Background: Ubiquitin ligase ATL31 and the target, 14-3-3 proteins, function in plant nutrient response.
Results: ATL31 binds to 14-3-3 proteins via phosphorylation of specific residues. These residues are essential for the function of ATL31.
Conclusion: ATL31 targets 14-3-3 proteins for degradation in a phosphorylation-dependent manner to regulate nutrient response.
Significance: Phosphorylation of ubiquitin ligase ATL31 controls plant nutrient response.
Keywords: E3 Ubiquitin Ligase, Plasma Membrane, Protein Phosphorylation, Protein Stability, Protein-Protein Interactions, Signal Transduction
Abstract
Ubiquitin ligase plays a fundamental role in regulating multiple cellular events in eukaryotes by fine-tuning the stability and activity of specific target proteins. We have previously shown that ubiquitin ligase ATL31 regulates plant growth in response to nutrient balance between carbon and nitrogen (C/N) in Arabidopsis. Subsequent study demonstrated that ATL31 targets 14-3-3 proteins for ubiquitination and modulates the protein abundance in response to C/N-nutrient status. However, the underlying mechanism for the targeting of ATL31 to 14-3-3 proteins remains unclear. Here, we show that ATL31 interacts with 14-3-3 proteins in a phosphorylation-dependent manner. We identified Thr209, Ser247, Ser270, and Ser303 as putative 14-3-3 binding sites on ATL31 by motif analysis. Mutation of these Ser/Thr residues to Ala in ATL31 inhibited the interaction with 14-3-3 proteins, as demonstrated by yeast two-hybrid and co-immunoprecipitation analyses. Additionally, we identified in vivo phosphorylation of Thr209 and Ser247 on ATL31 by MS analysis. A peptide competition assay showed that the application of synthetic phospho-Thr209 peptide, but not the corresponding unphosphorylated peptide, suppresses the interaction between ATL31 and 14-3-3 proteins. Moreover, Arabidopsis plants overexpressing mutated ATL31, which could not bind to 14-3-3 proteins, showed accumulation of 14-3-3 proteins and growth arrest in disrupted C/N-nutrient conditions similar to wild-type plants, although overexpression of intact ATL31 resulted in repression of 14-3-3 accumulation and tolerance to the conditions. Together, these results demonstrate that the physiological role of phosphorylation at 14-3-3 binding sites on ATL31 is to modulate the binding ability and stability of 14-3-3 proteins to control plant C/N-nutrient response.
Introduction
The ability to sense and respond to nutrient levels is critical for the growth of all living systems. Plants, in particular, have developed sophisticated mechanisms to robustly monitor and appropriately respond to the dynamic changes of nutrient availability due to plant immobility. Among many nutrients, carbon and nitrogen are crucial for plants to perform their routine and fundamental cellular activities. Carbon and nitrogen are not only important energy sources and structural components but are also signaling molecules that regulate gene expression, metabolism, physiology, and growth and development (1–4). Because carbon and nitrogen are tightly coordinated during metabolism, in addition to their independent utilization, the ratio of carbon to nitrogen metabolites in the cell is also important for regulation of plant growth and is referred to as the C/N-nutrient3 response (5, 6). Plants sense and adapt to changing carbon/nitrogen dynamics via precise partitioning of carbon and nitrogen sources and fine-tuning of complex cellular metabolic activity (7, 8). The typical plant C/N-nutrient response is observed in the early postgerminative growth of seedlings, at which stage the phase transition from heterotrophic to photo-autotrophic growth occurs in higher plants. Arabidopsis seedlings show purple anthocyanin pigmentation and stop postgerminative growth when grown on disrupted C/N-nutrient medium containing relatively high sugar and low nitrogen (e.g. 300 mm glucose and 0.3 mm nitrogen) (6, 9). The expression of photosynthetic genes, such as RBCS (ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit) and CAB (chlorophyll a/b-binding protein), is down-regulated by excess sugar in the medium and also repressed in nitrogen-deficient conditions (6). On the other hand, the expression of GS2 (glutamine synthase 2), which is involved in the assimilation of inorganic nitrogen in the glutamine synthetase-glutamate synthase cycle, is affected by the amount of sugar and/or light/dark treatment (10). Although the impact of C/N-nutrient status on plant growth has been recognized during the past decades, the detailed molecular mechanism mediating the C/N-nutrient signal are still unclear.
Our laboratory previously revealed that the RING-type ubiquitin ligase ATL31 is essential for correct C/N-nutrient response at an early postgerminative growth stage in Arabidopsis (9). ATL31 overexpression led to a carbon/nitrogen-insensitive phenotype of carbon/nitrogen insensitive 1-dominant (cni1-D) plants and resulted in expansion of green-colored cotyledon under high carbon/low nitrogen conditions, whereas the loss-of-function mutant of ATL31 showed a hypersensitive phenotype (9). ATL31 is a member of the plant-specific ATL ubiquitin ligase family, which comprises proteins that contain a transmembrane-like hydrophobic region at the N terminus, a GLD region, a RING-H2 type zinc finger domain, and a non-conserved C-terminal region (11). Biochemical and genetic studies demonstrated the membrane localization and ubiquitin ligase activity of ATL31, both of which are required for the regulation of C/N-nutrient response in plants (9). Subsequent proteomic analyses identified 14-3-3 proteins as interactors of ATL31 (12). 14-3-3 proteins bind to phosphorylated motifs and function in multiple developmental processes by regulating the activity of a wide variety of target proteins (13–15). In particular, 14-3-3 proteins have been reported to regulate primary carbon and nitrogen metabolism by directly interacting with key enzymes (16–18). Further analyses demonstrated that ATL31 directly ubiquitinates 14-3-3 proteins and regulates protein amounts in response to C/N-nutrient conditions, revealing a plant-specific regulatory mechanism of C/N-nutrient signaling via the ubiquitin-proteasome system (12).
The ubiquitin-proteasome system is one of the most well characterized protein degradation pathways and plays a fundamental role in eukaryotic cellular functions. In this system, ubiquitin ligases function as key regulators to recognize target proteins for proteasomal degradation (19, 20). The ubiquitin ligase ATL is a plant-specific multigene family and widely conserved in basal and higher plants, including mosses, monocots, and dicots (21). The Arabidopsis genome contains 91 members of the ATL proteins (22). Recently, several ATL proteins have been reported to function in multiple environmental stress adaptation (22). ATL2, ATL9, and ATL55/RING1 are involved in plant defense response, and ATL78 mediates adaptation to cold stress (23–26). Moreover, IDF1 (IRT1 (iron-regulated transporter 1) degradation factor 1)/ATL14 was recently identified as an ubiquitin ligase regulating iron uptake by the direct ubiquitination of the iron transporter IRT1 (27). However, less is understood about the molecular mechanism mediating target recognition by ATL proteins, including ATL31. The identification of the interaction domain of ATL31 and the signals mediating the interaction is required for further elucidation of the C/N-nutrient signaling mechanism.
In this study, we investigated the detailed molecular basis of the interaction between ATL31 and 14-3-3 proteins and demonstrated the phosphorylation signal on specific Ser/Thr residues in the C-terminal region of ATL31 that mediate the interaction with 14-3-3 proteins. Moreover, analysis of the C/N-nutrient response with transgenic Arabidopsis plants overexpressing mutated ATL31 that could not bind to 14-3-3 proteins indicated the physiological impact of these residues for correct growth regulation via 14-3-3 degradation in response to C/N-nutrient conditions.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the wild type and the genetic background for all transgenic lines. ATL31-overexpressing plants (35S-ATL31) were as described previously (9). Col-0 and all transgenic plants were grown under conditions as described previously (28). MM2d Arabidopsis cell suspension culture was maintained as described previously (29, 30). ATL31C143S-overexpressing MM2d cells (35S-ATL31C143S-FLAG) were as described previously (12). Nicotiana benthamiana was grown under conditions of 16 h light/8 h dark at 22 °C.
Plasmid Construction
The coding sequences of ATL6, ATL11, ATL31, 14-3-3χ, 14-3-3υ, 14-3-3λ, 14-3-3ι, 14-3-3ϵ, and FLS2 (flagellin-sensing 2) were amplified from Col-0 cDNA. The substituted forms of ATL31 and 14-3-3λ were generated by PCR-based site-directed mutagenesis. All primers used are listed in Table 1. Amplified fragments were cloned into pENTR/D-TOPO vector and then transferred into destination vectors using the Gateway system according to the manufacturer's protocol (Invitrogen). All amplified fragments and inserts were verified by DNA sequencing.
TABLE 1.
List of primers used in this study
Plant Transformation
To generate ATL31-4A-overexpressing Arabidopsis plants (35S-ATL31–4A) and ATL31C143S-4A-overexpressing MM2d cells, the full-length coding sequence of ATL31 with introduced T209A/S247A/S270A/S303A and C143S/T209A/S247A/S270A/S303A substitutions was subcloned into pGWB11 destination vector (31). The constructs were introduced into Agrobacterium tumefaciens strain GV3101 (pMP90) by electroporation. Arabidopsis plants were transformed using the floral dip method (32), and transformation of MM2d cells was performed as described previously (29, 30). Three independent T4 homozygous lines for 35S-ATL31–4A were selected for the carbon/nitrogen response assay.
Yeast Two-hybrid Assay
Yeast strain EGY48 was used for the two-hybrid assays. The truncated and substituted forms of ATL31, ATL6, and ATL11 were subcloned into pEG202gw destination vector as bait, and the full-length coding sequences of 14-3-3χ, 14-3-3υ, 14-3-3λ, 14-3-3ι, and 14-3-3ϵ as well as the truncated and substituted forms of 14-3-3λ were subcloned into pJG4–5gw destination vector as prey (Gateway-compatible versions of pEG202 and pJG4-5 vectors were kindly provided by Dr. Hironori Kaminaka, Tottori University). The pEG202 and pJG4-5 vectors were used as vector control. Yeast transformation was performed by the Frozen-EZ Yeast Transformation II Kit (Zymo Research) according to the manufacturer's protocol. Handling of yeast cultures and β-galactosidase assays was performed as described in the Yeast Protocols Handbook (Clontech). Amounts of expressed 14-3-3 proteins were verified by Western blotting using anti-HA antibody (Wako).
Transient Expression in N. benthamiana
For the co-immunoprecipitation assay, the truncated and substituted forms of ATL31 were subcloned into pGWB11 destination vector (31), and the full-length coding sequence of 14-3-3λ and the truncated and substituted forms of 14-3-3λ were subcloned into pEarleygate203 destination vector (33). For subcellular localization analysis, the full-length coding sequence of ATL31, which introduced C143S substitution (ATL31C143S), was subcloned into pMDC83 destination vector (34), and the full-length coding sequence of 14-3-3λ was subcloned into pB4GWmC destination vector. For the bimolecular fluorescence complementation (BiFC) assay, ATL31C143S was subcloned into pB4GWnG destination vector, the full-length coding sequence of 14-3-3λ was subcloned into pB4GWcG destination vector, and the full-length coding sequence of FLS2 was subcloned into pB4GWmC destination vector (pB4 destination vectors were kindly provided by Dr. Shoji Mano, National Institute for Basic Biology). All constructs were introduced into A. tumefaciens strain GV3101 (pMP90) by electroporation. Agrobacterium containing the above constructs was grown in 2× YT medium overnight at 28 °C with shaking. Cultured agrobacterium cells were collected and resuspended with infiltration buffer (10 mm MES, 10 mm MgCl2, and 450 μm acetosyringone, pH 5.6). To enhance transgene expression, agrobacterium expressing the p19 suppressor was resuspended together with all samples (35). Agrobacterium suspensions were mixed at equal ratios, and the mixture was infiltrated into the leaves of 3–5-week-old N. benthamiana plants using a needleless syringe.
Co-immunoprecipitation Assay
N. benthamiana leaves and MM2d cells were ground in liquid nitrogen and lysed with extraction buffer (50 mm Tris, 1% Triton X-100, 150 mm NaCl, 10% glycerol, 1 mm EDTA, pH 7.5) supplemented with 10 μm MG132 and Complete Protease Inhibitor Mixture (Roche Applied Science). After the removal of insoluble materials by centrifugation two times at 20,000 × g for 5 min at 4 °C, protein concentrations were determined and normalized by a protein assay (Bio-Rad) using bovine serum albumin (BSA) as a standard. Anti-FLAG M2 antibody conjugated to agarose beads (Sigma) was added to the extracts and gently shaken for 1 h at 4 °C with a rotary shaker. The beads were washed three times with extraction buffer, and bound proteins were eluted with SDS sample buffer (62.5 mm Tris, 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.01% bromphenol blue, pH 6.8) and heated for 30 min at 55 °C. Portions of the extracts prior to immunoprecipitation were also boiled with SDS sample buffer. The proteins were detected by Western blotting using anti-FLAG M2 antibody (Sigma), anti-Myc tag antibody (MBL), and anti-14-3-3 antibody (12).
Fluorescent Microscopy
Infiltrated N. benthamiana leaves were used for microscopic analysis at 2 days after infiltration. Fluorescent images were obtained using a Zeiss LSM510 laser scanning microscope equipped with a C-Apochromat (×40/1.2 numerical aperture water immersion) objective. Excitation/detection wavelengths were 488/505–550 nm for green fluorescent protein (GFP) and BiFC constructs and 561/575–615 nm for mCherry constructs. Image processing was carried out using ImageJ software (National Institutes of Health).
Identification of in Vivo Phosphorylation Sites on ATL31 Using Mass Spectrometry
ATL31C143S-FLAG-overexpressing MM2d cells were ground in liquid nitrogen and lysed with extraction buffer (50 mm Tris, 1% Triton X-100, 150 mm NaCl, 10% glycerol, 1 mm EDTA, pH 7.5) supplemented with 10 μm MG132, Complete Protease Inhibitor Mixture (Roche Applied Science), and PhosSTOP (Roche Applied Science). After the removal of insoluble materials by centrifugation two times at 20,000 × g for 5 min at 4 °C, protein concentrations were determined and normalized by a protein assay (Bio-Rad) using BSA as a standard. Anti-FLAG M2 antibody conjugated to agarose beads (Sigma) was added to the extracts and gently shaken for 1 h at 4 °C with a rotary shaker. The beads were washed three times with extraction buffer, and bound proteins were eluted with 150 μg/ml 3× FLAG peptide (Sigma) in extraction buffer. After precipitation by cold acetone, the proteins were resuspended with SDS sample buffer and heated to 55 °C for 30 min. The samples were subjected to SDS-PAGE followed by mass spectrometry analysis. For mass spectrometry analysis, a specific band containing ATL31 was excised after staining with Flamingo stain solution (Bio-Rad). Peptides for LC-MS/MS analysis were then prepared by in-gel digestion using sequencing grade modified trypsin (Promega) (36), and LC-MS/MS analysis was performed using a LTQ-orbitrap XL-HTC-PAL system (Thermo Fisher Scientific). The MS/MS spectra were compared against TAIR10 (Arabidopsis Information Resource), and MS scores were calculated using the MASCOT server as described previously (37).
Identification of in Vivo Phosphorylation of Thr209 on ATL31
Anti-ATL31 phospho-Thr209 antibody was generated by SCRUM Inc. (Tokyo, Japan) using a phosphorylated Thr209 peptide (CFPRSHpTTGHS). ATL31C143S-FLAG and ATL31C143S-T209A-FLAG were transiently expressed in N. benthamiana leaves and immunoprecipitated with anti-FLAG M2 antibody conjugated to agarose beads (Sigma) as described above. The proteins were detected by Western blotting using monoclonal anti-FLAG M2 antibody (Sigma) and anti-ATL31 phospho-Thr209 antibody.
Protein Expression and Purification
For glutathione S-transferase (GST)-KTA purification, the coding sequence of kinase targeted to ATL31 (KTA) was subcloned into pYU1274 destination vector (Gateway-compatible version of pGEX4T-1 kindly provided by Dr. Yoshihisa Ueno, National Institute of Agrobiological Sciences). For His-14-3-3λ purification, the full-length coding sequence of 14-3-3λ was subcloned into pDEST-his destination vector (38). These constructs were transformed into Escherichia coli strain BL21 (DE3) pLysS. The overnight cultures were diluted 1:100 into LB medium and grown at 37 °C with shaking. When the A600 reached ∼0.6, isopropyl β-d-thiogalactopyranoside was added to the cultures to a final concentration of 0.2 mm, and the incubation was continued for 16 h at 18 °C. The cells were collected and resuspended with lysis buffer (50 mm Tris, 0.1% Triton X-100, 100 mm NaCl, 1 mm MgCl2, 10% glycerol) supplemented with Complete Protease Inhibitor Mixture (Roche Applied Science). After three freeze-thaw cycles, the lysates were incubated with Benzonase nuclease (Novagen) for 10 min at 25 °C with shaking and clarified by centrifugation at 20,000 × g for 10 min at 4 °C. GST-KTA and His-14-3-3λ were purified using glutathione-Sepharose 4B (GE Healthcare) and Talon metal affinity resin (Clontech), respectively, according to the manufacturer's protocol. Protein concentrations were determined by a protein assay (Bio-Rad) using BSA as a standard. Maltose-binding protein (MBP)-ATL31 ΔTM was as described previously (9).
In Vitro Phosphorylation and Far Western Blotting
MBP-ATL31 ΔTM (1.5 μg) was incubated with GST (1.5 μg) or GST-KTA (1.5 μg) in 30 μl of reaction buffer (20 mm Tris, 2.5 mm MgCl2, 1 mm MnCl2, 0.5 mm CaCl2, 2 mm DTT, 1 mm ATP) for 60 min at 30 °C. The reaction was terminated by the addition of SDS sample buffer and heated for 5 min at 95 °C. Proteins were detected by Western blotting using anti-GST monoclonal antibody (Wako) and anti-MBP antiserum (New England Biolabs). To determine the phosphorylation of MBP-ATL31 ΔTM, the samples were subjected to Mn2+-Phos-tag SDS-PAGE (7.5% polyacrylamide gel, including 50 μm MnCl2 and 50 μm Phos-tag acrylamide (Wako)) and analyzed by Western blotting using anti-MBP antiserum (New England Biolabs).
For far Western blotting, the samples were subjected to SDS-PAGE and transferred to Immobilon-P membrane (Millipore). The membrane was incubated with blocking buffer (10 mm Tris, 150 mm NaCl, 0.1% Tween 20, 5% nonfat dry milk, pH 7.5) for 30 min at room temperature and then reacted with 0.1 μm His-14-3-3λ in blocking buffer for 1 h at room temperature. Bound His-14-3-3λ was subsequently detected using anti-His antibody (GE Healthcare).
Peptide Competition Assay
Thr209 peptide (CFPRSHTTGHS) and phosphorylated Thr209 peptide (CFPRSHpTTGHS) corresponding to the sequence surrounding Thr209 on ATL31 were synthesized by SCRUM Inc. (Tokyo, Japan). Anti-FLAG M2 antibody conjugated to agarose beads (Sigma) was added to N. benthamiana leaf extracts containing ATL31C143S-FLAG and Myc-14-3-3λ and gently shaken for 1 h at 4 °C with a rotary shaker. The beads were washed three times with extraction buffer and incubated with 1 mm Thr209 peptide or 1 mm phospho-Thr209 peptide in TBS buffer (10 mm Tris, 150 mm NaCl, pH 7.5) for 1 h at 4 °C. After centrifugation, the supernatants were collected, and the beads were washed three times with extraction buffer. Bound proteins were eluted with SDS sample buffer and heated to 55 °C for 30 min. Portions of the extracts prior to immunoprecipitation and collected supernatants were also boiled with SDS sample buffer. Proteins were detected by Western blotting using monoclonal anti-FLAG M2 antibody (Sigma) and monoclonal anti-Myc tag antibody (MBL).
RT-PCR Analysis
Total RNA was isolated from indicated plant materials using TRIzol reagent (Invitrogen) and treated with RQ1 RNase-free DNase (Promega) according to the manufacturer's protocol. First-strand cDNA was synthesized from 1 μg of total RNA using oligo(dT) primer and ReverTraAce reverse transcriptase (TOYOBO). First strand cDNA was then assayed for gene-specific DNA fragments using the primer pairs listed in Table 1. PCR amplification was performed using optimum cycles with each gene using the Ex Taq polymerase (TaKaRa). Amplified fragments were separated on agarose gels and visualized by ethidium bromide staining.
Carbon/Nitrogen Response Assay
Approximately 50 seeds of each genotype that were surface-sterilized and sown on Murashige and Skoog solid medium with no glucose, 20 mm KNO3, and 20 mm NH4NO3 (0 mm Glc, 60 mm nitrogen) or 300 mm glucose, 0.1 mm KNO3, 0.1 mm NH4NO3, and 19.9 mm KCl (300 mm Glc, 0.3 mm nitrogen). After cold treatment for 2 days to synchronize germination, seeds were transferred to 22 °C under a 16-h light/8-h dark photoperiod (this time point indicates 0 days old). The percentage of cotyledon greening was calculated at 7 days old from three independent experiments. To analyze the amount of 14-3-3 proteins, 7-day-old seedlings were ground in liquid nitrogen and lysed with extraction buffer (50 mm Tris, 1% Triton X-100, 150 mm NaCl, 10% glycerol, 1 mm EDTA, pH 7.5) supplemented with 10 μm MG132 and Complete Protease Inhibitor Mixture (Roche Applied Science). After the removal of insoluble materials by centrifugation two times at 20,000 × g for 5 min at 4 °C, protein concentrations were determined and normalized by a protein assay (Bio-Rad) using BSA as a standard. After heating with SDS sample buffer for 30 min at 55 °C, the proteins were analyzed by Western blotting using anti-14-3-3 antibody and anti-α-tubulin mouse mAb (Calbiochem).
RESULTS
The 14-3-3 Proteins Interact with the C-terminal Region of ATL31
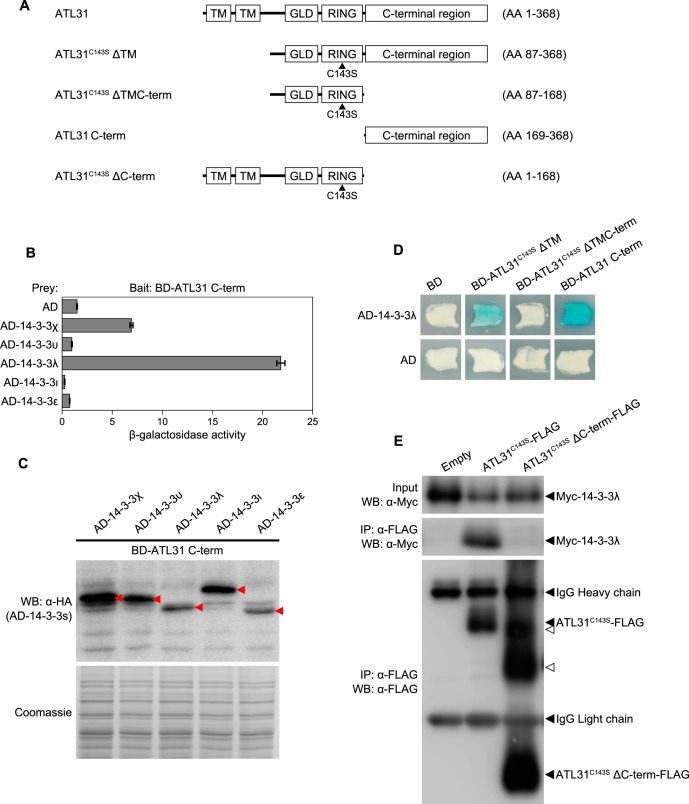
In Arabidopsis, 14-3-3 proteins consist of 13 isoforms, and each isoform can be separated into two major evolutionary branches, the ϵ group and the non-ϵ group (39). Each isoform has a different binding affinity to the client proteins (40, 41). Previously, we have shown that 14-3-3χ belonging to the non-ϵ group is directly associated and ubiquitinated by ATL31 (12). In addition to 14-3-3χ, immunoprecipitation and MS analysis revealed that several 14-3-3 isoforms are putative targets of ATL31, with the stability of each isoform being different in response to C/N-nutrient conditions (12). To evaluate the specificity of the interaction between ATL31 and each 14-3-3 isoform, we carried out yeast two-hybrid (Y2H) analysis. Among 13 isoforms, 14-3-3χ, -υ, and -λ were chosen as representative of the non-ϵ group, whereas 14-3-3ι and -ϵ were chosen as representative of the ϵ group. Because the C-terminal region of ATL31 (aa 169–368) was shown to interact with 14-3-3χ in Y2H analysis (12), we constructed the C-terminal region of ATL31 fused to the LexA DNA-binding domain (BD-ATL31 C-term) (Fig. 1A) and the full length of 14-3-3χ, -υ, -λ, -ι, or -ϵ fused to the B42 activation domain (AD-14-3-3). The constructs were co-transformed into yeast cells and the interactions were evaluated by β-galactosidase activity in a liquid assay using o-nitrophenyl-β-d-galactoside. Yeast cells harboring 14-3-3χ and -λ showed high β-galactosidase activity, whereas 14-3-3υ, -ι, and -ϵ had low activity (Fig. 1B). All AD-14-3-3 proteins were expressed in yeast, and the expression levels did not correlate with β-galactosidase activity (Fig. 1C). The 14-3-3λ showed higher activity than that of 14-3-3χ, although the expressed protein amount was lower (Fig. 1, B and C). These results indicate that 14-3-3λ has a higher binding affinity to ATL31 compared with the 14-3-3χ. Thus, the 14-3-3λ was chosen for further study.
FIGURE 1.
Binding domain of ATL31 and binding affinity to different 14-3-3 isoforms. A, drawings represent a schematic of ATL31 domains used in yeast two-hybrid and co-immunoprecipitation analyses. TM, transmembrane-like hydrophobic region; GLD, highly conserved region including Gly-Leu-Asp residues; RING, a RING-H2 type zinc finger domain. Mutated ATL31 with substitution of the conserved Cys143 residue in the RING domain to Ser (ATL31C143S), which lost ubiquitin ligase activity, was used to avoid the degradation of interacted 14-3-3 proteins. B, interactions between ATL31 and 14-3-3 isoforms in yeast. The C-terminal region of ATL31 fused to the LexA DNA binding domain (BD-ATL31 C-term) and the indicated 14-3-3 isoforms fused to the B42 activation domain (AD-14-3-3s) were co-transformed into yeast cells. Protein interactions were evaluated by β-galactosidase activity in a liquid assay using o-nitrophenyl-β-d-galactoside (ONPG). AD, no-fusion B42 activation domain. Data shown are means ± S.D. (error bars) from three independent assays. C, expression of Arabidopsis 14-3-3 proteins in yeast. Total proteins were extracted from yeast cells expressing the indicated proteins. AD-14-3-3 proteins were detected by Western blotting using anti-HA antibody. Coomassie staining was used as a loading control. Red arrowheads, AD-14-3-3 proteins. D, interactions between the truncated forms of ATL31 and 14-3-3λ in yeast. The indicated ATL31 constructs and AD-14-3-3λ were co-transformed into yeast cells. Yeast transformants were streaked onto solidified medium supplemented with 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal), and photographs were taken 2 days later. Blue patches indicate positive interaction. BD, no-fusion LexA DNA binding domain. E, effect of deletion of the C-terminal region of ATL31 on its association with 14-3-3λ in plant. ATL31C143S-FLAG or ATL31C143S ΔC-term-FLAG were transiently co-expressed with Myc-14-3-3λ in N. benthamiana leaves. Leaf extracts (Input) were subjected to immunoprecipitation with anti-FLAG beads (IP: α-FLAG), and proteins were analyzed by Western blotting using anti-FLAG (WB: α-FLAG) and anti-Myc antibodies (WB: α-Myc). Open arrowheads, position of ATL31C143S ΔC-term-FLAG after unknown post-translational modifications.
To further confirm the requirements of the C-terminal region of ATL31 binding to 14-3-3, we generated the truncated forms of ATL31 and tested the interaction with 14-3-3λ (Fig. 1A). Truncation of the C-terminal region of ATL31 abolished the interaction with 14-3-3λ in Y2H (Fig. 1D). This result was also confirmed by co-immunoprecipitation (co-IP) analysis following transient expression in N. benthamiana leaves. Myc-tagged 14-3-3λ (Myc-14-3-3λ) was co-immunoprecipitated with FLAG-tagged ATL31C143S (ATL31C143S-FLAG), which has no ubiquitin ligase activity, but not with the C-terminal region deletion mutant (ATL31C143S ΔC-term-FLAG) (Fig. 1, A and E). These results demonstrate that 14-3-3 proteins bind to the C-terminal region but not to other regions of ATL31. We therefore utilized ATL31C143S for interaction analysis, avoiding the possible degradation of the 14-3-3 proteins that interact with ATL31.
ATL31 Interacts with 14-3-3 Proteins at the Plasma Membrane
Previously, we have shown that ATL31 is localized at the plasma membrane, and the localization is essential for the regulation of C/N-nutrient response (9). This finding suggests that ATL31 interacts with 14-3-3 proteins and regulates their stability at the plasma membrane. To test this possibility, we explored the distribution of ATL31 and 14-3-3λ complex in plant cells. First, ATL31C143S fused to GFP at the C terminus (ATL31C143S-GFP) and 14-3-3λ fused to mCherry at the N terminus (mCherry-14-3-3λ) were transiently co-expressed in N. benthamiana leaf epidermal cells for co-localization analysis. The fluorescence of ATL31C143S-GFP was observed in the plasma membrane, as shown in a previous study (Fig. 2A) (9). By contrast, the fluorescence of mCherry-14-3-3λ was observed at the plasma membrane as well as in the cytoplasm and nucleus (Fig. 2A). The fluorescence of ATL31C143S-GFP overlapped with mCherry-14-3-3λ at the plasma membrane (Fig. 2A), indicating that ATL31 and 14-3-3λ were co-localized at the plasma membrane.
FIGURE 2.
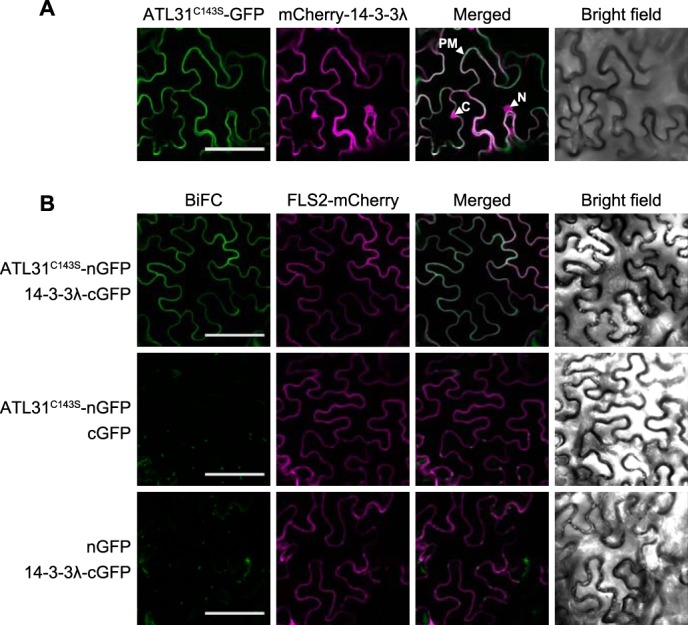
Subcellular distribution of ATL31 and 14-3-3λ complex in plant cells. A, subcellular localization of ATL31 and 14-3-3λ. ATL31C143S-GFP and mCherry-14-3-3λ were transiently co-expressed in N. benthamiana leaf epidermal cells. Confocal images were taken at 2 days after inoculation. PM, plasma membrane; C, cytoplasm; N, nucleus. B, BiFC analysis of ATL31 and 14-3-3λ. BiFC constructs to express ATL31C143S fused with the N-terminal half of GFP (ATL31C143S-nGFP) and 14-3-3λ fused with the C-terminal half of GFP (14-3-3λ-cGFP) were prepared, respectively. BiFC constructs were transiently co-expressed in N. benthamiana leaf epidermal cells. FLS2-mCherry was co-expressed as a transformation and plasma membrane marker. Confocal images were taken at 2 days after inoculation. Bars, 100 μm.
Next, we performed BiFC analysis to evaluate the direct interaction between ATL31 and 14-3-3λ at the plasma membrane. The N- or C-terminal parts of GFP were fused to ATL31C143S and 14-3-3λ, respectively (ATL31C143S-nGFP and 14-3-3λ-cGFP). The constructs were transiently co-expressed in N. benthamiana leaf epidermal cells with FLS2-mCherry used as a transformation and plasma membrane marker (42). As a result, reconstituted GFP fluorescence was observed at the cell surface when ATL31C143S-nGFP and 14-3-3λ-cGFP were co-expressed (Fig. 2B, top panels), whereas co-expression of ATL31C143S-nGFP with a no-fusion cGFP or 14-3-3λ-cGFP with a no-fusion nGFP yielded no GFP fluorescence (Fig. 2B, middle and bottom panels). The fluorescence of FLS2-mCherry was observed in all combinations and completely overlapped with the reconstituted GFP. Together, these results clearly demonstrate that ATL31 interacts with 14-3-3 proteins at the plasma membrane in plant cells.
14-3-3 Proteins Interact with ATL31 via the Phosphopeptide Binding Pocket
14-3-3 proteins form a homo- or heterodimer with the N-terminal region and are able to simultaneously bind to two distinct sites of a single target protein or two different target proteins (43). Each 14-3-3 monomer contains a conserved amphipathic groove that mediates the association of 14-3-3 proteins with phosphoserine- or threonine- containing motifs on their client proteins (44, 45). To test whether the dimerization and the conserved amphipathic groove are involved in the interaction with ATL31, we constructed mutated forms of 14-3-3λ in which the N-terminal region (aa 1–29) was deleted (ΔN) to inhibit dimer formation (46). Alternatively, a conserved Lys residue in the amphipathic groove was substituted by Glu (K56E) to abolish the ability to bind to the phosphopeptide (46). In Y2H analysis, the ΔN mutation did not affect the interaction with ATL31, whereas the K56E mutation significantly inhibited it (Fig. 3A). To further examine these results in plant cells, we carried out co-IP analysis. ATL31C143S-FLAG was transiently co-expressed with the native or mutated forms of Myc-14-3-3λ in N. benthamiana leaves and then subjected to co-IP and Western blotting analysis. The 14-3-3λ ΔN was co-immunoprecipitated with ATL31, indicating that 14-3-3 proteins can interact with ATL31 as monomers (Fig. 3B). The interaction between ATL31 and 14-3-3λ was only partially inhibited by the K56E mutation, which was inconsistent with the results of Y2H analysis that showed no interaction (Fig. 3, A and B). It is possible that Arabidopsis 14-3-3λ K56E could not interact with ATL31 but, rather, with the endogenous 14-3-3 isoforms of N. benthamiana which can form a heterodimer with Arabidopsis 14-3-3λ K56E protein and in turn mediate the interaction with ATL31. To evaluate this possibility, we used the 14-3-3λ protein with both N-terminal deletion and Lys substitution (14-3-3λ ΔN/K56E), a protein that showed a marked reduction in binding activity (Fig. 3B). These results indicate that the conserved Lys residue in the amphipathic groove of 14-3-3 proteins is essential for the binding to ATL31.
FIGURE 3.
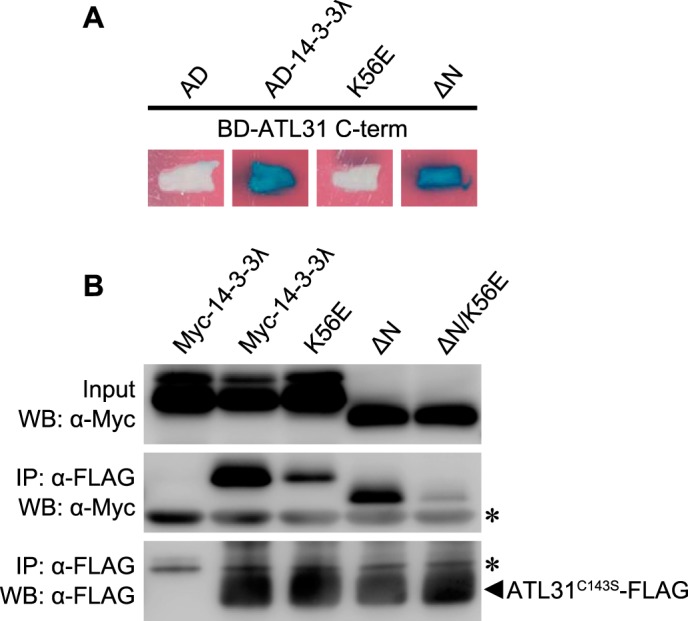
Effects of the mutation of 14-3-3λ on the interaction with ATL31. A, effects of the N-terminal deletion and the K56E mutation of 14-3-3λ on the interaction with ATL31 in yeast. The C-terminal region of ATL31 fused to the LexA DNA binding domain (BD-ATL31 C-term) and the native, N-terminus-deleted, and Lys-56 → Glu-substituted 14-3-3λ fused to the B42 activation domain (AD-14-3-3λ, AD-14-3-3λ ΔN, and AD-14-3-3λ K56E, respectively) were co-transformed into yeast cells. Yeast transformants were streaked onto solidified medium supplemented with X-Gal, and photographs were taken 2 days later. Blue patches indicate positive interaction. AD, no fusion B42 activation domain. B, effects of the N-terminal deletion and the K56E mutation of 14-3-3λ on the interaction with ATL31 in plant. ATL31C143S-FLAG and the indicated mutant variants of Myc-14-3-3λ were transiently co-expressed in N. benthamiana leaves. Leaf extracts (Input) were subjected to immunoprecipitation with anti-FLAG beads (IP: α-FLAG), and proteins were analyzed by Western blotting using anti-FLAG (WB: α-FLAG) and anti-Myc antibodies (WB: α-Myc). *, immunoglobulin chain.
ATL31 Interacts with 14-3-3 Proteins via Consensus 14-3-3 Binding Motif in the C-terminal Region
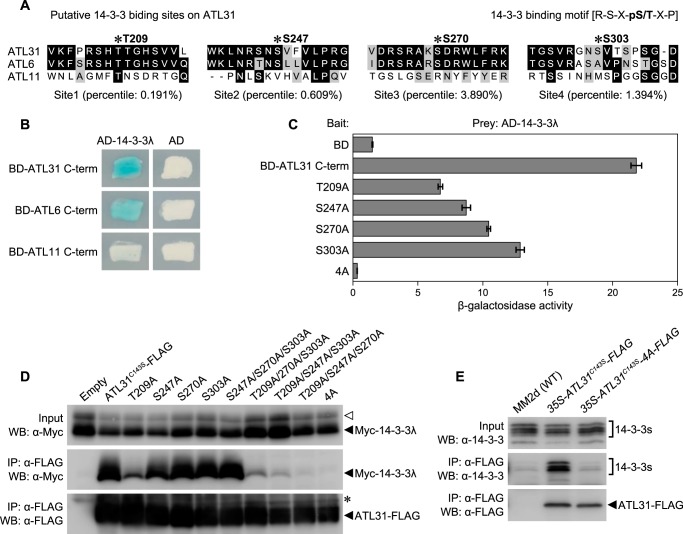
14-3-3 proteins are known to be mostly associated with two types of phosphopeptides: RSX(pS/T)XP (mode I) or RSXX(pS/T)XP (mode II), in which the phospho-Ser/Thr (pS/T) residue is required for 14-3-3 binding (45, 47). Scansite analysis of the C-terminal region of the ATL31 amino acid sequence revealed four putative 14-3-3 binding sites (Thr209, Ser247, Ser270, and Ser303) corresponding to the mode I 14-3-3 binding motif (Fig. 4A) (48). Among these sites, Thr209 showed the highest score (lowest percentile). These sites, except for Ser303, were conserved in ATL6, the most closely related homologue to ATL31, but not in ATL11 (Fig. 4A). Y2H analysis revealed that ATL6 also interacts with 14-3-3λ, whereas ATL11 did not show any interaction (Fig. 4B), suggesting that Thr209, Ser247, Ser270, and Ser303 residues in the C-terminal region of ATL31 may be involved in the interaction with 14-3-3 proteins.
FIGURE 4.
Identification of 14-3-3 protein binding sites on ATL31. A, putative 14-3-3 binding sites on the C-terminal region of ATL31 predicted by Scansite and the conservation in ATL6 and ATL11. *, possible phosphorylation sites. Scansite percentile scores are shown for each sequence. Identical residues conserved in two or three sequences are marked in black boxes, whereas similar residues are marked in gray. B, interactions of ATL31, ATL6, and ATL11 with 14-3-3λ in yeast. The C-terminal region of ATL31, ATL6, and ATL11 fused to the LexA DNA binding domain (BD-ATL31 C-term, BD-ATL6 C-term, and BD-ATL11 C-term) and 14-3-3λ fused to the B42 activation domain (AD-14-3-3λ) were co-transformed into yeast cells. Yeast transformants were streaked onto solidified medium supplemented with X-Gal, and photographs were taken 2 days later. Blue patches indicate positive interaction. AD, no-fusion B42 activation domain. C, effects of alanine substitution to putative 14-3-3 binding sites of ATL31 on the interaction with 14-3-3λ in yeast. BD-ATL31 C-term and the mutated forms with single or quadruple alanine substitution (T209A, S247A, S270A, S303A, or 4A) were co-transformed with AD-14-3-3λ into yeast cells. Protein interactions were evaluated by β-galactosidase activity in a liquid assay using o-nitrophenyl-β-d-galactoside. Data shown are means ± S.D. (error bars) from three independent assays. D, effects of alanine substitution to putative 14-3-3 binding sites of ATL31 on the interaction with 14-3-3λ in plant. ATL31C143S-FLAG and the indicated mutant variants were transiently co-expressed with Myc-14-3-3λ in N. benthamiana leaves. Leaf extracts (Input) were subjected to immunoprecipitation with anti-FLAG beads (IP: α-FLAG), and proteins were analyzed by Western blotting using anti-FLAG (WB: α-FLAG) and anti-Myc antibodies (WB: α-Myc). In addition to single and quadruple mutations, ATL31 proteins carrying triple Ala substitutions were tested (S247A/S270A/S303A, T209A/S270A/S303A, T209A/S247A/S303A, and T209A/S247A/S270A). Open arrowhead, nonspecific band. *, immunoglobulin heavy chain. E, interaction between ATL31–4A and endogenous 14-3-3 proteins in Arabidopsis cells. Extracts (Input) from WT and transgenic Arabidopsis cultured cells overexpressing ATL31C143S-FLAG (35S-ATL31C143S-FLAG) or ATL31C143S-4A-FLAG (35S-ATL31C143S-4A-FLAG) were immunoprecipitated with anti-FLAG beads (IP: α-FLAG). Proteins were analyzed by Western blotting using anti-FLAG (WB: α-FLAG) and anti-14-3-3 antibodies (WB: α-14-3-3).
To examine whether these residues on ATL31 are involved in the interaction with 14-3-3 proteins, we introduced a single or multiple alanine substitution of ATL31 at Thr209, Ser247, Ser270, and Ser303. In Y2H analysis, β-galactosidase activity was reduced by a single alanine substitution at one of four residues (T209A, S247A, S270A, or S303A) compared with the native form. T209A substitution showed the greatest reduction (Fig. 4C). Moreover, the β-galactosidase activity was significantly reduced by a quadruple alanine substitution (ATL31-4A) (Fig. 4C). Similar results were also demonstrated by co-IP and Western blotting analysis with N. benthamiana leaves (Fig. 4D). The amount of co-immunoprecipitated 14-3-3λ was dramatically decreased by T209A single substitution and triple substitution containing T209A (T209A/S270A/S303A, T209A/S247A/S303A, and T209A/S247A/S270A). On the contrary, triple substitution, except for Thr209 (S247A/S270A/S303A), did not have an obvious effect on the interaction; nor did a single substitution, except for T209A (Fig. 4D). ATL31-4A did not show the interaction with 14-3-3λ (Fig. 4D), suggesting that these residues are involved in the interaction with 14-3-3 proteins, and especially Thr209 plays the central role for 14-3-3 binding among the four residues. To evaluate these results in Arabidopsis, we prepared Arabidopsis cultured cells constitutively expressing ATL31C143S-4A-FLAG (35S-ATL31C143S-4A-FLAG) and tested the interaction with endogenous 14-3-3 proteins. Co-IP and Western blotting analysis demonstrated that ATL31C143S-4A-FLAG is not able to interact with endogenous 14-3-3 proteins, whereas intact ATL31C143S-FLAG did interact (Fig. 4E). Taken together, we conclude that Thr209, Ser247, Ser270, and Ser303 residues on ATL31 are essential for the interaction with 14-3-3 proteins.
Phosphorylation of 14-3-3 Binding Sites on ATL31 Is Critical for the Interaction
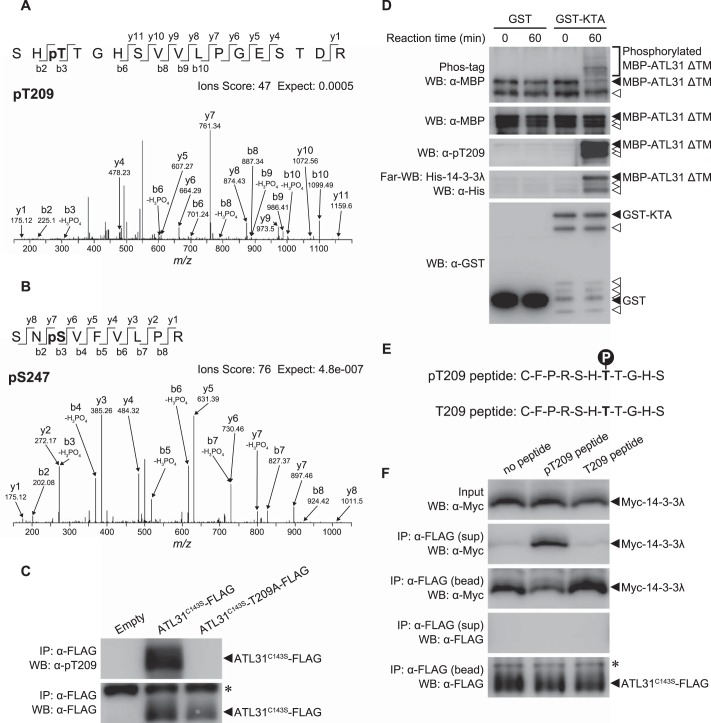
As shown in Fig. 3, the conserved amphipathic groove of 14-3-3λ is required for the interaction with ATL31, suggesting that 14-3-3 proteins bind to ATL31 in a phosphorylation-dependent manner. To evaluate this possibility, we first identified the in vivo phosphorylation sites of the ATL31 protein by mass spectrometry analysis. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis with immunoprecipitated ATL31C143S-FLAG from transgenic Arabidopsis cultured cells identified phosphorylation on Thr209 and Ser247 (Fig. 5, A and B), both of which are considered putative phosphorylation residues for the 14-3-3 binding sites on ATL31 (Fig. 4A). To further examine the phosphorylation, we generated the phospho-specific antibody (anti-Thr(P)209 antibody), raised against the Thr(P)209 peptide (for ATL31 consisting of amino acids Cys203–Ser213 with phosphorylation at Thr209; see Fig. 5E) and detected phosphorylation on the Thr209 residue in vivo. Immunoprecipitated ATL31C143S-FLAG was detected by anti-Thr(P)209 antibody, whereas ATL31 substituted with Ala at Thr209 (ATL31C143S-T209A-FLAG) was not detected (Fig. 5C). Together, these results demonstrate that 14-3-3 binding sites on ATL31 are phosphorylated in vivo.
FIGURE 5.
Effects of the phosphorylation of ATL31 on the interaction with 14-3-3λ. A and B, identification of in vivo phosphorylation sites of ATL31 by mass spectrometry. Extracts from transgenic Arabidopsis cultured cells overexpressing ATL31C143S-FLAG were subjected to immunoprecipitation with anti-FLAG beads. The proteins were separated by SDS-PAGE and in-gel digested with trypsin, and the resulting peptides were extracted and analyzed by LC-MS/MS. MS/MS spectra showing phosphorylation of Thr209 (A) and Ser247 (B) on ATL31. C, in vivo phosphorylation of Thr209 on ATL31. ATL31C143S-FLAG or ATL31C143S-T209A-FLAG was transiently expressed in N. benthamiana leaves. Leaf extracts were subjected to immunoprecipitation with anti-FLAG beads (IP: α-FLAG), and proteins were analyzed by Western blotting using anti-FLAG (WB: α-FLAG) and anti-ATL31 phospho-Thr209 antibodies (WB: α-pT209). *, immunoglobulin heavy chain. D, in vitro phosphorylation of ATL31 and far Western blotting analysis. Recombinant MBP-ATL31 ΔTM protein was incubated with GST or GST fused to the protein kinase targeting ATL31 (GST-KTA) in the reaction buffer at 30 °C for 60 min. After the reaction, proteins were subjected to Phos-tag SDS-PAGE (Phos-tag; top) or normal SDS-PAGE followed by Western blotting using anti-MBP (WB: α-MBP), anti-ATL31 phospho-Thr209 (WB: α-pT209), and anti-GST antibodies (WB: α-GST). Far Western blotting was carried out using His-14-3-3λ as a probe (Far-WB: His-14-3-3λ), and bound His-14-3-3λ was detected by Western blotting using anti-His antibody (WB: α-His). Open arrowheads, degraded fragments of MBP-ATL31 ΔTM and GST-KTA. E, amino acid sequences of synthetic peptides used for peptide competition assay. F, peptide competition assay. ATL31C143S-FLAG and Myc-14-3-3λ were transiently co-expressed in N. benthamiana leaves. Leaf extracts (Input) were incubated with anti-FLAG beads for 1 h. After incubation, beads were washed and then incubated with phosphorylated or unphosphorylated ATL31 Thr209 peptide (pT209 peptide and T209 peptide). After 1 h, the mixture was separated into supernatant (sup) and precipitated anti-FLAG beads (bead) by centrifugation. Proteins in each fraction were analyzed by Western blotting using anti-FLAG (WB: α-FLAG) and anti-Myc antibodies (WB: α-Myc). *, immunoglobulin heavy chain.
To evaluate the effect of ATL31 phosphorylation on interaction with 14-3-3 proteins, we performed an in vitro ATL31 phosphorylation assay followed by far Western blotting with recombinant His-14-3-3λ and anti-His antibody. Recently, we have identified the kinase targeted to ATL31 (KTA) using a proteomics approach (data not shown). Thus, we used KTA as the kinase for the in vitro phosphorylation assay against ATL31. The N-terminal hydrophobic region (aa 1–86) of ATL31 was deleted for sufficient expression in E. coli, and the remaining residues (aa 87–368) were fused to MBP to generate recombinant MBP-ATL31 fusion protein (MBP-ATL31 ΔTM) (9). MBP-ATL31 ΔTM was incubated with GST or GST-KTA in the reaction buffer for 60 min. After the reaction, the phosphorylation of MBP-ATL31 ΔTM was assessed by a Phos-tag mobility shift assay, in which binding of phosphoproteins to the Phos-tag reagent in the SDS-polyacrylamide gel matrix slows down their mobility (49). Western blotting with anti-MBP antibody demonstrated that MBP-ATL31 ΔTM incubated with GST-KTA, but not with GST, shows slow migration, indicating that ATL31 is phosphorylated by KTA in vitro (Fig. 5D). Furthermore, Western blotting analysis with anti-Thr(P)209 antibody revealed that KTA phosphorylates Thr209 at ATL31 (Fig. 5D). Far Western blotting analysis clearly demonstrated that only the phosphorylated MBP-ATL31 ΔTM can be associated with His-14-3-3λ (Fig. 5D). Together, these results indicate that the phosphorylation of ATL31 is required for the interaction with 14-3-3 proteins.
To determine whether phosphorylation of the 14-3-3 binding sites on ATL31 is critical for the interaction, we performed a competitive binding assay using the synthetic peptides Thr(P)209 and Thr209 (Fig. 5E). ATL31C143S-FLAG and Myc-14-3-3λ were transiently co-expressed in N. benthamiana leaves, and the extracts were immunoprecipitated with anti-FLAG beads in the presence of Thr(P)209 or Thr209 peptide. In the presence of Thr(P)209 peptide, co-immunoprecipitated Myc-14-3-3λ was decreased to some extent, and the dissociated Myc-14-3-3λ was recovered in the supernatant fraction from the precipitation, whereas T209 peptide did not affect the interaction (Fig. 5F). These results clearly demonstrate that the interaction of ATL31 with 14-3-3 proteins is mediated by the phosphorylation of the 14-3-3 binding sites on ATL31.
Mutation of 14-3-3 Binding Sites on ATL31 Affects the Function in C/N-nutrient Response via the Modulation of 14-3-3 Protein Stability
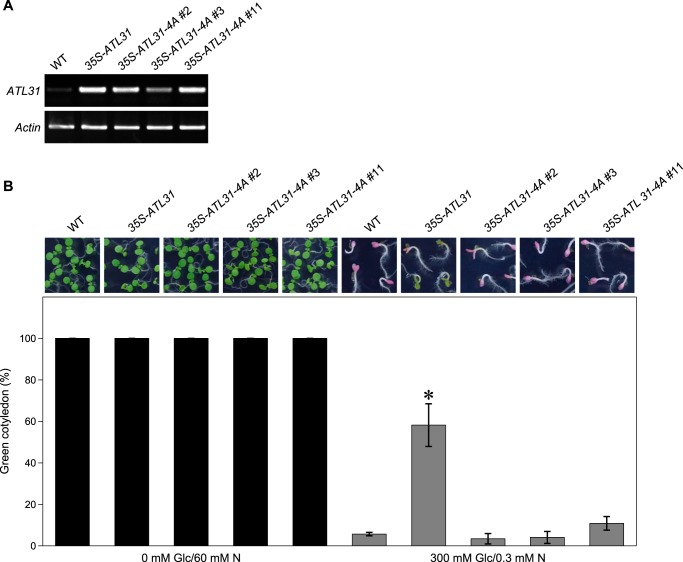
To examine the physiological requirements of phosphorylation at 14-3-3 binding sites for ATL31 function, the C/N-nutrient response of transgenic Arabidopsis overexpressing intact ATL31 (35S-ATL31) or quadruple-substituted ATL31 (35S-ATL31–4A) was analyzed (Fig. 6A). Wild-type (WT) and transgenic plants were grown on modified Murashige and Skoog medium under the indicated glucose and nitrogen conditions, and the percentage of 7-day-old seedlings with expanded green cotyledons was scored. All seedlings exhibited expanded green cotyledons under balanced C/N-nutrient conditions (0 mm glucose and 60 mm nitrogen; 0 mM Glc/60 mM N). In disrupted C/N-nutrient conditions (300 mm glucose and 0.3 mm nitrogen; 300 mM Glc/0.3 mM N), WT seedlings exhibited a strong purple pigmentation, and the percentage of cotyledon greening decreased significantly (Fig. 6B). On the other hand, 35S-ATL31 was less sensitive to the disruption of C/N-nutrient conditions and showed an increase in the percentage of cotyledon greening compared with the WT (Fig. 6B) (9). However, the percentage of cotyledon greening in all three independent lines for 35S-ATL31–4A decreased to a similar level as observed in the WT under disrupted C/N-nutrient conditions (Fig. 6B), indicating the physiological importance of these 14-3-3 binding sites for ATL31 function in plant C/N-nutrient response.
FIGURE 6.
Physiological impact of Thr209, Ser247, Ser270, and Ser303 on ATL31 in plant C/N-nutrient response. A, RT-PCR analysis of the transcript levels of ATL31 in WT and transgenic Arabidopsis plants overexpressing ATL31 (35S-ATL31) or ATL31–4A (35S-ATL31–4A, three independent lines). Total RNA was extracted from 7-day-old seedlings. Actin was used as an internal control. B, representative images and the percentage of seedlings with green expanded cotyledon under different C/N-nutrient conditions. WT, 35S-ATL31, and 35S-ATL31–4A were germinated and grown on Murashige and Skoog medium containing the indicated concentrations of glucose (Glc) and nitrogen (N). The percentage of cotyledon greening was scored at 7 days old. Data shown are means ± S.D. (error bars) from three independent assays. *, statistically significant difference from WT (two-tailed Student's t test, p < 0.05).
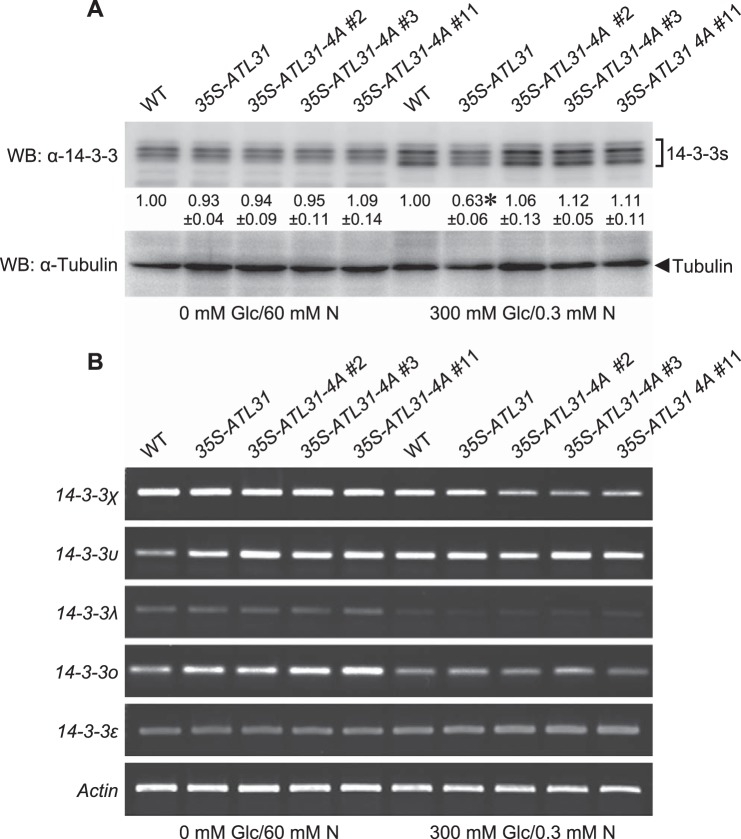
Previously, we demonstrated that ATL31 ubiquitinates and promotes the proteasomal degradation of a specific isoform, 14-3-3χ. Furthermore, we have shown that endogenous 14-3-3 proteins, as well as constitutively expressed FLAG-14-3-3χ, accumulate under disrupted C/N-nutrient conditions, an accumulation enhanced in ATL31/ATL6 double-knock-out mutants (12). Thus, we concluded that ATL31 negatively regulates the accumulation of 14-3-3 proteins. To examine the requirement of ATL31 phosphorylation for the regulation of 14-3-3 protein accumulation in response to C/N-nutrient conditions, we analyzed the amount of 14-3-3 proteins in plants for WT, 35S-ATL31, and 35S-ATL31–4A by Western blotting using the anti-14-3-3 antibody. 14-3-3 proteins in the WT were more accumulated under disrupted C/N-nutrient conditions compared with balanced C/N-nutrient conditions as reported previously (12), whereas the accumulation was repressed in 35S-ATL31 (Fig. 7A). Interestingly, such repression in 35S-ATL31 was released in 35S-ATL31–4A and restored to similar levels as in the WT (Fig. 7A). We also analyzed the transcriptional level of 14-3-3 genes used in the interaction analysis with ATL31 (Fig. 1B). Because we could not detect 14-3-3ι expression, we selected 14-3-3ο, which is located in the same subgroup as 14-3-3ι. Although the transcriptional level of some 14-3-3 genes was changed in these plants in different C/N-nutrient conditions (Fig. 7B), there was no correlation between mRNA levels and protein levels. Together, these results clearly demonstrate that the binding of 14-3-3 proteins to the Ser/Thr residues in the C terminus of ATL31 is essential for the mediation of C/N-nutrient response via 14-3-3 protein stability in plants.
FIGURE 7.
Effects of the mutation of 14-3-3 binding sites of ATL31 on the accumulation of 14-3-3 proteins. A, accumulation of 14-3-3 proteins in WT and transgenic Arabidopsis plants overexpressing ATL31 (35S-ATL31) or ATL31–4A (35S-ATL31–4A, three independent lines) under different C/N-nutrient conditions. Extracts of 7-day-old seedlings as indicated were subjected to SDS-PAGE followed by Western blotting using anti-14-3-3 (WB: α-14-3-3) and anti-tubulin antibodies (WB: α-Tubulin). Tubulin was used as a loading control. The numbers below the blot indicate the relative signal intensity of 14-3-3 proteins compared with WT (taken as 1.00) in the indicated C/N-nutrient condition. Data shown are means ± S.D. from three independent assays. *, statistically significant difference from WT (two-tailed Student's t test, p < 0.01). B, RT-PCR analysis of transcript levels of the indicated 14-3-3 isoforms in WT, 35S-ATL31, and 35S-ATL31–4A. Three independent 35S-ATL31–4A lines were tested. Total RNA was extracted from 7-day-old seedlings grown on Murashige and Skoog medium containing the indicated concentrations of glucose and nitrogen. Actin was used as an internal control.
DISCUSSION
In this study, we have shown the regulatory mechanism underlying the targeting of ubiquitin ligase ATL31 to 14-3-3 proteins. Quadruple Ala substitution at Thr209, Ser247, Ser270, and Ser303 on ATL31 (ATL31–4A), residues predicted to be putative 14-3-3 binding sites, clearly abolished the interaction with 14-3-3 proteins. Mass spectrometry analysis of purified ATL31 from Arabidopsis cells revealed that the 14-3-3 binding sites on ATL31 are phosphorylated. In vitro phosphorylation and peptide competition analyses demonstrated that phosphorylation of the 14-3-3 binding sites on ATL31 is required for the interaction with 14-3-3 proteins. Moreover, the physiological impact of these 14-3-3 binding sites was indicated because transgenic Arabidopsis plants overexpressing ATL31–4A (35S-ATL31–4A) lost tolerance under disrupted C/N-nutrient conditions, whereas transgenic Arabidopsis plants overexpressing intact ATL31 (35S-ATL31) retained tolerance. A similar phenotypic outcome was observed in a previous study (9), where transgenic Arabidopsis plants overexpressing ATL31 received a substitution of Cys143 to Ser residue in the RING domain, which led to a loss of ubiquitin ligase activity. Together, these results suggest that the phosphorylation of 14-3-3 binding sites on ATL31 is important in recruiting 14-3-3 proteins and in ubiquitinating the proteins to be degraded. Moreover, accumulation of 14-3-3 proteins under disrupted C/N-nutrient conditions was repressed in 35S-ATL31, whereas the accumulation was restored in 35S-ATL31–4A to a similar level as in WT plants. We have previously shown that 14-3-3 proteins accumulated more in ATL31/ATL6 double-knock-out mutants compared with WT, which resulted in a phenotype hypersensitive to disrupted C/N-nutrient conditions (12). In the present study, we further demonstrate that the ATL31 protein interacts with 14-3-3 proteins at phosphorylated Ser/Thr residues in the C-terminal region and regulates the C/N-nutrient response via controlling 14-3-3 stability in plants.
Phosphorylation modification plays a critical role in multiple biotic and abiotic stresses through transduction of signals that modulate the availability of nutrients and energy. The TOR (target of rapamycin) kinase complex regulates protein synthesis and starvation-induced autophagy via phosphorylation catalyzed by kinases for ribosomal protein S6 and ATG1/13 (autophagy-related 1/13) in response to amino acid and sugar status in yeast and mammals and is conserved in higher plants in a similar way (50). SnRK1 (sucrose non-fermenting protein-related kinase 1), the plant homologue of mammalian AMPK (AMP-activated protein kinase) and yeast Snf1 (sucrose non-fermenting protein 1), is alternatively a well characterized protein mediating nutrient signals in plants (51). Key enzymes in carbon and nitrogen metabolism, such as sucrose-6-phosphate synthase and nitrate reductase, are directly phosphorylated by SnRK1 to modulate enzymatic activity (51). In addition, a recent study revealed that NRT1.1 (nitrate transporter 1.1) functions as a nitrate sensor at the plasma membrane and is also phosphorylated in response to low nitrate concentrations by protein kinase CIPK23 (CBL-interacting protein kinase 23), a member of the SnRK3 subfamily (52). Whether or not the kinase mediates ATL31 phosphorylation is one of the most important questions to be addressed for further understanding of C/N-nutrient signaling. In this study, we used a kinase that targets ATL31 for in vitro phosphorylation followed by far Western blotting analysis with recombinant His-14-3-3λ to demonstrate that ATL31 can be phosphorylated by plant kinases in vitro and that the binding of 14-3-3 proteins to ATL31 is dependent on this phosphorylation. Furthermore, mutation of the phosphorylation sites of ATL31 affected the interaction with 14-3-3λ in yeast. It may be possible that ATL31 interacts with 14-3-3 proteins in a phosphorylation-dependent manner in yeast as well as in plants. These findings suggest that the kinase for ATL31 is broadly conserved in eukaryotes. It will be important to characterize the protein kinase of ATL31 to further understand C/N-nutrient signaling in plants.
On the other hand, the functional specificity of 14-3-3 isoforms to ATL31 should also be clarified. In this study, we used 14-3-3λ for biochemical interaction analyses because 14-3-3λ showed higher binding affinity than 14-3-3χ with ATL31 in Y2H analysis. However, it is unclear whether 14-3-3λ is the physiologically essential target. Because 14-3-3λ has a short C-terminal region, and the C-terminal region acts as a gate by negatively regulating the binding to target proteins in the plant 14-3-3 family (53, 54), 14-3-3λ is expected to bind many target proteins with high affinity. 14-3-3 interaction with ATL31 could be affected by many factors in plant cells, such as transcriptional level, subcellular localization, or post-translational modification. Moreover, co-IP analysis revealed that ATL31 interacts with multiple endogenous 14-3-3 isoforms, and ATL31 overexpression attenuated the accumulation of several 14-3-3 proteins under disrupted C/N-nutrient conditions. More detailed characterization of the physiological specificity of each 14-3-3 isoform should be performed in the future.
The C-terminal region of ATL proteins is predicted to recognize specific target proteins for ubiquitination because the region is not conserved among the ATL family, unlike the N-terminal hydrophobic region and RING domain (11). According to this hypothesis, our study demonstrates that the interaction of ATL31 with 14-3-3 proteins is mediated by the phosphorylation of specific Ser/Thr residues in the C-terminal region. ATL6 protein, the closest homologue of ATL31, shows a highly similar alignment (65% amino acid identity), including the C-terminal region. Moreover, transgenic Arabidopsis plants overexpressing ATL6 also exhibited insensitivity to disruption of C/N-nutrient conditions in the same way as did 35S-ATL31 (55). Y2H analysis showed that ATL6 also interacts with 14-3-3λ through the C-terminal region. Three residues (Thr209, Ser247, and Ser270) among the four 14-3-3 binding sites on ATL31, were conserved in ATL6. Also, Ser322 on ATL6 (which corresponds to Ser247 on ATL31) was reported to be phosphorylated by phosphoproteome analysis (56), suggesting that ATL6 can also interact with 14-3-3 proteins via phosphorylation at these conserved residues. In contrast to ATL6, the ATL11 protein, a close homologue of ATL31 next to ATL6 (35% amino acid identity), has no amino acid sequence corresponding to the 14-3-3 binding motif in the C-terminal region. Y2H analysis revealed no interaction of ATL11 with 14-3-3λ, suggesting that 14-3-3 proteins are not common targets for all ATL proteins and that the C-terminal region of the ATL proteins is critical for the specificity of target recognition.
Previously, we reported that ATL31 is localized at the plasma membrane with the N-terminal hydrophobic region and that this localization is essential for the function in C/N-nutrient response regulation (9). Other ATL proteins were also reported to localize to membrane compartments, including the plasma membrane and endoplasmic reticulum (24, 25). ATL55/RING1 was detected in detergent-resistant membranes, specialized microdomains in plasma membranes (24). In this study, we have shown that ATL31 interacts with 14-3-3λ at the plasma membrane by BiFC analysis. These findings suggest the possibility that ATL31 regulates the function of 14-3-3 client proteins, which are localized at the plasma membrane, via 14-3-3 protein degradation in C/N-nutrient response. 14-3-3 proteins directly target plasma membrane-localized proteins, such as plasma membrane H+-ATPase, phototropin receptor kinase, or putative glutamate receptor (57–59). Essential proteins involved in nutrient response, including receptors, transporters, and membrane trafficking factors, are localized at the plasma membrane, although their relationships with 14-3-3 proteins remain unclear. Interactome analysis with 14-3-3 proteins will provide significant information on the downstream molecular network involved in the C/N-nutrient response regulated by ATL31.
In conclusion, we propose a working model for the regulation of 14-3-3 protein stability mediated by ATL31 (Fig. 8). The in planta results on the accumulation of 14-3-3 proteins under different C/N-nutrient conditions suggest that phosphorylation of the 14-3-3 binding sites on ATL31 is regulated in response to C/N-nutrient conditions. Some protein kinases are activated in response to disrupted C/N-nutrient conditions (high carbon/low nitrogen), resulting in phosphorylation of the 14-3-3 binding sites on ATL31. Phosphorylated ATL31 binds to 14-3-3 proteins and subsequently mediates their ubiquitination and proteasomal degradation. Under balanced C/N-nutrient conditions (low carbon/high nitrogen), however, most protein kinases are inactivated, reducing ATL31 phosphorylation. Unphosphorylated ATL31 cannot bind to 14-3-3 proteins and mediate their ubiquitination and proteasomal degradation. In addition to C/N-nutrient conditions, other stimuli may also control the phosphorylation status on ATL31 because we detected phosphorylation in Arabidopsis cultured cells and in N. benthamiana in the absence of C/N-nutrient disruption.
FIGURE 8.
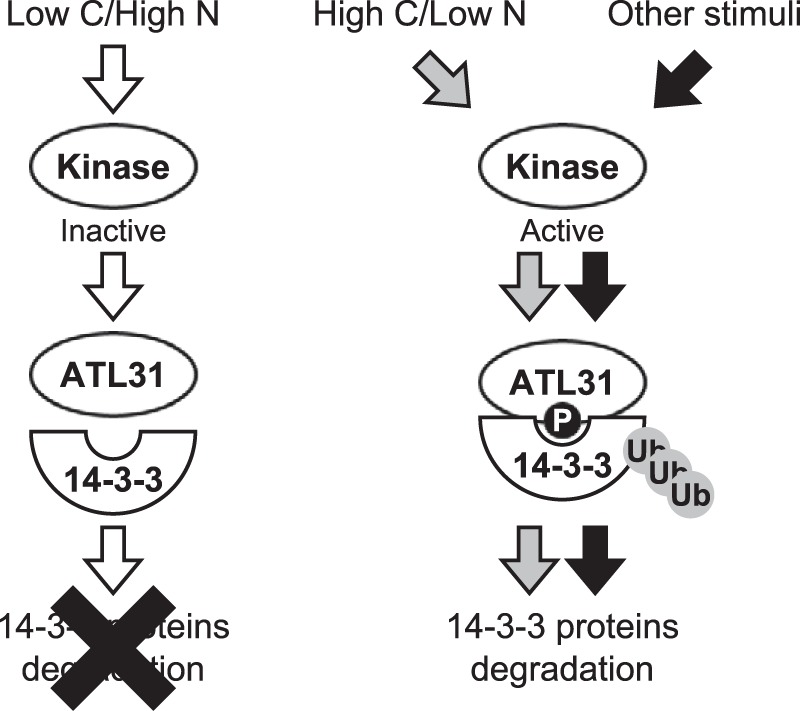
A working model for the regulation of 14-3-3 protein stability mediated by ATL31. Under balanced C/N-nutrient conditions (low carbon/high nitrogen), the protein kinases that catalyze phosphorylation to ATL31 are mostly inactivated. Unphosphorylated ATL31 cannot bind to 14-3-3 proteins. In contrast, the protein kinases become activated in response to disrupted C/N-nutrient conditions (high carbon/low nitrogen) or other stimuli, resulting in the phosphorylation of 14-3-3 binding sites on ATL31. Phosphorylated ATL31 binds 14-3-3 proteins, resulting in their ubiquitination and proteasomal degradation.
Acknowledgments
We thank Dr. Hironori Kaminaka (Tottori University, Japan), Dr. Shoji Mano (National Institute for Basic Biology, Japan), and Dr. Yoshihisa Ueno (National Institute of Agrobiological Sciences, Japan) for kindly providing vectors.
This work was supported by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research 24770035 (to T. S.) and on Innovation Areas 24114701 and 25112501 (to J. Y.) and in part by The Akiyama Foundation (to T. S.).
- C/N-nutrient
- carbon/nitrogen nutrient balance
- ATL
- Arabidopsis tóxicos en levadura
- Y2H
- yeast two-hybrid
- IP
- immunoprecipitation
- WB
- Western blot
- BiFC
- bimolecular fluorescence complementation
- MBP
- maltose-binding protein.
REFERENCES
- 1. Rolland F., Baena-Gonzalez E., Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709 [DOI] [PubMed] [Google Scholar]
- 2. Vidal E. A., Gutiérrez R. A. (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 11, 521–529 [DOI] [PubMed] [Google Scholar]
- 3. Gutiérrez R. A. (2012) Systems biology for enhanced plant nitrogen nutrition. Science 336, 1673–1675 [DOI] [PubMed] [Google Scholar]
- 4. Eveland A. L., Jackson D. P. (2012) Sugars, signalling, and plant development. J. Exp. Bot. 63, 3367–3377 [DOI] [PubMed] [Google Scholar]
- 5. Coruzzi G. M., Zhou L. (2001) Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects”. Curr. Opin. Plant Biol. 4, 247–253 [DOI] [PubMed] [Google Scholar]
- 6. Martin T., Oswald O., Graham I. A. (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 128, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sato T., Maekawa S., Yasuda S., Yamaguchi J. (2011) Carbon and nitrogen metabolism regulated by the ubiquitin-proteasome system. Plant Signal. Behav. 6, 1465–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sulpice R., Nikoloski Z., Tschoep H., Antonio C., Kleessen S., Larhlimi A., Selbig J., Ishihara H., Gibon Y., Fernie A. R., Stitt M. (2013) Impact of the carbon and nitrogen supply on relationships and connectivity between metabolism and biomass in a broad panel of Arabidopsis accessions. Plant Physiol. 162, 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sato T., Maekawa S., Yasuda S., Sonoda Y., Katoh E., Ichikawa T., Nakazawa M., Seki M., Shinozaki K., Matsui M., Goto D. B., Ikeda A., Yamaguchi J. (2009) CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J. 60, 852–864 [DOI] [PubMed] [Google Scholar]
- 10. Oliveira I. C., Coruzzi G. M. (1999) Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol. 121, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Serrano M., Parra S., Alcaraz L. D., Guzmán P. (2006) The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J. Mol. Evol. 62, 434–445 [DOI] [PubMed] [Google Scholar]
- 12. Sato T., Maekawa S., Yasuda S., Domeki Y., Sueyoshi K., Fujiwara M., Fukao Y., Goto D. B., Yamaguchi J. (2011) Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J. 68, 137–146 [DOI] [PubMed] [Google Scholar]
- 13. Mackintosh C. (2004) Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 381, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chevalier D., Morris E. R., Walker J. C. (2009) 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 60, 67–91 [DOI] [PubMed] [Google Scholar]
- 15. Denison F. C., Paul A. L., Zupanska A. K., Ferl R. J. (2011) 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22, 720–727 [DOI] [PubMed] [Google Scholar]
- 16. Bachmann M., Huber J. L., Liao P. C., Gage D. A., Huber S. C. (1996) The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Lett. 387, 127–131 [DOI] [PubMed] [Google Scholar]
- 17. Toroser D., Athwal G. S., Huber S. C. (1998) Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 435, 110–114 [DOI] [PubMed] [Google Scholar]
- 18. Comparot S., Lingiah G., Martin T. (2003) Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. J. Exp. Bot. 54, 595–604 [DOI] [PubMed] [Google Scholar]
- 19. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 20. Vierstra R. D. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397 [DOI] [PubMed] [Google Scholar]
- 21. Aguilar-Hernández V., Aguilar-Henonin L., Guzman P. (2011) Diversity in the architecture of ATLs, a family of plant ubiquitin-ligases, leads to recognition and targeting of substrates in different cellular environments. PLoS One 6, e23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guzmán P. (2012) The prolific ATL family of RING-H2 ubiquitin ligases. Plant Signal. Behav. 7, 1014–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Serrano M., Guzmán P. (2004) Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 167, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin S. S., Martin R., Mongrand S., Vandenabeele S., Chen K. C., Jang I. C., Chua N. H. (2008) RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J. 56, 550–561 [DOI] [PubMed] [Google Scholar]
- 25. Berrocal-Lobo M., Stone S., Yang X., Antico J., Callis J., Ramonell K. M., Somerville S. (2010) ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses. PLoS One 5, e14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S. J., Kim W. T. (2013) Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 587, 2584–2590 [DOI] [PubMed] [Google Scholar]
- 27. Shin L. J., Lo J. C., Chen G. H., Callis J., Fu H., Yeh K. C. (2013) IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25, 3039–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sako K., Maki Y., Kanai T., Kato E., Maekawa S., Yasuda S., Sato T., Watahiki M. K., Yamaguchi J. (2012) Arabidopsis RPT2a, 19S proteasome subunit, regulates gene silencing via DNA methylation. PLoS One 7, e37086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Menges M., Murray J. A. (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212 [DOI] [PubMed] [Google Scholar]
- 30. Menges M., Murray J. A. (2006) Synchronization, transformation, and cryopreservation of suspension-cultured cells. Methods Mol. Biol. 323, 45–61 [DOI] [PubMed] [Google Scholar]
- 31. Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 [DOI] [PubMed] [Google Scholar]
- 32. Clough S. J., Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 33. Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., Pikaard C. S. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 [DOI] [PubMed] [Google Scholar]
- 34. Curtis M. D., Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 [DOI] [PubMed] [Google Scholar]
- 36. Fujiwara M., Umemura K., Kawasaki T., Shimamoto K. (2006) Proteomics of Rac GTPase signaling reveals its predominant role in elicitor-induced defense response of cultured rice cells. Plant Physiol. 140, 734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fukao Y., Ferjani A., Fujiwara M., Nishimori Y., Ohtsu I. (2009) Identification of zinc-responsive proteins in the roots of Arabidopsis thaliana using a highly improved method of two-dimensional electrophoresis. Plant Cell Physiol. 50, 2234–2239 [DOI] [PubMed] [Google Scholar]
- 38. Tsunoda Y., Sakai N., Kikuchi K., Katoh S., Akagi K., Miura-Ohnuma J., Tashiro Y., Murata K., Shibuya N., Katoh E. (2005) Improving expression and solubility of rice proteins produced as fusion proteins in Escherichia coli. Protein Expr. Purif. 42, 268–277 [DOI] [PubMed] [Google Scholar]
- 39. DeLille J. M., Sehnke P. C., Ferl R. J. (2001) The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 126, 35–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bachmann M., Huber J. L., Athwal G. S., Wu K., Ferl R. J., Huber S. C. (1996) 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 398, 26–30 [DOI] [PubMed] [Google Scholar]
- 41. Rosenquist M., Sehnke P., Ferl R. J., Sommarin M., Larsson C. (2000) Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J. Mol. Evol. 51, 446–458 [DOI] [PubMed] [Google Scholar]
- 42. Robatzek S., Chinchilla D., Boller T. (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson C., Crowther S., Stafford M. J., Campbell D. G., Toth R., MacKintosh C. (2010) Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 427, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu D., Bienkowska J., Petosa C., Collier R. J., Fu H., Liddington R. (1995) Crystal structure of the ζ isoform of the 14-3-3 protein. Nature 376, 191–194 [DOI] [PubMed] [Google Scholar]
- 45. Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 [DOI] [PubMed] [Google Scholar]
- 46. Jaspert N., Oecking C. (2002) Regulatory 14-3-3 proteins bind the atypical motif within the C terminus of the plant plasma membrane H+-ATPase via their typical amphipathic groove. Planta 216, 136–139 [DOI] [PubMed] [Google Scholar]
- 47. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 48. Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteomics 5, 749–757 [DOI] [PubMed] [Google Scholar]
- 50. Robaglia C., Thomas M., Meyer C. (2012) Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15, 301–307 [DOI] [PubMed] [Google Scholar]
- 51. Coello P., Hey S. J., Halford N. G. (2011) The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 62, 883–893 [DOI] [PubMed] [Google Scholar]
- 52. Ho C. H., Lin S. H., Hu H. C., Tsay Y. F. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 [DOI] [PubMed] [Google Scholar]
- 53. Shen W., Clark A. C., Huber S. C. (2003) The C-terminal tail of Arabidopsis 14-3-3ω functions as an autoinhibitor and may contain a tenth α-helix. Plant J. 34, 473–484 [DOI] [PubMed] [Google Scholar]
- 54. Börnke F. (2005) The variable C-terminus of 14-3-3 proteins mediates isoform-specific interaction with sucrose-phosphate synthase in the yeast two-hybrid system. J. Plant Physiol. 162, 161–168 [DOI] [PubMed] [Google Scholar]
- 55. Maekawa S., Sato T., Asada Y., Yasuda S., Yoshida M., Chiba Y., Yamaguchi J. (2012) The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol. Biol. 79, 217–227 [DOI] [PubMed] [Google Scholar]
- 56. Nühse T. S., Bottrill A. R., Jones A. M., Peck S. C. (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fuglsang A. T., Visconti S., Drumm K., Jahn T., Stensballe A., Mattei B., Jensen O. N., Aducci P., Palmgren M. G. (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274, 36774–36780 [DOI] [PubMed] [Google Scholar]
- 58. Kinoshita T., Emi T., Tominaga M., Sakamoto K., Shigenaga A., Doi M., Shimazaki K. (2003) Blue-light- and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiol. 133, 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang I. F., Curran A., Woolsey R., Quilici D., Cushman J. C., Mittler R., Harmon A., Harper J. F. (2009) Proteomic profiling of tandem affinity-purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9, 2967–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]