Abstract
Cancer cells must integrate multiple biosynthetic demands to drive indefinite proliferation. An outstanding question is how these key cellular processes such as metabolism and protein synthesis cross-talk to fuel cancer cell growth. Here we uncover the mechanism by which the MYC oncogene coordinates the production of the two most abundant classes of cellular macromolecules, proteins and nucleic acids in cancer cells. We find that a single rate-limiting enzyme, phosphoribosyl-pyrophosphate synthetase 2 (PRPS2), promotes increased nucleotide biosynthesis in MYC-transformed cells. Remarkably, Prps2 couples protein and nucleotide biosynthesis through a specialized cis-regulatory element within the Prps2 5′UTR, which is controlled by the oncogene and translation initiation factor eIF4E downstream Myc activation. We demonstrate with a Prps2 knockout mouse that the nexus between protein and nucleotide biosynthesis controlled by PRPS2 is crucial for Myc-driven tumorigenesis. Together, these studies identify a translationally-anchored anabolic circuit critical for cancer cell survival and an unexpected vulnerability for “undruggable” oncogenes, such as Myc.
Introduction
The ability to alter metabolic output to fulfill the biosynthetic and bioenergetic demands of cell growth and proliferation is a defining feature of cancer cells (Cairns et al., 2011; Vander Heiden et al., 2012; Ward and Thompson, 2012). For example, the Myc oncogene reprograms several cellular machineries including those promoting protein synthesis, glycolysis, glutaminolysis, as well as nucleotide synthesis, vital for sustaining cancer cell survival (Dang, 2010; Gordan et al., 2007; Liu et al., 2008; Mannava et al., 2008; Morrish et al., 2008; Wise et al., 2008). An outstanding question is how cancer cells couple multiple macromolecular synthetic processes to sustain an enhanced bioenergetic homeostasis vital for cancer cell survival. For instance, cancer cells need to maintain a careful homeostasis between the rate of protein biosynthesis and the metabolic flux that supplies the biosynthetic precursors and energy necessary for cancer cell growth and survival. In particular, the increased rate of protein synthesis that sustains cancer cell growth upon Myc hyperactivation imposes an onerous biosynthetic and bioenergetic cost to cancer cells (Bywater et al., 2013; Granneman, 2004; Lempiäinen and Shore, 2009; White, 2008). Therefore, an outstanding question is how key cellular processes underlying cancer cell growth, such as metabolism and protein synthesis, become coordinated and are maintained, and whether this point of intersection reflects a unique vulnerability that could be targeted. This question is particularly critical, as the Myc oncogene, at present, remains “undruggable”.
In this work, we employed a multifaceted approach integrating metabolomics with a unique mouse genetic strategy to address how production of two of the most abundant classes of cellular macromolecules, proteins and nucleic acids, are integrated and coupled by the Myc oncogene. Surprisingly, we find that Myc-driven hyperactivation of protein synthesis stimulates the translational upregulation of one key rate-limiting enzyme within the nucleotide biosynthesis pathway, PRPS2. Many translationally-regulated transcripts downstream of mTOR hyperactivation harbor a Pyrimidine-Rich Translational Element (PRTE) positioned within their 5′ untranslated region (UTR) (Hsieh et al., 2012). Interestingly, we find that Prps2 contains a PRTE that enables translational regulation by Myc to directly increase nucleotide biosynthesis proportionately to the increased protein synthesis rates of cancer cells. Strikingly, PRPS1 a related isoform that is responsible for widespread effects on nucleotide metabolism in normal cells, lacks the PRTE cis-regulatory translational element within its 5′UTR, thereby revealing a distinguishing feature of an isoform-specific PRPS in cancer cells. Therefore, these findings delineate a self-regulating circuitry through which cancer cells ensure balanced coordination between the production of proteins and nucleic acids. Importantly, by specifically inhibiting expression of Prps2, we demonstrate synthetic lethality in Myc-overexpressing cells and dramatically decreased tumorigenic potential in vivo, illuminating an important vulnerability and therapeutic window for cancers driven by “undruggable” oncogenes, such as Myc.
Results
Elevated Rates of Protein Synthesis Sustain the Myc-Dependent Metabolic Program
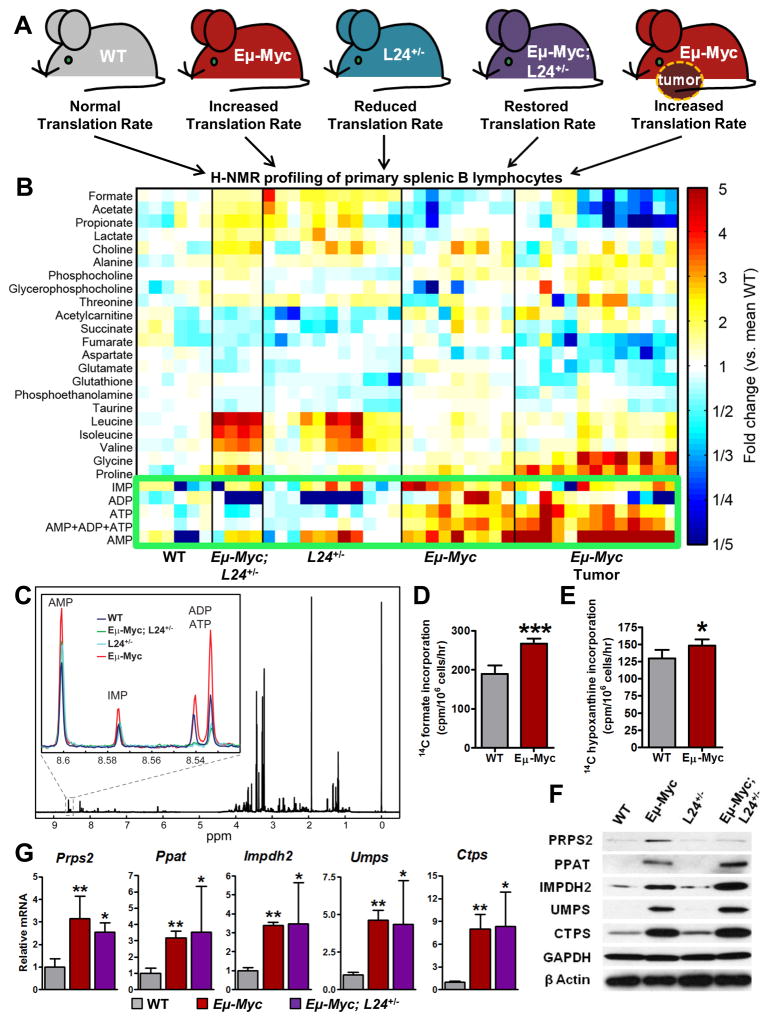
It remains unknown whether and how the elevated metabolic and protein synthesis program of cancer cells are sensed and coupled to promote cancer cell growth and development. We at first sought to genetically determine whether Myc’s ability to increase the protein synthetic capacity of cancer cells is directly linked to enhanced metabolism. The Eμ-Myc/+ transgenic mouse faithfully recapitulates the clinical features of human Burkitt’s lymphoma (Adams, 1985; Harris et al., 1988). B cells derived from these mice display a dramatic increase in Myc-dependent ribosome biogenesis and protein synthesis resulting in increased cell growth, which is a hallmark of Myc-driven cancers (Barna et al., 2008; Iritani and Eisenman, 1999). Previous studies have revealed that haploinsufficiency of a single ribosomal protein (RP), RPL24, leads to an overall decrease in protein synthesis, and that RPL24 haploinsufficiency in the Eμ-Myc/+ genetic background is sufficient to restrain Myc-dependent hyperactivation of protein synthesis to normal levels, dramatically thwarting Myc’s oncogenic activity (Barna et al., 2008). Therefore, we reasoned that Eμ-Myc/+;Rpl24BST/+ mice represent an ideal genetic tool to assess the functional relationship between Myc-dependent increases in protein synthesis rates and cancer cell metabolism.
We employed unbiased nuclear magnetic resonance (NMR)-based metabolomics to compare and measure the steady-state metabolite concentrations in freshly isolated Eμ-Myc/+ and Eμ-Myc/+;Rpl24BST/+ B cells, where elevated protein synthesis rates are restored to normal, as well as respective controls (Figure 1A). Consistent with an overall shift in metabolic processes to an anabolic state, we find that Myc-overexpressing cells, both in the pre-tumor and tumor setting, display an overall depletion in formate, acetate and propionate which are used for the construction of larger, more complex metabolites such as nucleotides, lipids, and amino acids (Figure 1B). We also find that in the pre-tumor setting, Myc broadly increases the levels of metabolites frequently found increased in cancer cells such as purine nucleotides, choline, phosphocholine, acetylcarnitine and several amino acids such as glycine and proline (Figure 1B). However, there also appears to be cell-type specificity. For example, the levels of glutamine or lactate previously linked to Myc-dependent metabolism are not altered in Eμ-Myc/+ B cells (Wise et al., 2008; Yuneva et al., 2012). The changes observed in the pre-tumor setting are maintained and, in most cases, amplified in the Myc-driven malignant lymphoma setting.
Figure 1. Unbiased Metabolomics Approach Reveals Myc-Dependent Translational Control of Purine Nucleotide Levels.
(A) Genetic approach to identify translationally regulated metabolites downstream of oncogenic Myc in B cells. (B) Intracellular steady state metabolite profiles of B cells isolated from wild-type (WT), Eμ-Myc/+ premalignancy, Rpl24BST/+, (L24+/-)Eμ-Myc/+;Rpl24BST/+, and Eμ-Myc/+ tumor-bearing mice. (C) Representative 1H NMR spectra of extracted metabolites from B cells with (inset) zoomed in view of region between 8.52–8.61ppm containing representative purine profiles from indicated genotypes. (D) Metabolic flux through de novo purine synthesis pathway measured by [14C] formate incorporation in WT and Eμ-Myc/+derived B cells. (E) Metabolic flux through purine salvage pathway measured by [8-14C] hypoxanthine incorporation in WT and Eμ-Myc/+-derived B cells. (F) Western blot analysis of indicated nucleotide biosynthesis enzymes from B cells derived from 5 week old WT, Eμ-Myc/+, Rpl24BST/+, and Eμ-Myc/+;Rpl24BST/+ mice. (G) qRT-PCR analysis of mRNA levels of indicated enzymes from mice as in (F). Error bars represent standard deviation, N=6 for (D) and (E), N=4 for (G)***P<0.001, **P<0.01, *P<0.05 by student’s t-test. See also Figure S1.
We directly ascertained how elevated protein synthesis rates in cancer influence the cancer metabolome. Strikingly, in Eμ-Myc/+;Rpl24BST/+ B cells we observed a specific rescue in a subset of metabolites normally found elevated in Eμ-Myc/+ cells. Unexpectedly, the most notable class of such metabolites is the nucleotides IMP, AMP, ADP and ATP (Figure 1B and Figure 1C, inset). The observed rescue in nucleotide metabolite production in Eμ-Myc/+ compared to Eμ-Myc/+;Rpl24BST/+ B cells was also further validated by HPLC (Figure S1A) and a PCR-based assay to measure free dNTP concentrations from a complex mixture of intracellular metabolites (Figure S1B). Together, these findings suggest that augmented protein biosynthesis may be directly coupled to the control of nucleotide metabolism downstream of Myc hyperactivation revealing an unexpected coordination between the production of the two most abundant classes of macromolecules in cancer cells – proteins and nucleic acids.
PRPS2 is a Critical Rate-limiting Enzyme Integrating Myc-dependent Protein Synthesis with Nucleotide Metabolism
Elevated nucleotide pools, through either de novo synthesis from carbohydrate and amino acid precursors or by salvage enzymes that rejoin recycled nucleobase and sugar moieties, are a defining feature of many cancer cells, required to carry out a diverse array of cellular functions (Tong et al., 2009). The effect of Myc on nucleotide levels is due, in large part, to increases in rates of de novo purine biosynthesis as revealed by the increased incorporation of radiolabeled precursors ([14C]-formate) for the de novo synthesis pathway (Figure 1D). Our findings also show that Myc increases nucleotide production by promoting flux through the purine salvage pathway (Figure 1E). Overall, these experiments suggest that Myc-dependent control of purine nucleotide concentrations relies on the increased biosynthetic production of purine precursors.
We next asked whether Myc-dependent increases in protein synthesis influence the expression of one or several key enzymes upstream or within the purine biosynthetic pathway. We identified a critical rate-limiting enzyme of purine biosynthesis, phosphoribosyl-pyrophosphate synthetase 2 (PRPS2), as a translationally-regulated mRNA whose protein levels are increased upon Myc overexpression and restored to wild-type levels in Eμ-Myc/+;Rpl24BST/+-derived B cells (Figure 1F and 1G). This effect on post-transcriptional control of Prps2 is highly specific as, notably, other important enzymes involved in nucleotide metabolism are expressed similarly in Eμ-Myc/+ and Eμ-Myc/+;Rpl24BST/+ B cells (Figure 1F, 1G and S1C). Therefore, these results unexpectedly suggest that one key enzyme within the purine biosynthesis pathway may be directly coupled to increased rates of protein synthesis elicited by the Myc oncogene to further regulate the flow of metabolic intermediates through the entire nucleotide pathway.
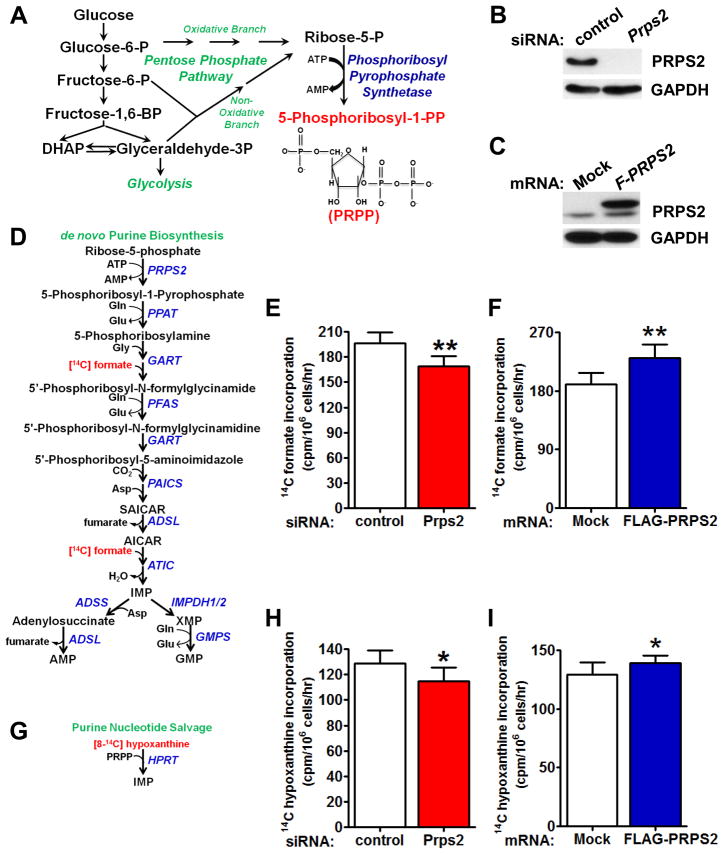
Phosphoribosyl pyrophosphate synthetase (PRPS) activity plays a central metabolic role in the production of nucleotides. The PRPS enzyme adds a pyrophosphate group donated from ATP to ribose-5-phosphate generated from the pentose phosphate pathway to produce 5-phosphoribosyl-1-pyrophosphate (PRPP) (Figure 2A). PRPP is a substrate for all nucleotide salvage pathway enzymes as well as the rate-limiting enzymes of purine and pyridine biosynthesis. Since the Km rate constants of nucleotide biosynthetic pathway enzymes that utilize PRPP as a substrate far exceed the physiological intracellular concentrations of this metabolite, an increase in PRPP levels may be sufficient to govern the overall nucleotide biosynthetic rate of cells. Therefore, we next addressed whether PRPS2 as a single enzyme is necessary and sufficient to control the overall purine biosynthesis and salvage rates of cells. To this end, we directly modulated Prps2 expression levels in freshly isolated primary B cells (Figure 2B, 2C and S2A). Upon knockdown of Prps2 expression in primary B cells using siRNA (Figure 2B and S2A), we observed a 14% decrease in the rate of [14C] formate incorporation into purines compared to control siRNA transfected cells (Figure 2D and E) demonstrating the requirement of PRPS2 enzymatic activity for the optimum rate of de novo purine biosynthesis. Conversely, overexpression of Prps2 in B cells caused a 21% increase in [14C] formate incorporated into purine nucleotides (Figure 2F). To assess the contribution of PRPS2 activity towards nucleotide salvage pathway function, we measured [8-14C] hypoxanthine labeling of purines upon knockdown or overexpression of Prps2 in B cells (Figure 2G). Although less dramatic than the effect observed on de novo purine biosynthesis, both knockdown (Figure 2H) as well as overexpression (Figure 2I) resulted in significant decreases or increases, respectively, in [14C] incorporation. Therefore, PRPS2 levels are rate limiting and important for controlling nucleotide biosynthesis rates, revealing a key enzyme that promotes nucleotide metabolism and is regulated at the post-transcriptional level by Myc hyperactivation.
Figure 2. PRPS2 is a Rate-limiting Enzyme for Purine Synthesis and Salvage Pathways.
(A) Schematic of the pathway of PRPP biosynthesis produced from glucose. (B) Western blot of primary wild-type (WT) B cells transduced with control or Prps2 siRNA. (C) Western blot of primary WT B cells mock transfected or transfected with capped, polyadenylated FLAG-Prps2 encoding mRNA. (D) Schematic illustrating [14C] formate incorporation into de novo purine biosynthesis pathway. (E) Measurement of [14C] formate incorporation into purines from cells treated as in (B). (F) Measurement of [14C] formate incorporation into purines from cells treated as in (C). (G) Schematic illustrating [8-14C] hypoxanthine incorporation into purines via HPRT nucleotide salvage enzyme. (H) Measurement of [8-14C] hypoxanthine incorporation into purines from cells treated as in (B). (I) Measurement of [8-14C] hypoxanthine incorporation into purines from cells treated as in (C). For all graphs, error bars represent standard deviation, N=6, **P<0.01, *P<0.05 by student’s t-test. See also Figure S2.
Prps2 but not Prps1 Gene Expression is Regulated Acutely at the Translational Level
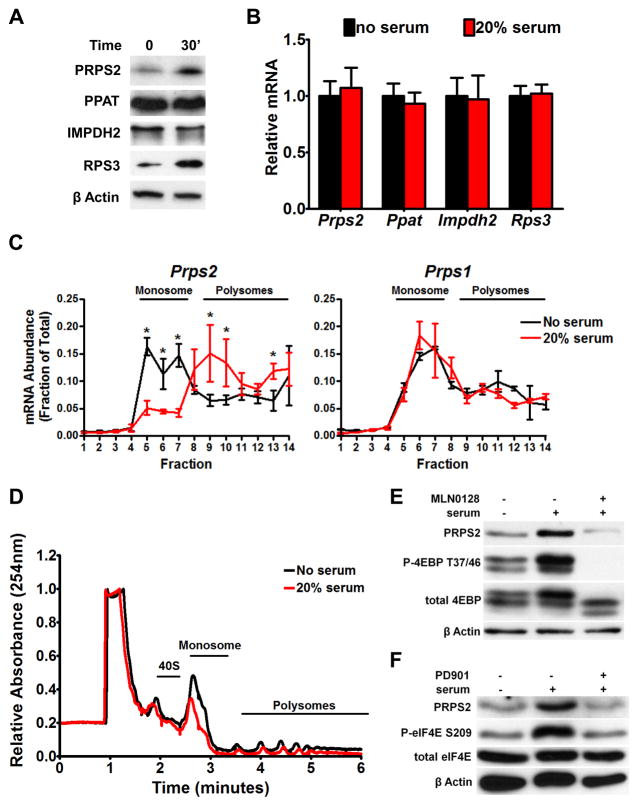
Given the critical importance of PRPS2 protein levels in purine metabolic flux, we hypothesized that translational control of Prps2 gene expression may serve at the interface of protein and nucleotide biosynthesis control. To address whether translational control of Prps2 may provide a rapid and dynamic mechanism to regulate purine synthesis, we acutely and rapidly induced mitogen-dependent protein synthesis in serum-starved cells by 30 minutes of serum stimulation (Geyer et al., 1982). Strikingly, the levels of PRPS2 protein, but not mRNA, increase dramatically upon addition of serum, suggesting that Prps2 may be regulated at the translation level (Figure 3A and B). To test this hypothesis, we employed ribosome sucrose-gradient fractionation. Indeed, upon 30 minutes of serum stimulation, we observe an increase in the amount of Prps2 mRNA associated with heavy poly-ribosomes fractions, containing highly translating mRNAs, compared to serum-starved cells (Figure 3C and D). Moreover, Prps2 showed the same acute translational activation as other growth factor responsive genes such as Ribosomal Protein S3 (Rps3), a bona fide translationally regulated mRNA, and c-Myc (Figure S3A, S3B, and S3C), that respond immediately and rapidly to acute serum stimulation.
Figure 3. Prps2 mRNA is Regulated Acutely at the Translational Level.
(A) Western blot (WB) of indicated purine biosynthesis enzymes from serum-starved or 30 minute fetal bovine serum (FBS) stimulated NIH3T3 cells. (B) qRT-PCR measurement of mRNA levels of inidcated genes from cells treated as in (A). Data are normalized to β-Actin mRNA levels and error bars represent standard deviation. (C) Polysomal analysis of Prps2 and Prps1 mRNA in cells treated as in (A). The polysomal association of Prps2 mRNA measuring mRNA abundance by qRT–PCR analysis in each fraction normalized to the corresponding 5S rRNA levels and expressed as a fraction of the total mRNA. Error bars represent standard deviation, N=4, *P<0.05 by student’s t-test. (D) Polysome profiles of serum starved and serum stimulated NIH3T3 cells treated as in (A) plotted as relative absorbance at 254nm versus elution time in minutes. (E) WB of serum-starved NIH3T3 cells treated for 30 minutes with 20% FBS or 20% FBS after a 10 minute pre-treatment with 100nM of the mTOR active site inhibitor MLN0128. (F) WB of serum-starved NIH3T3 cells treated for 30 minutes with 20% FBS or 20% FBS after a 10 minute pre-treatment with 100nM of the MEK1/2 inhibitor PD901. See also Figure S3.
Notably, while there are two isoforms of PRPP synthetase expressed in somatic tissues, only Prps2, and not Prps1, displays increased translation upon serum-stimulation (Figure 3C). Interestingly, the PRPS2 isoform is largely resistant to feedback inhibition by the nucleotide biosynthesis products ADP and GDP that is a feature of the PRPS1 enzyme (Nosal et al., 1993). This enzymatic property of PRPS2 may facilitate the unrestrained, elevated production of nucleotides observed in Myc-overexpressing cells as well as to explain why the levels of PRPS2 but not PRPS1 are increased in cancer cells. The translational regulation of Prps2 is highly specific, as other members of the purine biosynthetic pathway are not regulated in this manner (Figure 3A). Therefore, translational control of Prps2 is a rapid sensor of the total rate of protein biosynthesis within the cell, which precedes biomass accumulation.
Prps2 Translation is Controlled by the eIF4E Oncogene through a Conserved Cis-acting Regulatory Element that Directs Nucleotide Production
An outstanding question is how Prps2, but not other members of the purine biosynthesis pathway, is specifically and acutely regulated at the translation level downstream of Myc hyperactivation. Pro-growth signals such as serum stimulation and Myc hyperactivation are thought to regulate translation of specific subsets of mRNAs through increases in the activity of the major cap-binding protein, eIF4E (Hsieh and Ruggero, 2010; Ruggero et al., 2004; Topisirovic and Sonenberg, 2011). Importantly, eIF4E is also a direct transcriptional target of Myc (Jones et al., 1996) and is a master regulator of the rate-determining step in translation initiation. Upon forced overexpression of Myc, we also observe an increase in eIF4E levels that coincides with elevated Prps2 expression (Figure S3D and S3E). Therefore, we tested whether the effects of Prps2 translational control are mediated through eIF4E hyperactivation using specific inhibitors that target the activity of eIF4E. Specifically, we pretreated serum-starved fibroblasts with either an mTOR active site inhibitor (MLN0128) that blocks the phosphorylation of eIF4E binding protein 1 (4EBP1) as well as with a MEK1/2 inhibitor (PD901), which impairs eIF4E phosphorylation. Notably, in both of these treatments we observe that the increases in PRPS2 protein levels present upon acute serum stimulation are abolished (Figure 3E and 3F).
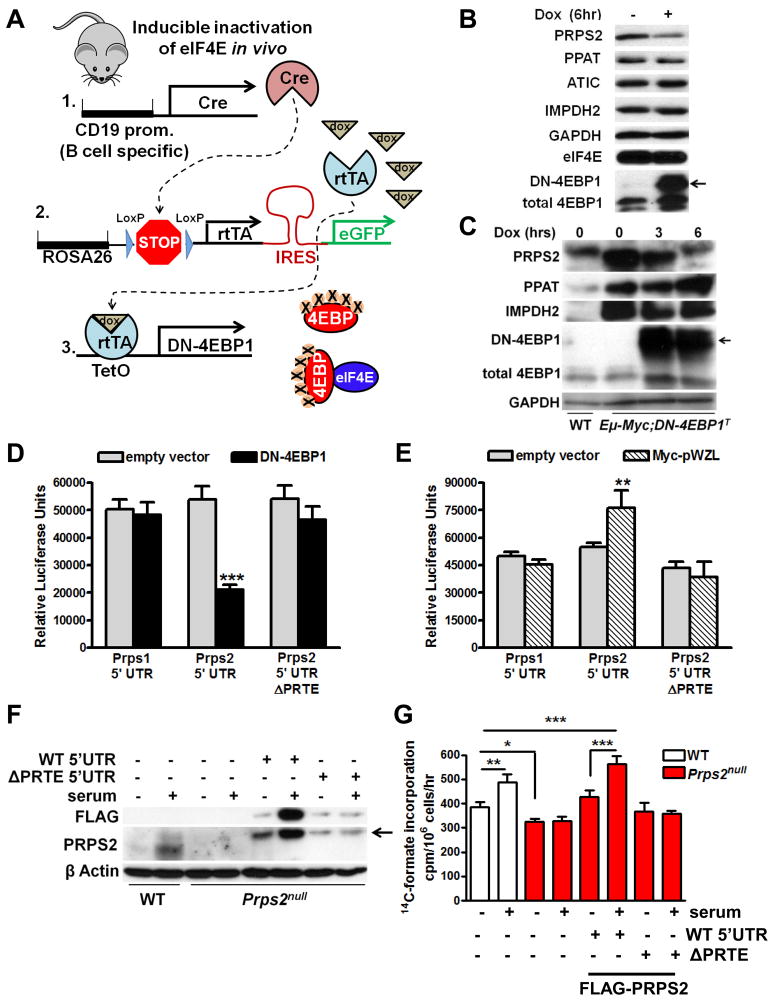
To extend our findings demonstrating eIF4E-mediated translational control of Prps2 mRNA in vivo, we employed a genetic system to specifically express a doxycycline-inducible dominant negative eIF4E binding protein 1 (DN-4EBP1) transgene in B cells (Figure 4A) (Hsieh et al., 2010; Pourdehnad et al., 2013). This genetic strategy does not perturb global protein synthesis nor cell viability, but rather only affects the translation of eIF4E rate-limiting target mRNAs (Hsieh et al., 2010). The inducible downregulation of eIF4E activity decreased expression of PRPS2 protein (Figure 4B) without affecting Prps2 mRNA levels (Figure S4A). Importantly, eIF4E-dependent translational control within the nucleotide biosynthesis pathway is selective for Prps2 as the expression levels of additional key enzymes in this pathway such as IMPDH2, PPAT and ATIC remain unchanged in DN-4EBP1 transgenic mice (Figure 4B). c-Myc expression is also unaffected by inhibition of eIF4E in Eμ-Myc/+-derived B cells (Pourdehnad et al., 2013). We next intercrossed Eμ-Myc/+ transgenic mice with inducible DN-4EBP1 transgenic mice. In Eμ-Myc/+;DN-4EBP1T mice PRPS2 abundance is restored to wild-type levels within 6 hours of DN-4EBP1 expression without affecting the Myc-dependent increases in Prps2 mRNA levels or the expression of other key nucleotide biosynthetic pathway enzymes (Figure 4C and S4B). These results demonstrate that eIF4E-mediated translational activity is selectively required for the Myc-dependent increase in total PRPS2 protein levels observed in Eμ-Myc/+ B cells.
Figure 4. Prps2 mRNA Translation, but not Prps1, is Regulated via a Cis Regulatory Motif in its 5’fUTR by the eIF4E Oncogene that Directs Increases in Nucleotide Metabolism.
(A) Compound transgenic mice to drive inducible expression of dominant-negative-4EBP1 (DN-4EBP1) specifically in B lymphocytes. (B) Western blot (WB) of indicated purine biosynthesis enzymes from purified B cells from mice described in (A) treated by i.p. injection with vehicle or doxycycline for 6hr. Arrow demarcates slower migrating transgenic DN-4EBP1 protein. (C) WB of indicated purine biosynthesis enzymes from purified B cells in (A) in wild-type (WT) or Eμ-Myc/+ transgenic mice treated by i.p. injection with vehicle or doxycycline for 3 or 6hr. Arrow demarcates slower migrating transgenic DN-4EBP1 protein. (D and E) Luciferase reporter assays of NIH3T3 with the indicated 5′UTR reporter constructs along with a DN-4EBP1 expression vector or control empty vector (D) or transfected into stable NIH3T3 cells expressing empty vector or Myc-pWZL (E). (F) WB using indicated antibodies of lysates prepared from WT or Prps2null mouse embryo fibroblasts (MEFs) mock transfected or transfected with the indicated FLAG-PRPS2 encoded mRNAs treated with or without 1 hour serum stimulation. Arrow denotes ectopic FLAG-tagged PRPS2. (G) [14C] formate incorporation into purine nucleotides was measured from cells in (F). Error bars represent standard deviation, N=4, ***P<0.001, **P<0.01, *P<0.05 by student’s t-test. See also Figure S4.
We next tested whether a specific sequence or motif could be responsible for the translational regulation of Prps2. A recently identified, important eIF4E cis-regulatory element that confers translational specificity is the pyrimidine-rich translational element (PRTE) (Hsieh et al., 2012). Consistent with our findings that Prps2, and not Prps1, is regulated at the level of translational control, only Prps2 contains a consensus PRTE motif within its 5′UTR (Figure S4C). To test the possibility that Prps2 displays differential sensitivity to translational regulation by eIF4E, we constructed luciferase reporter constructs fused to the 5′UTR of Prps1, Prps2, as well as deletion and transversion mutants of the PRTE domain within Prps2. Strikingly, transfection of these reporter constructs revealed that the PRTE motif is sufficient to direct translational control of Prps2 in an eIF4E-dependent manner and that the related PRPS1 isoform is not regulated by this mode of translational control (Figure 4D and S4D). Additionally, the wild-type Prps2 5′UTR luciferase reporter was selectively activated in Myc-overexpressing cells, whereas the Prps2 5′UTR PRTE deletion reporter and Prps1 5′UTR reporter were not (Figure 4E), therefore confirming the direct link between Myc-overexpression and increased Prps2 gene expression through a unique eIF4E-dependent cis-acting regulatory element. Importantly, other genes of the nucleotide biosynthesis pathway lack this functional motif within their 5′UTRs (data not shown and Figure S4C). Thus, Prps2 possesses a functional translational enhancer element within its 5′UTR that may act as a critical bottleneck in the production of nucleotides in cancer that is coupled to protein synthesis.
To further define the precise contribution of translational control of Prps2 towards nucleotide biosynthesis, we employed fibroblasts derived from Prps2null mice (see Figure 6 and Figure S6 for details regarding generation of Prps2null mice). We transfected mRNAs encoding FLAG-PRPS2 fused to either the wild-type 5′UTR of Prps2 or the 5′UTR of Prps2 engineered to lack the pyrimidine-rich translational element (ΔPRTE) which confers sensitivity to eIF4E-mediated translation control. Importantly, only Prps2null cells transfected with the Prps2 5′UTR, FLAG-PRPS2, but not the ΔPRTE 5′UTR mutant, displayed a serum-dependent increase in translation of FLAG-PRPS2 protein and increased purine nucleotide production (Figure 4F and 4G). Therefore, the PRTE sequence in Prps2 mRNA acts as a dynamic sensor that couples protein biosynthesis with nucleotide production.
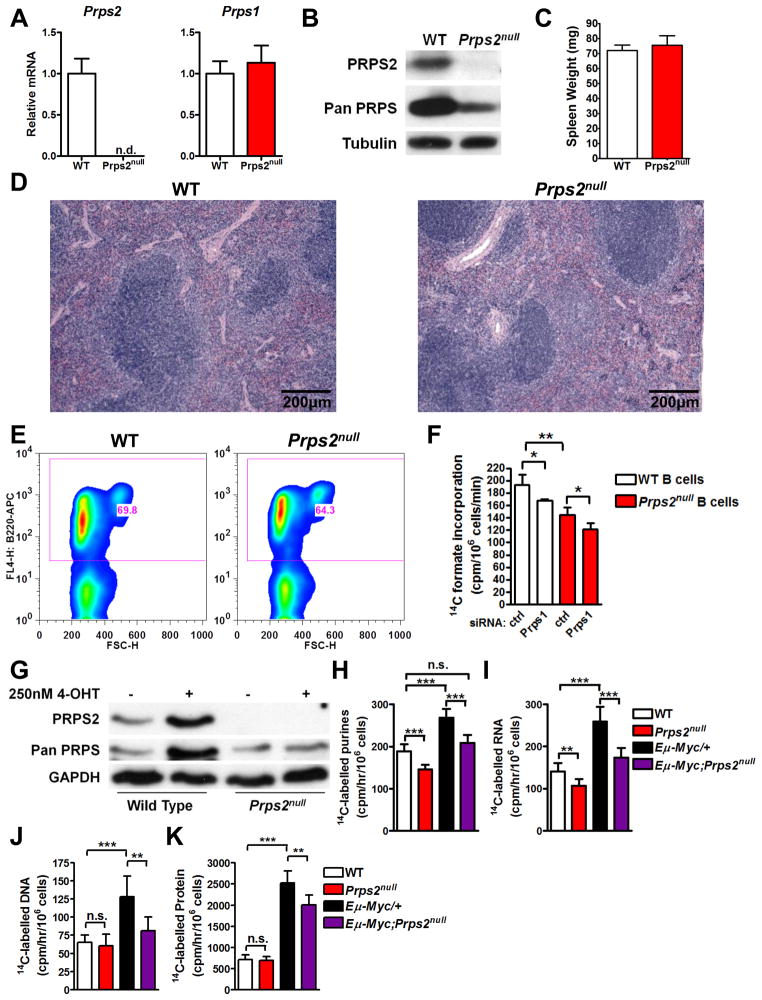
Figure 6. PRPS2 is Dispensable for Normal Physiology and Cellular Function, but Required for Myc-dependent Metabolic Reprogramming.
(A) RNA was isolated from purified B cells derived from wild-type (WT) or Prps2null mice and qRT-PCR was performed relative to β-Actin. (B) Western blot (WB) of B cell lysates from mice with indicated genotype. (C) Spleen weights of 8wk old WT or Prps2null mice. (D) Haematoxylin and Eosin stained tissue sections from WT or Prps2null spleens. (E) The percentage of splenic B220+ cells was assessed by FACS. (F) Measurement of [14C] formate incorporation into purine nucleotides from primary B lymphocytes transfected with control or Prps1-targeting siRNA. (G) WT or Prps2null MEFs were stably infected with retroviruses encoding the chimeric MycER fusion gene. WB of lysates prepared from cells treated with vehicle or 250nM 4-hydroxytamoxifen for 24hr. (H-K) Measurement of [14C] formate incorporation into purine nucleotides (H), RNA (I), DNA (J), and protein (K) from primary B lymphocytes isolated from the indicated genotypes. For all graphs, error bars represent standard deviation, ***P<0.001, **P<0.01, *P<0.05, and n.s.=not significant by student’s t-test. For (A) and (C), N=3. For (F), (H), (I), (J), and (K), N=6. n.d.=not detected. See also Figure S6.
PRPS2 Knockdown is Synthetically Lethal in Myc-Overexpressing Cells and Blocks Myc-dependent Tumor Initiation and Maintenance in vivo
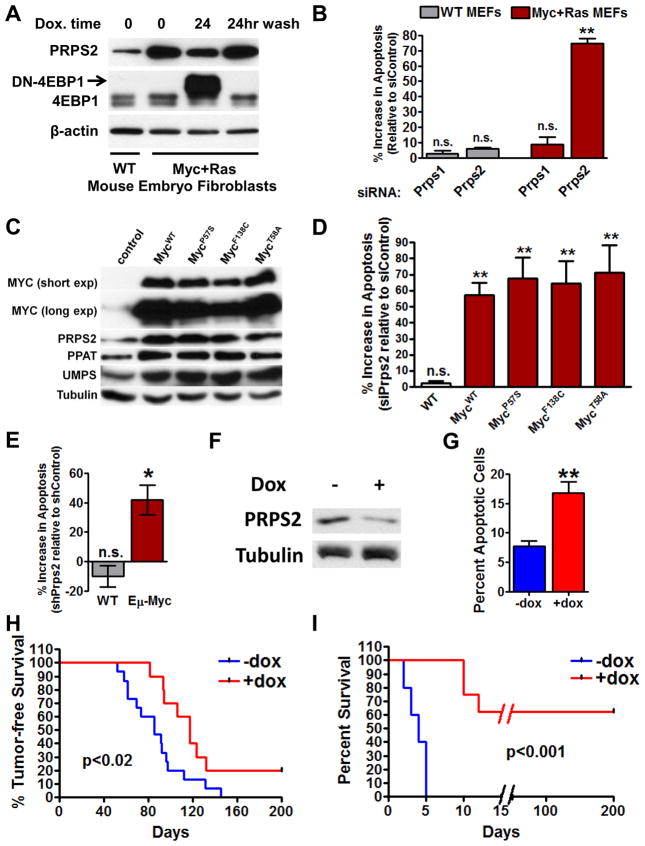
We tested whether inhibiting translational regulation of Prps2 mRNA could represent a potential synthetic lethal interaction with Myc hyperactivation. We observe a dramatic increase in Prps2 translation upon oncogenic cellular transformation driven by Myc and Ras that relies on eIF4E activity, demonstrating that translational regulation of Prps2 occurs as an early event during oncogenic transformation (Figure 5A). This is consistent with the upregulation in PRPS2 protein during the early pre-tumor stage of Myc-driven lymphomagenesis (Figure 1F). Next, to directly address the functional relevance of increased Prps2 expression in cellular transformation, we performed knockdown of Prps2 in Ras and Myc-transformed MEFs. Strikingly, we observed a 70% increase in apoptosis upon Prps2 knockdown in transformed, but not wild-type, cells (Figure 5B). Importantly, this increase in apoptosis was specific to the knockdown of the PRPS2 isozyme, as knockdown of PRPS1 did not affect cellular viability in normal or transformed MEFs (Figure 5B and S5A).
Figure 5. Loss-of-function of PRPS2 is Synthetically Lethal for Cancer Cells and Delays Myc-dependent Tumor Initiation and Maintenance in vivo.
(A) Western blot (WB) analysis of indicated proteins lysates from normal or transformed mouse embryo fibroblasts treated as specified. 24hr wash denotes sample with replenished media lacking doxycycline. (B) Indicated cells were transfected with control, Prps1, or Prps2 siRNA and apoptosis was measured after 48hr. Data in graph represents percent increase in the Annexin V+/Propidium iodide+ (PI) population in Prps1 or Prps2 siRNA transfected cells relative to control siRNA transfected cells. (C) WB of lysates from stable NIH3T3 cells expressing the indicated Myc WT or amino acid substitution alleles detecting the indicated nucleotide biosynthesis enzymes. (D) Cells from (C) were transfected with control or Prps2 siRNA and 48 hours later subjected to Annexin V/PI FACS analysis. (E) WT or Eμ-Myc/+-derived B lymphocytes were isolated and transduced with palmitoylated GFP (pGFP) expressing retrovirus co-expressing either control or Prps2 shRNA. 48 hours post-transduction, cells were stained for Annexin V and PI. (F) WB of lysates prepared from B cells isolated from mice transplanted with Eμ-Myc/+ derived hematopoietic stem cells harboring dox-inducible Prps2 shRNA. (G) Percent apoptotic cells represent Annexin V/PI+ percentage of GFP+ B lymphocytes prepared from mice treated as in (F). (H) Fetal liver-derived Eμ-Myc/+ hematopoietic stem cells were transduced with palmitoylated-GFP expressing retrovirus that co-express doxycycline (dox) inducible shRNA targeting Prps2 and subsequently transplanted to lethally-irradiated syngeneic mice. Survival curves measure days free of palpable lymph nodes between cohorts of mice. (I) Eμ-Myc/+ tumor cells were isolated from a lymphoma-bearing mouse, transduced with pGFP-expressing dox-inducible Prps2 shRNA, and subsequently transplanted into syngeneic mice. Survival curves begin at start of treatment regimen and monitor the time to sacrifice of tumor-bearing mice. For (B), (D) and (E), Data in graphs represent percent increase in Annexin V+/PI+ upon Prps1/2 siRNA or shRNA relative to control siRNA or shRNA transduced cells. For all experiments, error bars represent standard deviation and N=4 for (B), (D) and (E), N=3 for (G). **P<0.01, *P<0.05, n.s.=not significant by student’s t-test. See also Figure S5.
We next wanted to address why PRPS2 loss, but not PRPS1 loss, is deleterious for the viability of Myc-overexpressing transformed MEFs. Whereas siRNA-mediated knockdown of either PRPS2 or PRPS1 led to only a modest decrease in purine nucleotide production in wild-type MEFs, knockdown of PRPS2, but not PRPS1, resulted in a dramatic decrease in purine nucleotide production in MEFs transformed by Myc and Ras overexpression (Figure S5B). Additionally, knockdown of PRPS1 or PRPS2 in wild-type MEFs produced similar decreases in the rates of RNA and DNA production (Figure S5C and S5D). These results suggest that while PRPS2 and PRPS1 play interchangeable roles in normal cells, Myc-overexpressing cancer cells rely on the specific activity of PRPS2 to produce the nucleotides necessary to sustain their metabolic demands. Since Myc overexpression is known to sensitize cells to apoptosis, loss of function of PRPS2 may exacerbate the apoptotic program normally regulated by Myc. To test this possibility, we generated cells that overexpress apoptosis-resistant Myc amino acid substitution mutant alleles (Graves et al., 2010; Hemann et al., 2005). Each mutant tested (P57S, F138C, and T58A) was able to increase the expression of the nucleotide biosynthetic pathway enzymes PPAT, UMPS and PRPS2 to similar degrees (Figure 5C) and importantly, retained the sensitivity to programmed cell death induced by PRPS2 knockdown (Figure 5D). Taken together, these results suggest that loss of function of PRPS2 does not simply augment the apoptotic program already regulated by Myc, but rather disrupts the Myc-dependent metabolic program in cancer cells leading to synthetic lethality.
We next tested the potential therapeutic effects of PRPS2 knockdown in vivo. We observe a 40% induction of apoptosis (Figure 5E) in Myc-overexpressing B cells, but not normal cells upon PRPS2 knockdown in a pre-tumor setting. Upon induced expression of Prps2 shRNA in vivo, we also observed an increase in apoptosis of Myc-expressing cells within the tumor setting (Figure 5Fand 5G). Notably, mice induced to express shRNA directed toward Prps2 show a significant delay in Myc-driven tumor onset (Figure 5H). We next assessed the therapeutic efficacy of PRPS2 loss of function in established Myc-driven lymphomas. After tumor formation, Prps2 shRNA expression was induced and tumor progression was monitored (Figure 5I and Figure S5F). We observed a strong impairment in tumor progression upon knockdown of Prps2 and, remarkably, at least 30% of these mice displayed complete tumor regression and survival beyond 7 months of age revealing a critical oncogenic role of PRPS2.
Generation of a Prps2 Knock-out Mouse Reveals that PRPS2 is Dispensable for Normal Physiology and Cellular Function but Required for Metabolic Homeostasis in Myc-overexpressing Cells
An outstanding question is whether PRPS2 is normally essential for cell and organismal physiology in vivo. To address this outstanding question, we generated the first Prps2 knockout mouse (Figure S6A). Strikingly, mice homozygous null for the Prps2 gene (Prps2null) are viable and fertile and display no gross phenotypic abnormalities despite lacking Prps2 mRNA and protein expression (Figure S6B and S6C). Notably, there was no compensatory upregulation of Prps1 mRNA levels in tissues from Prps2null mice, suggesting that normal expression levels and enzymatic activity of PRPS1 are sufficient to maintain metabolic homeostasis (Figure S6C). As the critical nucleotide biosynthetic intermediate PRPP is only produced by PRPS enzymes, these findings are consistent with the continued activity of the PRPS1 isozyme, whose mRNA is interestingly normally found expressed at higher levels than Prps2 in all tissues we surveyed (Figure S6C and S6D).
We next investigated the function of PRPS2 in normal B cell homeostasis. In Prps2null mice, Prps1 expression in splenic B cells was maintained at the same levels observed in wild type cells (Figure 6A and 6B). Complete loss of Prps2 expression did not alter spleen weight, tissue architecture or morphology (Figure 6C, 6D, and S6E). Moreover, the percent of B lymphocytes present in the spleen (Figure 6E), as well as B cell size (Figure S6F), cell cycle distribution (Figure S6G) or cell viability (Figure S6H) were not altered. While Prps2null B lymphocytes display no overt phenotypic differences, they do have a minor decrease in rates of purine nucleotide biosynthesis which are further decreased upon siRNA-mediated knockdown of the PRPS1 isozyme (Figure 6F). Together, these results suggest that the activity of PRPS1 alone is sufficient to maintain the normal function of B lymphocytes and spleen development, whereas PRPS2 function is dispensable.
As PRPS2 shares approximately 95% amino acid identity with the PRPS1 isoform (Becker et al., 1990), it remains unknown whether cancer cells have evolved a mechanism to promote cell survival through only one of the two PRPS isoforms. To address this question, we utilized Prps2null MEFs engineered to overexpress a ligand-activatable MycER allele (Eilers et al., 1989). Notably, treatment with 4-hydroxytamoxifen to activate Myc increased only PRPS2, but not PRPS1, protein abundance (Figure 6G). Strikingly, our data demonstrate that the Myc-dependent increase in the purine nucleotide production rate can be completely restored to wild-type levels in Eμ-Myc/+;Prps2null B cells (Figure 6H). Therefore, these results suggest a specific requirement for PRPS2, but not PRPS1, function in maintaining the metabolic homeostasis of Myc-overexpressing cells.
We then sought to determine the most immediate effects of nucleotide production driven by PRPS2 downstream of Myc hyperactivation. An increased pool of RNA nucleotides is utilized by cancer cells for incorporation into rRNA and tRNA to generate increased numbers of ribosomes (Ben-Sahra et al., 2013). In addition, sufficient pools of deoxyribonucleotides are also required to maintain DNA fidelity during replication (Bester et al., 2011) and to bypass oncogene induced senescence (Aird et al., 2013). Importantly, in Eμ-Myc/+;Prps2null B cells the decrease in overall nucleotide production leads to overall decreases in the rate of ribo- and deoxyribo-nucleic acid production compared to Myc-overexpressing B cells (Figure 6I and 6J). As total cellular RNA is predominantly comprised of ribosomal RNA we next examined whether the augmented rates of protein synthesis evident upon Myc hyperactivation are altered in Eμ-Myc/+;Prps2null B cells. Indeed, our data demonstrate that the overall rate of protein synthesis in Eμ-Myc/+ B lymphocytes, but not normal B cells, is decreased towards normal levels in Prps2null mice (Figure 6K). Importantly, limiting the protein synthetic capacity in Eμ-Myc/+ B cells by genetic or pharmacological means has been previously shown to promote increased programmed cell death (Barna et al., 2008; Bywater et al., 2012). These findings suggest a possible mechanism for the synthetic lethality observed in Myc-overexpressing cells upon loss of PRPS2 function.
Interference with glucose metabolism has also been shown to limit oncogenic potential or promote apoptosis in Myc-overexpressing cells (Doherty et al., 2013; Faubert et al., 2013; Shim et al., 1998). Normal cells lacking PRPS2 display little to no difference in glucose uptake or the rate of lactate production, glucose oxidation, glucose conversion to purines, RNA and DNA (Figure S7A, S7B, S7C, S7D, S7E, S7F). On the contrary, Eμ-Myc/+;Prps2null B lymphocytes have increased glucose uptake compared to Eμ-Myc/+ counterparts (Figures S7A) and glucose oxidation occurs at a faster rate despite no alterations in overall mitochondrial mass or mitochondrial membrane potential (Figure S7C, S7G, S7H). Taken together, these experiments suggest that Myc-overexpressing cells lacking PRPS2 are thrown into “metabolic disarray” with perturbations in glucose utilization as well as the anabolic metabolism of nucleotides, nucleic acids and proteins, thereby hindering cancer cell survival.
PRPS2 is Critical for Myc-driven Cancer in Human and Mouse Models
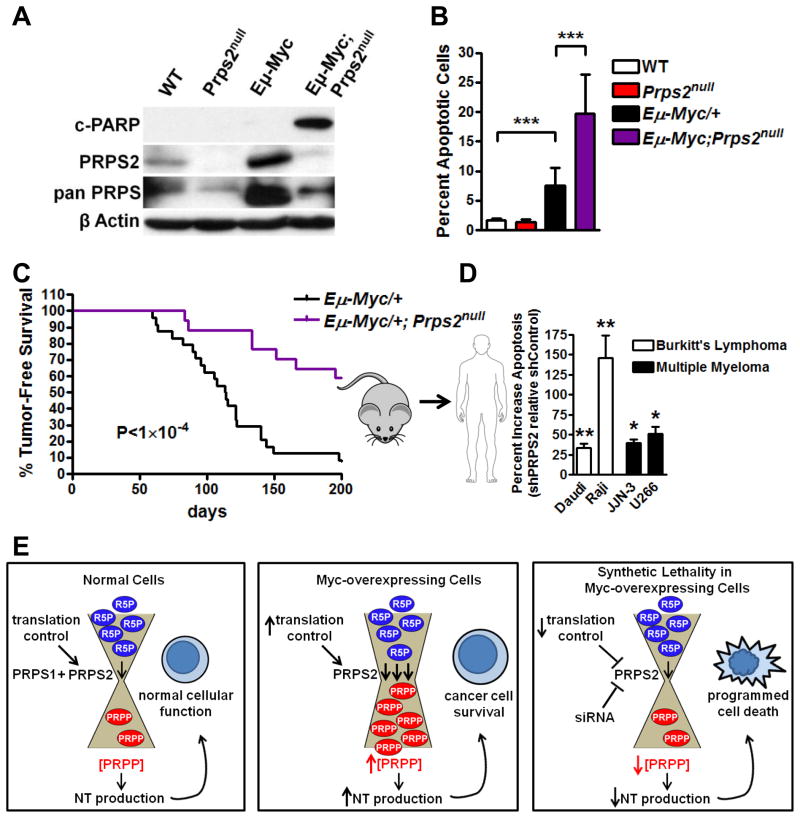
We employed Prps2null mice to directly genetically address the requirement of PRPS2 activity in Myc-driven lymphomagenesis. We first assessed the role of PRPS2 in the cellular function of premalignant Eμ-Myc/+ B lymphocytes. Consistent with an effect on Myc-overexpressing cell viability upon PRPS2 knockdown, we observed a dramatic increase in apoptosis in Eμ-Myc/+;Prps2null B lymphocytes (Figure 7A and 7B). To investigate the therapeutic benefit that loss of function of PRPS2 confers to Myc-driven lymphomagenesis, we carried out a survival study. Eμ-Myc/+;Prps2null mice display a remarkable delay in tumor initiation compared to their Eμ-Myc/+ littermates, with approximately 60% of Eμ-Myc/+;Prps2null mice that lived beyond 200 days, a time point when almost all Eμ-Myc/+ mice had died (Figure 7C). To extend our observations to human cancers, we employed two Burkitt’s lymphoma cell lines, Daudi and Raji, and two Multiple Myeloma cell lines, U266 and JJN-3, where Myc is found translocated and/or overexpressed. Knockdown of PRPS2 in these Myc-dependent cell lines resulted in a marked increase in apoptosis, revealing that PRPS2 expression is required to sustain cancer cell survival in both mouse and human cancers that rely on Myc hyperactivation (Figure 7D and S7I).
Figure 7. Loss of PRPS2 Function is Therapeutically Beneficial for Mice and Human Cancers Driven by Myc Hyperactivation.
(A) Representative western blot (WB) of primary B lymphocytes isolated from mice of the indicated genotypes. (B) Annexin V/Propidium Iodide staining and FACS analysis were performed on cells from (A). Error bars represent standard deviation, N=3 mice per condition, ***P<0.001 by student’s t-test. (C) Survival curves showing tumor-free survival between Eμ-Myc/+ (N=24) and Eμ-Myc/+;Prps2null (N=17) male mice. P value was calculated using the log rank test. (D) Apoptosis of human Myc-dependent cell lines assessed by Annexin V staining upon transduction with control or PRPS2 shRNA expressing retroviruses. Data in graph represents percent increase in Annexin V+ GFP-labeled PRPS2 shRNA transduced cells relative to Annexin V+ GFP-labeled control shRNA transduced cells. Error bars represent standard deviation, N=3 for Raji, U266 and JJN-3 cells, N=6 for Daudi cells, **P<0.01, *P<0.05 by student’s t-test. (E) Proposed model for PRPS2 regulation of nucleotide (NT) production in normal and cancer cells. See also Figure S7.
Discussion
The Role of Translational Control in Cellular Metabolism
Our findings point to a model whereby translational regulation of PRPS2 couples protein synthesis to metabolism and directly acts as a molecular rheostat for the nucleotide biosynthesis pathway in cancer cells, controlling the flow of ribose-5-phosphate from the pentose phosphate pathway into the nucleotide biosynthetic precursor PRPP (Figure 7E). Therapeutic strategies that interfere with this Myc-dependent translational control or direct inhibition of PRPS2 expression create a ‘bottleneck’ between the pentose phosphate pathway and nucleotide precursors to decrease the elevated nucleotide production that is specifically required for cancer but not normal cell survival, and consequently, tumor initiation and progression.
The ability to translationally up- or down-regulate mRNAs that encode enzymes capable of controlling metabolic flux ensures a quick response to the protein synthesis demands of cancer cells. Myc-overexpressing cells lacking PRPS2 have a reduced capacity to increase protein synthesis, likely as a consequence of the observed decrease in nucleotide production that is required to synthesize ribosomes. Therefore, Myc-overexpressing cells establish a feed-forward mechanism that tethers increased protein synthesis rates to nucleotide production in order to sustain their continued growth. In line with our findings, restriction of either protein synthesis capacity or ribosome biogenesis has been directly implicated as synthetically lethal in Myc-overexpressing cells (Barna et al., 2008; Bywater et al., 2012). The reliance of key oncogenes such as Myc on the post-transcriptional regulation of PRPS2 reveals a significant vulnerability to the maintenance of cancer cell homeostasis. In fact, our data demonstrate that growth factor-dependent increases in Prps2 translation can be blocked by kinase inhibitors of mTOR or MEK1/2 which converge on eIF4E activity. Therefore, the identification of PRPS2 as a key, translationally regulated enzyme may have broad implications for metabolic control in many additional cancer types driven by distinct oncogenic signals, such as mTOR, where the translation of PRTE-containing mRNAs are increased.
PRPS2 shares approximately 95% amino acid identity with the PRPS1 isoform (Becker et al., 1990). However, our data demonstrates that translational upregulation of PRPS expression by oncogenic signals occurs exclusively on the PRPS2 isozyme. Distinguishing biochemical features of the PRPS2 enzyme present intriguing insight as to why it may be favored to promote nucleotide biosynthesis in oncogenic cells. First, PRPS2 has been demonstrated to be more resistant to the allosteric feedback inhibition by nucleotide biosynthetic pathway byproducts ADP and GDP (Nosal et al., 1993). This enzymatic property may allow PRPS2-expressing cells to continue biosynthesis of nucleotides when their intracellular concentrations are elevated. Additionally, PRPS2 activity is more sensitive to fluctuations in the concentration of its substrate, ATP, which is present at elevated concentrations in Myc-overexpressing B cells (Figure 1B and S1A) (Nosal et al., 1993). These distinguishing enzymatic properties of the different PRPS isoforms suggest a biochemical basis for the development of specific inhibitors to selectively inhibit PRPS2.
PRPS2 as a Therapeutic Target
The Myc oncogene is currently undruggable. Because PRPS2 is an enzyme, it is considered part of the druggable genome (Hopkins and Groom, 2002). Surprisingly, although loss of expression of Prps2 resulted in a modest decrease in nucleotide production in normal cells, there are no apparent deleterious effects at the organismal level in Prps2null mice. Furthermore, we show a therapeutic benefit upon loss-of-function of PRPS2 from the earliest stage of pre-malignancy through the later stages of fully-formed tumors, indicating that PRPS2-dependent nucleotide production is an important, defining feature necessary for cancer cell survival. Therefore drugs capable of specifically inhibiting the PRPS2 isoform may possess a selective therapeutic window.
Inhibitors targeting the nucleotide biosynthetic pathway have long been utilized as chemotherapeutic agents in the clinic. However, many of these drugs produce significant toxicities in widespread organs. Antifolates, which interfere with the one carbon cycle, indiscriminately decrease nucleotide biosynthesis in all proliferating tissues. Other drugs that specifically inhibit the production of one of the five pyrimidine or purine nucleotides disrupt the delicate balance within the nucleotide pools which leads to incorporation of improper bases into the genetic code resulting in DNA damage and/or defective gene expression. Our data demonstrating that PRPS2 is a central integrator of the two most abundant classes of macromolecules comprising cells – proteins and nucleic acids-strongly suggests a distinct therapeutic window exploiting the synthetic lethal interactions important for metabolic reprogramming in cancer cells.
Experimental Procedures
Detailed experimental procedures can be found in the Extended Experimental Procedures. All experimental procedures performed on mice were approved by the UCSF Institutional Animal Care and Use Committees.
Magnetic Resonance Data Acquisition and Processing
One dimensional 1H MR spectra acquisition was performed on the aqueous fraction of freshly isolated B cell extracts described in Extended Experimental Procedures using a 600 MHz Bruker spectrometer equipped with a cryogenically cooled probe. 90° pulse and 4s relaxation delay were used and the water resonance was suppressed using excitation sculpting (Hwang, T. L. and Shaka, A. J., 1995).
All MRS datasets were processed using NMRLab in the MATLAB programming environment (The MathWorks, Inc.) (Günther et al., 2000). Following standard processing steps, selected signals arising from residual solvents and from TMSP were excluded and spectra were normalized according to the probabilistic quotient method (Dieterle et al., 2006). For all datasets, MRS resonances of metabolites were assigned by comparison with spectra of standard compounds (Ludwig et al., 2011). Peak integrals of selected metabolites were calculated using ACD/Spec Manager version 9.15 software (Advanced Chemistry Development) and normalized to the mean of WT samples for relative quantification. Statistical significance was determined using a Student’s t-test with p<0.05 considered significant (N=~8/genotype).
Metabolic Flux experiments using [14C] formate and [8-14C] hypoxanthine
Metabolic labeling experiments were performed as described (Boss and Erbe, 1982; White et al., 1975) with further details provided in Extended Experimental Procedures. [14C] formate and [8-14C] hypoxanthine were from American Radiolabeled Chemicals, (#ARC 0163A and #ARC 0364, respectively).
Sucrose Gradient Fractionation and Polysome Profiling
Sucrose gradient fractionation and polysome profiling were performed as described (Hsieh et al., 2012). Upon polysome fractionation, RNA was extracted and purified using Trizol reagent and PureLink RNA isolation kits (Invitrogen). RNA isolated from each fraction was reverse transcribed and qRT-PCR analysis of polysomal fractions was performed using primers listed in Extended Experimental Procedures. Data are normalized to 5S rRNA expression and subsequently expressed as a fraction of total mRNA for either Prps1 or Prps2.
Luciferase Reporter Assays
NIH3T3 cells were transfected with Prps2 or Prps1 5′UTR pGL3 promoter plasmids in presence of DN-4EBP1 pMSCV hygro or empty vector control in 6 well dishes using lipofectamine (Invitrogen). 24 hours post-transfection, cells were harvested, lysed in passive lysis buffer (Promega) and luciferase assays were performed using a Promega Glomax instrument as described (Hsieh et al., 2012).
Fetal Liver Hematopoietic Stem Cell Culture, Retroviral Preparation, Infection, and Transplantation
Fetal liver hematopoietic stem cell (HSC) culture, infection and transplantation were performed as described with slight variation (Zuber et al., 2010). Retroviral constructs used to transduce HSCs with GFP-expressing Tet-PRPS2 shRNA is described in Extended Experimental Procedures. Efficacy of infection was assessed by FACS analysis to determine the percentage of GFP positive cells. For all transplanted recipient mice, greater than 60% of donor HSCs were infected. Tumor-free survival was monitored twice weekly by palpating lymph nodes. Survival data was plotted using GraphPad Prism software. P values were calculated using the Logrank test.
Eμ-Myc/+ Tumor Cell Culture, Infection, Transplantation and Monitoring
Eμ-Myc tumor cells were harvested, cultured and infected with Tet-PRPS2 shRNA MSCV-pGFP as described (Schmitt et al., 2000). A detailed description is presented in Extended Experimental Procedures. After transplantation via tail vein of 5×106 live tumour cells into syngeneic recipients, tumors were allowed to engraft for several days. Between days 5 and 7 post transplantation, blood samples from tail bleeds were subjected to FACS analysis to assay for tumor take by measurement of GFP+ B220+ tumor cells. Upon detection of GFP+ population of circulating B220+ cells, mice were placed in either vehicle (water) or doxycycline (2mg/mL in water) treatment regimens representing day 0 of the survival curve. Survival was monitored daily and mice were sacrificed upon becoming moribund or developing tumors >1cm in diameter. Survival data was plotted using GraphPad Prism software. P values were calculated using the Logrank test.
Generation of Prps2null mice and Other Mouse Lines Used
PRPS2tm1a(KOMP)Wtsi ES cells used for this research project were generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository. PRPS2tm1a(KOMP)Wtsi ES cells contain a genetrap insertion in the first intron of the PRPS2 gene, which fuses the rst 40 amino acids of PRPS2 to a β-gal cassette flanked by a SV40 polyadenylation site that acts as a strong transcriptional termination signal. PRPS2tm1a(KOMP)Wtsi ES cells were propagated by the UCSF ES cell core and microinjections of these cells into C57Bl6 albino females were performed by the Gladstone Institute Transgenic Core Facility to generate chimeric founders. F1 mice were generated and named Prps2null in the text. A complete list of mouse lines used in this study is presented in Extended Experimental Procedures.
Supplementary Material
Research Highlights.
Protein synthesis and nucleotide metabolism are coupled downstream of Myc
Translational control of Prps2 integrates nucleotide metabolism and protein synthesis
Loss of function of PRPS2, but not PRPS1, is synthetic lethal upon Myc overexpression
Prps2 is dispensable for normal physiology, but essential for Myc-driven lymphoma
Acknowledgments
We thank Maria Barna and Kevan Shokat for helpful discussions and critical reading of the manuscript. D. Ruggero is a Leukemia and Lymphoma Society Scholar. This work was supported by NIH R01 CA130812 (to S. Ronen); NIH R01 CA154915 (to S. Ronen); American Cancer Society 121364-PF-11-184-01-TBG (to J.T. Cunningham); and NIH R01CA140456 (to D. Ruggero). NIH grants to Velocigene at Regeneron Inc (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8500 genes in the KOMP Program and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
John T. Cunningham, School of Medicine and Department of Urology, Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, CA 94158
Melissa V. Moreno, School of Medicine and Department of Urology, Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, CA 94158
Alessia Lodi, Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA 94158.
Sabrina M. Ronen, Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA 94158
Davide Ruggero, School of Medicine and Department of Urology, Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, CA 94158.
References
- Adams JM. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, Wu H, Wei Z, Wagner SN, Herlyn M, et al. Suppression of Nucleotide Metabolism Underlies the Establishment and Maintenance of Oncogene-Induced Senescence. Cell Rep. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MA, Heidler SA, Bell GI, Seino S, Le Beau MM, Westbrook CA, Neuman W, Shapiro LJ, Mohandas TK, Roessler BJ. Cloning of cDNAs for human phosphoribosylpyrophosphate synthetases 1 and 2 and X chromosome localization of< i> PRPS1</i> and< i> PRPS2</i> genes. Genomics. 1990;8:555–561. doi: 10.1016/0888-7543(90)90043-t. [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide Deficiency Promotes Genomic Instability in Early Stages of Cancer Development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss GR, Erbe RW. Decreased purine synthesis during amino acid starvation of human lymphoblasts. J Biol Chem. 1982;257:4242–4247. [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, et al. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Pearson RB, McArthur GA, Hannan RD. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat Rev Cancer. 2013;13:299–314. doi: 10.1038/nrc3496. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Dang CV. Enigmatic MYC Conducts an Unfolding Systems Biology Symphony. Genes Cancer. 2010;1:526–531. doi: 10.1177/1947601910378742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in 1 H NMR Metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- Doherty JR, Yang C, Scott KEN, Cameron MD, Fallahi M, Li W, Hall MA, Amelio AL, Mishra JK, Li F, et al. Blocking Lactate Export by Inhibiting the Myc Target MCT1 Disables Glycolysis and Glutathione Synthesis. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Picard D, Yamamoto KR, Bishop JM. Chimaeras of Myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. AMPK Is a Negative Regulator of the Warburg Effect and Suppresses Tumor Growth In Vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Meyuhas O, Perry RP, Johnson LF. Regulation of ribosomal protein mRNA content and translation in growth-stimulated mouse fibroblasts. Mol Cell Biol. 1982;2:685–693. doi: 10.1128/mcb.2.6.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: Sibling Rivals for Control of Cancer Cell Metabolism and Proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Graves JA, Rothermund K, Wang T, Qian W, Van Houten B, Prochownik EV. Point Mutations in c-Myc Uncouple Neoplastic Transformation from Multiple Other Phenotypes in Rat Fibroblasts. PLoS ONE. 2010;5:e13717. doi: 10.1371/journal.pone.0013717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther UL, Ludwig C, Rüterjans H. NMRLAB—Advanced NMR Data Processing in Matlab. J Magn Reson. 2000;145:201–208. doi: 10.1006/jmre.2000.2071. [DOI] [PubMed] [Google Scholar]
- Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Hsieh AC, Ruggero D. Targeting Eukaryotic Translation Initiation Factor 4E (eIF4E) in Cancer. Clin Cancer Res. 2010;16:4914–4920. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic Dissection of the Oncogenic mTOR Pathway Reveals Druggable Addiction to Translational Control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang TL, Shaka AJ. Water suppression that works - Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson. 1995;112:275–279. [Google Scholar]
- Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt EV. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Liu YC, Li F, Handler J, Huang CRL, Xiang Y, Neretti N, Sedivy JM, Zeller KI, Dang CV. Global Regulation of Nucleotide Biosynthetic Genes by c-Myc. PLoS ONE. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C, Easton JM, Lodi A, Tiziani S, Manzoor SE, Southam AD, Byrne JJ, Bishop LM, He S, Arvanitis TN, et al. Birmingham Metabolite Library: a publicly accessible database of 1-D 1H and 2-D 1H J-resolved NMR spectra of authentic metabolite standards (BML-NMR) Metabolomics. 2011;8:8–18. [Google Scholar]
- Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–1066. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosal JM, Switzer RL, Becker MA. Overexpression, purification, and characterization of recombinant human 5-phosphoribosyl-1-pyrophosphate synthetase isozymes I and II. J Biol Chem. 1993;268:10168–10175. [PubMed] [Google Scholar]
- Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci. 2013;110:11988–11993. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de Novo Pyrimidine Synthesis by Growth Signaling Through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Rosenthal CT, Lowe SW. Genetic analysis of chemoresistance in primary murine lymphomas. Nat Med. 2000;6:1029–1035. doi: 10.1038/79542. [DOI] [PubMed] [Google Scholar]
- Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci. 1998;95:1511–1516. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. mRNA Translation and Energy Metabolism in Cancer: The Role of the MAPK and mTORC1 Pathways. Cold Spring Harb Symp Quant Biol. 2011;76:355–367. doi: 10.1101/sqb.2011.76.010785. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Lunt SY, Dayton TL, Fiske BP, Israelsen WJ, Mattaini KR, Vokes NI, Stephanopoulos G, Cantley LC, Metallo CM, et al. Metabolic Pathway Alterations that Support Cell Proliferation. Cold Spring Harb Symp Quant Biol. 2012;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- White JC, Loftfield S, Goldman ID. The mechanism of action of methotrexate. III. Requirement of free intracellular methotrexate for maximal suppression of (14C)formate incorporation into nucleic acids and protein. Mol Pharmacol. 1975;11:287–297. [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang X-Y, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TWM, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Matés JM, Alonso FJ, Wang C, Seo Y, et al. The Metabolic Profile of Tumors Depends on Both the Responsible Genetic Lesion and Tissue Type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2010;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.