart and colleagues (15), has been coupled to the enzymatic synthesis reactions. We synthesized HS having specific biological activities including anticoagulant heparin, herpes simplex virus glycoproteinD(gD)-binding HS, and fibroblast growth factor 2 (FGF2)- binding HS. Each of these products had activities comparable with the heparin and HS. Furthermore, the amount of synthesized HS was sufficient to permit not only disaccharide analysis but also analysis using 1H NMR spectroscopy, resulting in the determination of the product structure with a high degree of confidence. The results from our study suggest that this is a general method for the large scale synthesis of a wide range of HS with specific activity required. In addition, this method provides a model system to better understand the biosynthesis of HS.
EXPERIMENTAL PROCEDURES
Preparation of Chemically Desulfated N-Sulfated Heparin (CDSNS Heparin)
Treatment of heparin with a dimethyl sulfoxide/methanol mixture (9:1, v/v) at 80 °C for 24 h resulted in nearly complete N,O-desulfation with <5% of the solvolytically resistant O-sulfo groups remaining. Chemical N-sulfation with sulfur trioxide-pyridine afforded CDSNS heparin, which was characterized by disaccharide analysis and 1H and two-dimensional COSY NMR.
Expression and Purification of 2-OST and 6-OST
The catalytic domains of 2-OST of Chinese hamster ovary (Arg51–Asn356) and mouse 6-OST-1 (His53–Trp401) were cloned into the pMAL-c2X vector (New England BioLabs) using the BamHI and HindIII sites to generate maltose- binding protein (MBP)-2-OST and MBP-6-OST fusion proteins. The full-length cDNAs of 2-OST and 6-OST-1 were gifts from Dr. Rosenberg (Massachusetts Institute of Technology, Cambridge, MA) and Dr. Kimata (Aichi University, Japan), respectively. Expression of 2-OST and 6-OST was achieved in Rosetta-gami B (DE3) cells (Novagen) using a standard procedure. Briefly, cells containing the plasmid expressing 2-OST or 6-OST were grown in LB medium supplemented with 2 mg/ml glucose, 15 μg/ml tetracycline, 15 μg/ml kenamycin, 35 μg/ml chloramphenicol, and 50 μg/ml carbenicillin at 37 °C. When the A600 reached 0.6–0.8, Isopropyl-β-D-thiogalactopyranoside (1mM)was added, and the cells were shaken overnight at 22 °C. The bacteria were harvested, and the proteins were purified by following a protocol from the manufacturer (New England Biolabs). The purified proteins migrated at 75 kDa on 12% SDS-PAGE with the purity greater than 80%.
Preparation of Immobilized HS Sulfotransferases
Dialyzed sulfotransferases (3 ml, 4 mg/ml) in phosphate-buffered saline buffer (3 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4, 138 mM NaCl, pH 7.2) were mixed with 1ml of Aminolink® plus beads (Pierce) following a protocol from the manufacturer. Immobilized enzyme was washed with 50 mM MES, 1% Triton X-100, 1 mM MgCl2, and 1 mM MnCl2, pH 7.0, and stored at 4 °C.
Measurement of Enzymatic Activities of Immobilized Proteins
To determine the activity of 3-OST-1 and 3-OST-3, HS from bovine kidney (ICN Biomedicals) was used as a substrate; to determine the activity of 6-OST and 2-OST, CDSNS heparin was used as a substrate. Immobilized proteins (100μl, ~300μg of immobilized enzyme) with 100μg of substrate (HS for 3-OST and CDSNS heparin for 2-OST or 6-OST) and 200 μM [35S]PAPS (1000 cpm/pmol) in 1 ml of 50 mM MES, pH 7.0, 1% Triton X-100, 1 mM MgCl2, and 1 mM MnCl2. After rotating at room temperature for 1 h, the supernatant was collected, and the beads were washed with 3 × 200 μl of 1 M NaCl in 25 mM MOPS (pH 7.0). The supernatant and washes were combined, diluted with 2ml of water, and subjected to DEAE chromatography to determine the amount of [35S]HS product used to determine the activities of the various HS O-STs.
PAPS Regeneration System
The reactions involved in the PAPS regeneration system are shown in Fig. 1B. N-terminal His6-tagged AST-IV was expressed in E. coli and purified as described by Burkat and colleagues (15) at a yield of ~50 mg/liter of bacterial culture. The fulllength cDNA of rabbit AST-IV was a generous gift of Dr. Michael Duffel (University of Iowa) (16).
Modification of Polysaccharides
All enzymatic modifications, including those catalyzed by 2-OST, 6-OST, 3-OST-1, and 3-OST-3 followed the same protocol. Briefly, 20 mg of purified AST-IV was incubated with 40 μM PAP and 1 mM PNPS in 20 ml of 50 mM MES, pH 7.0, 1% Triton X-100, 1 mM MgCl2, and 1 mM MnCl2 at 25 °C for 15 min. The reaction mixture was mixed with 4 ml of immobilized sulfotransferase, 2 mg of a polysaccharide substrate was added, and the mixture was rotated at 25 °C for 24 h. The supernatant was recovered, and the polysaccharide that bound to the beads was eluted by washing three times with 8ml of 1 M NaCl in 25mM MOPS. Both the supernatant and wash were combined and the product was precipitated by adding ethanol (3 volumes). After incubating overnight at 4 °C, polysaccharide was recovered by centrifugation.
Estimation of the Modification Level
The completion of 2-OST modification was monitored by incubating 10 μg of polysaccharide (2) with 20 μg of soluble 2-OST in the presence of 100 μM of [35S]PAPS at 37 °C for 30 min. In a control experiment, 10 μg of compound 1 replaced the previously modified polysaccharide in the otherwise identical reaction mixture. By comparing the amount of 35S incorporation, we could estimate the extent of the original 2-O-sulfation reaction. For monitoring the completion of 6-OST, compound 1 or 2 was used as a substrate; for 3-OST-1, compound 4 was used.
Using [35S]PAPS as a Sulfate Donor
Antithrombin (AT)-binding and gD-binding experiments utilized 35S-labeled polysaccharides prepared using [35S]PAPS. In a typical reaction, 2 mg of HS substrate was incubated with 4 ml of beads with immobilized sulfotransferase (~12 mg of immobilized enzyme) at 25 °C in 20 ml of 50mM MES, pH 7.0, 1% Triton X-100, 1 mM MgCl2, and 1 mM MnCl2, 200 μM [35S]PAPS (1000 cpm/pmol) for 1 h. The resultant polysaccharide was recovered using DEAE chromatography.
Disaccharide Analysis
Synthesized polysaccharides (100 μg) were degraded by amixture of heparin lyases as previously described (17) and desalted on BioGel P-2 column (0.5 × 200 cm) in 0.1 M NH4HCO3. Disaccharides were analyzed by a C18 reversed phase column (0.45×25 cm; Vydac) with UV 232 detection and identified by coelution with appropriate standards (18). The overall recovery yield of the disaccharide analysis was estimated by using 2-O-[35S]heparin (100,000 cpm/70 ng; compound 2) as an internal control.
NMR Analysis
Polysaccharide sample (1–2 mg)was dissolved in 0.5 ml of 2H2O(99.9%), freeze dried to remove exchangeable protons, redissolved in 75 μl of 2H2O (100.00%), and transferred to a Shigemi NMR microtubes. NMR spectra were referenced relative to the HO2H at 4.80 ppm, and in COSY water was suppressed by presaturation of the HO2H resonance.
Determination of the Binding to AT and FGF2 Using Surface Plasmon Resonance Spectroscopy (SPR)
Heparin and the synthesized polysaccharides were biotinylated following the published procedure (19). A solution of the biotinylated polysaccharides (0.1 mg/ml) in Hepes (10 mM), 0.15 M NaCl, 3 mM EDTA, and 0.005% surfactant P20 (HBS-EP) was flowed over the cells of the streptavidin chip at 5 μl/min. To determine the KD values, different concentrations (25–1600 nM in HBS-EP buffer) of FGF2 andATwere individually injected at 30μl/min (running buffer: HBS-EP). Kinetic injection was used, leading the protein to flow for 180 s and dissociated for the next 180 s, and the surface was regenerated by a 60-s injection of 30 μl of 2 M NaCl. Response units were monitored as a function of time to afford sensorgrams. The SPR curves for FGF2 fit well at individual concentrations, but the global fit suggested significant binding heterogeneity. Thus, the equilibrium response unit RU(eq) values from the sensorgrams of FGF2 binding to polysaccharide-containing surfaces were used to construct Scatchard plots, RU(eq)/C versus RU(eq), where C is the free protein concentration, resulting in linear, first degree polynomial functions confirming the one-to-one binding of FGF-polysaccharide and to estimate binding affinity. A two-state reaction model was applied to the AT-polysaccharide interactions measured by SPR using curve fitting to estimate the association and dissociation rate constants and affinity constant.
FGF2/FGFR1C-mediated Proliferation Assay
The BaF3 cells ectopically expressing FGFR1C have been previously described (20). The BaF3-FGFR1c cells were maintained in RPMI 1640 medium (Sigma) supplemented with 10% fetal bovine serum, 0.5 ng/ml interleukin (IL)-3 (PeproTech Inc., Rocky Hill, NJ), 2mML-glutamine, penicillin (50 IU/ml) and streptomycin (50 μg/ml), and 50 μM β-mercaptoethanol. For mitogenic assays, BaF3 FGFR1c cells were washed three times with RPMI 1640 medium to remove IL-3 and resuspended in the growth medium lacking IL-3. About 30,000 cells were plated per well in a 96-well plate in medium containing 1μg/ml heparin, compound 1, 2, 3, 4a, or 4b, and 2 nM FGF-2 (PeproTech) in a total volume of 200 μl. The cells were then incubated at 37 °C for 40 h. To each well, an additional 50 μl of growth medium containing 1 μCi of [3H]thymidine was added. Cells were harvested after 4–5 h by filtration through glass fiber paper. The incorporation of [3H]thymidine into the DNA was determined by scintillation counting.
Determination of the Binding of Compounds 5–8 to AT
Approximately 1 × 105 cpm of 35S-labeled compound was incubated with 5 μg of humanAT(Cutter Biological) in 50μl of binding buffer containing 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Mn2+, 1 mM Mg2+, 1 mM Ca2+, 10 μM dextran sulfate, 0.0004% Triton X-100, and 0.02% sodium azide for 30 min at room temperature. Concanavalin A-Sepharose (Sigma; 50μl of 1:1 slurry) was then added, and the reaction was shaken at room temperature for 1 h. The beads were then washed by 3 × 1 ml of binding buffer, and the bound polysaccharide was eluted with 1 M NaCl.
Inhibition Effect of the Polysaccharides on the Activities of Factor Xa and Thrombin
Assays were based on two previous methods (21, 22). Briefly, factor Xa (Enzyme Research Laboratories, South Bend, IN) and thrombin (Sigma) were diluted to 20 and 8 units/ml with phosphate-buffered saline containing 1mg/ml bovine serum albumin, respectively. AT was diluted with phosphate-buffered saline containing 1 mg/ml bovine serum albumin to give a stock solution at the concentration of 27 μM. The chromogenic substrates, S-2765 (for factor Xa assay) and S-2238 (for thrombin assay) were from Diapharma and made up at 1mM with 1 mg/ml Polybrene in water. The synthesized polysaccharide (compounds 5, 7, and 8) or heparin was dissolved in a buffer containing 50 mM Tris-HCl, pH 8.4, 7.5 mM Na2EDTA, 175 mM NaCl at various concentrations (1–10,000 ng/ml). The reaction mixture, which consisted of 25μl of AT stock solution and 25μl of the solution containing polysaccharide, was incubated at 37 °C for 2 min. Factor Xa (25 μl) or thrombin (25 μl) was added. After incubating 37 °C for 4 min, 25 μl of S-2765 or S-2238 was added. The absorbance of the reaction mixture was measured at 405 nm continuously for 10 min. The absorbance values were plotted against the reaction time. The absorbance of the reaction mixture was measured at 405 nm continuously for 10 min. The initial reaction rates as a function of concentration were used to calculate the IC50 values. The concentrations of the synthesized polysaccharides were determined using Alcian blue as described by Bjornsson (23) and quantitative disaccharide analysis as described above.
The Binding to Herpes Simplex Virus gD
The assay for determining binding of 35S-labeled polysaccharides (compounds 5 and 6) to gD was carried out by an immunoprecipitation procedure using anti-gD monoclonal antibody (DL6) (24).
RESULTS
Development of an Efficient Enzymatic Synthesis of Sulfated Polysaccharides
Expression of HS Sulfotransferases in E. coli
Our goal is to develop an approach to synthesize FGF-binding HS (compound 4, including 4a and 4b) AT-binding HS (compound 5), and herpes simplex virus gD-binding HS (compound 6). Four enzymes, including 2-OST, 6-OST, 3-OST-1, and 3-OST-3, were required for the syntheses of these targets. We recently demonstrated that bacterial expressed 3-OST-1 and 3-OST-3 exhibit substrate specificity and specific enzymatic activity comparable with those of their counterparts expressed in insect cells (17, 25). Expression of the catalytic domains of 2-OST and 6-OST was also achieved in relatively high yield by preparing a fusion protein with MBP and 2-OST or 6-OST in Rosetta-gami B cells. Because 2-OST and 6-OST fusion proteins were enzymatically active and highly soluble, we decided to retain the MBP domain in the current study.
Immobilized Enzymes Are Reusable
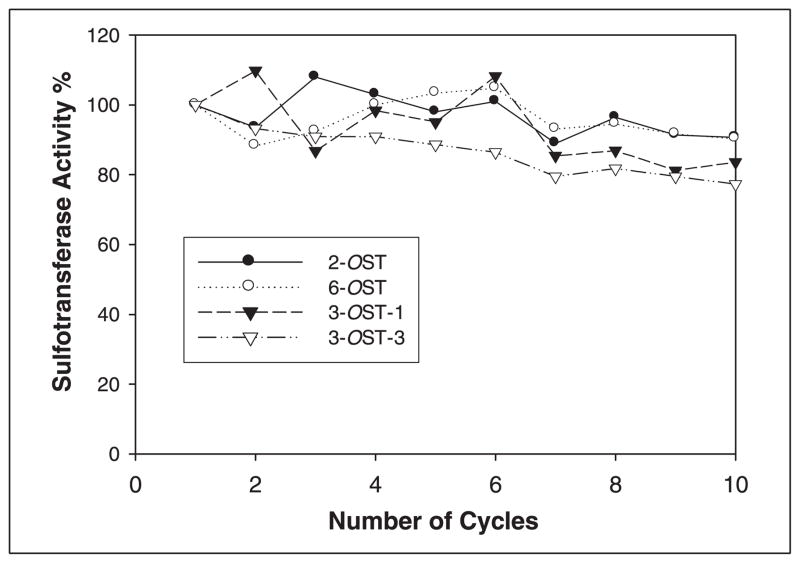
2-OST, 6-OST, 3-OST-1, and 3-OST-3 were immobilized on agarose to be reusable and to enhance the thermal stability. We incubated immobilized enzymes with polysaccharide substrate and PAPS for 1 h at room temperature. The sulfated polysaccharide product was separated from the immobilized enzyme by washing the beads with 1MNaCl followed by centrifugation, making the immobilized enzymes ready for the next synthetic cycle. The cycle was repeated 10 times, after which each of the immobilized enzymes were assayed and showed >80% of their catalytic activity (Fig. 2). We have also found that the immobilized enzymes also maintained>65% of their catalytic activity after 2 months of storage at 8 °C (data not shown).
FIGURE 2. Activities of recycled immobilized sulfotransferases.
The immobilized enzymes were utilized multiple cycles. The activities of the immobilized enzymes after each were determined as described under “Experimental Procedures.” A total of 10 cycles were conducted.
Introduction of PAPS Regeneration System
PAP inhibits sulfotransferase- catalyzed reactions. A PAPS regeneration system can be used to convert PAP to PAPS by relying on AST-IV to catalyze the transfer of the sulfo group from PNPS to PAP, as illustrated in Fig. 1B (15). We observed that the PAPS regeneration system performed very well with 2-OST, 6-OST, and 3-OST-1. Complete modification of the substrate could be demonstrated by the low susceptibility of polysaccharide product to undergo additional sulfation using [35S]PAPS with soluble enzymes, as described under “Experimental Procedures.” Under the standard conditions, 2-OST, 6-OST, and 3-OST-1 afforded 98, 97, and 98% complete modification, respectively, using the PAPS regeneration system. These results demonstrate that the PAPS regeneration system functioned properly, providing sufficient PAPS for the complete sulfotransferase- catalyzed modification of polysaccharide substrates. This conclusion was further supported by characterizing the structures of the newly synthesized polysaccharide products 2, 3, 4a, 4b, and 5 (described below). In a parallel experiment, we used [35S]PAPS as the sulfo donor to sulfate the polysaccharides in order to estimate the number of sulfo groups that were incorporated into the product. Six 2-O-sulfo and 6-O-sulfo groups and three 3-O-sulfo groups were estimated to be transferred to one polysaccharide molecule, respectively, assuming that the length of the polysaccharide is 25 disaccharide units. Approximately 1.5 mg of anticoagulant HS (5) was synthesized from 10 mg of CDSNS heparin (1) using immobilized sulfotransferases and the PAPS regeneration system.
Structural Characterization of Synthesized Polysaccharides
Disaccharide Analysis of the Polysaccharides
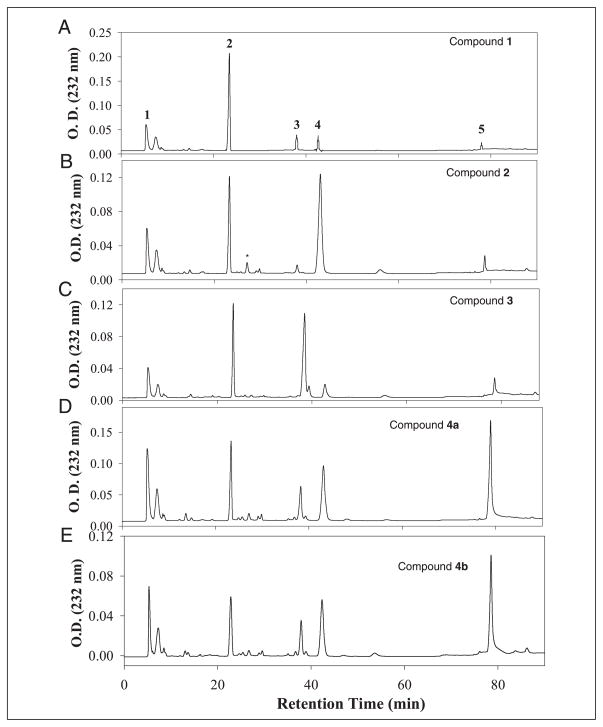
Synthesized polysaccharide intermediates were digested with a mixture of heparin lyases, and the resulting disaccharides were analyzed using RPIP-HPLC (Fig. 3). As expected, analysis of compound 1 showed a disaccharide component of the structure of ΔUA-GlcNS (disaccharide 2), residual unsulfated disaccharide, ΔUA-GlcNAc (disaccharide 1), and small amounts of sulfated disaccharides (disaccharides 3–5) due to incomplete chemical desulfation (Fig. 3A and TABLE ONE). The modification by 2-OST elevated the level of ΔUA2S-GlcNS (disaccharide 4) by about 5-fold (Fig. 3B and TABLE ONE), confirming the structure of compound 2. Analysis of compound 3 afforded a 7-fold increase in the level of the disaccharide ΔUA-GlcNS6S (disaccharide 3) compared with compound 1, consistent with 6-OST-catalyzed modification (Fig. 3C and TABLE ONE). The level of trisulfodisaccharide, ΔUA2S-GlcNS6S (disaccharide 5), in compounds 4a and 4b was increased by about 10-fold compared with that of compound 1 (Fig. 3, D and E, and TABLE ONE). The results from the disaccharide analysis clearly establish that the expected enzymatic modifications took place at each step. It is interesting to note that 6-O-sulfation occurs at N-sulfoglucosamine, consistent with the substrate specificity of 6-OST in vitro (26). The 2-O-sulfation predominantly occurs at the uronic acid with an N-sulfoglucosamine residue at the reducing end. We also note that substantial amounts of ΔUA-GlcNS (disaccharide 2) remain in compounds 4a and 4b. This observation is not unexpected, since the HS from various tissues also afford a ΔUA-GlcNS disaccharide unit, suggesting that the structures of compounds 4a and 4b are similar to HS from natural sources (27).
FIGURE 3. RPIP-HPLC chromatograms of the disaccharide analysis of synthesized polysaccharides.
The synthesized polysaccharides were digested with a mixture of heparinases, including heparin lyase I, II, III, and heparitinase IV. The resultant disaccharides were purified by BioGel P-2 and resolved on RPIP-HPLC. A–E, the chromatogram of the disaccharide analysis of compound 1, 2, 3, 4a, and 4b, respectively. The arrows indicate the eluted positions of authentic disaccharide standards, where 1 represents ΔUA-GlcNAc, 2 represents ΔUA-GlcNS, 3 represents ΔUA-GlcNS6S, 4 represents ΔUA2S-GlcNS, and 5 represents ΔUA2S-GlcNS6S. *, the eluted position of ΔUA2SGlcNAc. Because we do not have this disaccharide standard, we could not determine the quantity of ΔUA2S-GlcNAc.
TABLE ONE. Summary of the disaccharide compositions of the synthetic polysaccharides.
Each synthesized polysaccharide (100 μg) was digested with a mixture of heparin lyases. The resultant disaccharides were purified by a BioGel P-2 column and resolved by RPIP-HPLC.
| Compound | ΔUA-GlcNAc | ΔUA-GlcNS | ΔUA2S-GlcNS | ΔUA-GlcNS6S | ΔUA2S-GlcNS6S | Total recovery |
|---|---|---|---|---|---|---|
| nmol a % | nmol % | nmol % | nmol % | nmol % | %b | |
| 1 | 12.5 (16.1%) | 50.0 (64.4%) | 6.3 (8.1%) | 6.3 (8.1%) | 2.5 (3.2%) | 54.0 |
| 2 | 12.5 (16.6%) | 27.5 (36.6%) | 30.0 (40.0%) | 2.5 (3.3%) | 2.5 (3.3%) | 49.2 |
| 3 | 10.0 (14.0%) | 27.5 (38.5%) | 5.0 (7.0%) | 25.0 (35.0%) | 3.8 (5.3%) | 51.0 |
| 4a | 12.5 (18.8%) | 17.5 (26.2%) | 7.5 (11.2%) | 10.0 (15.0%) | 20.0 (30.0%) | 50.0 |
| 4b | 17.5 (21.7%) | 15.0 (18.6%) | 8.0 (9.9%) | 15.0 (18.6%) | 25.0 (31.0%) | 48.0 |
The amount of each disaccharide was estimated by determining its peak area with a standard curve generated with a known amount of the disaccharide standard.
A recovery yield was calculated by using 2-O-[35S]sulfoheparin as an internal standard.
NMR Analysis of the Polysaccharides
The 1H NMR analysis was conducted on the synthesized polysaccharides to allow us to assess their structures at the polymer levels as well as to estimate the composition. The assignment of each nonexchangable proton was made using COSY, and their chemical shifts are reported in TABLE TWO.
TABLE TWO.
1H chemical shift data (in ppm) for the synthesized polysaccharides
| Sample | GlcN H1 | GlcN H2 | GlcN H3 | GlcN H4 | GlcN H5 | GlcN H6a/b | IdoUA H1 | IdoUA H2 | IdoUA H3 | IdoUA H4 | IdoUA H5 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Heparin | 5.41 | 3.26 | 3.69 | 3.79 | 4.04 | 4.29 | 5.22 | 4.35 | 4.22 | 4.12 | 4.82 |
| 4.42 | |||||||||||
| Compound 1 | 5.40 | 3.23 | 3.67 | 3.72 | 3.79 | 3.84 | 4.87 | 3.70 | 3.93 | 4.17 | 4.72 |
| Compound 2 | 5.33 | 3.24 | 3.70 | 3.74 | 3.75–3.84 | 3.85 | 5.23 | 4.34 | 4.23 | 4.08 | 4.87 |
| Compound 3 | 5.38 | 3.24 | 3.63 | 3.78 | 4.13 | 4.30 | 4.95 | 3.72 | 4.10 | 4.05 | 4.74 |
| Compound 4a | 5.38 | 3.25 | 3.70 | 3.83 | 4.19 | 4.25 | 5.23 | 4.33 | 4.21 | 4.13 | 4.90 |
| 4.33 | |||||||||||
| Compound 4b | 5.37 | 3.25 | 3.67 | 3.83 | 4.18 | 4.26 | 5.22 | 4.35 | 4.21 | 4.15 | 4.90 |
| 4.33 | |||||||||||
| Compound 5 | 5.36 | 3.22 | 4.10 | 3.71 | 4.17 | 4.32 | 5.23 | 4.33 | 4.22 | 4.08 | 4.94 |
| 4.38 |
The integration of the N-acetyl signal in the 1H NMR of heparin was compared with the integration of the GlcNS-H1 signal, allowing us to estimate the level of modifications. The ratio of these integrals showed that ~15% of the glucosamine residues contained N-acetyl groups. Using this information, we examined the incorporation of 2-O-, 3-O-, and 6-O-sulfo groups. Comparison of the N-acetyl peak with IdoUA2S-H2 in compound 2 demonstrates incorporation of a 2-O-sulfo group into 35% of the IdoUA residues. Based on this incorporation into compound 2, the integral of GlcNS-H6a and IdoUA2S-H2 (overlapping signal) was compared with the integral of the N-acetyl methyl group in compound 4a to estimate the level of incorporation of the 6-O-sulfo group. Integration showed that the 6-O-sulfo group was incorporated into 25% of the GlcNS residues. The reduced level of incorporation is not surprising, since only the 6-OST-1, and not 6-OST-2 and -3, was used in this synthesis. In compound 5, the incorporation of the 3-O-sulfo group was calculated by comparing the integral of the N-acetyl methyl group with GlcN-H3 and IdoUA-H4 (overlapping signal). Based on these calculations, the 3-O-sulfo group was incorporated into 32% of the GlcNS6S residues. According to 1HNMR and COSY experiments (TABLE TWO), the structures of the compounds 4a, 4b, and 5 were all found to be similar to heparin and contained no unusual signals. Compound 5 showed a slightly greater heterogeneity, as evidenced by additional minor signals corresponding to additional 3-O-sulfo group-containing sequences in the 1H NMR, when compared with heparin. This is not unexpected, since heparin contains a lower level of 3-O-sulfo groups/chain than the content of 3-O-sulfo groups/chain observed in compound 5 as determined by 35S incorporation.
The composition estimated by NMR is consistent with the results of disaccharide analysis. NMR analysis suggests that about 15% of the glucosamine unit is N-acetylated, which is similar to the results of disaccharide analysis for ΔUA-GlcNAc (14–22%; TABLE ONE). NMR analysis demonstrates that compound 2 consists of 35% of IdoUA2S-GlcNS, and compound 4a consists of 25% of IdoUA2S-GlcNS6S, and disaccharide analysis demonstrates that compounds 2 and 4a contain 40% of ΔUA2S-GlcNS and 30% of ΔUA2S-GlcNS6S, respectively. Because of the signal overlap, we were unable to calculate the degree of 6-O-sulfation in compounds 3 and 4b using 1HNMR.
Determination of the Biological Activities of the Synthesized Polysaccharides
The Binding of the Polysaccharides to AT and FGF2
Characterization of the affinities of AT to heparin and enzymatically modified heparin derivatives were performed by SPR. A two-state reaction model was applied to the SPR study of AT and polysaccharide interactions using BIA evaluation Software for curve fitting analysis. None of the derivatives with the exception of the 2,6,3-O-sulfopolysaccharide (5) and heparin had high affinity to AT. The binding constant (KD) for the binding of compound 5 to AT was determined to be 170 nM, which is very similar to that of heparin (75 nM).We also estimated the binding affinity of FGF2 to the synthesized polysaccharides. Compound 1 showed no interaction with FGF2, whereas compounds 4a and 4b showed the identical binding affinity to FGF2 at a Kd of 35 nM, which is similar to heparin (22 nM) and is consistent with the value reported in the literature (28).
Synthetic Polysaccharides Promote Cell Proliferation
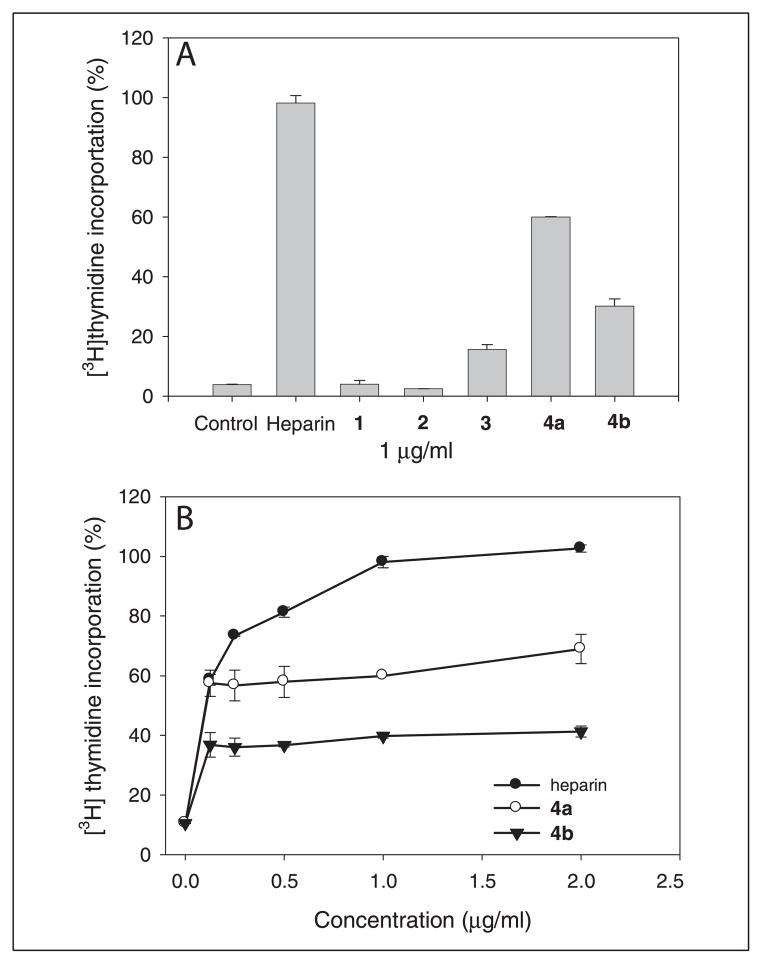
The BaF3 FGFR1c cells normally depend on IL-3 for growth. In the absence of IL-3, the cell proliferation depends on the addition of both FGF and heparin or HS (20). We measured the activity of compounds 1, 2, 3, 4a, and 4b and heparin in promoting cell mitogenesis using the FGF-2/ FGFR1c system in BaF3 cells as described under “Experimental Procedures.” The cells receiving compounds 4a and 4b (at 1 μg/ml) showed an increase in [3H]thymidine incorporation, which was about 60 and 40% of that of heparin, respectively, suggesting that the combinations of 2-O- and 6-O-sulfations confer the activity in promoting cell proliferation (Fig. 4A). We also compared the activity of 4a and 4b with that of heparin at different concentrations (Fig. 4B). We found that compound 3 had moderate activity in promoting cell proliferation, whereas compounds 1 and 2 did not exhibit activity. The results are consistent with previously reported data on the contributions of the sulfo groups of HS to the mitogenic activity (29, 30). We noted that 4a appears to have stronger activity in promoting cell proliferation than 4b, suggesting that different sequences are generated by the different order of 2-O- and 6-O- sulfation as suggested by Jacobsson and Lindahl (31). However, such sequence difference could not be detected by the disaccharide or NMR analysis.
FIGURE 4. The effect of the synthesized polysaccharides on FGF-2-dependent BaF3 FGFR1c cell proliferation.
BaF3 FGFR1c cells were seeded in 96-well plates as described with 2 nM FGF2 for control and 2 nM FGF2 plus a 1μg/ml concentration of the following compounds: heparin, 1, 2, 3, 4a, and 4b (A). B, dose-response curves of heparin, 4a, and 4b for their activities in stimulating cell proliferation. Cells were cultured for 40 h, followed by incubating in the media containing [3H]thymidine for 4 h. The cellular proliferation was determined by [3H]thymidine incorporation into the DNA. Data are mean ± range of duplicates.
The Anticoagulant Activity of the Synthesized Polysaccharides
Heparin achieves its anticoagulant activity by forming a 1:1 complex with AT, which inhibits the activities of factor Xa and thrombin (32). Because it is known that the introduction of the 3-O-sulfo group by 3-OST-1 is essential for the synthesis of anticoagulant HS, we prepared different types of 3-O-sulfo group-containing polysaccharides, compounds 5, 7, and 8, and tested their activities in inhibiting factor Xa and thrombin (TABLE THREE). As expected, heparin is a potent activator for AT-mediated inhibition of factor Xa and thrombin, whereas Arixtra specifically activates the AT-mediated inhibition of factor Xa (5). Compound 5 has very similar potency to heparin, inhibiting the activities of both factor Xa and thrombin, suggesting that our enzyme-based approach is indeed capable of synthesizing the anticoagulant polysaccharide. It has been reported that the presence of 2-O-sulfo groups is not essential for HS binding to AT and its resulting anticoagulant activity (33). Indeed, our polysaccharide intermediate compound 8 lacks 2-O-sulfo groups but still exhibits anticoagulant activity, consistent with this previous report (33). In contrast, compound 7 lacks 6-O-sulfo groups and, thus, has no anticoagulant activity, since 6-O-sulfo groups are critical in AT binding (34). Another 3-O-sulfated polysaccharide (compound 6) was also prepared to test for its anti-Xa and antithrombin activities. It is important to note that both compounds 5 and 6 carry a 3-O-sulfoglucosamine unit, although it is located in different saccharide sequences (Fig. 1A). It is known that 3-OST-3-modified HS does not bind to AT (35). As expected, compound 6 does not exhibit any anti-Xa and antithrombin activities. We also measured the binding of AT to the synthesized compounds (TABLE THREE). It is clear that the anticoagulant activities of the compounds correlated to their binding affinity to AT. Taken together, these results demonstrate that the anticoagulant activities of these enzymatically synthesized polysaccharides are consistent with the known structure activity relationship of HS.
TABLE THREE.
Anti-factor Xa and antithrombin activities of synthesized polysaccharide intermediates
| Sample | Factor Xa inhibition (IC50)a | Thrombin inhibition (IC50)a | Binding to ATb |
|---|---|---|---|
| ng/ml | ng/ml | % | |
| Heparinc | 20 | 10 | NDd |
| Heparan sulfatee | >5000 | >3000 | ND |
| Arixtraf | 58 | >3000 | ND |
| Compound 7 | >2000 | >3000 | 5 |
| Compound 8 | 126 | 96 | 31 |
| Compound 5 | 40 | 32 | 38 |
| Compound 6 | >2000 | >2000 | 2 |
The procedures for measuring the activities of factor Xa and thrombin are described under “Experimental Procedures.”
3-O-[35S]sulfo compounds were used to determine their bindings to AT as described under “Experimental Procedures.” 3H-Labeled HS from CHO cells was used as a negative control for the AT binding. About 0.3% of 3H-labeled HS bound to AT.
Heparin was from Sigma.
ND, not determined.
Heparan sulfate was isolated from bovine kidney.
Arixtra is the chemically synthesized antithrombin-binding pentasaccharide, which was obtained from a local pharmacy.
Preparation of the Polysaccharide That Binds to Herpes Simplex Virus gD
Herpes simplex virus utilizes HS as a receptor to infect the target cells (36). A specific 3-O-sulfo group-containing HS, generated by 3-OST-3, -5, or -6, serves as an entry receptor for herpes simplex virus type 1 through HS binding of gD (24, 37, 38). Structural characterization of the gD-binding site revealed a unique octasaccharide sequence carrying a 3-O-sulfo glucosamine residue (39). To test if our enzymatic approach synthesizes gD-binding HS, we incubated compound 4 with immobilized 3-OST-3 to generate compound 6. We have found that 12% of the resultant polysaccharide (compound 6) bound to gD, whereas only 2.6% of compound 5, a 3-OST-1-modified polysaccharide, bound to gD as determined by immunoprecipitation (TABLE FOUR). Comparison with the appropriate controls showed that the percentage binding for 3-OST-3-modified HS closely resembled that of compound 6. In conclusion, our results suggest that our enzymatic approach is capable of effectively synthesizing gD-specific binding HS. It should be noted that commercial heparin does not bind to gD, and heparin is not a substrate for 3-OST-3 (40, 41). Therefore, our results also demonstrate that it is possible to redesign the sulfation patterns by first solvolytically removing all O-sulfo groups and then selectively enzymatically replacing sulfo groups required for specific interactions.
TABLE FOUR.
The binding of synthesized polysaccharides to gD
| Sample | gD binding |
|---|---|
| % | |
| Unmodified HSa | 1.0 |
| 3-OST-1-modified HSb | 2.6 |
| 3-OST-3-modified HSb | 12.0 |
| Compound 5 | 2.6 |
| Compound 6 | 12.1 |
[3H]HS (from CHO cells) was prepared by metabolically labeling the cells with [3H]glucosamine.
3-OST-1- and 3-OST-3-modified HS were prepared by incubating [3H]HS with purified 3-OST-1 or 3-OST-3 enzymes.
DISCUSSION
Heparin and HS have a wide range of biological activities, including anticoagulation, antiviral, and anticancer activities. Sulfo group-containing saccharide sequences dominate the specificity of the functions of heparin and HS. Thus, the synthesis of a polysaccharide with the appropriate positioning of these critical functional groups to carry out its unique biological activity represents an important goal. In this study, we report an approach to synthesize sulfo group-containing HS polysaccharides that bind to FGF2, herpes simplex virus gD, and AT. These HS polysaccharides can also demonstrate appropriate biological activity, such as anticoagulant activity mediated through AT binding and the activity in promoting cell proliferation. More importantly, this approach permits the synthesis of greater than 1mg amounts of specific sulfo group-containing polysaccharides, sufficient for testing their activities in biochemical and biological assays. The quantities synthesized are also sufficient for extensive structural analysis, including disaccharide analysis and one- and two-dimensional NMR analysis.
Two approaches were used to increase the scale of enzymatic synthesis. First, HS sulfotransferases were expressed in E. coli, readily affording 20–50 mg of purified enzymes. We had previously shown that both 3-OST-1 and 3-OST-3 were expressed in E. coli in relatively high yield. In this study, both 2-OST and 6-OST were similarly expressed in E. coli as the soluble 2-OST and 6-OST-MBP fusion proteins. Second, we improved efficiency of enzymatic sulfation by utilizing immobilized O-sulfotransferases and coupling their use to a PAPS regeneration system. When this PAPS regeneration system was applied for preparing N-sulfoheparosan from heparosan using N-deacetylase/N-sulfotransferase, the N-sulfation yield was lower than that obtained using added exogenous PAPS (42). We considered the application of this system essential to our study for the following reasons. First, PAP inhibits HS O-sulfotransferase activities with IC50 values of ~100 μM under the reaction conditions used in our syntheses (data not shown), making milligram-scale synthesis difficult without continuously removing PAP. Second, a PAPS regeneration system would permit the PNPS as the sulfo donor and require only catalytic amounts of PAP, significantly reducing the cost of synthesis. The listed price of PNPS in the Aldrich catalog is 1300-fold less than PAPS. The immobilized enzyme format allowed us to use the enzymes repeatedly, making the method amendable to a large scale synthesis, since scale-up of immobilized enzyme columns is relatively simple.
Developing an effective approach to synthesize HS is also essential for understanding the mechanism of its biosynthesis. Unlike the protein biosynthesis, polysaccharide biosynthesis is not a template-driven process. Although the cDNAs encoding the HS biosynthetic enzymes have been cloned, the genetic regulation mechanism for the synthesis of the HS with defined biological functions is not fully understood. We now know that the unique and often remote sequence features in substrate can influence the action of HS O-sulfotransferases. Furthermore, whether additional factors or the formation of complexes of biosynthetic enzymes takes part in controlling the structure of HS remains to be elucidated. It is interesting to note that a complex of HS epimerase and 2-OST in vivo has been reported (43). Our approach could provide a tool to answer these questions, since it is capable of generating a sufficient amount of HS product required for extensive structural biochemical and biological analysis.
HS is believed to be present in block structures, consisting of highly sulfated and nonsulfated domains (44), and the HS with the biological activities largely contain the highly sulfated domains. We believe that the modifications in our system indeed happen in a block fashion based on the following facts. First, the results of the disaccharide analysis concluded that both 6-O-sulfation and 2-O-sulfation are carried out predominantly in the sulfated region (Fig. 3 and TABLE ONE). Second, the enzymatically modified products exhibit the anticipated biological functions, including the activation of FGF/FGF receptor signaling (compounds 4a and 4b), carrying anticoagulant activity (compound 5), and binding to herpes simplex virus glycoprotein D (compound 6). Because the HS carrying these functions must contain the domain structures with the size larger than pentasaccharide (5, 34, 39, 45), the synthetic products with the desired functions suggest that the modifications indeed occur in a block fashion. Further investigation is needed to determine whether the enzymatic modifications are identical to those in vivo. We note that the GlcNAc6S unit is a common saccharide residue in the HS from natural sources; however, it cannot be generated by 6-OST modification in vitro, suggesting the difference between in vitro and in vivo synthesis (46).
In summary, we have described a method for enzymatic sulfation to prepare the HS with three distinct biological activities. Unique sulfated saccharide sequences play a dominant role in the function and specificity of HS/heparin. Our method demonstrates the feasibility of using a collection of HS biosynthetic enzymes to synthesize HS/heparin with selected biological activities. The synthetic scale with this method can be easily increased for large scale synthesis, provided that both the enzymes and the sulfo donor are easily accessible. Since the synthesis of HS polysaccharide is not a template-driven process, it is still not possible to design an approach to synthesize a “sequence-pure” polysaccharide. Nevertheless, the current method clearly demonstrates that HS/heparin having specific biological activities can be synthesized by subjecting a backbone saccharide polymer to different enzymatic modifications. This enzymatic selectivity is currently not accessible by chemical sulfation approaches. Further improvement of this method will significantly aid the exploration of new potential therapeutic applications for HS. In addition, enzymatic synthesis of anticoagulant heparin could lead to a better anticoagulant drug by reducing its side effects.
Acknowledgments
We thank Drs. Gary Cohen and Roselyn Eisenberg (University of Pennsylvania) for providing gD and anti-gD antibody and Dr. Michael Duffel (University of Iowa) for providing rat AST-IV cDNA. We also thank Dr. Rosenberg (Massachusetts Institute of Technology) and Dr. Koji Kimata (Aichi University) for providing hamster 2-OST cDNA and mouse 6-OST-1 cDNA and Dr. David Baker (University of Tennessee) for helping with nomenclature.
Footnotes
This work is supported in part by National Institutes of Health Grants AI50050 (to J. L.), HL52622 and GM38060 (to R. J. L.), and GM069968 (to L. Z.) and American Heart Association, Mid-Atlantic Affiliate, Grant-in-Aid 0355800U (to J. L.).
The abbreviations used are: HS, heparan sulfate(s); RPIP-HPLC, reversed-phase ion pairing- high performance liquid chromatography; GlcNS, N-sulfoglucosamine; IdoUA, L-iduronic acid; 2-, 3-, and 6-OST, 2-, 3-, and 6-O-sulfotransferase, respectively; FGF, fibroblast growth factor; CDSNS, chemically desulfated N-sulfated heparin; MBP, maltose- binding protein; MES, 4-morpholineethanesulfonic acid; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; MOPS, 4-morpholinepropanesulfonic acid; gD, glycoprotein D; SPR, surface plasmon resonance spectroscopy; IL, interleukin; AT, Antithrombin.
References
- 1.Esko JD, Selleck SB. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Thorp SC. Med Res Rev. 2002;22:1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 3.Capila I, Linhardt RJ. Angew Chem Int Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Linhardt RJ. J Med Chem. 2003;46:2551–2564. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 5.Petitou M, van Boeckel CAA. Angew Chem Int Ed. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 6.Petitou M, Herault LP, Bernat A, Driguez PA, Duchaussoy P, Lormeau JC, Herbert JM. Nature. 1999;398:417–422. doi: 10.1038/18877. [DOI] [PubMed] [Google Scholar]
- 7.Dementiev A, Petitou M, Herbert JM, Gettins PG. Nat Struct Biol. 2004;11:867–863. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
- 8.Avci FY, Karst NA, Linhardt RJ. Curr Pharm Des. 2003;9:2323–2335. doi: 10.2174/1381612033453929. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl U, Kusche-Gullberg M, Kjellen L. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 10.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 11.Balagurunathan K, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Nat Biotechnol. 2003;21:1343–1346. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- 12.Kuberan B, Beeler DL, Lawrence R, Lech M, Rosenberg R. J Am Chem Soc. 2003;125:12424–12425. doi: 10.1021/ja036737g. [DOI] [PubMed] [Google Scholar]
- 13.Balagurunathan K, Beeler DL, Lech M, Wu ZL, Rosenberg RD. J Biol Chem. 2003;278:52613–52621. doi: 10.1074/jbc.M305029200. [DOI] [PubMed] [Google Scholar]
- 14.Lindahl U, Li J, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T, Emies J, Roberts I, Taylor C, Oreste P, Zoppetti G, Naggi A, Torri G, Casu B. J Med Chem. 2005;48:349–352. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
- 15.Burkart MD, Izumi M, Chapman E, Lin C, Wong C. J Org Chem. 2000;65:5565–5574. doi: 10.1021/jo000266o. [DOI] [PubMed] [Google Scholar]
- 16.Sheng JJ, Saxena A, Duffel MW. Drug Metabol Dispos. 2004;32:559–565. doi: 10.1124/dmd.32.5.559. [DOI] [PubMed] [Google Scholar]
- 17.Moon A, Edavettal SC, Krahn JX, Munoz EM, Negishi M, Linhardt RJ, Liu J, Pedersen LC. J Biol Chem. 2004;279:45185–45193. doi: 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Duncan MB, Carrick K, Pope M, Liu J. Glycobiology. 2003;13:785–794. doi: 10.1093/glycob/cwg101. [DOI] [PubMed] [Google Scholar]
- 19.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Biochem Biophys Res Commun. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 20.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Beeler DL, Lawrence R, Lech M, Liu J, Davis JC, Shriver Z, Sasisekharan R, Rosenberg RD. J Biol Chem. 2001;276:42311–42321. doi: 10.1074/jbc.M101441200. [DOI] [PubMed] [Google Scholar]
- 22.Duncan MB, Chen J, Krise JP, Liu J. Biochim Biophys Acta. 2004;1671:34–43. doi: 10.1016/j.bbagen.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Bjornsson S. Anal Biochem. 1993;210:282–291. doi: 10.1006/abio.1993.1197. [DOI] [PubMed] [Google Scholar]
- 24.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 25.Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. J Biol Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- 26.Smeds E, Habuchi H, Do AT, Hjertson E, Grundberg H, Kimata K, Lindahl U, Kusche-Gullberg M. Biochem J. 2003;372:371–380. doi: 10.1042/BJ20021666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ledin J, Staatz W, Li JP, Gotte M, Selleck SB, Kjellen L, Spillmann D. J Biol Chem. 2004;279:42732–42741. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- 28.Ibrahimi OA, Zhang F, Lang S, Hrstka C, Mohammadi M, Linhardt RJ. Biochemistry. 2004;43:4724–4730. doi: 10.1021/bi0352320. [DOI] [PubMed] [Google Scholar]
- 29.Guimond SE, Turnbull JE. Curr Biol. 1999;9:1343–1346. doi: 10.1016/s0960-9822(00)80060-3. [DOI] [PubMed] [Google Scholar]
- 30.Pye DA, Vives RR, Turnbull JE, Hyde P, Gallagher JT. J Biol Chem. 1998;273:22936–22942. doi: 10.1074/jbc.273.36.22936. [DOI] [PubMed] [Google Scholar]
- 31.Jacobsson I, Lindahl U. J Biol Chem. 1980;255:5094–5100. [PubMed] [Google Scholar]
- 32.Rosenberg RD, Showrak NW, Liu J, Schwartz JJ, Zhang L. J Clin Invest. 1997;99:2062–2070. doi: 10.1172/JCI119377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Lawrence R, Schwartz JJ, Bai X, Wei G, Esko JD, Rosenberg RD. J Biol Chem. 2001;276:28806–28813. doi: 10.1074/jbc.M100204200. [DOI] [PubMed] [Google Scholar]
- 34.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Shworak NW, Sinaÿ P, Schwartz JJ, Zhang L, Fritze LMS, Rosenberg RD. J Biol Chem. 1999;274:5185–5192. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 36.Shukla D, Spear PG. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia G, Chen J, Tiwari V, Ju W, Li JP, Malmström A, Shukla D, Liu J. J Biol Chem. 2002;277:37912–37919. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 38.Xu D, Tiwari V, Xia G, Clement C, Shukla D, Liu J. Biochem J. 2005;385:451–459. doi: 10.1042/BJ20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Shriver Z, Pope RM, Thorp SC, Duncan MB, Copeland RJ, Raska CS, Yoshida K, Eisenberg RJ, Cohen G, Linhardt RJ, Sasisekharan R. J Biol Chem. 2002;277:33456–33467. doi: 10.1074/jbc.M202034200. [DOI] [PubMed] [Google Scholar]
- 40.Nicola AV, Willis SH, Naidoo NN, Eisenberg RJ, Cohen GH. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Shriver Z, Blaiklock P, Yoshida K, Sasisekharan R, Rosenberg RD. J Biol Chem. 1999;274:38155–38162. doi: 10.1074/jbc.274.53.38155. [DOI] [PubMed] [Google Scholar]
- 42.Saribas AS, Mobasseri A, Pristatsky P, Chen X, Barthelson R, Hakes D, Wang J. Glycobiology. 2004;14:1217–1228. doi: 10.1093/glycob/cwh129. [DOI] [PubMed] [Google Scholar]
- 43.Pinhal MA, Smith B, Olson S, Aikawa J, Kimata K, Esko JD. Proc Natl Acad Sci U S A. 2001;98:12984–12989. doi: 10.1073/pnas.241175798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallagher JT. J Clin Invest. 2001;108:357–361. doi: 10.1172/JCI13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maccarana M, Casu B, Lindahl U. J Biol Chem. 1993;268:23898–23905. [PubMed] [Google Scholar]
- 46.Holmborn K, Ledin J, Smeds E, Eriksson I, Kusche-Gullberg M, Kjellen L. J Biol Chem. 2004;279:42355–42358. doi: 10.1074/jbc.C400373200. [DOI] [PubMed] [Google Scholar]