Abstract
HIV sensory neuropathy and distal neuropathic pain (DNP) are common, disabling complications associated with combination antiretroviral therapy (cART). We previously associated iron-regulatory genetic polymorphisms with a reduced risk of HIV sensory neuropathy during more neurotoxic types of cART. We here evaluated the impact of polymorphisms in 19 iron-regulatory genes on DNP in 560 HIV-infected subjects from a prospective, observational study, who underwent neurological examinations to ascertain peripheral neuropathy and structured interviews to ascertain DNP. Genotype-DNP associations were explored by logistic regression and permutation-based analytical methods. Among 559 evaluable subjects, 331 (59%) developed HIV-SN, and 168 (30%) reported DNP. Fifteen polymorphisms in 8 genes (p<0.05) and 5 variants in 4 genes (p<0.01) were nominally associated with DNP: polymorphisms in TF, TFRC, BMP6, ACO1, SLC11A2, and FXN conferred reduced risk (adjusted odds ratios [ORs] ranging from 0.2 to 0.7, all p<0.05); other variants in TF, CP, ACO1, BMP6, and B2M conferred increased risk (ORs ranging from 1.3 to 3.1, all p<0.05). Risks associated with some variants were statistically significant either in black or white subgroups but were consistent in direction. ACO1 rs2026739 remained significantly associated with DNP in whites (permutation p<0.0001) after correction for multiple tests. Several of the same iron-regulatory-gene polymorphisms, including ACO1 rs2026739, were also associated with severity of DNP (all p<0.05). Common polymorphisms in iron-management genes are associated with DNP and with DNP severity in HIV-infected persons receiving cART. Consistent risk estimates across population subgroups and persistence of the ACO1 rs2026739 association after adjustment for multiple testing suggest that genetic variation in iron-regulation and transport modulates susceptibility to DNP.
Introduction
The success of combination antiretroviral therapy (cART) in human immunodeficiency virus (HIV) infection has focused attention on addressing long-term complications that reduce quality of life, such as peripheral neuropathy [1]. Peripheral neuropathy in HIV infection is a distal symmetric, often painful, sensory polyneuropathy associated with the virus itself and/or to toxic effects of certain antiretroviral drugs, particularly the dideoxy-nucleoside reverse-transcriptase inhibitors (so-called “D-drug” NRTIs) such as didanosine and stavudine [2], [3]. Clinically and histopathologically, neuropathy due to HIV infection in the pre-cART era and the neuropathy associated with cART toxicity are usually indistinguishable; both are encompassed by the term “HIV sensory neuropathy” (HIV-SN). HIV-SN may first manifest or worsen upon initiation of cART and is associated with sensory loss, paresthesias, and distal neuropathic pain (DNP) or dysesthesias. The known mitochondrial toxicity of D-drugs and dysregulated inflammation have both been implicated in its pathogenesis, but the pathophysiology of HIV-SN remains incompletely understood [2], [4]–[8].
Development of DNP is particularly disabling [9]–[11], because the symptoms respond poorly to analgesic medications that are used to treat other types of neuropathic pain [1], [12]–[15]. HIV-SN therefore has an adverse impact on quality of life, social and emotional functioning, and neuropsychological test performance, and DNP in particular contributes to depression, physical deconditioning, and non-adherence to treatment [10], [11], [16]–[18]. Although substitution of less toxic antiretroviral drugs lowers the risk of HIV-SN, older D-drugs like stavudine are likely to remain in use in low-resource settings for some time as part of generic, fixed-dose cART regimens, and the study of HIV-SN and DNP remains very relevant [19], [20]. Improved understanding of the biological mechanisms underlying painful HIV-SN may lead to better ways of categorizing and therapeutically targeting DNP [21]–[23].
Iron is a critical micronutrient for metabolically active cells such as neurons, and a carefully controlled supply of iron is essential for mitochondrial function, axonal transport, and myelination [24]–[26]. Iron transport also plays an important role in regulating macrophage-mediated inflammation via the hepcidin pathway [27]–[30]. Hepcidin, a peptide hormone which is synthesized in the liver in response to inflammatory stimuli, leads to iron sequestration within the reticuloendothelial cell (macrophage-monocyte) compartment, potentiating macrophage oxidative killing via Fenton chemistry while reducing availability of iron to micro-organisms. We previously reported associations between iron-loading, single-nucleotide polymorphisms (SNPs) in the hemochromatosis (HFE) gene and reduced risk of HIV-SN in HIV-infected individuals exposed to D-drug-containing cART [31]. While improved cellular and mitochondrial iron delivery might play a protective role in individuals with HFE variants, in whom the action of hepcidin is reduced, these associations could also be explained by the known linkage of HFE to the HLA Class I locus and thereby to other immunomodulatory haplotypes [31], [32]. Studies of non-HLA-linked genetic variants that are prevalent in racially diverse populations, unlike HFE gene variants, are needed to determine whether iron-transport is of fundamental importance in susceptibility to HIV-SN and neuropathic symptoms like DNP.
We hypothesized that like HFE variants, common variants in other iron-management but non-HLA-Class I-linked genes modulate susceptibility to cART-associated HIV-SN and DNP in HIV-infected individuals, and that these genes impact neuropathy phenotypes independent of race/ethnicity and disease-related factors. This study assessed the role of variants in key iron-transport and iron-regulatory genes (henceforth termed “the ferrome” for brevity) in susceptibility to neuropathy phenotypes in a racially diverse, HIV-infected population, the CNS HIV Antiretroviral Therapy Effects Research (CHARTER) Cohort. CHARTER is the largest U.S.-based prospective study of neurological complications in HIV/AIDS in the cART era [33]. Particular strengths of CHARTER include its clear definition of neurological phenotypes and sensitive and specific methods of ascertaining HIV-SN. This analysis specifically targeted genes that encode components of the nuclear and mitochondrial ferrome, for which SNP genotypes were available from a genomic study conducted previously in CHARTER. In addition to several SNPs that were nominally associated with DNP, a common variant (rs2026739) in the cytoplasmic aconitase gene (ACO1) was identified to be statistically associated with DNP after multiple-testing correction.
Patients and Methods
Study population
The CHARTER Study is a prospective, observational study conducted at six outpatient centers within the U.S.: Johns Hopkins University, Baltimore, Maryland; Mount Sinai School of Medicine, New York, NY; the University of California, San Diego, CA; the University of Texas Medical Branch, Galveston, TX; the University of Washington, Seattle, WA; and Washington University, St Louis, MO. Data collection was standardized across centers and employed a protocol of comprehensive neuromedical, neurobehavioral, and laboratory assessments, including: nadir CD4+ T-cell count, cART history, D-drug exposure, history of a major depressive disorder, and demographic information (from structured interviews), as well as current CD4+ T-cell count, HIV plasma viral load, and hepatitis C viral serology (from laboratory data). Further details regarding CHARTER Study eligibility and enrollment procedures have been reported previously [1]. A subset of CHARTER Study subjects (N = 579) was recruited between 2003 and 2007 to a cross-sectional genomic sub-study of peripheral neuropathy. Genetic study eligibility criteria included the ability to provide details of cART use and to undergo a structured interview and examination for signs/symptoms of HIV-SN; individuals with active opportunistic infections, uncontrolled psychiatric disorders, or who were unable to cooperate with detailed evaluations over the course of a full day were excluded.
Ethics Statement
The CHARTER Study and this study were approved by the University of California-San Diego Human Research Protections Program (Biomedical Institutional Review Board) and by the Institutional Review Boards (IRBs) of the following participating CHARTER institutions: the Johns Hopkins University School of Medicine IRB, Icahn School of Medicine at Mount Sinai IRB, University of Texas (Medical Branch) IRB, University of Washington School of Medicine IRB, and Washington University -St. Louis School of Medicine IRB. The present study was conducted according to the principles expressed in the Declaration of Helsinki. All study subjects whose data are reported herein provided written informed consent.
Ascertainment of neuropathy and distal neuropathic pain
Standardized, targeted neurological examinations were performed by physicians and nurses trained in neurological AIDS disorders. HIV-SN and DNP were identified using criteria published previously [1], [10]. Briefly, participants were queried at baseline and each follow-up visit about symptoms of HIV-SN and DNP, specifically, tingling, or burning, aching, shooting, or stabbing pain in a bilateral, predominantly distal distribution (e.g., in the fingers and toes only, extending to the ankles or wrists, or extending to the knees or elbows). If subjects endorsed tingling in this distribution, they were considered to have paresthesias; if they endorsed any of the pain symptoms in this distribution, they were considered to have a positive self-report of DNP; loss of sensation in the lower extremities was also assessed on examination. HIV-SN cases were defined by the bilateral presence of at least one of the following signs at any visit: diminished vibration sense, reduced sharp-dull discrimination in the feet and toes, reduced ankle reflexes, and DNP cases by the above-mentioned pain symptoms; controls did not have any of these signs or symptoms of peripheral nerve disease. DNP was also classified into the following 5 severity levels by structured survey; none, slight (occasional, fleeting), mild (frequent), moderate (frequent, disabling) or severe (constant, daily, disabling, and requiring treatment with analgesics or other medications).
Genomic DNA isolation and gene selection
Genomic DNA was isolated from whole blood samples using PUREGENE (Gentra Systems Inc., Minneapolis, MN, USA). Samples were subjected to whole-genome nuclear genotyping using the Affymetrix Genome-Wide Human SNP Array 6.0 platform (Affymetrix, Inc., Santa Clara, CA). Among 579 extracted samples, 560 yielded analyzable genotypes that passed quality-control filters using the Platform for the Analysis, Translation, and Organization of large scale data (PLATO) [34]. Sample genotyping efficiency was 95%. Variants with less than 95% genotyping efficiency or minor allele frequencies (MAFs) less than 1% in the study population were excluded from analysis.
Genes reported in the published literature to be associated with iron metabolism and/or neurological phenotypes were searched in the publicly available Online Mendelian Inheritance in Man database (http://www.ncbi.nlm.nih.gov/omim). We then identified 20 genes based on their well-recognized direct or indirect involvement in iron metabolism, transport, storage, or regulation (e.g., the hepcidin pathway). SNPs mapping to the following iron-regulatory and transport genes were selected for this exploratory analysis based on their inclusion in the Affymetrix Genome-Wide Human SNP Array 6.0: HFE, HFE2, SLC40A1, SLC11A1, HAMP, TF, TFRC, TFR2, BMP2, BMP6, CP, SLC11A2, FXN, FTMT, FTH1, ACO1, ACO2, B2M and ATP13A2. Nineteen of these genes were covered by the Affymetrix 6.0 chip, and the platform provided evaluable genotypic information for 192 candidate SNPs (Table S1). Many of these iron-related genes/SNPs have also been previously reported to play a role in iron transport within the nervous system, and genes previously linked to neurodegenerative diseases and neural differentiation were included [35]. Based on the common disease-common variant hypothesis, this list was further narrowed to those SNPs with MAFs of at least 5% in one or both of the largest subsets of the CHARTER study population (non-Hispanic blacks or non-Hispanic whites, henceforth referred to as blacks or whites, respectively) [36]. Using a typical analysis strategy that has been applied in numerous previous studies, we used all available genotyped SNPs in those genes, though some of them may be correlated due to LD structure.
Statistical methods
General approach
The role of iron-related genes in susceptibility to cART-associated HIV-SN and DNP was evaluated by both multivariable logistic regression and permutation analysis. Since the SNPs analyzed could not be assumed to be independent, Bonferroni adjustment of statistical tests would almost certainly overcorrect and result in unacceptable Type II error; permutation-based analysis is a complementary statistical method that accounts for multiple statistical tests, gene size, and linkage disequilibrium [37], [38]. Specifically, the observed p-value from the real dataset is compared to the distribution of p-values generated from 1000 permutated sample sets which randomly switch case/control labels while maintaining the same case/control ratio [37]. An empirical p-value is generated by the number of permutation sets whose p-values are less than the observed p-value divided by 1000.
Population stratification was also evaluated in our analyses by adjustment for four principal components variables (PCs) representing ancestry-related genetic information, which were generated from genome-wide genotype data in CHARTER and used as covariates in multivariate logistic regression models, in addition to adjustment for self-reported race/ethnicity. Logistic regression models stratified by self-reported race/ethnicity were also explored (black or white; other categories were omitted due to insufficient numbers). The degree of potential overlap between individual PCs was also evaluated by plotting these variables against one another (Figure S1).
The following neuropathy outcomes were analyzed: presence of at least 1 sign of HIV-SN, at least 2 signs of HIV-SN, and DNP. Each outcome was analyzed as a dichotomous variable. Potential confounders were identified by univariable logistic regression to calculate unadjusted odds ratios (ORs) and by prior published associations of these variables with HIV-SN, neuropathy symptoms (loss of sensation, paresthesias, and/or dysesthesias/DNP), or DNP alone in CHARTER.[1], [9], [33]. Significant variables were then included in all multivariable logistic models with or without race-stratification. These phenotype-specific covariates were as follows: age (years), cumulative D-drug exposure (months), CD4+ T-cell nadir, log10[plasma HIV RNA], hepatitis C virus (HCV) serologic status (positive/negative), protease inhibitor use (0 or ≥1 protease inhibitor), current or prior major depressive disorder (yes/no), self-reported race (in non-stratified models), and the 4 genomic sequence-derived ancestry PC variables in HIV-SN analyses; and age, cumulative D-drug exposure, CD4+ T-cell nadir, log10[plasma HIV RNA], self-reported race (in non-stratified analyses), and the 4 PC variables in analyses of neuropathy symptoms and DNP. In order to account for the effects of diabetes in some subjects who might also be on treatment for this condition, we used a more stringent blood glucose criterion for ascertaining diabetes than is normally used for non-fasting blood glucose levels (≥126 mg/dL, rather than ≥200 mg/dL). By this criterion, 66 subjects (11.8%) were coded as diabetic. Since additional adjustment for diabetes by this method did not alter p-values appreciably for any of the studied SNPs, we did not include this covariate in final regression models. (Hemoglobin A1c levels, which would have allowed for more accurate diabetes diagnosis, were available in <10% subjects, and blood glucose values were missing in 6 subjects.)
The reported associations of genetic variants with presence or absence of HIV-SN or DNP made no genetic model assumptions regarding dominant or recessive allelic effects (genotypes were coded as 0, 1, or 2, based on the number of minor alleles present); all SNPs in the genes studied were bi-allelic.
For analysis of DNP severity, genotypes were dichotomized (variant alleles present or absent) to optimize power, and p-values were obtained using Pearson's chi-square test or, for those SNPs with fewer than 5 observations per cell, Fisher's exact test.
Multivariable single-SNP analyses
Statistical tests were performed using publicly available R software (http://www.r-project.org/). For each phenotype, logistic regression was used for single-marker association tests, while adjusting for phenotype-specific covariates in addition to race as a categorical covariate and ancestry PCs, as discussed above [1], [9]. Additional adjustment for cART-naïve status beyond inclusion of D-drug exposure and HIV viral load did not significantly alter results; hence, this variable was not included in multivariable models. PC variables were computed in advance based on genome-wide genotype data available in all 560 CHARTER study participants. Permutation tests were conducted by randomizing case/control labels in multivariable models while keeping the same numbers of cases and controls as in the original dataset. We generated 1000 permutation datasets. An empirical p-value was computed for each SNP in each phenotype, according to p emp = #{P(π) <P(real)/1000, where P(π) is the p-value in the πth permutation [38], [39].
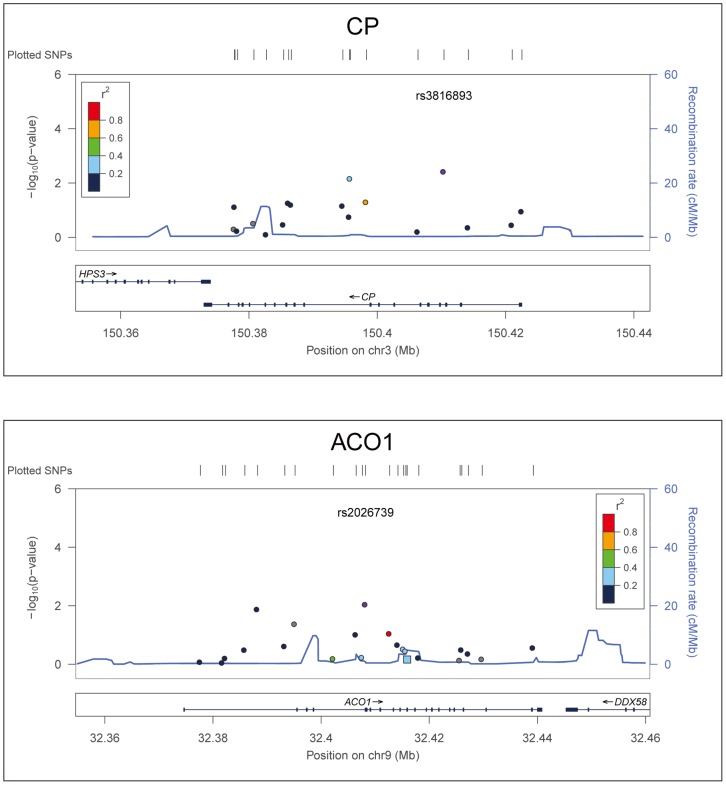
Results of association tests for two key genes, CP and ACO1, with neuropathic pain were depicted using the LocusZoom tool ( Figure 1 , generated using HapMap Phase II CEU) [40].
Figure 1. Representative Linkage Disequilibrium (LD) plot.
The LD plot for two representative SNPs that were statistically significantly associated with neuropathic pain in the CHARTER cohort is displayed: CP rs3816893 and ACO1 rs2026739. Each plot shows the degree of LD between the SNP of interest (rs number) and all other SNPs analyzed in the same gene, color-coded according to the r2 value (correlation of frequencies in this sample). The LD is based on HapMap Phase II CEU data. Association p-values are displayed on the y-axis and chromosomal distance (Mb) on the x-axis.
The p-values for all statistical tests were two-tailed, and for this exploratory analysis, the value of α (statistical significance) was set at 0.05 to identify SNPs of potential interest in this exploratory study. A Bonferroni correction for multiple statistical tests was also applied by multiplying p-values obtained for each association test by 192, the number of SNPs evaluated to identify more robust associations.
Results
Genotypes from the CHARTER genome study were available for 192 SNPs encompassing 19 iron-regulatory or iron-transport-related (ferrome) genes queried (Table S1): HFE, HJV, FPN1, SLC11A1, HAMP, TF, TFRC, TFR2, BMP2, BMP6, CP, SLC11A2, FXN, FTMT, FTH1, ACO1, ACO2, B2M, and ATP13A2. Genotypes at the HEPH locus on the X chromosome, which encodes a ceruloplasmin-like, membrane-bound ferroxidase that is important in transmembrane iron transport, and genotypes at the HFE C187G locus (previously studied by the authors in HIV-infected subjects) were not covered by the Affymetrix Human SNP Array 6.0 platform. The CHARTER genomic study provided genotypes at all other loci of interest in >99% of subjects; of 559 evaluable individuals, 21% were women, and the mean age of the population was 44 years (range 21–68). Self-reported race/ethnicity was 43% (n = 242) black, 44% (n = 247) white, 11% (n = 58) Hispanic, and 2% Asian or “Other” (n = 11). Due to insufficient power for analyses in subjects of Hispanic, Asian or “Other” ancestry, only analyses performed in non-Hispanic blacks, whites, or the entire CHARTER genetic study population are reported. As shown in Figure S1, the 4 genome-derived PC variables in multivariable models of neuropathic pain accurately represented distinct ancestral strata in this study sample.
Demographic and HIV disease characteristics of subjects with and without HIV-SN are presented in Table 1 . Among 559 evaluable subjects, 331 (59%) had at least one sign of HIV-SN, and 160 (29%) exhibited more severe HIV-SN (at least two signs). Fifty percent of the study population (281 subjects) reported at least one neuropathic symptom, including paresthesias, loss of sensation, dysesthesias, and/or DNP in both lower extremities. Ascertainment of dysesthesias correlated so closely with DNP that only results for DNP will be discussed henceforth. A total of 168 subjects (30% of this study sample) experienced DNP of some degree of severity. CHARTER study subjects with HIV-SN (at least one sign) and those who reported neuropathic symptoms and/or DNP were older than subjects without these complications (p<0.01). No significant differences were noted with regard to educational level (data not shown), self-reported race/ethnicity, or sex, but subjects reporting DNP tended to be female (26% vs. 19%, p = 0.06). Individuals with HIV-SN of any severity and those who reported neuropathy symptoms other than DNP had a lower CD4 nadir than corresponding controls [median (IQR) for cases vs. controls, respectively, were 112 (31, 246) vs. 242 (125, 391) cells/µL, p<0.01 for at least one HIV-SN sign; 107 (23, 214) vs. 200 (70, 350), p<0.01 for at least two signs; and 151 (37, 275) vs. 198 (60, 348), p<0.01 for neuropathy symptoms]. Median viral load was lower among HIV-SN cases than controls [median (IQR) 1.7 (1.7, 3.3) vs. 2.6 (1.7, 4.2) log10 (HIV RNA copies/mL), respectively, p<0.01] but not statistically different between subjects with or without neuropathy symptoms and DNP. Substantially fewer individuals with HIV-SN and DNP were cART-naïve as compared to controls (6% of cases with at least one sign of HIV-SN vs. 29% of controls; 8% of DNP cases vs. 19% of controls, p<0.01 for both). Cumulative D-drug exposure was also higher among cases than controls in all outcome categories, including DNP [e.g., median (IQR) 14 (0, 53) vs. 0 (0, 21) months for DNP]; all p-values<0.01]. HIV-SN of any severity was associated with the use of protease inhibitors (53% current use in subjects with at least one sign vs. 31% of controls, p<0.01. Co-infection with hepatitis C virus (HCV) was more common among HIV-SN cases (28% of cases with at least one sign vs. 19% of controls; 31% of cases with at least two signs vs. 22% of controls, p<0.05 for both). History of a major depressive disorder was more frequently reported by subjects without HIV-SN (46%) than among HIV-SN cases (36%, p<0.05) and was slightly more common among individuals with DNP.
Table 1. Characteristics of neuropathy cases and controls in the CHARTER genetic study population.
| Baseline Characteristic | HIV-SN, ≥1 sign | p-value* | HIV-SN, ≥2 signs | p-value* | Neuropathy Symptoms† | p- value* | Neuropathic Pain | p-value* | ||||
| Cases N = 331 | Controls N = 228 | Cases N = 160 | Controls N = 399 | Cases N = 281 | Controls N = 274 | Cases N = 168 | Controls N = 390 | |||||
| Age, mean±SD | 46±8 | 40±8 | <0.01 | 42±8 | 47±8 | <0.01 | 45±8 | 42±8 | <0.01 | 46±8 | 43±9 | <0.01 |
| Race/ethnicity, % | 0.10 | 0.80 | 0.23 | 0..30 | ||||||||
| Black | 46 | 40 | 43 | 43 | 40 | 47 | 38 | 46 | ||||
| White | 44 | 44 | 46 | 44 | 48 | 40 | 49 | 42 | ||||
| Hispanic | 9 | 13 | 10 | 11 | 10 | 11 | 11 | 10 | ||||
| Other | 1 | 3 | 1 | 2 | 2 | 2 | 2 | 2 | ||||
| Sex, % women | 19 | 23 | 0.23 | 18 | 22 | 0.23 | 23 | 18 | 0.13 | 26 | 19 | 0.06 |
| CD4+ T-cell nadir,cells/uL, median(IQR) | 112 (31, 246) | 242 (125, 391) | <0.01 | 107 (23, 214) | 200 (70, 350) | <0.01 | 151 (37, 275) | 198 (60, 348) | <0.01 | 139 (56, 267) | 187 (54, 325) | 0.09 |
| Log10 [HIV RNA copies/mL], median (IQR) | 1.7 (1.7, 3.3) | 2.6 (1.7, 4.2) | <0.01 | 1.7 (1.7, 3.0) | 2.3 (1.7, 4.1) | <0.01 | 1.7 (1.7, 3.4) | 2.2 (1.7, 4.0) | 0.09 | 1.7 (1.7, 3.6) | 2.0 (1.7, 3.9) | 0.31 |
| cART-naïve, % | 6 | 29 | <0.01 | 3 | 20 | <0.01 | 11 | 20 | <0.01 | 8 | 19 | <0.01 |
| Current PI use, % yes | 53 | 31 | <0.01 | 56 | 40 | <0.01 | 48 | 40 | 0.14 | 46 | 44 | 0.49 |
| Cumulative D-drug use, median mos.,(IQR) | 14 (0, 53) | 0 (0, 21) | <0.01 | 17 (0, 48) | 0 (0, 36) | <0.01 | 11 (0, 41) | 0 (0, 43) | 0.01 | 12 (0, 48) | 0 (0, 36) | <0.01 |
| HCV positive, % | 28 | 19 | <0.05 | 31 | 22 | <0.05 | 24 | 25 | 0.73 | 24 | 25 | 0.78 |
| Current/prior major depression, % | 36 | 46 | <0.05 | 32 | 43 | <0.05 | 42 | 39 | 0.43 | 43 | 39 | 0.39 |
Abbreviations: SD, standard deviation; HIV-SN, HIV sensory neuropathy; cART, combination antiretroviral therapy; PI, protease inhibitor; HCV, hepatitis C virus; mos., months; IQR, interquartile range.
Neuropathy symptoms included bilateral paresthesias, dysesthesias/neuropathic pain, and/or loss of sensation in the lower extremities.
*p-values<0.05 are statistically significant.
Results of multivariable logistic regression and permutation analyses evaluating associations of selected SNPs with HIV-SN phenotypes, including DNP, are summarized in Table 2 , adjusted for potential confounders, including self-reported race and 4 PC variables that captured racial admixture. Twelve SNPs in 8 iron-related genes were associated with at least one sign of HIV-SN (p<0.05) by permutation analyses, as compared with 13 SNPs in the same 8 genes analyzed by logistic regression. One variant (rs1049296 in the gene TF) was also associated with HIV-SN with a p-value<0.01. One variant in the HFE gene was also associated with reduced risk of HIV-SN in this population, replicating a prior association of this gene with reduced risk of HIV-SN [31]. More severe HIV-SN was associated with 10 SNPs in 7 iron-related genes (p<0.05) and with 4 SNPs in 4 genes (p<0.01) by permutation analysis. Fourteen SNPs in 8 iron-regulatory genes were significantly associated with DNP (p<0.05) by logistic regression and 15 SNPs by permutation analysis, including two variants (rs3816893 in the gene CP and rs2026739 in the gene ACO1). These SNPs were also associated with more severe HIV-SN (at least two signs, data not shown). Eighteen SNPs in 6 genes were associated in permutation analysis with the presence of one or a combination of uncomfortable neuropathy symptoms (paresthesias, loss of sensation, or dysesthesias/DNP, p<0.05). Four variants in 2 of these genes, including CP rs3816893 and CP rs13072552, were strongly associated with neuropathy symptoms by logistic regression (all p-values<0.01) as well as by permutation analysis (p<0.0001 for both SNPs mentioned), thereby also meeting criteria for significance after correction for multiple testing.
Table 2. Summary of significant results from multivariable analyses of peripheral neuropathy in CHARTER subjects.
| Neuropathy Phenotype | p-values from Logistic regression* | Empirical p-values from Permutation* | ||
| No. of Significant genes (SNPs) | ||||
| p<0.05 | p<0.01 | pemp<0.05 | pemp<0.01 | |
| HIV-SN, ≥1 sign | 8 (13) | 1 (1) | 8 (12) | (0) |
| HIV-SN, ≥2 signs | 7 (12) | 4 (4) | 7 (10) | 4 (4) |
| Neuropathic pain | 8 (14) | 4 (5) | 8 (15) | 4 (5) |
| ≥1 Neuropathy symptom(s)¥ | 7 (18) | 2 (4) | 6 (18) | 2 (4) |
*Models shown were adjusted for the following covariates: age, cumulative D-drug exposure, CD4+ T-cell nadir, plasma HIV RNA concentration, self-reported race, protease inhibitor use, HCV status, history of major depression, and 4 ancestry by principal components (for HIV-SN); first five of the same factors plus principal components (for neuropathic pain or neuropathy symptoms).
Includes presence of paresthesias, loss of sensation, and/or dysesthesias/neuropathic pain.
Analyses were performed with no genetic model assumptions (genotypes at each locus were coded as 0, 1, or 2 minor alleles).
Abbreviations: HIV-SN, HIV-sensory neuropathy.
A summary of iron-regulatory gene variants significantly associated in multivariable-adjusted analyses with DNP in CHARTER subjects stratified by self-reported race, with estimated ORs and 95% confidence intervals (CIs), is presented in Table 3 . Eight iron-regulatory or iron-transport-related genes, TF, CP, TFRC, BMP6, ACO1, FXN, SLC11A2, and B2M were associated with DNP. Five SNPs in 4 iron regulatory genes, TF rs2718796, CP rs13072552 and rs3816893, ACO1 rs2026739, and B2M rs16966334, were associated with increased risk in the combined CHARTER population by both logistic regression and permutation analysis in the entire population, with ORs ranging from 1.5 to 3.1 (all p<0.01). Race-stratified analyses revealed, however, that statistically significant associations for some of these SNPs occurred primarily within black or white subgroups. Point estimates for the associations of CP rs3816893 and B2M rs16966334 with DNP were greater in blacks than in whites and statistically significant only in blacks [OR 2.6 (95% CI 1.3–5.0) and 2.8 (95% CI 1.3–6.0), respectively, both p<0.01], while those for ACO1 rs2026739 and B2M rs1901531 were statistically significant among whites only [OR 2.3 (95% CI 1.4–3.7, p<0.0001) and OR 2.2 (95% CI 1.3–3.8, p<0.01), respectively]. With the exception of B2M rs1901531, point estimates for ORs associating SNPs with DNP in this population were consistent in direction and very similar in magnitude across race-stratified analyses (black and white subgroups). Several additional SNPs were nominally associated with DNP by logistic regression as well as permutation-based methods of analysis, including FXN rs3793451 (all p<0.05). Two variants, TF rs8177306 and ACO1 rs4495514, could not be analyzed in whites due to insufficient numbers of variant alleles.
Table 3. Genetic variants associated with neuropathic pain.
| SNP | Gene | All Subjects (390 Cases/168 Controls) | White (163 Cases/83 Controls) | Black (179 Cases/63 Controls) | ||||||
| OR (95% CI) | p | p emp | OR (95% CI) | p | p emp | OR (95% CI) | p | p emp | ||
| rs2718796_g | TF | 3.1 (1.4–7.3) | 0.007 | 0.003 | 5.0 (1.1–28.1) | 0.045 | 0.026 | 3.0 (0.7–12.3) | 0.118 | 0.055 |
| rs8177306_g | TF | 0.4 (0.2–0.9) | 0.023 | 0.017 | – | – | – | 0.5 (0.2–1.0) | 0.060 | 0.059 |
| rs13072552_t | CP | 1.6 (1.1–2.4) | 0.007 | 0.008 | 1.5 (0.7–3.1) | 0.292 | 0.280 | 1.5 (1.0–2.4) | 0.054 | 0.071 |
| rs13075921_c | CP | 1.6 (1.0–2.4) | 0.048 | 0.035 | 1.4 (0.7–2.8) | 0.343 | 0.358 | 2.1 (1.0–4.0) | 0.037 | 0.049 |
| rs3816893_t | CP | 1.9 (1.2–3.0) | 0.004 | 0.002 | 1.6 (0.8–3.3) | 0.165 | 0.151 | 2.6 (1.3–5.0) | 0.005 | 0.006 |
| rs480760_t | TFRC | 0.6 (0.4–0.9) | 0.042 | 0.042 | 0.8 (0.2–2.1) | 0.660 | 0.678 | 0.7 (0.0–1.1) | 0.098 | 0.094 |
| rs270388_t | BMP6 | 1.3 (1.0–1.8) | 0.057 | 0.049 | 1.2 (0.7–2.2) | 0.430 | 0.448 | 1.2 (0.2–1.9) | 0.323 | 0.314 |
| rs267202_a | BMP6 | 0.8 (0.6–1.0) | 0.050 | 0.066 | 0.8 (0.5–1.3) | 0.414 | 0.424 | 0.7 (0.5–1.1) | 0.162 | 0.182 |
| rs267206_c | BMP6 | 1.4 (1.0–2.0) | 0.029 | 0.034 | 1.5 (0.9–2.6) | 0.098 | 0.118 | 1.4 (0.9–2.2) | 0.173 | 0.196 |
| rs7033149_g | ACO1 | 1.6 (1.1–2.4) | 0.013 | 0.012 | 1.7 (0.9–3.2) | 0.118 | 0.128 | 1.7 (1.0–2.9) | 0.044 | 0.046 |
| rs4495514_t | ACO1 | 0.4 (0.2–0.9) | 0.040 | 0.036 | – | – | – | 0.5 (0.2–1.1) | 0.118 | 0.112 |
| rs2026739_g | ACO1 | 1.5 (1.1–2.0) | 0.009 | 0.007 | 2.3 (1.4–3.7) | 0.001 | <0.0001 | 1.3 (0.8–2.1) | 0.233 | 0.228 |
| rs3793451_t | FXN | 0.4 (0.2–0.9) | 0.046 | 0.047 | 0.3 (0.1–0.9) | 0.047 | 0.054 | 0.6 (0.1–2.1) | 0.513 | 0.526 |
| rs224446_t | SLC11A2 | 0.7 (0.4–1.0) | 0.047 | 0.047 | 0.7 (0.4–1.2) | 0.222 | 0.233 | 0.9 (0.4–2.0) | 0.899 | 0.897 |
| rs16966334_g | B2M | 2.4 (1.3–4.2) | 0.003 | 0.006 | 1.3 (0.3–4.7) | 0.708 | 0.726 | 2.8 (1.3–6.0) | 0.008 | 0.004 |
| rs1901531_c | B2M | 1.6 (1.1–2.5) | 0.028 | 0.023 | 2.2 (1.3–3.8) | 0.004 | 0.006 | 0.5 (0.1–1.3) | 0.262 | 0.265 |
The p-values (p) shown were obtained by multivariable logistic regression, adjusting for age, total D-drug exposure, CD4+ T-cell nadir, plasma HIV RNA concentration, all 4 principal component ancestry variables, and self-reported race (if not race-stratified) and by permutation analysis (empiric p-value, p emp). Abbreviations: OR, odds ratio; 95% CI, 95% Confidence Interval; TF, transferrin; CP, ceruloplasmin; TFRC, transferrin receptor 1; BMP6, bone morphogenetic protein 6; ACO1, cytoplasmic aconitase; SLC11A2, divalent metal transporter 1; B2M, beta-2 microglobulin.
The strong association of SNP rs2026739 in ACO1 with DNP in whites by both logistic regression and permutation analysis remained statistically significant after adjustment for multiple testing (pemp<0.0001). Notably, no linkage disequilibrium was observed between statistically significant SNPs and other SNPs within the same genes, as exemplified in the LD maps for CP and ACO1 (SNPs rs3816893 and rs2026739), shown in Figure 1 .
The prevalence of ferromic variants was compared across three categories of severity of DNP, which was graded as none (0); slight or mild (1–2); and moderate to severe (3–4) in race-stratified analyses ( Table 4 ). Only associations with p-values≤0.05 are presented. Polymorphisms in ACO1 (rs2026739), B2M (rs16966334 and rs1901531), CP (rs3816893), TF (rs2718796 and rs8177306), and TFRC rs480760, most of which are very common among both blacks and whites, showed statistically significant associations with DNP severity in one or both subgroups as well as in the entire study population.
Table 4. Iron-transport-related genes associated with severity of distal neuropathic pain in CHARTER.
| Gene/SNP | Neuropathic Pain Severity (n,%)1 | All subjects N = 558 | Blacks N = 242 | Whites N = 247 | Allele Frequency3 Overall (B/W) | ||
| None | Slight/Mild | Mod/Severe | p-value for trend2 | ||||
| ACO1 | |||||||
| rs2026739_g | 209 (54) | 76 (66) | 38 (73) | 0.013 | 0.148 | 0.002 | 0.46 (0.54/0.36) |
| rs7033149_g | 133 (34) | 52 (44) | 24 (46) | 0.044 | 0.089 | 0.222 | 0.20 (0.30/0.14) |
| rs4495514_t | 39 (10) | 3 (3) | 4 (8) | 0.027 | 0.111 | ---4 | 0.05 (0.11/---) |
| B2M | |||||||
| rs16966334_g | 37 (10) | 20 (17) | 7 (13) | 0.027 | 0.009 | 0.618 | 0.06 (0.08/0.02) |
| rs1901531_c | 71 (18) | 29 (25) | 16 (31) | 0.009 | 0.316 | 0.005 | 0.12 (0.04/0.21) |
| BMP6 | |||||||
| rs267202_a | 258 (67) | 68 (59) | 24 (47) | 0.008 | 0.078 | 0.206 | 0.57 (0.57/0.50) |
| rs376308_g | 243 (63) | 59 (51) | 23 (44) | 0.014 | 0.203 | 0.315 | 0.49 (0.58/0.35) |
| CP | |||||||
| rs9853335_c | 38 (10) | 16 (15) | 10 (20) | 0.034 | 0.758 | 0.124 | 0.06 (0.02/0.11) |
| rs3816893_t | 65 (17) | 32 (27) | 10 (19) | 0.024 | 0.011 | 0.210 | 0.10 (0.12/0.10) |
| FXN | |||||||
| rs3793451_t | 46 (12) | 7 (6) | 4 (8) | 0.051 | 0.289 | 0.094 | 0.06 (0.04/0.04) |
| TF | |||||||
| rs2718796_g | 14 (3) | 5 (4) | 9 (17) | 0.004 | 0.154 | 0.009 | 0.03 (0.02/0.02) |
| rs8177306_g | 54 (14) | 7 (6) | 4 (8) | 0.009 | 0.026 | ---4 | 0.08 (0.18/---) |
| TFRC | |||||||
| rs480760_t | 110 (28) | 22 (19) | 8 (15) | 0.005 | 0.038 | 0.476 | 0.19 (0.37/0.05) |
Numbers of subjects shown are for entire study population, including 10.4% non-Hispanic Black and 2.1% “Other” self-reported race/ethnicity.
P-values presented were obtained using a non-parametric test for trend.
Allele frequency in entire CHARTER study population.
Allele not present in whites.
Abbreviations: Mod/Sev, moderate to severe neuropathic pain; B/W, Non-Hispanic Blacks/Whites; ACO1, cytoplasmic aconitase (iron-regulatory protein 1); B2M, beta-2 microglobulin; BMP6, bone morphogenetic protein-6; CP, ceruloplasmin; TF, transferrin; TFRC, transferrin receptor 1.
Discussion
This represents the first study to associate key iron-regulatory and iron-transport-pathway genes, which make up the human ferrome and are critical for maintenance of neuronal metabolism and mitochondrial function, with painful neuropathy in HIV-infected subjects. Our results particularly support the concept that iron metabolism plays a role in the pathophysiology of DNP for the following reasons: 1) the same genomic variants showed associations with DNP by conventional regression and by permutation-based analytical methods, which account for the effects of multiple statistical tests, gene size, and linkage disequilibrium; 2) thirteen of the same SNPs (in 7 genes) were also associated with the severity of DNP in this population; and 3) point estimates for ORs were consistent in direction and very similar in magnitude in both blacks and whites, as might be anticipated for valid, fundamental biological associations. Furthermore, ACO1 SNP rs2026739 retained its statistically significant association with DNP after adjustment for multiple testing. Our observation that associations of certain SNPs (e.g., in CP, ACO1, TF, and B2M genes) with DNP and with DNP severity predominate in either blacks or whites may in part explain reported population differences with regard to pain threshold and pain-related disability, albeit in non-HIV-infected populations [41], [42]. Interaction with race-specific genomic and lifestyle factors within these population subgroups is also possible. With the exception of BMP6, none of the genes we associated with DNP occurs on chromosome 6p, indicating that genomic variation in iron metabolism and transport per se, not linkage disequilibrium with HLA Class I-linked haplotypes that modulate inflammation, is likely to be responsible for the observed associations.
Several lines of evidence support a role for iron metabolism in the development of HIV-SN and/or DNP, which remain significant problems even in the era of newer-generation cART [43], [44]. Common variants in the iron-loading HFE gene were associated with reduced risk of HIV-SN during mitochondrial-toxic cART regimens in AIDS Clinical Trial Group Study 384, and the protective association with HFE was replicated in this study [31]. HIV infection itself induces cellular iron dysregulation, possibly via downregulation of macrophage Hfe expression by the viral Nef protein and induction of the pro-inflammatory, iron-regulatory hormone hepcidin [45]–[47]. Restless legs syndrome (RLS), a disorder characterized by unpleasant sensations in the lower extremities that are relieved by movement, has been reported in some studies to be more prevalent in HIV-infected individuals with painful neuropathy and has been likened to a unique form of neuropathic pain [48], [49]. Systemic iron deficiency, brain iron deficiency, and activation of the hypoxia-inducible factor pathway have been linked to the development of RLS, and in a significant subset of patients, iron supplementation alleviates symptoms [50]–[52]. The biological plausibility of a role for iron metabolism in the pathophysiology of inflammatory peripheral neuropathy and DNP is also suggested by prior experimental studies and growing appreciation within the field of iron metabolism of the importance of iron transport and its tight regulation to maintenance of energy metabolism in highly metabolically active cells such as neurons, and to immune regulation [27], [47], [53]–[57]. Iron transport is critical to neuronal energy metabolism and axoplasmic transport [58]–[60]. Inhibitors of heme oxygenase, which catalyzes the breakdown of heme to iron, biliverdin and carbon monoxide, have been shown to have analgesic effects in some models of inflammatory or neuropathic pain [61]. Genetic variation in iron transport has also been associated with altered inflammatory cytokine levels, which at least in part mediate neuropathic pain [62]–[66]. Involvement of bone morphogenetic proteins in pain perception and sensitivity has also been reported in rodent models [67].
The ACO1 and FXN genes are important for cellular energy metabolism and mitochondrial iron transport: ACO1 encodes a bifunctional protein containing an iron-sulfur cluster that reversibly binds to regulatory elements in transferrin receptor (Tfr) and ferritin mRNAs depending on ambient cellular iron levels [55]. This protein functions as a cytoplasmic aconitase under low-iron conditions, upregulating synthesis of Tfr (the major cellular iron importer) by stabilizing Tfr mRNA, while preventing translation of ferritin (the principal iron storage protein in the cell); the reverse occurs when ambient iron levels are high [68]. The FXN gene, which is mutated in Friedreich's ataxia, regulates mitochondrial iron utilization and export; a defect in the encoded frataxin molecule in this disease results in progressive mitochondrial iron accumulation and oxidative injury in a variety of tissues with high metabolic demand, including the heart and central nervous system [69]–[71]. Beta-2 microglobulin is a ubiquitous iron- and Hfe-stabilizing protein which also promotes nerve repair following injury, and it is a marker of disease progression in a variety of diseases, but it has not previously been associated with DNP [72]. The CP rs3816893 and rs1302552 SNPs, which were associated with DNP alone in this population, also showed strong associations with the presence of neuropathy symptoms in general, which included paresthesias and loss of sensation. These SNP associations also met multiple-testing criteria for significance after permutation analysis, lending support to a role for CP in DNP. Ceruloplasmin is a ferroxidase of critical importance to neuronal copper and iron regulation and in neuronal protection against iron-mediated oxidative injury in the central and peripheral nervous systems [73]. The non-synonymous CP SNP rs13072552 has been associated in genome-wide association studies with higher serum ceruloplasmin levels [74]. While serum ceruloplasmin is an acute-phase reactant (levels rise during acute inflammation), serum ceruloplasmin levels may also be a marker of oxidative neuronal injury, as in diabetic neuropathy.[75]. Disturbances in ceruloplasmin homeostasis have been linked to other neurodegenerative diseases, and to neuropathic pain in the constriction injury rat model of neuropathic pain.[76]. Similarly, transferrin levels have been associated with peripheral neuropathy after bariatric surgery, which has been shown to disrupt iron homeostasis [77].
Iron transport in all tissues is intricately balanced with cellular and mitochondrial iron demand; indeed, recent studies including our own suggest that iron levels regulate mitochondrial biogenesis, a determinant of mitochondrial function [78]. (Kallianpur et al, unpublished data) Therefore, it should not be surprising if altered iron transport increases susceptibility to HIV-SN and DNP, especially in individuals exposed to mitochondria-toxic forms of cART. Mitochondrial injury, as well as activation of glia in the spinal cord and dorsal root ganglia due to HIV envelope glycoprotein gp120, with consequent release of inflammatory cytokines, may be involved in the pathogenesis of DNP [79]. Finally, others have reported that dysregulated iron transport also alters glutamate metabolism, which is implicated in both drug-induced and non-toxic peripheral neuropathies [80]–[82].
This study has some acknowledged limitations. The functional effects of these SNPs in iron-related genes are currently unknown. Since regulation of iron metabolism occurs largely at the post-transcriptional level, we speculate that many of these variants alter gene regulation and protein levels rather than protein structure or function. Assay of serum protein levels was not possible as part of this study, but CP rs13072552 has been linked to increased ceruloplasmin levels [74], and several of the genes identified here encode measurable serum proteins that may be investigated in future studies. The functional relatedness of the genes we selected in iron metabolism, in addition to tightly coordinated regulation of some of these genes (e.g., ACO1 and TFRC) in order to fine tune cellular iron access, storage, and utilization, argue against reliance on genome-wide levels of significance or stringent multiple-testing corrections in an exploratory study of this type; individual iron-regulatory SNP genotypes are highly unlikely to be independent, as represented in Figure 1 . In this situation, correction for the number of genes analyzed may be a more logical approach: with this type of correction, TF rs2718796, CP rs3816893, and B2M rs16966334, as well as ACO1 rs2026739 in whites, remain statistically significant or nominally significant. (As noted, the ACO1 SNP also remained significant after Bonferroni correction for multiple tests.) We did not adjust in our analyses for height or alcohol consumption, factors which can influence susceptibility to neuropathy, but not necessarily DNP. However, alcohol consumption was not associated with DNP on univariable analysis. Information about hair color, which has been correlated with neuropathic pain sensitivity, was not available [83]. We also attempted to address the potential effect of diabetes on genetic associations with peripheral neuropathy and DNP in this population. Glycosylated hemoglobin values were not available for analysis on most subjects, but additional adjustment for the possible presence of treated or untreated diabetes, ascertained using a more stringent-than-normal cut-off for non-fasting blood glucose values, did not significantly impact our results. Finally, the purpose of this study was to build on our prior findings of iron-related genetic associations with HIV-SN and to test the hypothesis that iron metabolism, rather than inflammation alone, plays a role in HIV-associated DNP; we intentionally did not study an exhaustive list of iron-regulatory genes. Much of the ferrome therefore remains to be studied. Keltner et al recently reported that HIV-associated DNP is associated in CHARTER subjects with reduced total cortical gray matter volumes on neuroimaging; cortical gray matter volumes have also been linked to R2* relaxation times, a sensitive measure of brain iron content [84], [85].
In summary, SNPs in eight iron-related genes selected for analysis showed significant and biologically plausible associations with DNP, independent of known potential confounders; SNPs in 7 of these genes also predicted DNP severity, adding to the biological plausibility argument. Most importantly, the common ACO1 rs2026739 variant remained significantly associated with DNP and DNP severity after stringent correction for multiple tests. Furthermore, associations with DNP were relatively robust, and point estimates for SNP associations were similar in direction and magnitude in blacks and whites, if not statistically significant in both groups. These results support the concept that the ferrome may be involved in the pathogenesis of HIV-SN and DNP in HIV-infected individuals on cART. These new genetic associations will require replication in other population samples as well as further functional characterization to determine underlying biological mechanisms that may be therapeutically targeted.
Supporting Information
Minimal overlap between ancestry principal components (PCs). PCs plotted against one another demonstrate appropriate clustering, with few outliers. Black dots: black; blue dots: Hispanic; red dots: white; cyan dots: other.
(TIF)
Iron-related genes and variants evaluated in relation to neuropathy in CHARTER.
(PDF)
Acknowledgments
The authors would like to express gratitude to all CHARTER study participants. We also thank all CHARTER investigators and site PIs, who assisted in subject recruitment:
The CNS HIV Antiretroviral Therapy Effects Research (CHARTER) Study is affiliated with the University of California-San Diego, Johns Hopkins University School of Medicine, Mount Sinai School of Medicine, Washington University- Saint Louis, University of Texas- Galveston, and the University of Washington-Seattle and is headquarted at the University of California-San Diego. Members of the CHARTER Study Group include: Director, Igor Grant, M.D.; Co-Directors, Scott Letendre, M.D., Ronald Ellis, M.D., Ph.D., Thomas Marcotte, Ph.D.; Center Manager, Donald Franklin, Jr.; Neuromedical component: Ronald Ellis, M.D., Ph.D. (PI); J. Allen McCutchan, M.D., Terry Alexander, R.N.; Laboratory, Pharmacology and Immunology Component: Scott Letendre, M.D. (P.I.), Edmund Capparelli, Pharm.D.; Neurobehavioral Component: Robert K. Heaton, Ph.D. (P.I.), J. Hampton Atkinson, M.D., Steven Paul Woods, Psy.D., Matthew Dawson; Virology Component: Davey M. Smith, M.D. (P.I.), Joseph K. Wong, M.D.; Imaging Component: Christine Fennema-Notestine, Ph.D. (Co-P.I.), Michael J. Taylor, Ph.D. (Co-P.I.), Rebecca Theilmann, Ph.D.; Data Management Component: Anthony C. Gamst, Ph.D. (P.I.), Clint Cushman, Ph.D; Statistics Component: Ian Abramson, Ph.D. (P.I.), Florin Vaida, Ph.D.; Johns Hopkins University Site: Justin McArthur (P.I.), Mary Smith; Mount Sinai School of Medicine Site: Susan Morgello, M.D. (Co-P.I.) and David Simpson, M.D. (Co-P.I.), Letty Mintz, N.P.; University of California, San Diego Site: J. Allen McCutchan, M.D. (P.I.), Will Toperoff, N.P.; University of Washington, Seattle Site: Ann Collier, M.D. (Co-P.I.) and Christina Marra, M.D. (Co-P.I.), Trudy Jones, M.N., A.R.N.P.; University of Texas, Galveston Site: Benjamin Gelman, M.D., Ph.D. (P.I.), Eleanor Head, R.N., B.S.N.; and Washington University, St. Louis Site: David Clifford, M.D. (P.I.), Muhammad Al-Lozi, M.D., Mengesha Teshome, M.D.
The views expressed in this article are those of the authors and do not reflect the official policy or position of the United States Government.
Funding Statement
The co-authors of this manuscript have no pertinent financial disclosures. Funding support for this work was provided by the following grants: National Institutes of Health contracts N01 MH22005 and HHSN271201000036C from the National Institutes of Health (to CHARTER PI Igor Grant) and 1R01 MH 095621 (to PIs Asha Kallianpur and Todd Hulgan). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ellis RJ, Rosario D, Clifford DB, McArthur JC, Simpson D, et al. (2010) Continued high prevalence and adverse clinical impact of human immunodeficiency virus-associated sensory neuropathy in the era of combination antiretroviral therapy: the CHARTER Study. Arch Neurol 67: 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kallianpur AR, Hulgan T (2009) Pharmacogenetics of nucleoside reverse-transcriptase inhibitor-associated peripheral neuropathy. Pharmacogenomics 10: 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hitchcock SA, Meyer HP, Gwyther E (2008) Neuropathic pain in AIDS patients prior to antiretroviral therapy. S Afr Med J 98: 889–892. [PubMed] [Google Scholar]
- 4. Apostolova N, Blas-Garcia A, Esplugues JV (2011) Mitochondrial interference by anti-HIV drugs: mechanisms beyond Pol-gamma inhibition. Trends in Pharmacological Sciences 32: 715–725. [DOI] [PubMed] [Google Scholar]
- 5. Hulgan T, Haas DW, Haines JL, Ritchie MD, Robbins GK, et al. (2005) Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: an adult AIDS clinical trials group study. AIDS 19: 1341–1349. [DOI] [PubMed] [Google Scholar]
- 6. Hulgan T, Tebas P, Canter JA, Mulligan K, Haas DW, et al. (2008) Hemochromatosis gene polymorphisms, mitochondrial haplogroups, and peripheral lipoatrophy during antiretroviral therapy. J Infect Dis 197: 858–866. [DOI] [PubMed] [Google Scholar]
- 7. Kamerman PR, Moss PJ, Weber J, Wallace VC, Rice AS, et al. (2012) Pathogenesis of HIV-associated sensory neuropathy: evidence from in vivo and in vitro experimental models. J Peripher Nerv Syst 17: 19–31. [DOI] [PubMed] [Google Scholar]
- 8. Keswani SC, Jack C, Zhou C, Hoke A (2006) Establishment of a rodent model of HIV-associated sensory neuropathy. J Neurosci 26: 10299–10304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellis RJ, Marquie-Beck J, Delaney P, Alexander T, Clifford DB, et al. (2008) Human immunodeficiency virus protease inhibitors and risk for peripheral neuropathy. Ann Neurol 64: 566–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robinson-Papp J, Morgello S, Vaida F, Fitzsimons C, Simpson DM, et al. (2010) Association of self-reported painful symptoms with clinical and neurophysiologic signs in HIV-associated sensory neuropathy. Pain 151: 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lucey BP, Clifford DB, Creighton J, Edwards RR, McArthur JC, et al. (2011) Relationship of depression and catastrophizing to pain, disability, and medication adherence in patients with HIV-associated sensory neuropathy. Aids Care-Psychological and Socio-Medical Aspects of Aids/Hiv 23: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phillips TJ, Cherry CL, Cox S, Marshall SJ, Rice AS (2010) Pharmacological treatment of painful HIV-associated sensory neuropathy: a systematic review and meta-analysis of randomised controlled trials. PLoS One 5: e14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chetty S, Baalbergen E, Bhigjee AI, Kamerman P, Ouma J, et al. (2012) Clinical practice guidelines for management of neuropathic pain: expert panel recommendations for South Africa. Samj South African Medical Journal 102: 312–325. [DOI] [PubMed] [Google Scholar]
- 14. Derry S, Moore RA (2012) Topical capsaicin (low concentration) for chronic neuropathic pain in adults. Cochrane Database Syst Rev 9: CD010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evans SR, Simpson DM, Kitch DW, King A, Clifford DB, et al. (2007) A Randomized Trial Evaluating Prosaptide (TM) for HIV-Associated Sensory Neuropathies: Use of an Electronic Diary to Record Neuropathic Pain. PLoS One 2: e551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keltner JR, Vaida F, Ellis RJ, Moeller-Bertram T, Fitzsimmons C, et al. (2012) Health-related quality of life ‘well-being’ in HIV distal neuropathic pain is more strongly associated with depression severity than with pain intensity. Psychosomatics 53: 380–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fellows RP, Byrd DA, Elliott K, Robinson-Papp J, Mindt MR, et al. (2012) Distal sensory polyneuropathy is associated with neuropsychological test performance among persons with HIV. J Int Neuropsychol Soc 18: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis L, Evans S, Fishman B, Haley A, Spielman LA (2004) Predictors of attrition in HIV-positive subjects with peripheral neuropathic pain. Aids Care-Psychological and Socio-Medical Aspects of Aids/Hiv 16: 395–402. [DOI] [PubMed] [Google Scholar]
- 19. Oshinaike O, Akinbami A, Ojo O, Ogbera A, Okubadejo N, et al. (2012) Influence of Age and Neurotoxic HAART Use on Frequency of HIV Sensory Neuropathy. AIDS Res Treat 2012: 961510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pahuja M, Grobler A, Glesby MJ, Karim F, Parker G, et al. (2012) Effects of a reduced dose of stavudine on the incidence and severity of peripheral neuropathy in HIV-infected adults in South Africa. Antiviral Therapy 17: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Backonja M, Woolf CJ (2010) Future directions in neuropathic pain therapy: closing the translational loop. Oncologist 15 Suppl 2 24–29. [DOI] [PubMed] [Google Scholar]
- 22. Maag R, Baron R (2006) Neuropathic pain: translational research and impact for patient care. Curr Pain Headache Rep 10: 191–198. [DOI] [PubMed] [Google Scholar]
- 23. Max MB, Stewart WF (2008) The molecular epidemiology of pain: a new discipline for drug discovery. Nature Reviews Drug Discovery 7: 647–658. [DOI] [PubMed] [Google Scholar]
- 24. Galy B, Ferring-Appel D, Sauer SW, Kaden S, Lyoumi S, et al. (2010) Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab 12: 194–201. [DOI] [PubMed] [Google Scholar]
- 25. Millecamps S, Julien JP (2013) Axonal transport deficits and neurodegenerative diseases. Nature Reviews Neuroscience 14: 161–176. [DOI] [PubMed] [Google Scholar]
- 26. Sheftel AD, Lill R (2009) The power plant of the cell is also a smithy: the emerging role of mitochondria in cellular iron homeostasis. Ann Med 41: 82–99. [DOI] [PubMed] [Google Scholar]
- 27. Drakesmith H, Prentice AM (2012) Hepcidin and the Iron-Infection Axis. Science 338: 768–772. [DOI] [PubMed] [Google Scholar]
- 28. Wu X, Yung LM, Cheng WH, Yu PB, Babitt JL, et al. (2012) Hepcidin regulation by BMP signaling in macrophages is lipopolysaccharide dependent. PLoS One 7: e44622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang X, Rovin BH (2013) Beyond anemia: hepcidin, monocytes and inflammation. Biol Chem 394: 231–238. [DOI] [PubMed] [Google Scholar]
- 30. Johnson EE, Sandgren A, Cherayil BJ, Murray M, Wessling-Resnick M (2010) Role of ferroportin in macrophage-mediated immunity. Infect Immun 78: 5099–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kallianpur AR, Hulgan T, Canter JA, Ritchie MD, Haines JL, et al. (2006) Hemochromatosis (HFE) gene mutations and peripheral neuropathy during antiretroviral therapy. AIDS 20: 1503–1513. [DOI] [PubMed] [Google Scholar]
- 32. Bulaj ZJ, Griffen LM, Jorde LB, Edwards CQ, Kushner JP (1996) Clinical and biochemical abnormalities in people heterozygous for hemochromatosis. N Engl J Med 335: 1799–1805. [DOI] [PubMed] [Google Scholar]
- 33. Holzinger ER, Hulgan T, Ellis RJ, Samuels DC, Ritchie MD, et al. (2012) Mitochondrial DNA variation and HIV-associated sensory neuropathy in CHARTER. J Neurovirol 18: 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grady BJ, Dudek S, Ritchie MD (2009) Finding Unique Filter Sets in PLATO: Preparation for Efficient Interaction Analysis. Genetic Epidemiology 33: 788–788. [Google Scholar]
- 35. Kumar KR, Djarmati-Westenberger A, Grunewald A (2011) Genetics of Parkinson's disease. Semin Neurol 31: 433–440. [DOI] [PubMed] [Google Scholar]
- 36. Reich DE, Lander ES (2001) On the allelic spectrum of human disease. Trends Genet 17: 502–510. [DOI] [PubMed] [Google Scholar]
- 37. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jia P, Wang L, Fanous AH, Chen X, Kendler KS, et al. (2012) A bias-reducing pathway enrichment analysis of genome-wide association data confirmed association of the MHC region with schizophrenia. J Med Genet 49: 96–103. [DOI] [PubMed] [Google Scholar]
- 39. Bush WS, Moore JH (2012) Chapter 11: Genome-wide association studies. PLoS Comput Biol 8: e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cano A, Mayo A, Ventimiglia M (2006) Coping, pain severity, interference, and disability: the potential mediating and moderating roles of race and education. Journal of Pain 7: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wandner LD, Scipio CD, Hirsh AT, Torres CA, Robinson ME (2012) The perception of pain in others: how gender, race, and age influence pain expectations. Journal of Pain 13: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lorber M, Haddad S (2013) Hypersensitivity and desensitization to darunavir in a case of HIV infection with triple-class drug resistance: case description and review of the literature. J Int Assoc Provid AIDS Care 12: 378–379. [DOI] [PubMed] [Google Scholar]
- 44. Kranick SM, Nath A (2012) Neurologic complications of HIV-1 infection and its treatment in the era of antiretroviral therapy. Continuum (Minneap Minn) 18: 1319–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boelaert JR, Weinberg GA, Weinberg ED (1996) Altered iron metabolism in HIV infection: mechanisms, possible consequences, and proposals for management. Infect Agents Dis 5: 36–46. [PubMed] [Google Scholar]
- 46. Drakesmith H, Chen N, Ledermann H, Screaton G, Townsend A, et al. (2005) HIV-1 Nef down-regulates the hemochromatosis protein HFE, manipulating cellular iron homeostasis. Proc Natl Acad Sci U S A 102: 11017–11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finberg KE (2013) Regulation of systemic iron homeostasis. Curr Opin Hematol 20: 208–214. [DOI] [PubMed] [Google Scholar]
- 48. Bentley AJ, Rosman KD, Mitchell D (2007) Can the sensory symptoms of restless legs syndrome be assessed using a qualitative pain questionnaire? Clin J Pain 23: 62–66. [DOI] [PubMed] [Google Scholar]
- 49. Gemignani F, Vitetta F, Brindani F, Contini M, Negrotti A (2013) Painful polyneuropathy associated with restless legs syndrome. Clinical features and sensory profile. Sleep Med 14: 79–84. [DOI] [PubMed] [Google Scholar]
- 50. Connor JR, Ponnuru P, Wang XS, Patton SM, Allen RP, et al. (2011) Profile of altered brain iron acquisition in restless legs syndrome. Brain 134: 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Patton SM, Ponnuru P, Snyder AM, Podskalny GD, Connor JR (2011) Hypoxia-inducible factor pathway activation in restless legs syndrome patients. Eur J Neurol 18: 1329–1335. [DOI] [PubMed] [Google Scholar]
- 52. Hare D, Ayton S, Bush A, Lei P (2013) A delicate balance: Iron metabolism and diseases of the brain. Front Aging Neurosci 5: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dowling P, Klinker F, Amaya F, Paulus W, Liebetanz D (2009) Iron-deficiency sensitizes mice to acute pain stimuli and formalin-induced nociception. J Nutr 139: 2087–2092. [DOI] [PubMed] [Google Scholar]
- 54. Dubovy P (2011) Wallerian degeneration and peripheral nerve conditions for both axonal regeneration and neuropathic pain induction. Ann Anat 193: 267–275. [DOI] [PubMed] [Google Scholar]
- 55. Gille G, Reichmann H (2011) Iron-dependent functions of mitochondria–relation to neurodegeneration. J Neural Transm 118: 349–359. [DOI] [PubMed] [Google Scholar]
- 56. Joseph EK, Levine JD (2006) Mitochondrial electron transport in models of neuropathic and inflammatory pain. Pain 121: 105–114. [DOI] [PubMed] [Google Scholar]
- 57. Richardson DR, Lane DJ, Becker EM, Huang ML, Whitnall M, et al. (2010) Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci U S A 107: 10775–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Carlson ES, Tkac I, Magid R, O'Connor MB, Andrews NC, et al. (2009) Iron is essential for neuron development and memory function in mouse hippocampus. J Nutr 139: 672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moos T, Rosengren Nielsen T (2006) Ferroportin in the postnatal rat brain: implications for axonal transport and neuronal export of iron. Seminars in Pediatric Neurology 13: 149–157. [DOI] [PubMed] [Google Scholar]
- 60. Moos T, Bernth N, Courtois Y, Morgan EH (2011) Developmental iron uptake and axonal transport in the retina of the rat. Mol Cell Neurosci 46: 607–613. [DOI] [PubMed] [Google Scholar]
- 61. Li X, Clark JD (2003) Heme oxygenase type 2 participates in the development of chronic inflammatory and neuropathic pain. Journal of Pain 4: 101–107. [DOI] [PubMed] [Google Scholar]
- 62. Mitchell RM, Lee SY, Randazzo WT, Simmons Z, Connor JR (2009) Influence of HFE variants and cellular iron on monocyte chemoattractant protein-1. J Neuroinflammation 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lawless MW, White M, Mankan AK, O'Dwyer MJ, Norris S (2007) Elevated MCP-1 serum levels are associated with the H63D mutation and not the C282Y mutation in hereditary hemochromatosis. Tissue Antigens 70: 294–300. [DOI] [PubMed] [Google Scholar]
- 64. Uceyler N, Rogausch JP, Toyka KV, Sommer C (2007) Differential expression of cytokines in painful and painless neuropathies. Neurology 69: 42–49. [DOI] [PubMed] [Google Scholar]
- 65. Leung L, Cahill CM (2010) TNF-alpha and neuropathic pain - a review. Journal of Neuroinflammation 7: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Foulkes T, Wood JN (2008) Pain Genes. Plos Genetics 4: e1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tramullas M, Lantero A, Diaz A, Morchon N, Merino D, et al. (2010) BAMBI (bone morphogenetic protein and activin membrane-bound inhibitor) reveals the involvement of the transforming growth factor-beta family in pain modulation. J Neurosci 30: 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang J, Pantopoulos K (2011) Regulation of cellular iron metabolism. Biochem J 434: 365–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Calabrese V, Lodi R, Tonon C, D'Agata V, Sapienza M, et al. (2005) Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich's ataxia. J Neurol Sci 233: 145–162. [DOI] [PubMed] [Google Scholar]
- 70. Stemmler TL, Lesuisse E, Pain D, Dancis A (2010) Frataxin and mitochondrial FeS cluster biogenesis. J Biol Chem 285: 26737–26743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ye H, Rouault TA (2010) Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry 49: 4945–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Staats KA, Schonefeldt S, Van Rillaer M, Van Hoecke A, Van Damme P, et al. (2013) Beta-2 microglobulin is important for disease progression in a murine model for amyotrophic lateral sclerosis. Front Cell Neurosci 7: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ayton S, Zhang M, Roberts BR, Lam LQ, Lind M, et al. (2014) Ceruloplasmin and beta-amyloid precursor protein confer neuroprotection in traumatic brain injury and lower neuronal iron. Free Radic Biol Med Feb 7 (Epub ahead of print).. [DOI] [PubMed] [Google Scholar]
- 74. Tang WH, Wu Y, Hartiala J, Fan Y, Stewart AF, et al. (2012) Clinical and genetic association of serum ceruloplasmin with cardiovascular risk. Arterioscler Thromb Vasc Biol 32: 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. El Boghdady NA, Badr GA (2012) Evaluation of oxidative stress markers and vascular risk factors in patients with diabetic peripheral neuropathy. Cell Biochem Funct 30: 328–334. [DOI] [PubMed] [Google Scholar]
- 76. Tan EC, Bahrami S, Kozlov AV, Kurvers HA, Ter Laak HJ, et al. (2009) The oxidative response in the chronic constriction injury model of neuropathic pain. J Surg Res 152: 84–88. [DOI] [PubMed] [Google Scholar]
- 77. Thaisetthawatkul P, Collazo-Clavell ML, Sarr MG, Norell JE, Dyck PJ (2004) A controlled study of peripheral neuropathy after bariatric surgery. Neurology 63: 1462–1470. [DOI] [PubMed] [Google Scholar]
- 78. Rensvold JW, Ong SE, Jeevananthan A, Carr SA, Mootha VK, et al. (2013) Complementary RNA and Protein Profiling Identifies Iron as a Key Regulator of Mitochondrial Biogenesis. Cell Rep 3: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hao S (2013) The Molecular and Pharmacological Mechanisms of HIV-Related Neuropathic Pain. Curr Neuropharmacol 11: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li JY, Wang X, Ji PT, Li XF, Guan GH, et al. (2012) Peripheral nerve injury decreases the expression of metabolic glutamate receptor 7 in dorsal root ganglion neurons. Neurosci Lett 531: 52–56. [DOI] [PubMed] [Google Scholar]
- 81. Yan X, Jiang E, Gao M, Weng HR (2013) Endogenous activation of presynaptic NMDA receptors enhances glutamate release from the primary afferents in the spinal dorsal horn in a rat model of neuropathic pain. J Physiol 591: 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mitchell RM, Lee SY, Simmons Z, Connor JR (2011) HFE polymorphisms affect cellular glutamate regulation. Neurobiol Aging 32: 1114–1123. [DOI] [PubMed] [Google Scholar]
- 83. Belfer I, Dai F (2010) Phenotyping and Genotyping Neuropathic Pain. Curr Pain Headache Rep 14: 203–212. [DOI] [PubMed] [Google Scholar]
- 84. Keltner JR, Fennema-Notestine C, Vaida F, Wang D, Franklin DR, for the CHARTER Group, et al. (2014) HIV-associated distal neuropathic pain is associated with smaller total cerebral cortical gray matter. J Neurovirol Feb 19 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ropele S, Wattjes MP, Langkammer C, Kilsdonk ID, de Graaf WL, et al. (2013) Multicenter R2* mapping in the healthy brain. Magn Reson Med May 8 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Minimal overlap between ancestry principal components (PCs). PCs plotted against one another demonstrate appropriate clustering, with few outliers. Black dots: black; blue dots: Hispanic; red dots: white; cyan dots: other.
(TIF)
Iron-related genes and variants evaluated in relation to neuropathy in CHARTER.
(PDF)