Abstract
Background
Previous genomic analysis of pathogenic Leptospira has identified two circular chromosomes but no plasmid. This study aims to investigate potential extrachromosomal elements of L.interrogans serovar Canicola strain Gui44.
Methodology
Two novel plasmids, pGui1 and pGui2, were isolated from the pathogenic strain Gui44, using a modified alkaline lysis method. Southern blotting was performed to determine the presence and size of them. Then, 454 and Hiseq sequencing were applied to obtain and analyze the complete sequences of the two plasmids. Furthermore, real-time quantitative PCR and next-generation sequencing were used to compare relative copy numbers of the two plasmids with that of the chromosomes. Finally, after serial passages in vitro for more than 2 years, the strain Gui44 was subsequently re-sequenced to estimate stability of the two plasmids.
Principal Findings
The larger plasmid, pGui1, 74,981 base pairs (bp) in length with GC content of 34.63%, possesses 62 open reading frames (ORFs). The smaller plasmid, pGui2, is 66,851 bp in length with GC content of 33.33%, and contains 63 ORFs. The replication initiation proteins encoded by pGui1 and pGui2 demonstrate significant sequence similarity with LA1839 (86% and 88%), a well-known replication protein in another pathogenic L.interrogans serovar Lai strain Lai, suggesting the ability for autonomous plasmid replication. Quantitative PCR and next-generation sequencing confirms a single copy of both plasmids and their stable presence in the strain Gui44 with in vitro serial passages after more than 2 years. Interestingly, the two plasmids both contain a significant number of novel genes (35 in pGui1 and 52 in pGui2).
Conclusions
This report confirms the presence of two separate circular plasmids in serovar Canicola strain Gui44 and provides a new understanding of genomic organization, adaptation, evolution and pathogenesis of Leptospira, which will aid in the development of in vivo genetic manipulation systems in pathogenic Leptospira species.
Author Summary
Leptospira species are the causative agent of leptospirosis, one of the most common animal to human transmitted diseases. Previous genomic analysis of L.interrogans serovar Lai and Copenhageni has identified the presence of large (4.33 mega base) and small (350 kilo base) circular chromosomes without evidence of any plasmids. Detailed understanding of Leptospira and its pathogenicity was delayed by the lack of available genetic tools. In this study we confirm the existence of two novel plasmids in L.interrogans serovar Canicola strain Gui44, an epidemic strain in China. Some novel genes identified in the two plasmids may play important roles in the characterization of the strain. The two plasmids will provide useful information in understanding the diversity of Leptospira genome and markedly improve our understanding of the evolution and pathogenesis of L.interrogans. In particular, it will contribute to the development of genetic manipulation systems in pathogenic Leptospira species.
Introduction
Leptospirosis is caused by spirochetes belonging to the genus Leptospira, which is comprised of both saprophytic and pathogenic species [1]. The spectrum of clinical symptoms of human leptospirosis ranges from asymptomatic to severe manifestation causing multi-organs dysfunction and even death [2]. There are eight known pathogenic species of Leptospira, and over 200 pathogenic serovars [3], [4]. Humans and animals become infected either through direct or indirect exposure to Leptospira spp. shed from the host organism via urine [5]. More than 500,000 cases of severe leptospirosis are reported each year, with mortality rates up to 23% in some outbreaks [6]. In addition, Leptospirosis remains recognized as a major veterinary disease worldwide as it infects a broad range of domestic animals, incurring large economic losses [5], [7].
Until recently, the lack of available genetic tools delayed the full characterization and understanding of pathogenic Leptospira and its pathogenicity [6]. Plasmid vectors have been successfully generated using the 54 kilo base (kb) genomic island LaiGI from the large chromosome of L.interrogans strain Lai, which has the ability to excise itself from the chromosome [8]. The plasmid vector can autonomously replicate in both L.biflexa and E. coli. In addition, DNA has been introduced into Leptospira by electroporation [8]. However, none of this work has led to the construction of a replicative plasmid vector for pathogenic Leptospira, and absence of naturally occurring plasmids in pathogenic Leptospira further impedes investigation of its biology and virulence mechanisms by classic genetic studies.
The recent whole genome sequencing of seven pathogenic Leptospira strains including L.interrogans serovar Lai strain Lai and its virulence-attenuated strain IPAV, serovar Copenhageni strains Fiocruz L1-130 and two virulent strains of L.borgpetersenii serovar Hardjo (strain L550 and JB197) [9]–[12], as well as two saprophytic, L.biflexa serovar Patoc strains Paris and Ames [13], has provided a better understanding of evolution, metabolism and pathogenicity of the organism. Based on these genomic data, L.biflexa is the first reported species in which a 74 kb extrachromosomal plasmid named p74 was identified [13]. Previous evidence had showed that all of the pathogenic Leptospira species sequenced only contained two chromosomes as well as one special 54 kb genomic island LaiGI in the large chromosome [14].
L.interrogans is the most frequently reported agent of leptospirosis and many serovars have adapted to a biased mammalian reservoir host, such as serovar Lai and Copenhageni (rodents), serovar Canicola (dogs) and serovar Hardjo (cattle) [15], [16]. Interestingly, focusing on one pathogenic serovar Lai strain 56601, our recent study re-analyzed the previously reported mobile, phage-related genomic island LaiGI in its large chromosome and first confirmed the presence of one separate circular extrachromosomal plasmid named Laicp in strain 56601 by a combination of various experimental methods [17]. Based on the finding, we continued to perform isolation and characterization of potential circular extrachromosomal elements in another pathogenic L.interrogans serovar Canicola strain Gui44, which was originally isolated from a patient with leptospirosis and was also one of the main epidemic strains in China (Institute for Infectious Disease Control and Prevention, Beijing, China). Surprisingly, in the present study, strain Gui44 was proved to possess two circular plasmids which were distinct from the simple one plasmid Laicp found in strain 56601. This report first described the two novel plasmids, pGui1 and pGui2, isolated from the pathogenic strain Gui44. Sequence analysis determined that the two plasmids contained novel genes, which may provide strain Gui44 with unique properties involved in adaptation and pathogenesis and give insight into the genome dynamics of this organism. To best of our knowledge, so far, except the pathogenic strain 56601 and Gui44, there was no other report of circular plasmid isolated from pathogenic Leptospira. Finally, these results may significantly contribute to our comprehensive understanding of the complicated diversity of pathogenic Leptospira genome and enable construction of a new shuttle vector and molecular tool to facilitate the investigation of its virulence mechanisms.
Methods
Bacterial strains and growth conditions
L.interrogans serogroup Canicola serovar Canicola strain Gui44 was obtained from the Institute for Infectious Disease Control and Prevention, Beijing, China. The strain is grown at 28°C in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium under aerobic conditions and collected at mid-log phase [18].
Isolation of plasmids
Plasmids of strain Gui44 were isolated using a previously published alkaline lysis method with some modifications [19], [20]. During mid-log growth phase, 500 mL of cells were collected by centrifugation (8000 rpm for 20 min at 4°C) and washed twice in 100 mL Tris-EDTA (TE) buffer (10 mM Tris–Cl, 1 mM EDTA, pH 8.0). Cells were then resuspended in 10 mL of solution I (2 mg/ml lysozyme, 10% sucrose, 20 mM Tris–Cl, 1 mM EDTA, RNase 100 µg/ml, pH 8.0) and incubated at 37°C for 2 h. Cells in solution I were then mixed with 20 ml of solution II (0.2 M NaOH, 1% SLS) and incubated for 3 min on ice. Cells were treated with 15 mL of solution III (sodium acetate, pH 4.8) and incubated for 5 min on ice followed by centrifugation at 12,000 rpm for 30 min. DNA was precipitated with 38% isopropanol, washed twice with 70% ethanol, and dissolved in sterile water. Total genomic DNA of strain Gui44 was extracted by the phenol-chloroform method as previously described [21].
Sequencing and sequence analysis of total genomic and plasmid DNA of strain Gui44
In March 2009, the genome of L.interrogans serovar Canicola strain Gui44 was first sequenced using 454 GS 20 sequencing system (454 Life Science Corporation, Branford, CT, USA) at the Chinese National Human Genome Center (CHGC, Shanghai, China). After physical gap closing, the complete genome of the strain contained two circular chromosomes (unpublished data) and two novel extrachomosomal elements. Based on this finding, plasmid DNA and total genomic DNA of strain Gui44, respectively, were obtained to further perform whole genome re-sequencing with Hiseq 2000 sequencing system (Illumina, San Diego, CA, USA) in September 2011 at CHGC. According to the complete sequences of the two plasmids by 454 sequencing, the nucleotide sequences of two plasmids were assembled into 16 contigs larger than 1 kb. The physical gaps were closed by sequencing gap-spanning PCR products or clones using ABI 3730×l DNA Analyzer. PCRs were performed under the following conditions: 5 min at 95°C, followed by 35 cycles of 30 sec at 95°C, 30 sec at 56°C, and 30 sec at 72°C. Sequence assembly was accomplished by using the Phred/Phrap/Consed software package. Sequencing in this study was performed at CHGC. In addition, the restriction sites were analyzed by BioEdit software (http://bioedit.software.informer.com/). The sequence alignments were performed using BLAST (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi). Glimmer (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi) was used to predict open reading frames (ORFs). Transmembrane helices in proteins were predicted by TMHMM Server v.2.0. SecretomeP 2.0 Server was used to predict the secretion of proteins. Conserved domains were determined via Conserved Domain Database (CDD) and Clusters of Orthologous Groups of proteins (COGs) in the NCBI database and InterPro (http://www.ebi.ac.uk/interpro). Orthologous proteins were identified by performing BLAST searches against the NCBI non-redundant protein database and subsequently checked manually.
Pulsed-Field Gel Electrophoresis (PFGE)
Either 2×109 cells or 20 µL (130 ng/µL) of extracted plasmid DNA were embedded in agarose plugs (1%) (SeaKem Gold agarose; Cambrex, Rockland, ME) and incubated with 0.1 mg/mL proteinase K overnight at 50°C. The plugs were then washed twice with pre-warmed TE buffer, and then digested with either restriction enzyme NotI or BamHI at 37°C or 30°C, respectively, following manufacturer's instructions (New England Biolabs, Ipswich, MA, USA). DNA fragments were separated using a 1% pulse-field agarose gel, which was prepared and ran in 0.5%TBE buffer on a contour-clamped homogenous electric field machine (CHEF-DR III; Bio-Rad, Richmond, CA). The lysates were electrophoresed at 5 V at room temperature for either 5–65 sec for 24 h or for 5–25 sec for 24 h based on the fragment size. Gels were stained in 20 µg/mL ethidium bromide (Sigma, St Louis, MO, USA) for 15 min and destained with distilled water for 30 min. Salmonella Braenderup (H9812) from the Institute for Infectious Disease Control and Prevention (Beijing, China) was digested with XbaI (New England Biolabs) and used as the molecular weight size standard.
Southern hybridization
Total genomic and plasmid DNA were digested with either NotI or BamHI and electrophoresed in a 1% agarose gel. Southern blot hybridization was performed using a DIG luminescent Detection Kit (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer's instructions with primers listed in Table 1.
Table 1. Primers used in this study.
| Probe | Primer ID | Primer sequence (5′-3′) | Product size (bp) | Location |
| SpGui1 | S pGui1-F | TCGTTGTGCCCAACTCTCAA | 486 | pGui1 |
| S pGui1-R | TCAAATCCGAAAACCCCA | |||
| SpGui2 | S pGui2-F | AAGATCTCCTTTATATAGTGATTTC | 496 | pGui2 |
| S pGui1-R | GTATTTTTATCAATCACAAATAATG | |||
| dnaA | dnaA-F | GCTCCATCTGCCACAATC | 92 | large chromosome |
| dnaA-F | GGAATCTTATCGCCACAAGTT | |||
| P1 | pGui1-F | CATCGGTATTAGCGTAAGAG | 94 | pGui1 |
| pGui1-R | AGGAATCATCGGAGTTGT | |||
| P2 | pGui2-F | AATCGCACCACCTAATATC | 100 | pGui2 |
| pGui2-R | TCATCGCAATCGTAGAAC |
Relative copy number determination by quantitative real-time PCR (qPCR) and next-generation sequencing (NGS)
As mentioned above, total genomic DNA of strain Gui44 including both chromosome and plasmid DNA was sequenced with Hiseq 2000 system. Sequenced reads were aligned to the reference genome (including the two chromosomes and two plasmids) of the strain Gui44 by Bowtie2 using the default parameters [22]. When the reads were exclusively mapped to the simple location of the genome, they were counted by using the idxstats module in SAMtools software [23]. The relative copy number was calculated by: aligned reads numbers× average read length/(corresponding chromosome or plasmid size), as well as by qPCR on an ABI 7500 fast real-time PCR system (Applied Biosystems of Thermo Fisher Scientific Inc., Waltham, MA, USA) with a FastStart Universal SYBER Green Master mix (Roche Diagnostics., Indianapolis, USA) according to manufacturer's instructions. PCR primers were designed using Beacon Designer 7 software (http://www.premierbiosoft.com/molecular_beacons/). Eight-fold serial dilutions of the plasmids from Peasy-T1 (Beijing Transgen Biotech Co., LTD, Beijing, China) and template DNA were used to construct standard curves of all plasmids. Reactions were performed in duplicate in a total reaction volume of 20 µL. A 92 bp fragment of a replication initiation factor (dnaA), a single-copy gene in the larger chromosome, was used as a reference gene. A 94 bp fragment designated P1 that is unique to pGui1 and a 100 bp fragment designated P2 which is specific to pGui2 were amplified (all primers used for qPCR are listed in Table 1). The reactions were performed under the following conditions: 10 min at 95°C, followed by 40 cycles of 30 sec at 95°C, 30 sec at 56°C, and 30 sec at 72°C. The experiment was carried out in triplicate and averages were reported. The relative copy number of plasmids was calculated using Nrelatives = (1+E)−ΔC T, where E stands for the PCR amplification efficiency and ΔCT was for the difference in threshold cycle number between the reference gene and target gene.
Nucleotide sequence accession number
The complete nucleotide sequences of pGui1 and pGui2 plasmids have been deposited in GenBank under the following accession number: KF648557 and KF648558.
Results and Discussion
Analysis of pGui1 and pGui2 isolated from L. interrogans strain Gui44
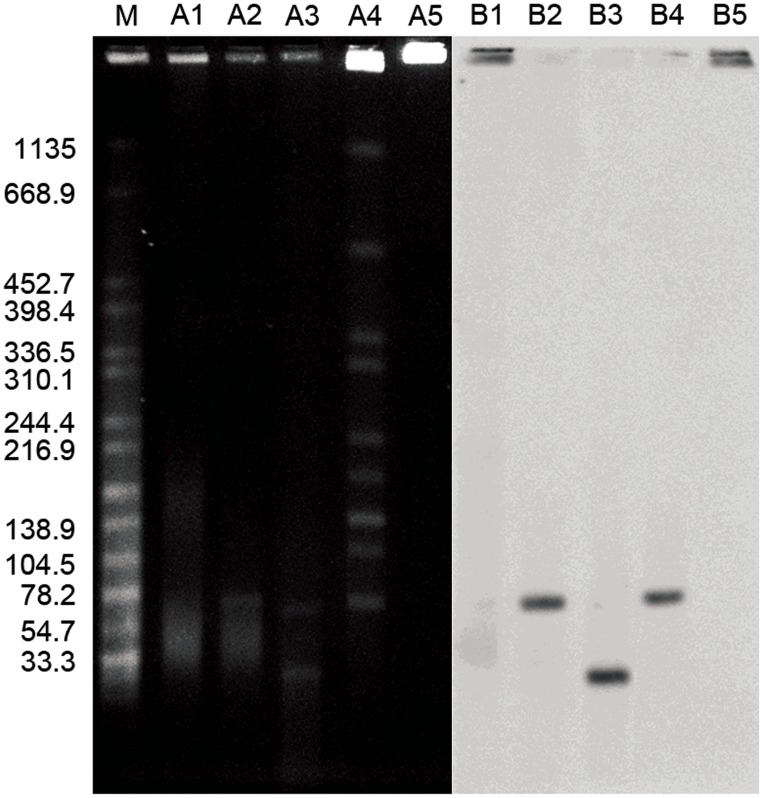
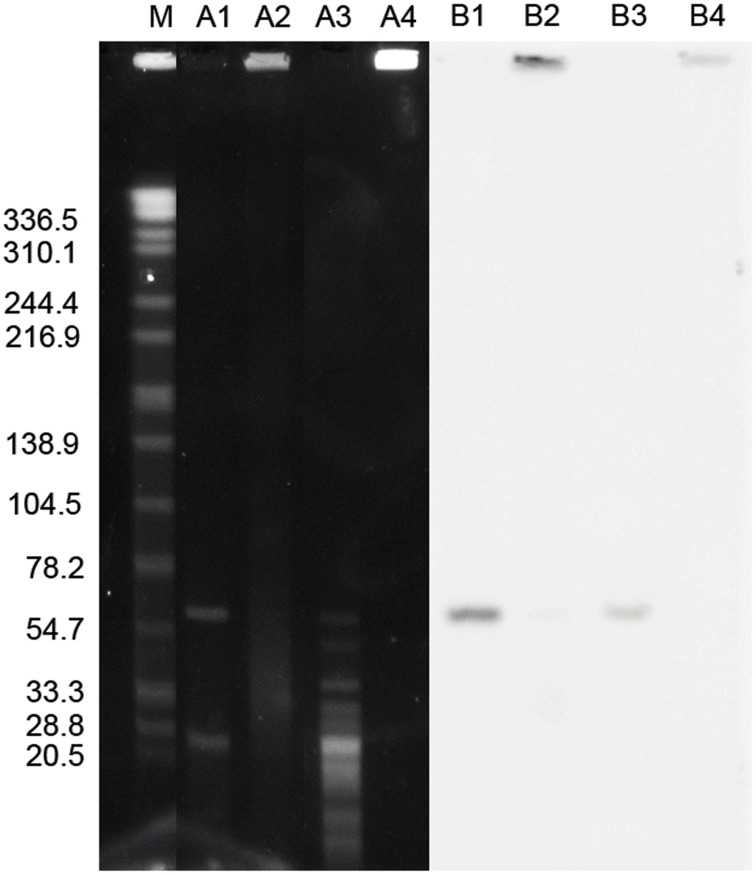
In silico analysis revealed that the pGui1 has one NotI restriction site and nine BamHI restriction sites, whereas pGui2 only contains a BamHI restriction site but no NotI restriction site. Therefore, following plasmid digestion with BamHI, pGui1 has eight specific fragments of 217, 1903, 2328, 3134, 3947, 19971, 21277, and 22104 bp. Digestion of genomic and plasmid DNA with NotI both identified the presence of pGui1 by a fragment of the expected 78 kb size (Fig. 1). Southern hybridization results using a specific probe SpGui1 suggested that pGui1 was circular and independent of the chromosome (Fig. 1). Digestion of genomic and plasmid DNA using BamHI and Southern hybridization with SpGui2 probe revealed a ∼65 kb fragment (Fig. 2). BamHI digested plasmids hybridized with probe SpGui1 revealed a ∼23 kb fragment (Fig. 1). Collectively, these results confirmed the presence of two circular plasmids in the L.interrogans serovar Canicola strain Gui44 with sizes of approximately 78 kb and 65 kb.
Figure 1. Characterization of pGui1 and pGui2 in the pathogenic L.interrogans strain Gui44 by PFGE and Southern blot.
PFGE and Southern Blot (using radiolabeled spGui1 probe) of undigested plasmid DNA (lanes A1 and B1); plasmid DNA digested with NotI (A2 and B2); plasmid DNA digested with BamHI (A3 and B3); total genomic DNA digested with NotI (A4 and B4); and undigested total genomic DNA (A5 and B5). The fragment in A2 and B2 indicates the linearized pGui1 (∼78 kb). The corresponding fragment presents in A4 and B4. The larger fragment in A3 indicates the linearized pGui2. The smaller fragment in A4 is one fragment of pGui1 digested with BamHI. Salmonella Braenderup (H9812) digested with XbaI and is used as the molecular weight size standard (M).
Figure 2. Characterization of pGui2 in the pathogenic L.interrogans strain Gui44 by PFGE and Southern blot.
PFGE and Southern Blot (using radiolabeled spGui2 probe) of plasmid DNA digested with BamHI (lanes A1 and B1); undigested plasmid DNA (A2 and B2); total genomic DNA digested with BamHI (A3 and B3); and undigested total genomic DNA (A4 and B4). Salmonella Braenderup (H9812) digested with XbaI was used as the molecular weight size standard (M). The larger fragment in A1 indicates the linearized plasmid pGui2. The corresponding fragment presents in A3. The smaller fragment in A1 is a fragment of the plasmid pGui1.
General characteristics of pGui1 and pGui2
pGui1 and pGui2, were found to be 74,981 and 66,851 bp in length with overall GC content of 34.63% and 33.33%, respectively. A summary of the general characteristics is presented in Table 2, and detailed genomic maps are provided in Supplementary Fig.S1. The G+C content of the two plasmids was lower in comparison with the chromosomes of L.interrogans strain Gui44 (35.05%, unpublished data), as well as with the third circular replicon p74 (37.47%) previously found in the L.biflexa and the extrachromosomal circular plasmid Laicp in strain 56601 (34.64%) [17]. The pGui1 contains 62 protein-coding genes representing a coding percentage of 78.3%, with the majority of the identified genes (56%) encoding hypothetical proteins. In addition, mobile elements (15%), genes related to heavy-metal and drug resistance (10%), and regulatory genes (6%) were also identified. A total of 63 protein-coding genes, comprising 72% of the plasmid, were identified in pGui2 (Table 2). The percentage of genes encoding hypothetical proteins (82%) on pGui2 was higher than on pGui1 (56%). In addition, mobile elements (6%) and regulatory genes (2%) were also identified. Identified genes and their putative functions are listed in Supplementary Table S2 and S3. The high percentage of mobile elements suggests the presence of gene rearrangement. The differences between pGui1 and pGui2 suggest they have different roles in parasite cell function.
Table 2. Genome features of pGui1 and pGui2.
| Genomic Features | pGui1 | pGui2 |
| Size (bp) | 74981 | 66851 |
| G+C content (%) | 34.63 | 33.33 |
| Protein coding (%) | 78.32 | 72.497 |
| Protein-coding genes | 62 | 63 |
| Hypothetical genes | 35 | 52 |
| Gene density (bp per gene) | 947 | 769 |
| Transposases | 9 | 4 |
Plasmid stability
As extrachromosomal DNA, plasmids must meet the following criteria if they are to be stably maintained over time in the host cells: (i) execute functions that ensure accurate segregation, (ii) contain effective systems that result in inherited stability, and (iii) be capable of replicating autonomously within a suitable host. Accurate segregation and inherited stability are especially important for the low copy number plasmids [24]. Sequence analysis of pGui1 and pGui2 shows that both plasmids meet these criteria.
In the present study, since we sequenced the genome of strain Gui44 by 454 sequencing and firstly detected the existence of the two novel extrachromosomal elements in 2009, the Gui44 strain has been cultured in EMJH and maintained by serial passages at least once a month in hamsters for the preservation of virulence for more than two years, in 2011, whole genome re-sequencing of plasmid DNA and total genomic DNA of the strain Gui44 were independently performed using Hiseq 2000 system. Although we do not have a precise number of total passages, the two independent sequencing results of plasmid DNA and genomic DNA both confirmed that the two plasmids, pGui1 and pGui2, are stably maintained in the strain Gui44, representing their potential application as novel shuttle vectors in pathogenic Leptospira.
Furthermore, the PFGE profiles in the present study following NotI digestion are fully consistent with a previous report in 2012 [25]. Particularly, the same 75 kb fragment of pGui1 was observed, further confirming the stable presence of pGui1. However, due to the absence of NotI restriction site, pGui2 did not appear as a linearized fragment when digested with NotI performed by Tang GP, et al and our study [25].
The partition system
Low-copy-number plasmids require a segregation mechanism to ensure that equivalent plasmid copies are inherited by each daughter cell during cell division. Several specific mechanisms may be involved in DNA segregation [26], [27]. One such mechanism involves par loci, which ensures receipt by every daughter cell, thereby enhancing the inheritance and stability of the plasmid DNA [27], [28]. Genes that may be involved in plasmid segregation were also identified in pGui1 and pGui2. pGui10067, identified in pGui1, encodes a protein that shares high similarity with LA1838, the chromosome partitioning protein parB locating on the newly confirmed plasmid Laicp of L.interrogans serovar Lai str.56601 in our recent study (98/227 aa, 43% identity) [17] and LEP1GSC075_0005, ParB-like protein of L.interrogans str.Kito (261/261 aa, 100%) in the PATRIC database [29]. Another gene found in pGui1, pGui10068, encodes a protein that exhibits similarities with LA1837, one parA family protein encoded by the extrachromosomal plasmid of str.56601 (116/258 aa, 45%) [17] and LEPBI_p0001, putative chromosome partitioning protein ParA in the plasmid p74 of L.biflexa serovar Patoc strain Patoc 1 (Paris) (117/257 aa, 46%) [29]. Similarly, pGui20067 identified in pGui2, shares high similarity with LEP1GSC069_1546, ParA nucleotide binding domain protein of L.interrogans serovar Canicola str. Fiocruz LV133 (229/247 aa, 92%), and its downstream pGui20066, exhibits similarity with LEP1GSC051_0353, ParB-like protein in Leptospira sp.P2653 (114/235 aa, 49%) [29]. Therefore, as ParB is known to be less conserved [14], pGui20066 may constitute the parB within the Gui44 strain. These partition systems in pGui1 and pGui2 may play an important role in the plasmids maintenance in the L.interrogans serovar Canicola strain Gui44.
The postsegregational killing system (PSK)
One way organisms maintain a plasmid in a population is through the addiction system also termed plasmid PSK [24]. Although there are several types of plasmid addiction systems, the one found in L.interrogans serovar Canicola strain Gui44 consists of toxin and antitoxin genes, most likely encoding a labile antitoxin and a stable toxin. In the toxin/antitoxin system (TA), the daughter cells that contain the addiction system, are able to survive due to the presence of the antitoxin whereas, the daughter cells that lack the plasmid either die or exhibit reduced growth-rate due to the presence of the toxin being produced by the parent cell [24], [26]. The addiction system in cells plays an essential role in the execution of postsegregational killing. In pGui1, pGui10006 and pGui10007 are identical to ppGpp-regulated growth inhibitor MazF of L.interrogans serovar Lai str.Lai (99/99 aa, 100%) and the transcriptional regulator/antitoxin MazE of L.interrogans serovar Lai str.Lai (99/99 aa, 100%), respectively [29]. An additional Toxin-Antitoxin system was identified in pGui1, encoded by pGui100011 and pGui10012, which show high similarity with MazF and MazE in L.borgpetersenii serovar Hardjo-bovis L550 (103/107, 96% and 77/82, 94%, respectively). MazF is a sequence-specific mRNA endoribonuclease that initiates a programmed cell death pathway, whereas MazE acts as its antagonist [30]. In pGui2, no obvious addiction system was found. However, hypothetical protein pGui20047, displays some similarity with the antitoxin component of repressor protein CI in Burkholderia multivorans CGD2M (22/62 aa, 35%) and antitoxin component in Algoriphagus spp. PR1 (36/121 aa, 30%). These homologs involved in the plasmid addiction system may contribute to the stable inheritance of pGui1 and pGui2 in the strain Gui44.
The replication region
Proper plasmid replication depends upon the replication region and replication origin [31]. Replication origins of pGui1 and pGui2 were predicted by GC skew [32] (Supplementary Fig.S1). The most important replication protein (Rep) of low-copy-number plasmid is often found in the vicinity of ParAB system [8]. Analysis of pGui1 and pGui2 showed that pGui10066 and pGui20065, both located downstream of ParAB-system, share high similarity with LA1839 in the plasmid Laicp as well as in the previously identified genomic island LaiGI which has the ability for autonomous replication in L.interrogans serovar Lai strain Lai and serovar Canicola strain HondUtrech as well as L.biflexa [8]. The C-terminal regions of pGui10066 and pGui20065 (272–478 aa) share 86% and 88% similarity with the LA1839 protein, respectively. Analysis of the conserved domains in the N-terminal region of the three replication proteins (1–75 aa) revealed a common helix-turn-helix motif, a sequence-specific DNA binding domain characteristic of Rep proteins [31]. Hence, these findings suggest that pGui10066 and pGui20065 are potential replication proteins for pGui1 and pGui2.
Genes with putative role in virulence
Genes involved in pathogenicity were also identified in both pGui1 and pGui2. pGui10044 encodes one protein that contains six tandem repeat bacterial immunoglobulin-like (Big) domains, which are classical characteristic domain of the leptospiral Ig-like (Lig) proteins [33]. Lig proteins have been shown to contribute to microbial adhesion of host mammalian cell and initiate infection [34]. Bioinformatic analysis indicates that pGui10044 is a secretory protein with a transmembrane helix, suggesting that it may be involved in Leptospira-host interaction. pGui20059 possesses a rearrangement hotspot domain (RHS), which appears to be involved in ligand binding and activates the inflammasome [35], [36]. Based on Interpro analysis, pGui20061 shows features related to VCBS domain (Repeat domain in Vibrio, Colwellia, Bradyrhizobium and Shewanella) that was found in some pathogenic bacteria and played a very important role in pathogen adhesion to host cells. The exact function of these proteins deserves further investigations.
Relative plasmid copy numbers
Whole genome sequencing using total genomic DNA of strain Gui44 generated a total of 26,589,548 reads and nearly 2 Gb high-quality filtered sequences. The numbers of aligned reads and genomic coverage are 23,612,793 and 558.0599 (larger chromosome), 2,014,331 and 564.8649 (small chromosome), 392,726 and 523.7674 (pGui1), 322,523 and 482.4505 (pGui2), respectively. The relative ratio of reads to length of the large chromosome, small chromosome, pGui1 and pGui2 sequenced are 5.5806, 5.6486, 5.2377, and 4.8245, respectively, indicating the closely similar relative copy number.
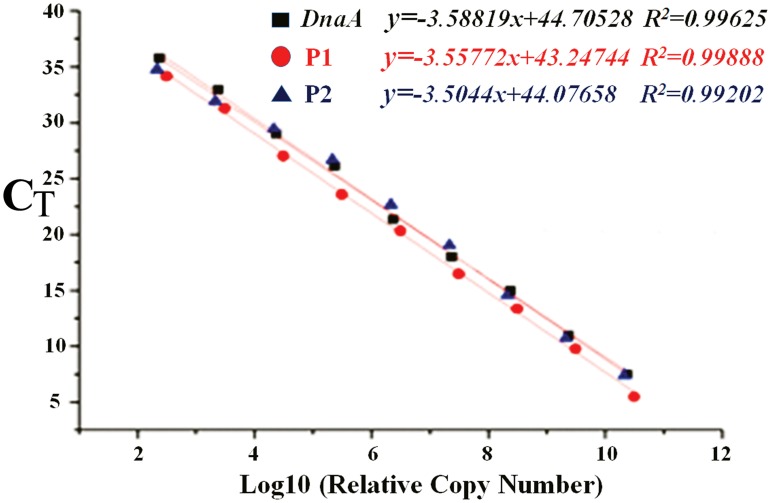
In order to more precisely determine the relative copy numbers of the two plasmids, the qPCR method was applied. As shown in Figure 3, the standard curves obtained for DnaA (larger chromosome), P1 (pGui1) and P2 (pGui2) are linear (R2>0.99) in the range tested with slopes of 3.58819, 3.55772, and 3.50440, respectively. In addition, the intercepts of dnaA, P1 and P2 are 44.70528, 43.24744 and 44.07658, with Ct values of 13.71492, 15.20904 and 14.09529, respectively. Taken together, these results indicate that the relative copy number of pGui1 and pGui2 is about one copy per chromosome. Hence, we speculate that it is difficult to isolate and extract plasmid DNA in pathogenic Leptospira because of the low plasmid copy number.
Figure 3. Relative copy number of large chromosome (DnaA), pGui1 (P1) and pGui2 (P2) as determined by real-time quantitative PCR.
The standard curves obtained from eight-fold DNA dilutions were linear (R2>0.99) in the range tested.
Comparison of the two plasmids against the extrachromosomal plasmid Laicp of L. interrogans str.56601
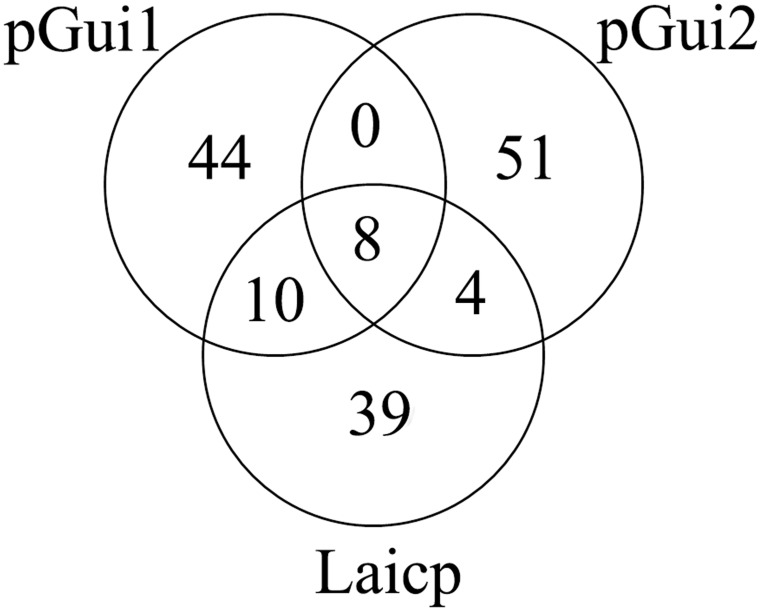
Sequence comparisons showed that the Laicp has 18 orthologs in pGui1 (Fig. 4). Six (33%) of these have no known function and six (33%) are transposases. In addition, Toxin-antitoxin proteins (two), transcriptional regulator (one), plasmid partition proteins (two) and replication origin protein (one) were identified on pGui1. Additionally, twelve common orthologs between pGui2 and Laicp were identified. The majority (8/12) of them are hypothetical proteins, whereas two are transposases, as well as one replication origin protein and one AraC family transcriptional regulator. The number of orthologs found in pGui1 is larger than in pGui2. Eight genes were common to all three plasmids (Supplementary Table S1). The transposase, which belong to the transposase 31 superfamily, is distributed widely in Leptospira species as confirmed by BLAST searches against the NCBI non-redundant protein database. Orthologs of LA1827 contain the PDDEXK domain, which belongs to the nuclease superfamily, and is identified in several Leptospira species. Global sequence comparisons demonstrate that the similarity between pGui1 and Laicp is greater than similarity between pGui2 and Laicp.
Figure 4. Venn diagram detailing the unique and common gene complements among the three plasmids pGui1, pGui2 and Laicp.
Eighteen orthologs in pGui1 and twelve orthologs in pGui2 are common with Laicp. Eight genes are common in the three plasmids. pGui2 possesses the largest number of unique genes (51) among the three plasmids. And 44 genes are unique to pGui1. (Supplementary Tables S1, S2, S3).
Conclusion
In this report, we present two novel plasmids, pGui1 and pGui2, found in one pathogenic L.interrogans strain Gui44. The sequence analysis shows that the two plasmids possess a high percentage of novel genes (56% and 82%, respectively), transposases, integrases and so on which may contribute to the strain's virulence and survival mechanism necessary for persistent infection in different ecological environments and mammalian hosts. Knowledge of the different genomic composition among pathogenic Leptospira may deepen our understanding of the discrepancies in their adaptation and pathogenesis. Thus far, at genome level, there was no other report on the successful isolation and characterization of two circular plasmids from pathogenic Leptospira. We speculate that this is most likely due to high genomic diversity, low copy number, fastidious growth requirements, slow bacterial growth rate in both solid and liquid media and the lack of suitable plasmid extraction methods. Until recently, the absence of available genetic tools has severely impeded our understanding of pathogenic Leptospira species. The demonstrated stability of these two plasmids suggests their potential use for future genetic studies on Leptospira, leading to an improved understanding of virulence and gene dissemination in Leptospira spp.
Supporting Information
Genomic maps of pGui1 and pGui2 plasmids. Functional information for pGui1 and pGui2 is presented (replicon sizes are shown in bp). The outside circle (circle 1) represents the genomic sequence. The arrows (red) identify the replication protein. OriCs were predicted by GC skew method. Inner circles (circle 2 and circle 3) represent predicted protein coding regions (forward and reverse strands, respectively). Functional COG categories are delineated by default color. GC content is depicted in circle 4, and GC skew is depicted in circle 5. GC content deviations from the genomic average were calculated by using a window of 500 bp in steps of 100 bp.
(TIF)
Common genes among pGui1, pGui2 and Laicp.
(XLSX)
Unique genes of pGui1 compare to pGui2 and Laicp.
(XLSX)
Unique genes of pGui2 compare to pGui1 and Laicp.
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files including the sequences of plasmid pGui1 and pGui2 which is available from Genbank under the accession number KF648557 and KF648558.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (81101264, 81171587, 81271793, 81201334, and 81261160321), the National High Technology Research and Development Program of China (2011AA02A119), the Research Fund for the Doctoral Program of Higher Education of China (20120073110071), the research project of Shanghai Municipal Health Bureau (20124162 and 2012Y4083). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Evangelista KV, Coburn J (2010) Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol 5: 1413–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Victoriano AF, Smythe LD, Gloriani-Barzaga N, Cavinta LL, Kasai T, et al. (2009) Leptospirosis in the Asia Pacific region. BMC Infect Dis 9: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, et al. (2003) Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 4. Vijayachari P, Sugunan AP, Shriram AN (2008) Leptospirosis: an emerging global public health problem. J Biosci 33: 557–569. [DOI] [PubMed] [Google Scholar]
- 5. Monahan AM, Miller IS, Nally JE (2009) Leptospirosis: risks during recreational activities. J Appl Microbiol 107: 707–716. [DOI] [PubMed] [Google Scholar]
- 6. Ko AI, Goarant C, Picardeau M (2009) Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Valverde Mde L, Goris MG, Gonzalez V, Anchia ME, Diaz P, et al. (2013) New serovars of Leptospira isolated from patients in Costa Rica: implications for public health. J Med Microbiol 62: 1263–1271. [DOI] [PubMed] [Google Scholar]
- 8. Bourhy P, Salaun L, Lajus A, Medigue C, Boursaux-Eude C, et al. (2007) A genomic island of the pathogen Leptospira interrogans serovar Lai can excise from its chromosome. Infect Immun 75: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, et al. (2003) Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422: 888–893. [DOI] [PubMed] [Google Scholar]
- 10. Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, et al. (2006) Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci U S A 103: 14560–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fraga TR, Chura-Chambi RM, Goncales AP, Morais ZM, Vasconcellos SA, et al. (2010) Refolding of the recombinant protein OmpA70 from Leptospira interrogans from inclusion bodies using high hydrostatic pressure and partial characterization of its immunological properties. J Biotechnol 148: 156–162. [DOI] [PubMed] [Google Scholar]
- 12. Zhong Y, Chang X, Cao XJ, Zhang Y, Zheng H, et al. (2011) Comparative proteogenomic analysis of the Leptospira interrogans virulence-attenuated strain IPAV against the pathogenic strain 56601. Cell Res 21: 1210–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, et al. (2008) Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3: e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bourhy P, Frangeul L, Couve E, Glaser P, Saint Girons I, et al. (2005) Complete nucleotide sequence of the LE1 prophage from the spirochete Leptospira biflexa and characterization of its replication and partition functions. J Bacteriol 187: 3931–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adler B, de la Pena Moctezuma A (2010) Leptospira and leptospirosis. Vet Microbiol 140: 287–296. [DOI] [PubMed] [Google Scholar]
- 16. Adler B, Lo M, Seemann T, Murray GL (2011) Pathogenesis of leptospirosis: the influence of genomics. Vet Microbiol 153: 73–81. [DOI] [PubMed] [Google Scholar]
- 17. Huang L, Zhu W, He P, Zhang Y, Zhuang X, et al. (2014) Re-characterization of an extrachromosomal circular plasmid in the pathogenic Leptospira interrogans serovar Lai strain 56601. Acta Biochim Biophys Sin (Shanghai). [DOI] [PubMed] [Google Scholar]
- 18. Zuerner RL (2005) Laboratory maintenance of pathogenic Leptospira. Curr Protoc Microbiol Chapter 12: Unit 12E 11. [DOI] [PubMed] [Google Scholar]
- 19. Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7: 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Birnboim HC (1983) A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol 100: 243–255. [DOI] [PubMed] [Google Scholar]
- 21. Picardeau M, Brenot A, Saint Girons I (2001) First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol Microbiol 40: 189–199. [DOI] [PubMed] [Google Scholar]
- 22. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Engelberg-Kulka H, Glaser G (1999) Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol 53: 43–70. [DOI] [PubMed] [Google Scholar]
- 25. Li SJ, Zhang CC, Li XW, Tian KC, Tang GP, et al. (2012) Molecular typing of Leptospira interrogans strains isolated from Rattus tanezumi in Guizhou Province, Southwest of China. Biomed Environ Sci 25: 542–548. [DOI] [PubMed] [Google Scholar]
- 26. Picardeau M, Ren S, Saint Girons I (2001) Killing effect and antitoxic activity of the Leptospira interrogans toxin-antitoxin system in Escherichia coli. J Bacteriol 183: 6494–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bignell C, Thomas CM (2001) The bacterial ParA-ParB partitioning proteins. J Biotechnol 91: 1–34. [DOI] [PubMed] [Google Scholar]
- 28. Mierzejewska J, Jagura-Burdzy G (2012) Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid 67: 1–14. [DOI] [PubMed] [Google Scholar]
- 29. Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, et al. (2014) PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42: D581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Christensen SK, Pedersen K, Hansen FG, Gerdes K (2003) Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol 332: 809–819. [DOI] [PubMed] [Google Scholar]
- 31. del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R (1998) Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev 62: 434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arakawa K, Tomita M (2007) The GC skew index: a measure of genomic compositional asymmetry and the degree of replicational selection. Evol Bioinform Online 3: 159–168. [PMC free article] [PubMed] [Google Scholar]
- 33. Lin YP, McDonough SP, Sharma Y, Chang YF (2010) The terminal immunoglobulin-like repeats of LigA and LigB of Leptospira enhance their binding to gelatin binding domain of fibronectin and host cells. PLoS One 5: e11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, et al. (2003) Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol Microbiol 49: 929–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Minet AD, Rubin BP, Tucker RP, Baumgartner S, Chiquet-Ehrismann R (1999) Teneurin-1, a vertebrate homologue of the Drosophila pair-rule gene ten-m, is a neuronal protein with a novel type of heparin-binding domain. J Cell Sci 112 Pt 12: 2019–2032. [DOI] [PubMed] [Google Scholar]
- 36. Kung VL, Khare S, Stehlik C, Bacon EM, Hughes AJ, et al. (2012) An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proc Natl Acad Sci U S A 109: 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic maps of pGui1 and pGui2 plasmids. Functional information for pGui1 and pGui2 is presented (replicon sizes are shown in bp). The outside circle (circle 1) represents the genomic sequence. The arrows (red) identify the replication protein. OriCs were predicted by GC skew method. Inner circles (circle 2 and circle 3) represent predicted protein coding regions (forward and reverse strands, respectively). Functional COG categories are delineated by default color. GC content is depicted in circle 4, and GC skew is depicted in circle 5. GC content deviations from the genomic average were calculated by using a window of 500 bp in steps of 100 bp.
(TIF)
Common genes among pGui1, pGui2 and Laicp.
(XLSX)
Unique genes of pGui1 compare to pGui2 and Laicp.
(XLSX)
Unique genes of pGui2 compare to pGui1 and Laicp.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files including the sequences of plasmid pGui1 and pGui2 which is available from Genbank under the accession number KF648557 and KF648558.