Abstract
Antibodies capable of effectively neutralizing HIV-1 generally exhibit very high levels of somatic hypermutation, both in their complementarity-determining and framework-variable regions. In many cases, full reversion of the antibody framework mutations back to germline has been shown to result in substantial to complete loss of HIV-1 neutralizing activity. However, it is currently not known whether all or most of the observed framework mutations are necessary, or whether a small subset of these mutations may be sufficient for broad and potent neutralization. To address this issue and to explore the dependence of neutralization activity on the level of somatic hypermutation in antibody framework, we applied a computationally guided framework reversion procedure to two broadly neutralizing anti-HIV-1 antibodies, VRC01 and 10E8, which target two different HIV-1 sites of vulnerability. Antibody variants in which up to 78% (38 out of 49 for VRC01) and 89% (31 out of 35 for 10E8) of framework mutations were reverted to germline retained breadth and potency within 3-fold of the mature antibodies when evaluated on a panel of 21-diverse viral strains. Further, a VRC01 variant with a ~50% framework-reverted light chain showed a 2-fold improvement in potency over the mature antibody. Our results indicate that only a small number of antibody-framework mutations may be sufficient for high breadth and potency of HIV-1 neutralization by antibodies VRC01 and 10E8. Partial framework revertants of HIV-1 broadly neutralizing antibodies may present advantages over their highly mutated counterparts as antibody therapeutics and as targets for immunogen design.
Introduction
Recent years have seen an explosion in the number of broadly neutralizing antibodies (bNAbs) against HIV-1 (1-10). Many of these bNAbs have been shown to protect from or to provide control of infection (11-13), and are therefore of interest for passive immunization approaches (14). An underlying characteristic of anti-HIV-1 antibodies is the substantially increased levels of somatic hypermutation (15). Somatic hypermutation is part of the diversification of antibodies that occurs during affinity maturation: this process occurs in activated B cells exposed to antigen within germinal centers where high affinity antibodies are selected over their low affinity counterparts (16). Generally, chronic viral infections are associated with the generation of antibodies with increased numbers of mutations compared to acute viral infections, suggesting that persistent antigen exposure plays a role in stimulating repeated rounds of somatic hypermutation and selection (17, 18). In the case of HIV-1, bNAbs mostly show higher mutation levels compared to weakly neutralizing antibodies. Moreover, the inferred germline antibodies of several anti-HIV bNAbs lack neutralization activity (19, 20), indicating that somatic hypermutation is important for neutralizing breadth and potency (18).
While somatic mutations occur preferentially within the CDR regions of antibodies (21), large numbers of mutations in anti-HIV-1 bNAbs are also found within the antibody framework regions (18, 19). Klein et al. (18) analyzed a set of anti-HIV-1 bNAbs targeting diverse epitopes on the HIV-1 envelope glycoprotein and found that full framework reversions to germline residues substantially reduced or completely abrogated neutralization activity for many of these antibodies. Function was only minimally restored in some antibodies by allowing framework mature mutations in positions that were in direct contact with the antigen (18). The results in (18) underline the importance of framework maturation for broad and potent neutralization by anti-HIV-1 antibodies. However, it is currently unknown whether most or all of the framework mutations are necessary for retention of antibody function or whether some of these mutations can be reverted to germline with minimal effects on function.
To investigate this question, we selected two bNAbs that target different sites of vulnerability on the HIV-1 Env glycoprotein: the CD4-binding-site (CD4bs) antibody VRC01 and the membrane-proximal external region (MPER) antibody 10E8. These antibodies neutralize approximately 90% and 98% of HIV-1 strains at average potency of 0.25 and 0.22 μg/ml, respectively (4, 10). The variable regions of both of these antibodies exhibit high degrees of amino acid mutation: VRC01 V-gene, 42% heavy/28% light; 10E8 V-gene, 22% heavy/17% light. The putative germline-reverted versions of these antibodies have been shown to be incapable of neutralizing HIV-1 viral strains (19 and unpublished data). For VRC01, the mature CDRs alone or in combination with the antigen-contacting framework residues are not sufficient for potent neutralization, as they only weakly neutralize 0 and 3 out of 10 strains, respectively (18). These results confirm the importance of framework mutations in VRC01. However, we conjectured that not all mutations from germline are necessary for retention of antibody neutralization activity. To test this conjecture, we created a series of VRC01 and 10E8 variants with partial framework reversions to germline in both heavy and light chains and compared their neutralization activity to that of the mature antibodies. This approach allowed us to explore the relationship between neutralization activity and number of framework mutations in anti-HIV-1 bNAbs. Our results challenge the notion that extensive framework mutations are necessary for broad and potent neutralization by anti-HIV-1 bNAbs and suggest strategies for antibody redesign in the context of antibody-product optimization or immunogen design.
Materials and Methods
Design and analysis of partial framework revertants to germline
Different approaches for various degrees of antibody germline reversion, ranging from full V-gene reversion to selective point mutations, have been described previously, with applications to a number of viruses (18, 19, 22-24). In our study, a CDR-grafting procedure typically used for humanization of non-human antibodies (25) was used as the basis for our designs, in combination with structural modeling and mutation analysis. Specifically, the CDR regions (Kabat definition) of the mature antibodies (VRC01 and 10E8) were grafted onto homologous human germline genes (in effect, this replaces the germline CDRs with their mature counterparts) (Figure 1). Additionally, series of germline framework residues were selected for mutation to their mature counterparts. Structural modeling, analysis, and visualization were used to determine which germline framework resides to mutate to mature. Specifically, mutations were selected based on a combination of several factors: (a) whether the position is implicated in supporting CDR regions (25); (b) the differences between germline and mature amino acid types (e.g., change in amino acid charge, hydrophobicity, or structural properties); and (c) proximity to antigen or CDR regions as determined by the antigen-bound crystal structure. The antigen-bound crystal structure of VRC01 was obtained from PDB id: 3ngb (19). The antigen-bound crystal structure of 10E8 (PDB id: 4G6F (10)) was not available at the time of the design, and was thus only used for retrospective analysis. Structural modeling, analysis, and visualization were performed using OSPREY (26, 27), MolProbity (28), and PyMOL (29). Throughout this paper, the mAb-nfH/mfL nomenclature was used for designed partial framework revertants, where n is the number of framework mutations in the heavy chain and m is the number of framework mutations in the light chain; for example, VRC01-5fH/6fL refers to the VRC01 variant with grafted mature CDRs and 5 heavy chain and 6 light chain mature framework mutations. Designed heavy and light chains were paired in a sparse matrix that was deemed to sufficiently represent the diversity of the possible heavy-light chain combinations.
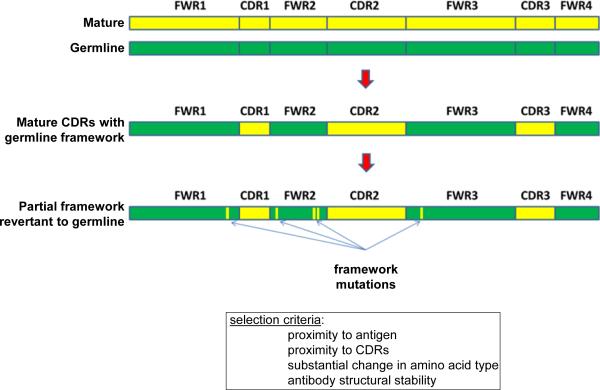
Figure 1. Procedure for partial reversion of antibody framework to germline.
The CDR regions of the mature target antibody (top, yellow) were grafted onto the backbone of the putative germline precursor, prior to somatic hypermutation (top, green) to obtain a chimeric CDR-grafted antibody (middle). Additionally, based on sequence and structural considerations, germline framework residues were selected for mutation to their mature counterparts, to obtain a partial framework reversion to germline of the target antibody with generally retained function.
Protein expression and purification
Site-directed mutagenesis or gene synthesis was used to generate the CMV/R plasmids encoding the heavy or light chain framework revertants. Full length IgG proteins were expressed by co-transfection of heavy and light chain plasmids into 293F cells, and antibodies were purified with affinity chromatography using either ProteinA Fast Flow Resin (GE Healthcare) or HiTrap ProteinA HP Columns (GE Healthcare).
Neutralization assays
Neutralization was measured using single-round-of-infection HIV-1 Env-pseudoviruses and TZM-bl target cells, as described previously (30-32). Neutralization curves were fit by nonlinear regression using a 5-parameter hill slope equation as previously described (31). The 50% and 80% inhibitory concentrations (IC50 and IC80) were reported as the antibody concentrations required to inhibit infection by 50% and 80%, respectively. IC50 values were used for the computation of neutralization breadth and potency for the different antibody variants (Figure 2). Average neutralization potency for a given antibody was computed as the geometric mean of the IC50 values for a set of 21 diverse viral strains (7), where values >50 μg/ml were set to 50. Neutralization breadth for a given antibody was computed as the percentage of strains with IC50 values of <50 μg/ml.
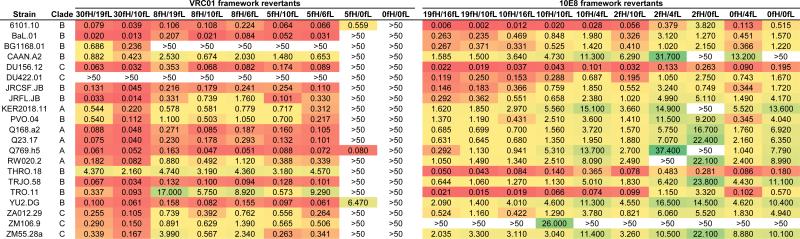
Figure 2. HIV-1 neutralization by VRC01 and 10E8 antibody variants.
For each antibody, shown are IC50 values (in μg/ml) for the wildtype mature antibody, a set of partial framework revertants, and a CDR-grafted variant where the entire framework regions have been reverted to germline (columns; antibody names are based on the number of framework mutations in the heavy and light chains). Neutralization data was obtained for 21 diverse HIV-1 strains (rows) from clades A, B, and C. Values are colored according to potency, on a scale from green (least potent) to red (most potent), with white corresponding to a neutralization potency of >50 μg/ml.
Results
Partial framework reversion of VRC01
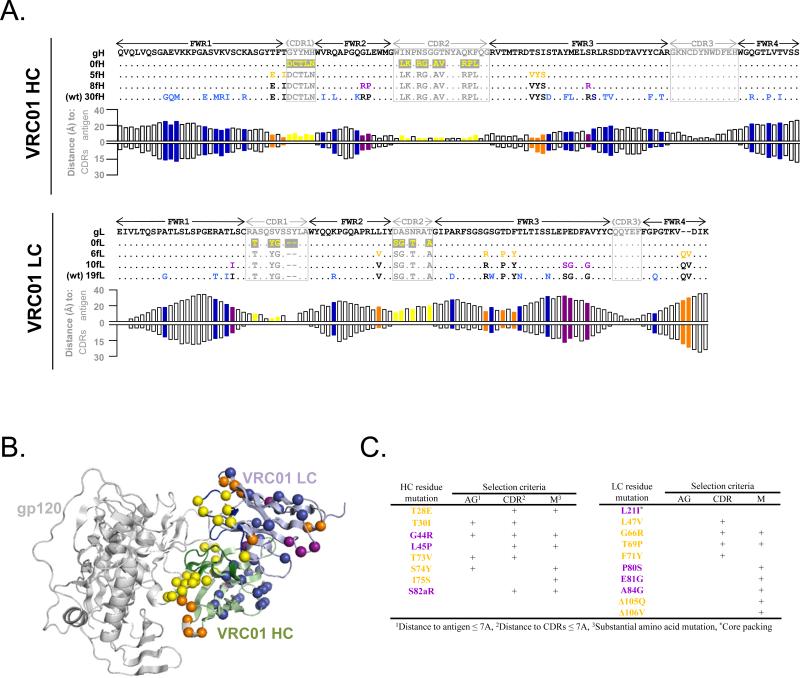
The heavy chain of the mature VRC01 antibody contains 30 mutations within framework regions 1-4 (Figure 3A). To test whether a subset of these mutations can be reverted back to germline without substantially affecting antibody activity, we created two partial framework revertants of the heavy chain, with 5 and 8 mature framework mutations each (in the context of mature CDR regions), as well as the CDR-grafted version of the heavy chain (with mature CDR regions and 0 framework mutations) (Figure 3A-C). The selected mutations in the VRC01-5fH variant were in proximity to the antigen (distances of 3.6 - 7.7 Å) (Figure 3A,B). The additional 3 mutations in the VRC01-8fH variant were deemed to represent substantial changes in amino acid type that were still in relative proximity to the antigen (distances of 6.7 - 8.8 Å). The remaining 22 residue positions of maturation were more distal to the antigen (mean distance of 16.3 ± 4.7 Å; range 8.3 - 24.9 Å) and/or represented more conserved amino acid changes, and thus were not selected for study. Similarly, the mature VRC01 light chain contains 19 framework mutations, so we created partial framework revertants with mature CDR regions and 0, 6, and 10 mature framework mutations (Figure 3A-C). None of the framework mutations in the VRC01 light chain were in substantial proximity to the antigen (mean distance of 21.7 ± 6.9 Å; range 9.8 - 32.3 Å), so the reversion analysis was focused on framework interactions with the CDR regions and/or within the overall antibody structure. The VRC01-6fL light chain variant included mutations in proximity to the CDR regions (distances of 2.9 - 5.5 Å), as well as a 2-residue FWR4 insertion that is not found in most other HIV-1 bNAbs, including other antibodies from the VRC01 class (9). Structural modeling identified an additional 4 mutations that could be of importance for the overall structure of VRC01, and the addition of these mutations formed the VRC01-10fL variant (Figure 3A-C).
Figure 3. Framework revertants of antibody VRC01.
(A) Sequence alignment (top: heavy chain; bottom: light chain) of a putative germline version, designed partial framework revertants, and mature VRC01. Residues that are identical to germline are shown as dots. CDR mutations are shown in yellow on grey background; framework mutations are colored according to the variant in which they are first introduced (orange – closest to germline; purple; blue – mature). For each residue position, the respective distance to antigen and CDRs is shown. (B) Mapping of framework mutations from germline backbone onto a VRC01 complex structure (coloring same as in A). (C) Selection criteria for framework mutations.
HIV-1 Neutralization by VRC01 partial framework revertants
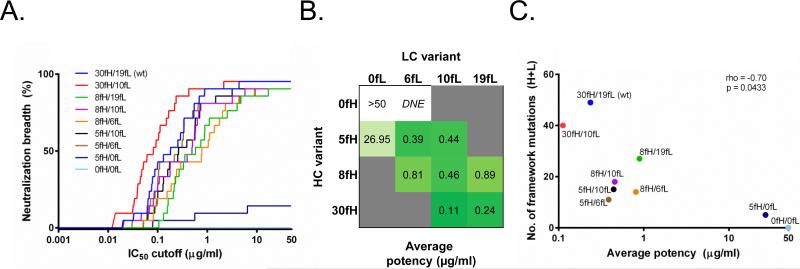
A total of 10 heavy-light chain combinations using the 4 heavy and 4 light chain VRC01 variants were tested for neutralization activity. The mature VRC01 (VRC01-30fH/19fL, consisting of 30 heavy/19 light chain framework mutations) was among the best in terms of neutralization function (Figure 2); however, the VRC01-30fH/10fL variant (with mature heavy and partially reverted light chain) consistently outperformed all other constructs when using neutralization potency-breadth curves as a metric (the percent of neutralized viral strains at different IC50 potency cutoffs) (Figure 4A). Six of the nine partial framework revertants maintained good neutralization activity. In fact, with the exception of VRC01-0fH/6fL (which did not express), VRC01-5fH/0fL, and VRC01-0fH/0fL, the revertants generally had an average potency within a ~10-fold range from mature (Figure 4B). The VRC01-0fH/0fL CDR-grafted construct showed complete lack of neutralization activity, in concordance with previous findings (18). Generally, whereas a significant correlation (p = 0.0433) was found between the number of framework mutations and the neutralization potency for VRC01 variants (Figure 4C), a small number of framework mutations appeared to be sufficient for retention of antibody function: for example, the VRC01-5fH/6fL construct had a <2-fold decrease in potency compared to the mature VRC01 (Figure 4A,B).
Figure 4. HIV-1 neutralization by framework revertants of antibody VRC01.
(A) Neutralization breadth at different IC50 neutralization cutoff values. (B) Average (geometric mean) neutralization potency for the different combinations of heavy and light chain variants; grey – combination not tested; DNE – variant did not express. (C) Spearman correlation of neutralization potency vs. number of framework mutations (heavy + light); colors same as in (A).
Partial framework reversion of antibody 10E8
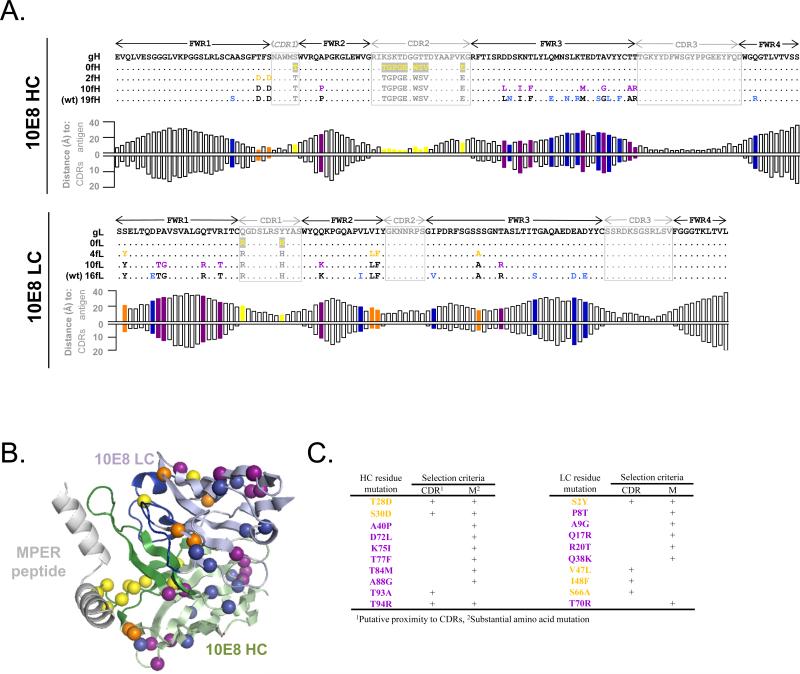
The crystal structure of the 10E8-MPER complex was not available at the time of the designs, and was thus used only for retrospective analysis. The heavy and light chains of mature 10E8 contain 19 and 16 mutations within the framework regions 1-4, respectively (Figure 5A). We created 3 heavy chain variants with mature CDR regions and 0, 2, and 10 framework mutations and 3 light chain variants with mature CDR regions and 0, 4, and 10 framework mutations (Figure 5A-C). The selected mutations in the 10E8-2fH and 10E8-10fH heavy chain variants and 10E8-4fL and 10E8-10fL light chain variants were deemed to represent more substantial changes in amino acid type and/or were found at positions that have been implicated in supporting CDR regions (Figure 5C). Post-hoc structural analysis revealed that the mutations in the 10E8-2fH heavy chain variant were in proximity to the antigen (distances of 3.6 and 6.9 Å), but the distances to the antigen or the CDR regions for the additional 8 mutations in the 10E8-10fH heavy chain variant vs. the remaining 9 heavy chain mutations were not substantially different (respectively, 18.1 ± 6.6 Å vs. 20.6 ± 4.9 Å for antigen, 7.5 ± 3.4 Å vs. 6.6 ± 2.1 Å for CDR regions) (Figure 5A,B). None of the framework mutations in the 10E8-light chain were in substantial proximity to the antigen (mean distance of 22.4 ± 6.8 Å; range 14.2 - 33.0 Å; Figure 5A). The 10E8-4fL light chain variant included mutations in proximity to the CDR regions (distances of 3.1 - 5.9 Å), but the distances to the CDR regions for the additional 6 mutations in the 10E8-10fL light chain variant versus the six remaining residue positions of maturation were not substantially different (10.3 ± 3.2 Å vs. 8.3 ± 4.0) (Figure 5A,B).
Figure 5. Framework revertants of antibody 10E8.
(A) Sequence alignment (top: heavy chain; bottom: light chain) of a putative germline version, designed partial framework revertants, and mature 10E8. Residues that are identical to germline are shown as dots. CDR mutations are shown in yellow on grey background; framework mutations are colored according to the variant in which they are first introduced (orange – closest to germline; purple; blue – mature). For each residue position, the respective distance to antigen and CDRs is shown. (B) Mapping of framework mutations from germline backbone onto a 10E8 complex structure (coloring same as in A). (C) Selection criteria for framework mutations.
HIV-1 Neutralization by 10E8 partial framework revertants
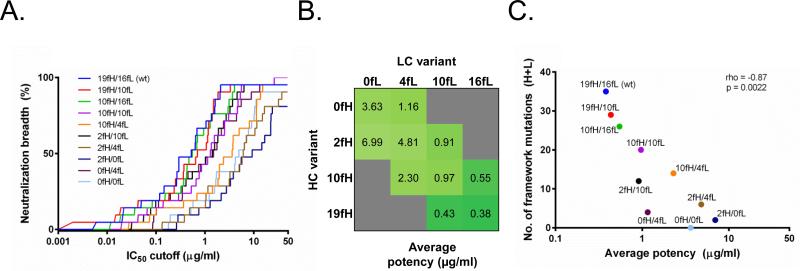
A total of 10 heavy-light chain combinations using the 4 heavy and 4 light chain 10E8 variants were tested for neutralization activity. The mature 10E8 (10E8-19fH/16fL) along with the 10E8-19fH/10fL and 10E8-10fH/16fL variants displayed the best neutralization potency and breadth (Figures 2 and 6A). Unlike VRC01, the 10E8 variant with 0 framework mutations (10E8-0fH/0fL) retained significant, but reduced, neutralization activity (Figure 6B). As with VRC01, a significant correlation was found between the number of framework mutations and the neutralization potency for 10E8 variants (Figure 6C; p=0.0022). Interestingly, however, the 10E8-0fH/4fL variant that contained no heavy chain and only 4 light chain framework mutations had substantially improved neutralization potency compared to 10E8-0fH/0fL, with only a ~3-fold decrease in potency relative to mature 10E8 (Figure 6A,B).
Figure 6. HIV-1 neutralization by framework revertants of antibody 10E8.
(A) Neutralization breadth at different IC50 neutralization cutoff values. (B) Average (geometric mean) neutralization potency for the different combinations of heavy and light chain variants; grey – combination not tested. (C) Spearman correlation of neutralization potency vs. number of framework mutations (heavy + light); colors same as in (A).
Discussion
The high rate of mutation and the numerous other immune-evasion mechanisms of HIV-1 serve as a constant stimulus for HIV-1-reactive antibodies to continuously evolve and mature as they target the ever-changing viral envelope. Perhaps as a consequence, broadly neutralizing anti-HIV-1 antibodies typically have extreme numbers of variable domain (CDR and framework region) mutations. In the case of VRC01, the importance of framework mutations is underscored by the fact that, while VRC01-5fH/0fL showed virtually no neutralization activity, the addition of 6 framework mutations in the light chain resulted in VRC01-5fH/6fL being within 2-fold potency of the mature antibody. The precise role of each of these 6 mutations is not clear; however three of these mutations (G66R, T69P, and F71Y) clustered closely to CDR L1 (Figure 3B,C), suggesting that these mutations may help support the CDR L1 loop in a conformation appropriate for interaction with antigen. In the case of antibody 10E8, a variant with no mature framework mutations showed reasonable, albeit ~10-fold lower, neutralization activity. Thus, framework maturation in anti-HIV-1 antibodies does not appear to be universally required for potent and broad neutralization activity.
While the overall number of framework mutations can affect neutralization activity, some mutations appeared to have a more substantial effect on antibody activity, so the specific selection of residues for germline reversion is also important. For example, the 10E8-2fH/4fL and 10E8-2fH/0fL variants had a ~4-fold and ~2-fold decrease in potency, respectively, compared to the analogous variants without the 2 heavy chain framework mutations (10E8-0fH/4fL and 10E8-0fH/0fL). Similarly, the VRC01-5fH/6fL variant had a ~2-fold better potency than the substantially more mutated VRC01-8fH/19fL. In the case of 10E8-0fH/4fL, post-hoc analysis of the 10E8-MPER peptide structure indicated that additional reversions of mutations that do not directly interact with the antigen or CDR regions may be possible (e.g., S2Y, Figure 5). Further, the reversion analysis here was focused solely on framework residues; however, it should also be possible to selectively revert CDR mutations as not all mutations within the CDR regions will be important for antigen recognition or overall antibody structure (33, 34). This process could allow for the generation of partially reverted antibody variants with even lower degrees of mutation and with similar, if not better, neutralization activity as their mature bNAb counterparts. Interestingly, we also found that reversion of selected mutations resulted in improved antibody variants compared to the mature antibodies. In the case of VRC01, a combination of the mature heavy chain and a ~50% reverted light chain (VRC01-30fH/10fL) resulted in 2-fold improved potency over the mature antibody. For 10E8, two variants, 10E8-19fH/10fL (mature heavy/~40% reverted light chain) and 10E8-10fH/16fL (~50% reverted heavy/mature light chain), exhibited similar neutralization breadth and potency to the mature antibody. These results show the promise of partial framework reversion for antibody optimization.
Anti-HIV-1 bNAbs with lowered mutation levels may be of utility both in vaccine design and as therapeutic agents. An effective prophylactic HIV-1 vaccine must be able to elicit broadly neutralizing antibodies. The high mutation levels of most anti-HIV-1 bNAbs thus present a challenge for vaccine development and immunogen design since the vaccine immunogen (or perhaps a series of different immunogens) would need to guide the antibody affinity maturation process from germline activation through the appropriate intermediate stages of antibody development to the mature broadly neutralizing antibody (35-37). However, bNAbs with lower levels of mutations, such as those presented here or combined with antibody ontogeny data from next-generation sequencing (1, 9), may be more easily inducible and can be suitable targets for immunogen design, alleviating the requirement for guiding the affinity maturation process to extreme levels of hypermutation. Antibodies with high mutation rates could also potentially be problematic as clinical products since the likelihood of immunogenicity may be increased. While, to our knowledge, no highly somatically mutated antibodies have been evaluated for immunogenicity in humans, antibodies that are closer to their germline ancestors (the genes that are commonly found in most individuals) may be more easily tolerated in the general population. Partial germline reversion may thus be a useful addition to the armamentarium of technologies for translating anti-HIV-1 bNAbs into clinical products. Overall, our results suggest that at least some of the anti-HIV-1 antibodies can retain broad and potent neutralization activity even when their framework regions are substantially reverted back to germline. These results should aid in efforts for immunogen design targeting the elicitation of bnAbs and for antibody-based therapeutics for the treatment or prevention of HIV-1.
Acknowledgments
We thank the Structural Biology Section, Structural Bioinformatics Core Section, Virology Laboratory, Vector Core Section, Humoral Immunology Section, and Humoral Immunology Core at the NIH Vaccine Research Center for helpful discussions and comments on the manuscript. The authors are grateful to J. Stuckey for assistance with graphics, and thank J. Baalwa, D. Ellenberger, D. Gabuzda, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in our neutralization panels.
This work was supported by the Intramural Research Program of the Vaccine Research Center, NIAID, and the Office of AIDS Research, NIH.
Abbreviations
- bNAbs
broadly neutralizing antibodies
- wt
wildtype
- LC
light chain
- HC
heavy chain
References
- 1.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O'Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- 8.Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev Ivelin S., Altae-Tran Han R., Chuang G-Y, Joyce MG, Do Kwon Y, Longo Nancy S., Louder Mark K., Luongo T, McKee K, Schramm Chaim A., Skinner J, Yang Y, Yang Z, Zhang Z, Zheng A, Bonsignori M, Haynes Barton F., Scheid Johannes F., Nussenzweig Michel C., Simek M, Burton Dennis R., Koff Wayne C., Mullikin James C., Connors M, Shapiro L, Nabel Gary J., Mascola John R., Kwong Peter D. Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders KO, Rudicell RS, Nabel GJ. The design and evaluation of HIV-1 vaccines. AIDS. 2012;26:1293–1302. doi: 10.1097/QAD.0b013e32835474d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 16.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breden F, Lepik C, Longo NS, Montero M, Lipsky PE, Scott JK. Comparison of Antibody Repertoires Produced by HIV-1 Infection, Other Chronic and Acute Infections, and Systemic Autoimmune Disease. PLoS One. 2011;6:e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev I, Pancera M, Zhou T, Incesu RB, Fu BZ, Gnanapragasam PN, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, Nussenzweig MC. Somatic mutations of the immunoglobulin framework are generaly required for broad and potent HIV-1 neutralizing activity. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid J, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, Diskin R, Scheid JF, Sather DN, Burton DR, Stamatatos L. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS pathogens. 2013;9:e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annual review of biochemistry. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 22.Bates JT, Keefer CJ, Utley TJ, Correia BE, Schief WR, Crowe JE. Reversion of Somatic Mutations of the Respiratory Syncytial Virus–Specific Human Monoclonal Antibody Fab19 Reveal a Direct Relationship between Association Rate and Neutralizing Potency. The Journal of Immunology. 2013;190:3732–3739. doi: 10.4049/jimmunol.1202964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingwood D, McTamney PM, Yassine HM, Whittle JRR, Guo X, Boyington JC, Wei C-J, Nabel GJ. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber M, Le KM, Doores KJ, Fulton Z, Stanfield RL, Wilson IA, Burton DR. Very Few Substitutions in a Germ Line Antibody Are Required To Initiate Significant Domain Exchange. Journal of virology. 2010;84:10700–10707. doi: 10.1128/JVI.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo BK. Antibody humanization by CDR grafting. Methods Mol Biol. 2004;248:135–159. doi: 10.1385/1-59259-666-5:135. [DOI] [PubMed] [Google Scholar]
- 26.Gainza P, Roberts KE, Georgiev I, Lilien RH, Keedy DA, Chen C-Y, Reza F, Anderson AC, Richardson DC, Richardson JS, Donald BR. Chapter Five - osprey: Protein Design with Ensembles, Flexibility, and Provable Algorithms. In: Keating AE, editor. Methods in Enzymology. Academic Press; 2013. pp. 87–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C-Y, Georgiev I, Anderson AC, Donald BR. Computational structure-based redesign of enzyme activity. Proceedings of the National Academy of Sciences. 2009;106:3764–3769. doi: 10.1073/pnas.0900266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrodinger L. The PyMOL Molecular Graphics System, Version 1.3. 2010:r1. [Google Scholar]
- 30.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human Immunodeficiency Virus Type 1 env Clones from Acute and Early Subtype B Infections for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. Journal of virology. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for neutralizing antibody assessment. Journal of virology. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Zhou T, O'Dell S, Wyatt RT, Kwong PD, Mascola JR. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. Journal of virology. 2009;83:10892–10907. doi: 10.1128/JVI.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Pascalis R, Iwahashi M, Tamura M, Padlan EA, Gonzales NR, Santos AD, Giuliano M, Schuck P, Schlom J, Kashmiri SV. Grafting of “abbreviated” complementarity-determining regions containing specificity-determining residues essential for ligand contact to engineer a less immunogenic humanized monoclonal antibody. J Immunol. 2002;169:3076–3084. doi: 10.4049/jimmunol.169.6.3076. [DOI] [PubMed] [Google Scholar]
- 34.Kim JH, Hong HJ. Humanization by CDR grafting and specificity-determining residue grafting. Methods Mol Biol. 2012;907:237–245. doi: 10.1007/978-1-61779-974-7_13. [DOI] [PubMed] [Google Scholar]
- 35.Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, Kwong PD. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. Journal of virology. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georgiev IS, Joyce MG, Zhou T, Kwong PD. Elicitation of HIV-1-neutralizing antibodies against the CD4-binding site. Current opinion in HIV and AIDS. 2013;8:382–391. doi: 10.1097/COH.0b013e328363a90e. [DOI] [PMC free article] [PubMed] [Google Scholar]