Background: The function and biosynthetic mechanism of GlcNH3+ in heparan sulfate remain unclear.
Results: GlcNH3+-containing tetrasaccharides inhibited heparanase activity. Exostosin-like 3 regulated the expression of GlcNH3+ structures.
Conclusion: GlcNH3+-containing tetrasaccharides suppressed cancer cell invasion in vitro. Production of GlcNH3+ was controlled by EXTL3 (exostosin-like 3) and NDST-1 (N-deacetylase/N-sulfotransferase-1).
Significance: Tumor metastasis might be suppressed by utilizing GlcNH3+-containing oligosaccharides. Additionally, a biosynthetic mechanism of GlcNH3+-containing heparan sulfates was proposed.
Keywords: Glycobiology, Glycosaminoglycan, Glycosidase, Heparan Sulfate, Heparanase, Proteoglycan, Proteoglycan Synthesis
Abstract
Degradation of heparan sulfate (HS) in the extracellular matrix by heparanase is linked to the processes of tumor invasion and metastasis. Thus, a heparanase inhibitor can be a potential anticancer drug. Because HS with unsubstituted glucosamine residues accumulates in heparanase-expressing breast cancer cells, we assumed that these HS structures are resistant to heparanase and can therefore be utilized as a heparanase inhibitor. As expected, chemically synthetic HS-tetrasaccharides containing unsubstituted glucosamine residues, GlcAβ1–4GlcNH3+(6-O-sulfate)α1–4GlcAβ1–4GlcNH3+(6-O-sulfate), inhibited heparanase activity and suppressed invasion of breast cancer cells in vitro. Bifunctional NDST-1 (N-deacetylase/N-sulfotransferase-1) catalyzes the modification of N-acetylglucosamine residues within HS chains, and the balance of N-deacetylase and N-sulfotransferase activities of NDST-1 is thought to be a determinant of the generation of unsubstituted glucosamine. We also report here that EXTL3 (exostosin-like 3) controls N-sulfotransferase activity of NDST-1 by forming a complex with NDST-1 and contributes to generation of unsubstituted glucosamine residues.
Introduction
Heparan sulfate proteoglycans (HS-PGs)3 are universally distributed glycoproteins consisting of HS chains substituted on core proteins and are located in the extracellular matrices and on cell surfaces of various kinds of human tissues. Some HS-PGs modulate cell adhesion, cell proliferation, and morphogenesis by serving as receptors. These functions are often focused at specific sequence domains within HS chains, which are characterized by modifications such as sulfation. Generation of these sequence domains is regulated during HS biosynthesis in the Golgi apparatus. The HS chain consists of up to 200 repeating disaccharide units of d-glucuronic acid (GlcA) β1→4 linked to N-acetylglucosamine (GlcNAc) α1→4. Biosynthesis of the HS chain backbone occurs by alternate additions of GlcA and GlcNAc by HS polymerase. Subsequently, the elongating linear polysaccharide chains are modified through a series of reactions involving the N-deacetylation and N-sulfation of GlcNAc, epimerization of GlcA, and O-sulfation at various positions of both residues. The first modification step, N-deacetylation and N-sulfation of GlcNAc, is catalyzed by glucosaminyl N-deacetylase/N-sulfotransferase (NDST), which removes acetyl groups from GlcNAc residues and then sulfates the N-position of glucosamine (GlcN) residues. Further modification steps include C5 epimerization of GlcA, 2-O-sulfation of GlcA/iduronic acid, 6-O-sulfation of GlcN, and 3-O-sulfation of GlcN units. In this study, we focused on the biosynthetic mechanism and biological function of N-unsubstituted glucosamine (GlcNH3+) generated during modification of GlcN by NDST-1.
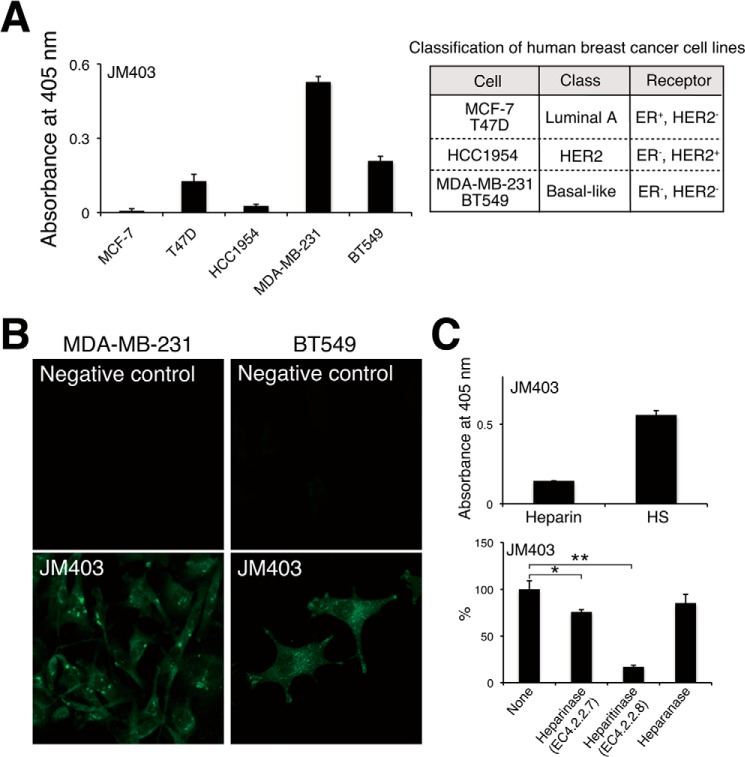
Although rare, naturally occurring N-unsubstituted GlcNH3+ residues are reported to have a biological function. The presence of GlcNH3+ residues in native HS chains has been documented using amine-reactive fluorescence labeling (1) and by deaminative cleavage at pH 3.9 (2–4). In addition, it has been shown that GlcNH3+ residues occur more commonly in preparations of purified bovine HS using the monoclonal antibody (mAb) JM403, which recognizes an HS structure with one or more GlcNH3+ structures (5). In addition, it has been reported that GlcNH3+ structures are expressed as the predominant disaccharides in male mouse liver in a gender-specific manner (6). Regarding the function of GlcNH3+, it has been reported that HS chains containing GlcNH3+ structures form the binding site for L-selectin in endothelial cells (2). Furthermore, cytoplasmic expression of GlcNH3+ structures was detected in mammary carcinomas at high frequency and in normal crypt epithelial cells in the small intestine, where these cells actively proliferate (7). Thus, it is suggested that GlcNH3+ structures are involved in cellular proliferation and associated with an increased degree of malignancy. From the finding that HS chains bearing unsubstituted glucosamine residues accumulate in heparanase-expressing malignant breast cancer cell lines (Figs. 1 and 2) (7), we assumed that these HS structures are resistant to heparanase and can therefore be utilized as a heparanase inhibitor. Here, we examined the effects of HS-oligosaccharides containing unsubstituted glucosamine structures on heparanase activity.
FIGURE 1.
Measurement of GlcNH3+ structures in various human breast cancer cell lines. A, HS chains were isolated and purified from human breast cancer cells, and GlcNH3+ structures contained in the HS chains were detected by ELISAs using monoclonal antibody JM403, which recognizes GlcNH3+ structures. Five breast cancer cell lines used in this study are classified according to the expression profile of cell surface receptors (46). B, cellular localization of JM403 epitopes in MDA-MB-231 and BT549 cells was examined by immunofluorescence. Mouse control IgG (non-immune) was used as the negative control. C, the expression levels of JM403 epitopes in heparin and HS were measured by ELISA. After biotinylated HS chains immobilized onto a streptavidin plate were completely digested with either heparinase, heparitinase, or heparanase, residual JM403 epitopes were detected by ELISA. This assay was performed in triplicate, and error bars represent the S.D. *, p < 0.05; **, p < 0.01.
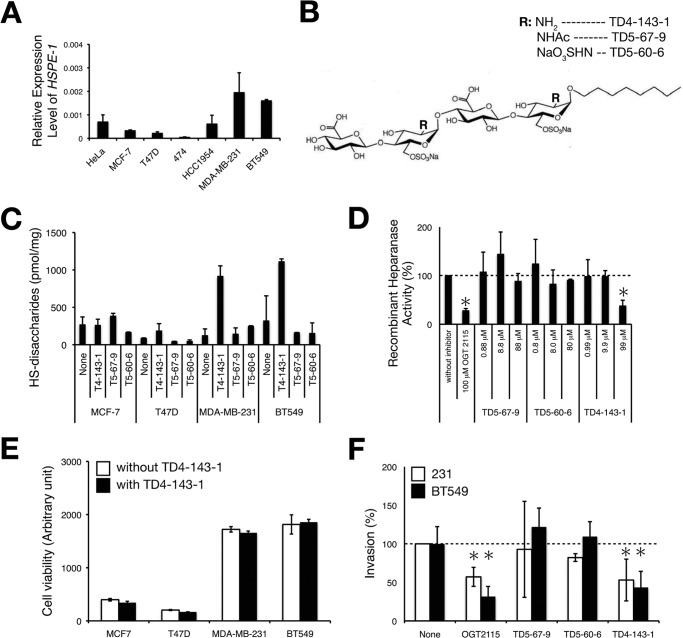
FIGURE 2.
HS tetrasaccharides containing GlcNH3+ structures inhibit heparanase activity and tumor invasion in vitro. A, the expression levels of HSPE-1 in various human breast cancer cell lines were analyzed by real-time PCR. The expression levels of HSPE-1 were normalized to G3PDH, and the means were obtained from three independent experiments. Error bars, S.D. B, structures of chemically synthesized HS tetrasaccharides. R indicates the modification of the N-position (-Ac, -SO3H, or -H) of the GlcN residue. C, four kinds of human breast cancer cell lines were treated with either TD4-143-1, T5-67-9, or T5-60-6, and HS expression level in each cell type was analyzed as described under “Experimental Procedures.” D, the heparanase inhibitory activities of various chemically synthesized HS tetrasaccharides were compared. Heparanase activity was measured as described under “Experimental Procedures.” OGT2115 is a commercially available heparanase inhibitor. This assay was performed in triplicate, and the means were obtained from two independent experiments. Error bars, S.D. E, cell viability was measured after cells were treated with (solid bar) or without (open bar) 100 μg/ml TD4-143-1 for 22 h. F, the invasion potential of MDA-MB-231 (open bar) and BT549 cells (solid bar) treated either with 1 μm OGT2115, 100 μg/ml TD5-67-9, TD5-60-6, or TD4-143-1, was tested by a Matrigel in vitro invasion assay. This assay was performed in triplicate, and error bars represent the S.D. *, p < 0.05.
The NDST family responsible for the N-deacetylation/N-sulfation reaction consists of four homologues (NDST-1, -2, -3, and -4). In vertebrates, Ndst1 and Ndst2 mRNA are abundantly and ubiquitously expressed in all embryonic and adult tissues. In addition, analyses of either NDST-1 or NDST-2 knock-out mice have revealed that NDST-1 is essential for development (8), whereas NDST-2 plays an important role only in mast cell production (9). Compared with Ndst1 and Ndst2, Ndst3 and Ndst4 transcripts are predominantly expressed during embryonic development and in the adult brain (10). Comparison of the enzymatic properties of the four murine NDSTs revealed striking differences in N-deacetylation and N-sulfation activities; NDST-1 and NDST-2 had both activities, NDST-3 had high deacetylase activity but weak sulfotransferase activity, and NDST-4 had the opposite properties (10). It has been thought that NDST-3 is involved in the synthesis of GlcNH3+ structures, because NDST-3 showed high deacetylase activity and weak sulfotransferase activity. However, the expression level of GlcNH3+ was not affected by the lack of NDST-3 (11). Thus, the synthetic mechanism of GlcNH3+ structures remained unclear. We postulated that GlcNH3+ structures are generated through the control of NDST activities by an unidentified molecule. Fortuitously, when we analyzed HS chains produced by EXTL3-knockdown cells, we perceived that EXTL3 (exostosin-like 3) is a candidate regulation factor of NDST-1 (Fig. 4A) (12). Here, we also report an EXTL3-dependent mechanism involved in the generation of GlcNH3+ structures. EXTL3 is a member of the EXT (hereditary multiple exostoses) gene family. Current knowledge indicates that EXTL3 has both N-acetylglucosaminyltransferase-I and -II activities, which are involved in the initiation of HS biosynthesis and elongation of HS chains, respectively. In addition, the analysis of EXTL3-deficient mice showed that EXTL3 plays an important role in HS biosynthesis in vivo, because the amounts of HS were dramatically decreased by lack of EXTL3. In this study, we report a novel function of EXTL3, its involvement in the production of GlcNH3+ structures through the control of N-sulfotransferase activity of NDST-1.
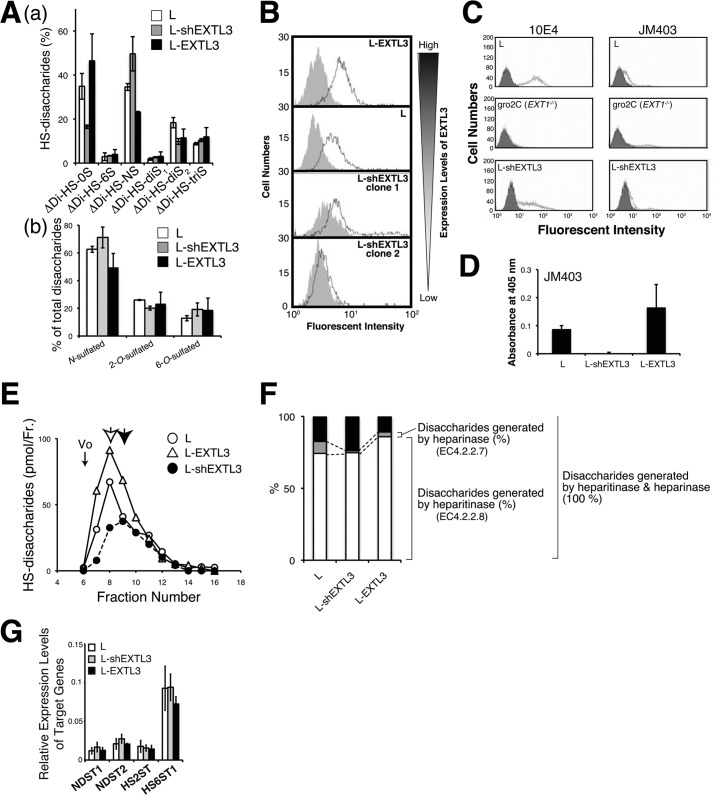
FIGURE 4.
GlcNH3+ structures were controlled by the expression level of EXTL3. A, a, HS disaccharide composition analysis of the cells expressing EXTL3 at various levels was carried out. White bar, L cells; gray bar, EXTL3-knockdown L cells; black bar, EXTL3-overexpressing L cells. Means were obtained from three independent experiments. Error bars, S.D. *, p < 0.05. ΔDiHS-0S, ΔHexAα1–4GlcNAc; ΔDiHS-6S, ΔHexAα1–4GlcNAc(6S); ΔDiHS-NS, ΔHexAα1–4GlcNS; ΔDiHS-diS1, ΔHexAα1–4Glc(NS,6S); ΔDi-diS2, ΔHexA(2S)α1–4GlcNS; ΔDi-triS, ΔHexA(2S)α1–4Glc(NS,6S). b, percentages of N-sulfated, 2-O-sulfated, and 6-O-sulfated disaccharides. B, the expression levels of JM403 epitopes of the cells expressing EXTL3 at various levels were examined by flow cytometry analysis. Shaded area, negative control staining (without the primary antibody); solid line, staining with mAb JM403. C, the expression levels of 10E4 and JM403 epitopes in the cells expressing EXTL3 at various levels were examined by flow cytometry analysis. gro2C cells, a mouse L cell mutant, are deficient in the expression of EXT1, which encodes a glycosyltransferase related to the formation of the HS backbone, and thereby synthesize marginal HS chains (12). Shaded area, negative control staining (without the primary antibody); solid line, staining with mAb 10E4 or JM403. D, biotinylated HS chains (2 nmol as GlcA) isolated from L, L-shEXTL3, and L-EXTL3 cells were immobilized onto a streptavidin plate, and the expression levels of JM403 epitopes were examined by ELISA. E, the length of HS chains produced in the cells expressing EXTL3 at various levels was measured by gel filtration chromatography as described under “Experimental Procedures.” Open circle, L cells; open triangle, EXTL3-overexpressed L cells; closed circle, EXTL3-knockdown L cells. An open arrow indicates the peak top of HS chains isolated from L and EXTL3-overexpressing L cells, whereas a closed arrow represents the peak of HS chains isolated from EXTL3-knockdown L cells. Vo, void volume. F, HS chains purified from the three kinds of cells indicated were completely digested with either heparitinase, heparinase, or both heparitinase and heparinase. The percentage of disaccharides generated by heparitinase (or heparinase) was calculated as 100 × disaccharides generated by heparitinase (or heparinase)/disaccharides generated by both heparitinase and heparinase. G, the expression levels of sulfotransferase genes involving sulfation modification of HS were investigated by real-time PCR. The expression levels of sulfotransferase genes were normalized to G3PDH, and the means were obtained from three independent experiments. Error bars, S.D.
EXPERIMENTAL PROCEDURES
Materials
Monoclonal antibodies JM403 (catalog no. 370730) and F58-10E4 (catalog no. 370255), heparitinase from Flavobacterium heparinum (EC 4.2.2.8), and heparinase from F. heparinum (EC 4.2.2.7) were provided by Seikagaku Corp. (Tokyo, Japan). Mouse L fibroblasts and their derivatives, gro2C, were kindly provided by Dr. Frank Tufaro (Allera Health Products Inc., St. Petersburg, FL), and human breast cancer MCF7 (ATCC® HTB-22TM), T47D (ATCC® HTB-133TM), HCC1954 (ATCC® CRL-2338TM), and BT549 (ATCC® HTB-122TM) were purchased from the American Type Culture Collection (ATCC) (Manassas, VA). MDA-MB-231 (catalog no. 92020424) was obtained from the European Collection of Cell Cultures (Salisbury, UK). Monoclonal antibodies against GM130 (catalog no. 610822) and Lamp-1 (catalog no. 555798) were purchased from BD Transduction Laboratories and BD Pharmingen, respectively. Anti-DYKDDDDK tag antibody beads (catalog no. 012-22781) and Ni-NTA-agarose were obtained from WAKO Pure Chemicals (Osaka, Japan) and Qiagen (Venlo, Netherlands), respectively. Anti-FLAG® polyclonal antibody (catalog no. F7425) was purchased from Sigma-Aldrich.
Plasmid Construction
Human NDST-1 cDNA was obtained by reverse transcription-coupled polymerase chain reaction using HeLa cDNA library and the following primers: fwP, 5′-GCGGCCGCGCCACCATGGCTGCCCTGGCATGC-3′ (underline, NotI site; double underline, start codon; boldface type, Kozak sequence) and rvP, 5′-GGATCCCCTGGTGTTCTGGAGGTCCTCTCGTAGCCGG (underline, BamHI site). The resultant fragments were cloned in a TA cloning vector, pT7Blue vector (Novagen). The 2.7-kb fragments obtained by digestion of pT7Blue-hNDST-1 with NotI and BamHI were inserted into the NotI and BamHI sites of p3xFLAG CMV14 (Sigma) to express hNDST-1(1–882) full-length protein tagged with the FLAG epitope at the C terminus. pEF-BOS/IP-hNDST1(42–882), pEF-BOS/IP-hNDST1(42–557), and pEF-BOS/IP-hNDST1(558–882) were constructed as follows. The BamHI-BamHI fragment of a PCR product was amplified using pT7Blue-hNDST-1 as a template with the following primers: fwP-hNDST1(42–557 or 42–882), 5′-CGGGATCCAAGCGAGGCCTGGAGCCCTC-3′ (underline, BamHI site); fwP-hNDST1(558–882), CGGGATCCCTGCAGACACTGCCCCCTGTGC (underline, BamHI site); rvP-hNDST1(42–557), CGGGATCCCTACCGGAGGTTCGTCCAGGAGTGCAG (underline, BamHI site; double underline, stop codon); and rvP-hNDST1(42–882 or 558–882), CGGGATCCCTACCTGGTGTTCTGGAGGTCC (underline, BamHI site; doublet, stop codon). The resultant fragments digested with BamHI were inserted into the BamHI site of pEF-BOS/IP vector (13).
pCMVscript-hEXTL3 was generated as follows. Full-length hEXTL3 was amplified using the previously cloned EXTL3 cDNA (14) as a template and the following primers: fwP, 5′-GAAGATCTGCCACCATGACAGGCTATAC-3′ (underline, BglII site; double underline, start codon; boldface type, Kozak sequence); rvP, 5′-GGGGTACCAGCCATCTCCTCCCTCT-3′ (underline, KpnI site). After the resulting fragments were subcloned into pGEM-T(Easy) vector (Promega), pGEM-T(Easy)-hEXTL3 was digested with BglII and KpnI. The 2.8-kb BglII-KpnI fragments were inserted into the BamHI and KpnI site of pCMVscript vector (Agilent Technologies). To construct an expression vector for Myc- and His-tagged EXTL3 proteins, the sequence corresponding to full-length hEXTL3 was amplified by PCR using pGEM-T(Easy)-hEXTL3 as a template and the following primers: fwP, 5′-GAAGATCTGCCACCATGACAGGCTATAC-3′ (underline, BglII site; double underline, start codon; boldface type, Kozak sequence); rvP, 5′-AAGCTTGATGAACTTGAAGCACT-3′ (underline, HindIII site). The fragments were cloned in a TA cloning vector and then digested with BglII and HindIII. The BglII-HindIII fragments were inserted into the BamHI and HindIII site of pcDNATM3.1/Myc-His A vector.
Isolation and Purification of HS from Cells
Glycosaminoglycans were isolated as described previously (12). After the glycosaminoglycan fraction was digested with chondroitinase ABC to remove CS chains, intact HS chains were isolated using a PD MiniTrap G-10 column (GE Healthcare). Isolated HS chains were quantified by the carbazole method or HPLC method (12).
Measurement of JM403 Epitope by ELISA
About 10 μg of HS was biotinylated to be immobilized on a Nunc ImmobilizerTM Streptavidin plate (Nalge Nunc International, Rochester, NY) as described previously (15). Briefly, HS isolated from cells was dissolved in 100 mm MES-NaOH, pH 5.5, at a concentration of 1 mg/ml. To this solution were added 2.5 μg of biotin-LC-hydrazide freshly dissolved in dimethyl sulfoxide and 0.25 μl of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (Pierce). The mixture was incubated overnight at room temperature with continuous shaking. Excess biotinylating reagents were removed using Ultrafree®-MC (5000 NMWL filter unit) (Millipore, Billerica, MA) against several changes of phosphate-buffered saline.
After Nunc ImmobilizerTM Streptavidin was washed with PBS containing 0.1% Tween 20 (PBST), 40 μg/ml biotinylated HS was added to each well and incubated for 1 h at room temperature. After washing with PBST, each well was blocked with PBS containing 2% BSA for 30 min at room temperature. HS immobilized to the plate was reacted with JM403 (dilution 1:100) overnight at 4 °C. After washing with PBST, horseradish peroxidase-conjugated anti-mouse IgM antibody (dilution 1:1,000) was added to each well and incubated for 2 h. After washing with PBST, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) was added to each well as a peroxidase substrate. The blue-green-colored materials produced by peroxidase were measured at 405 nm.
Immunofluorescence
Cells cultured on a 3.5-cm glass-bottomed dish (AGC Techno Glass Co. Ltd., Shizuoka, Japan) were fixed with PBS containing 4% paraformaldehyde for 20 min on ice. After washing with PBS, cells were permeabilized with PBS containing 0.2% Triton X-100 for 10 min at room temperature. After blocking with PBS containing 2% BSA and 0.1% Tween 20 for 1 h at room temperature, cells were incubated overnight at 4 °C with the following antibodies: JM403 (dilution 1:500), anti-GM130 monoclonal antibody (dilution 1:50), or anti-Lamp-1 monoclonal antibody (dilution 1:200). After washing with PBST, Dylight488-conjugated anti-mouse IgM antibody (dilution 1:1,000) (Rockland Immunochemicals, Gilbertsville, PA) for detection of JM403 or Alexa568-conjugated anti-mouse IgG (dilution 1:1,000) (Molecular Probes, Inc., Eugene, OR) for GM130 and Lamp-1 was employed. Hoechst33342 was used as a nuclear counterstain. Images were acquired with a Zeiss LSM 700 confocal laser scanning system (Carl Zeiss Inc., Oberkochen, Germany) equipped with an inverted Axio observer Z1 microscope. To investigate the cellular localization of incorporated chemically synthesized tetrasaccharides containing glucosamine residues, TD4-143-1, cells were incubated overnight in the presence or absence of 50 μm TD4-143-1 and double-stained with JM403 and anti-GM130 antibody or with JM403 and anti-Lamp-1 antibody as described above.
Real-time PCR Analysis
Total RNA was extracted from cells by the guanidine phenol method using TRIzol reagent (Invitrogen) according to the manufacturer's protocols. Aliquots (1 μg) of total RNA were digested with 2 IU of RQ1 RNase-free DNase (Promega) for 30 min at 37 °C and then incubated for 10 min at 65 °C with stop solution (Promega). For reverse transcription, total RNA (0.75 μg) was treated with Moloney murine leukemia virus reverse transcriptase (Invitrogen) using random primers (nonadeoxyribonucleotide mixture; pd(N)9) (Takara Bio Inc., Shiga, Japan). Quantitative real-time PCR was conducted using FastStart DNA Master plus SYBR Green I and a LightCycler 1.5 (Roche Applied Science) according to the manufacturer's protocols. The housekeeping gene GAPDH was used as an internal control for quantification. The primers used for real-time PCR are shown in Table 1.
TABLE 1.
Primers used for real-time PCR
| Gene name | 5′-primer | 3′-primer |
|---|---|---|
| Human HSPE-1 | 5′-TTGCTATCCGACACCTTTGC-3′ | 5′-CACGCTTGCCATTAACACCT-3′ |
| Human G3PDH | 5′-ATGGGTGTGAACCATGAGAAGTA-3′ | 5′-GGCAGTGATGGCATGGAC-3′ |
| Mouse Ndst1 | 5′-TCCTATCAGCATCTTCATGACAC-3′ | 5′-TCTTCTCCTTAGACCAGATGTCC-3′ |
| Mouse Ndst2 | 5′-AGACCGGTACTGCGTGGAGT-3′ | 5′-GGCTGTGCTGCTGGGCT-3′ |
| Mouse Hs2st | 5′-GACCCTATTGAGAGGCTAGTT-3′ | 5′-CCAGAGCTTCTCTGGAGCA-3′ |
| Mouse Hs6st1 | 5′-CGCCCAGAAAGTTCTACTACATC-3′ | 5′-GGTTGTTAGCCAGGTTATAGGG-3′ |
| Mouse G3pdh | 5′-CATCTGAGGGCCCACTG-3′ | 5′-GAGGCCATGTAGGCCATGA-3′ |
Disaccharide Analysis of HS from Cells
Disaccharide analysis of HS from cells was carried out as described previously (12). Human breast cancer cells were cultured for 2 days in the presence or absence of 50 μm TD4-143-1, and glycosaminoglycans subsequently isolated from the cells were subjected to disaccharide analysis of HS.
Measurement of Heparanase Inhibitory Activity of Chemically Synthesized Tetrasaccharides
Heparanase activity was measured using a HepActivTM heparanase activity assay kit (InSight Biopharmaceuticals Ltd., Rehovot, Israel), and the heparanase inhibitory activity of chemically synthesized tetrasaccharides (16) was examined using 10 ng of recombinant human heparanase-1 (InSight Biopharmaceuticals Ltd.).
Cell Viability Assay
After washing cells with PBS, cells were incubated with 1 ml of 1% (v/v) Triton X-100 to release cellular lactate dehydrogenase. The enzyme activity of cellular lactate dehydrogenase was measured as an indicator of cell viability using the Cyto Tox-ONE homogeneous membrane integrity assay (Promega).
Matrigel Invasion Assay
An invasion assay was carried out using BD BioCoatTM MatrigelTM invasion chambers according to the manufacturer's instructions. Cells (5 × 104 cells/ml) were incubated in the presence or absence of 100 μg/ml chemically synthesized tetrasaccharides for 20 min at room temperature and then added to the upper chamber and allowed to invade for 22 h at 37 °C in a CO2 incubator. After the non-invading cells and Matrigel were removed from the upper surface of the membrane by scrubbing with a cotton swab, invading cells (those adhering to the bottom surface of the membrane) were subjected to Giemsa staining (WAKO Pure Chemicals, Osaka, Japan). All migrated cells on a membrane were photographed using a Keyence BZ-8000 fluorescence microscope and counted.
Flow Cytometry
After cells were detached with PBS containing 1 mm EDTA, 0.5 × 106 cells were incubated with PBS containing 0.1% NaN3 for 10 min at room temperature. After washing with PBS, cells were incubated with JM403 (dilution 1:50) or F58-10E4 (dilution 1:50) for 1 h on ice. After washing with PBS containing 0.2% BSA, cells were incubated with FITC-conjugated anti-mouse IgM (dilution 1:100) for 1 h on ice, washed again with PBS containing 0.2% BSA, and then analyzed by BD FACSCaliburTM flow cytometry.
Determination of Molecular Size of Glycosaminoglycan Chains
Isolated glycosaminoglycan chains were subjected to gel filtration chromatography using a Superdex 200 HR 10/30 (10 × 300-mm) column (GE Healthcare) (17). The column was eluted with 0.2 m NH4HCO3 at a flow rate of 0.4 ml/min. Fractions were collected, lyophilized, and digested with a mixture of 1 mIU heparinase and 1 mIU heparitinase in 20 mm sodium acetate (pH 7.0) containing 2 mm calcium acetate at 37 °C for 4 h. The resultant digests were labeled with 2-aminobenzamide and analyzed by anion exchange HPLC as described previously (12).
Interactions between NDST-1 and EXTL3 Proteins
p3xFLAG CMV14-hNDST-1(1–882) and/or pcDNA3.1-EXTL3(1–919)-Myc-His expression plasmids were transfected into COS-1 cells on 60-mm dishes using Lipofectamine 2000 (Invitrogen). Two days after transfection, cells were solubilized with lysis buffer (1% Nonidet P-40, 20 mm Tris-HCl (pH 7.5), 0.15 m NaCl, 1 mm EDTA, 10% glycerol, and protease inhibitor mixture), and cellular FLAG-tagged NDST-1 proteins and histidine-tagged EXTL3 were purified by incubation overnight at 4 °C with 10 μl of anti-DYKDDDDK tag antibody beads and Ni-NTA-agarose, respectively. The anti-DYKDDDDK tag antibody beads (WAKO Pure Chemicals, Osaka, Japan) were washed with PBS containing 0.1% Tween 20, and bound proteins were eluted by 100 μg/ml DYKDDDDK peptides (WAKO Pure Chemicals, Osaka, Japan). The Ni-NTA-agarose beads recovered by centrifugation were washed four times with the lysis buffer. Each sample was resolved on 7.5% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and incubated overnight with anti-FLAG antibody (Sigma-Aldrich). The bound antibody was detected with anti-mouse IgG conjugated to horseradish peroxidase.
N-Sulfotransferase Activity
To compare between sulfotransferase activity of NDST-1 alone and sulfotransferase activity of a complex of NDST-1 with EXTL3, full-length or deletion mutants of NDST-1 were expressed in COS-1 cells with or without EXTL3. Two days after transfection, cells were solubilized as above and then incubated with 10 μl of IgG-Sepharose (GE Healthcare) overnight at 4 °C. The beads recovered by centrifugation were washed with lysis buffer and then resuspended in the assay buffer and tested for sulfotransferase activity. N-Sulfotransferase activity was analyzed as described previously (18) by measuring incorporation of 35S from the sulfate donor [35S]PAPS into Escherichia coli capsular K5 polysaccharide (Iduron, Manchester, UK) as a substrate.
N-Deacetylase Activity
N-Deacetylase activity was analyzed using an ELISA-based assay as described previously (15, 19). Briefly, a Corning® 96-well immunoassay/radioimmunoassay clear flat bottom polystyrene high bind microplate was coated with 50 μg/ml E. coli K5 capsular polysaccharide in PBS overnight at room temperature. After washing with TBS containing 0.1% Tween 20 (TBST), an N-deacetylase assay mix containing 50 mm MES-NaOH (pH 6.3), 10 mm MnCl2, and 1% Triton X-100 was added to the wells together with the samples (recombinant enzyme proteins bound to beads) and incubated for 30 min at 37 °C. After washing with TBST, the wells were then blocked with 1% BSA in TBST for 2 h at room temperature. Incubation with JM403 (dilution 1:100) in TBST overnight at 4 °C was followed by washing and incubation with peroxidase-conjugated anti-mouse IgM in TBST (dilution 1:1,000) for 2 h at room temperature. After washing with TBST, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) was added to each well as a peroxidase substrate. The blue-green-colored materials produced by peroxidase were measured at 405 nm.
Statistical Analysis
Data are expressed as mean ± S.D. Statistical significance was determined by Student's t test.
RESULTS
HS Structures Possessing N-Unsubstituted Glucosamine Are Effective as a Heparanase Inhibitor
We found that the highly invasive cell lines MDA-MB-231 and BT549 express JM403-recognizing antigens. The JM403 epitope was previously reported to be a sequence of GlcA-GlcNH3+ in HS chains (20). Immunofluorescence analysis indicated that JM403-recognizing epitopes were localized within cells (Fig. 1B). In addition, these two cell lines, classified as a basal-like breast cancer, expressed JM403 epitopes at a comparatively higher level than other breast cancer cell lines, MCF-7, T47D (luminal A-type breast cancer), and HCC1954 (HER2-type breast cancer) (Fig. 1A). We examined the susceptibility of JM403 epitopes to HS-degrading enzymes using HS as a substrate, because JM403 epitopes were more abundant in HS than in heparin (Fig. 1C). Immobilized biotinylated HS to the streptavidin-coated plate was digested with either heparinase, heparitinase, or heparanase. JM403 epitopes were decreased by the digestion with heparitinase and slightly decreased by the digestion with heparinase. In contrast, the JM403 epitope was resistant to mammalian heparanase (Fig. 1C). Fig. 2A shows the expression level of each cell line; the expression level of heparanase in MDA-MB-231 and BT549 cells was higher than in other cell lines. These results prompted us to speculate that the JM403 epitope is concentrated within cells due to the presence of heparanase-resistant structures. To test this hypothesis, we chemically synthesized three kinds of HS-tetrasaccharides with different modifications of the amino group in GlcN residues (16) (Fig. 2B). When MDA-MB-231 and BT549 cells were treated with TD4-143-1, the amounts of HS were significantly higher than before treatment, whereas TD5-67-9 and TD5-60-6 hardly affected the amount of HS (Fig. 2C). In addition, TD4-143-1 did not increase the amounts of HS in MCF-7 and T47D cells, where heparanase was expressed at a lower level compared with MDA-MB-231 and BT549 cells (Fig. 2, A and C). This result implies that TD4-143-1 inhibits degradation of HS chains by heparanase in MDA-MB-231 and BT549 cells. Thus, we investigated whether TD4-143-1 could inhibit heparanase activities in vitro. As expected, TD4-143-1 significantly inhibited recombinant heparanase activities, whereas TD5-67-9 and TD5-60-6 did not (Fig. 2D). Therefore, it is thought that the structure possessing N-unsubstituted glucosamine (GlcNH3+) exhibits more effective inhibition of heparanase activity than N-acetylated and N-sulfated glucosamine. Then we examined the effects of TD4-143-1 on the cell viability and invasion activity of breast cancer cells. Although TD4-143-1 did not affect the cell viability of four breast cancer cell lines (Fig. 2E), invasion of highly metastatic breast cancer cells, MDA-MB-231 and BT549 cells, was significantly inhibited by TD4-143-1 in vitro (Fig. 2F). In addition, the inhibitory activity of TD4-143-1 was comparable with that of a commercially available heparanase inhibitor, OGT2115 (Fig. 2F). TD4-143-1 is supposed to suppress tumor invasion mediated by inhibition of heparanase activity. In contrast, invasion of MDA-MB-231 and BT549 cells was hardly inhibited by TD5-67-9 and TD5-60-6 (Fig. 2F), both of which exert no heparanase inhibitory activity (Fig. 2D). These results indicate that TD4-143-1, an HS structure possessing N-unsubstituted glucosamine (GlcNH3+), is effective as a heparanase inhibitor; thus, TD4-143-1 can inhibit breast cancer cell invasion.
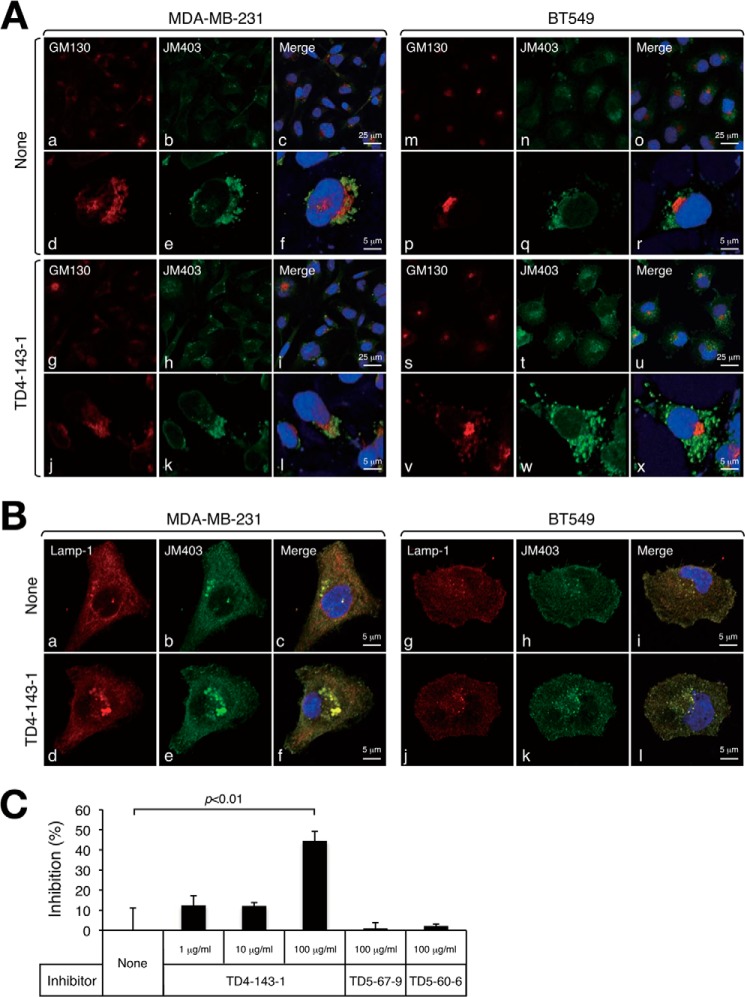
Furthermore, we investigated the localization of TD4-143-1 incorporated into cells. TD4-143-1 has N-unsubstituted glucosamine (GlcNH3+) structures and can thus be detected by JM403. The JM403-recognizing structure was endogenously expressed (Fig. 3A, b and n). The addition of TD4-143-1 to cells increased staining signals by JM403 within the cells (Fig. 3A, h and t). We confirmed whether JM403 could recognize TD4-143-1 by competitive inhibition ELISA (Fig. 3C). TD4-143-1 (100 μg/ml) inhibited the binding of JM403 to HS containing JM403 epitopes (see Fig. 1C) by 50%, whereas TD5-67-9 and TD5-60-6 could not inhibit (Fig. 3C). These results suggest that TD4-143-1 was incorporated into the cells. Both endogenous JM403-recognizing GlcNH3+ structures and exogenously added TD4-143-1 were partially co-localized with the cis-Golgi marker GM130 in both breast cancer cells (Fig. 3A, f, l, r, and x). In contrast, localization of TD4-143-1 was considerably merged with the lysosomal marker Lamp-1 (Fig. 3B, c, f, i, and l). These results indicate that endogenous JM403-recognizing GlcNH3+ structures reside partially in the Golgi and mostly in the lysosome. Generally, cell surface HS-PGs are taken up by endocytosis, transported to the lysosome, where HS chains are degraded by heparanase. However, HS oligosaccharides containing GlcNH3+ structures evade degradation by heparanase, accumulate during turnover of HS, and might have an inhibitory effect on heparanase activities.
FIGURE 3.
Cellular localization of endogenously expressed JM403 epitopes and exogenously added TD4-143-1. Human breast cancer cells, MDA-MB-231 and BT549, were incubated overnight in the presence (TD4-143-1) or absence (None) of 50 μm TD4-143-1. A, Golgi localization of JM403 epitopes and TD4-143-1 incorporated into the cells was examined as an indicator of the localization of GM130, which is a member of the Golgi family of proteins. a, b, c, g, h, i, m, n, o, s, t, and u, low magnification; d, e, f, j, k, l, p, q, r, v, w, and x, high magnification. B, lysosome localization of JM403 epitopes and TD4-143-1 incorporated into the cells was examined as an indicator of the localization of the lysosome-associated membrane protein Lamp-1. C, binding of JM403 to HS was examined by the inhibition assay using three kinds of chemically synthesized tetrasaccharides as inhibitors.
EXTL3 Contributes to Generation of Unsubstituted Glucosamine Residues
We next examined the synthetic mechanism of endogenous GlcNH3+ structures. To date, we have investigated the role of three EXTLs (EXTL1, EXTL2, and EXTL3) in HS biosynthesis (21–24). During the course of these studies, we noted that the expression level of EXTL3 controls N-sulfation of HS. As shown in Fig. 4A, knockdown of EXTL3 in mouse L fibroblasts increased the amount of ΔDi-HS-NS, whereas the amount of ΔDi-HS-0S was decreased. These results prompted us to ask if EXTL3 might have other functions, although it has been reported that EXTL3 has N-acetylglucosaminyltransferase-I and -II activities, which are involved in the initiation and elongation of HS chains, respectively. Because the N-sulfation level of glucosamine was affected by the expression level of EXTL3, we examined whether EXTL3 could regulate the expression level of the JM403 epitope by the generation of cells expressing EXTL3 at various levels. The cells with a higher expression level of EXTL3 express larger amounts of the JM403 epitope (Fig. 4B). However, there is the possibility that the expression level of the JM403 epitope might increase with the increasing level of HS, because EXTL3 is reported to regulate the amounts of HS (21, 22). Thus, we examined the expression level of HS in the cells expressing EXTL3 at various levels using another anti-HS antibody, 10E4. Although knockdown of EXTL3 decreased the expression of both of 10E4 and JM403 epitopes, the effect of EXTL3 knockdown on the 10E4 epitope was milder than that on the JM403 epitope (Fig. 4C). Furthermore, we measured the proportion of the JM403 epitope within HS chains isolated from the cells expressing EXTL3 at various levels. Knockdown of EXTL3 remarkably decreased the proportion of JM403 epitopes in HS chains (Fig. 4D). These results indicate that EXTL3 can control N-sulfation of glucosamine via an unknown mechanism. From the results of heparanase inhibition assays (Fig. 2D), we assumed that JM403 epitope is resistant to heparanase. Thus, the length of HS chains produced by cells with various expression levels of EXTL3 was measured (Fig. 4E). Expectedly, knockdown of EXTL3 caused a decrease in the amount and length of HS chains, probably because of the lower level of JM403 epitope. Overexpression of EXTL3 increased the amount of HS but had little or no effect on the length of HS chains. These results raise the possibility that EXTL3 controls modifications at the N-position (-Ac, -SO3H, or -H) of glucosamine and the catabolism of HS by heparanase. We next investigated whether the expression level of EXTL3 could affect C5 epimerization of GlcA by taking advantage of the substrate specificities of heparinase and heparitinase, because modifications, such as epimerization and O-sulfation, depend on prior N-sulfation of glucosamine residues. Heparinase (EC 4.2.2.7) catalyzes the cleavage of the α-glucosaminidic linkage to 2-O-sulfated iduronic acid, whereas heparitinase (EC 4.2.2.8) cleaves the α-glucosaminidic linkage to GlcA. As shown in Fig. 4F, disaccharides generated by heparinase considerably decreased in L-shEXTL3 cells compared with L cells, suggesting that epimerization and 2-O-sulfation might be affected by knockdown of EXTL3. Both knockdown and overexpression of EXTL3 had little effect on the expression levels of sulfotransferases involved in sulfation of HS chains, NDST1, NDST2, HS2ST, and HS6ST1 (Fig. 4G). These results indicate that EXTL3 can control N-sulfation of glucosamine at a post-transcriptional level. So far, four NDSTs (NDST-1, -2, -3, and -4) have been identified as enzymes involved in the modification of the N-position of glucosamine, with the expression of NDST-1 and NDST-2 being widely distributed (25). The most predominant N-sulfotransferase responsible for the HS biosynthesis is NDST-1, whereas NDST-2 is a key enzyme in heparin synthesis. Therefore, we investigated whether EXTL3 could modulate enzymatic activities of NDST-1. First, we examined the interaction of EXTL3 with NDST-1.
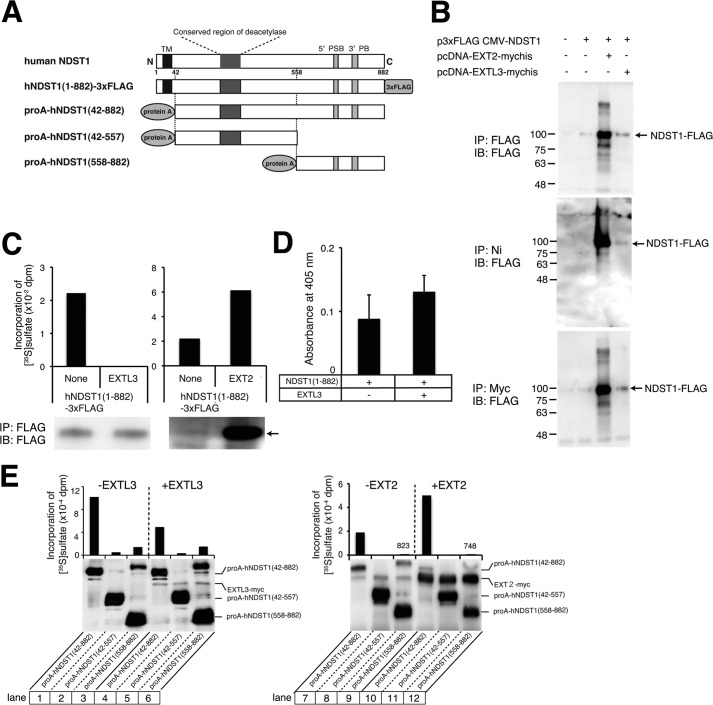
NDST-1 and EXTL3 proteins were expressed as full-length proteins tagged with a FLAG peptide (Fig. 5A) or a C-terminal polyhistidine metal-binding peptide, respectively. EXTL3-Myc-His6 and NDST-1-FLAG were expressed either individually or in combination in COS1 cells. Upon purification of EXTL3-Myc-His6 from the cell lysates of transfected cells, using nickel-agarose, the NDST-1-FLAG proteins could be detected in the precipitates only when both NDST-1-FLAG proteins and EXTL3-Myc-His6 were co-expressed (Fig. 5B). These results suggest that EXTL3 and NDST-1 are capable of forming complexes. As reported previously (15), interaction of NDST-1 with EXT2 was observed in our experimental system (Fig. 5B). In addition, we confirmed that the protein expression of NDST-1 was increased by co-expression of EXT2 for an increase in protein stability, as reported (15). We next explored the possibility that N-sulfotransferase activities of NDST-1 may be affected by the forming of a complex with EXTL3. When NDST-1 and EXTL3 were co-expressed in COS1 cells, N-sulfotransferase activities were strongly inhibited compared with the expression of NDST-1 alone. In contrast, EXT2 increased N-sulfotransferase activities because EXT2 could enhance the stability of NDST-1 (Fig. 5C). Because NDST-1 possesses N-deacetylase activity in addition to N-sulfotransferase activity, we investigated whether or not N-deacetylase activity was affected by the formation of a complex with EXTL3. NDST-1 N-deacetylase activity was slightly enhanced by co-expression with EXTL3; however, there was no significant difference between the two conditions (Fig. 5D).
FIGURE 5.
N-Sulfotransferase activity of NDST-1 was controlled by interaction of NDST-1 with EXTL3. A, schematic representations of expressed human NDST-1 proteins. Full-length NDST-1 was expressed as a FLAG-tagged protein, and NDST-1 deletion mutants were expressed as a protein A fusion protein. TM, transmembrane domain. The conserved region of deacetylase is located at the N terminus of NDST-1. 5′PSB and 3′PB, 5′-phosphosulfate of PAPS binding region and 3′-phosphate of PAPS binding region, respectively. B, FLAG-tagged NDST-1 was expressed with or without Myc- and His-tagged EXTL3 in COS-1 cells. Cell lysates were pulled down using anti-FLAG-agarose or nickel-agarose. In addition, cellular proteins were incubated with an anti-Myc antibody, followed by pull-down using protein G-Sepharose. Precipitated proteins were subjected to electrophoresis and immunoblotting using anti-FLAG antibodies. Arrow, FLAG-tagged NDST-1. +, the expression plasmids encoding the indicated proteins were transfected into COS-1 cells; −, the expression plasmids encoding the indicated proteins were not transfected into COS-1 cells. C, after FLAG-tagged NDST-1 was expressed alone (None) or co-expressed with EXTL3 or EXT2 in COS-1 cells, N-sulfotransferase activities were measured as described under “Experimental Procedures.” An arrow indicates FLAG-tagged NDST-1. D, COS-1 cells were co-transfected with a plasmid encoding FLAG-tagged NDST-1 together with or without EXTL3, as indicated. N-Deacetylase activities were measured as described under “Experimental Procedures.” E, protein A-fused NDST-1 deletion mutants were co-expressed with EXTL3 or EXT2 (+EXTL3 or +EXT2) or without EXTL3 or EXT2 (−EXTL3 or −EXT2) in COS-1 cells and precipitated using IgG-Sepharose, and N-sulfotransferase activities were measured. In addition, precipitated protein A fused NDST-1 and Myc-tagged EXTL3 were analyzed by immunoblotting using anti-Myc antibodies. IP, immunoprecipitation; IB, immunoblotting.
It has been reported that the two functional domains responsible for N-sulfotransferase (NST) and N-deacetylase (NDA) are located at the C terminus (amino acids 558–882) and the N terminus (amino acids 1–557) of NDST-1 protein, respectively (26). Thus, we examined the binding of EXTL3 to the C-terminal NST and N-terminal NDA domains of NDST-1. It was found that both domains of NDST-1 bound to EXTL3 (Fig. 5E, lanes 5 and 6). However, N-sulfotransferase activities of NST domain were not affected by EXTL3 (Fig. 5E, lanes 3 and 6). In addition, EXT2 can bind to NST and NDA domains of NDST-1 (Fig. 5E, lanes 11 and 12), although EXT2 had little effect on N-sulfotransferase activities of NST domain (Fig. 5E, lanes 9 and 12). Taken together, these results suggest that binding of EXTL3 to both NDA and NST domains is needed to regulate N-sulfotransferase activities of NDST-1.
DISCUSSION
In this study, we found that HS chains containing GlcA-GlcNH3+ accumulate within the highly invasive human breast cancer cell lines MDA-MB-231 and BT549 (Fig. 1), which is consistent with previous reports that GlcA-GlcNH3+ is expressed in highly malignant breast cancers (27). These two cell lines express high levels of heparanase (Fig. 2). Previous reports showed that cleavage of HS on glypican by heparanase yielded an HS fragment containing GlcA-GlcNH3+ (28). These results suggest that heparanase cannot cleave at glucuronidic acid linkages of GlcA-GlcNH3+. Thus, we tested whether or not HS containing GlcA-GlcNH3+ could inhibit heparanase activities. We first examined inhibitory activity of a series of chemically synthesized tetrasaccharides, because it has been reported that the inhibitory activity of heparin is lost upon a decrease in chain length below tetrasaccharidic sizes (29). As reported, tetrasaccharides carrying N-sulfates and N-acetyl groups, TD5-60-6 and TD5-67-9, had little effect on heparanase activities in vitro. In contrast, tetrasaccharides carrying free glucosamine groups, TD4-143-1, inhibited heparanase activities in vitro to the same extent as the commercial heparanase inhibitor OGT2115 (Fig. 2D). Furthermore, we indicated that TD4-143-1 inhibited invasion of breast cancer cells by a Matrigel invasion assay (Fig. 2F). When TD4-143-1 was added to cells, TD4-143-1 was incorporated into the cells and localized in the lysosome (Fig. 3). This result is consistent with the previous report that GlcNH3+-containing HS was present at the cell surface and in cytoplasmic vesicles, where it colocalized extensively with LysoTracker Red, a marker for acidic endosomes and lysosomes. These results shown in Fig. 3 suggest that exogenous TD4-143-1 might inhibit heparanase activity in extracellular matrices as well as the conversion of the precursor form into the active form of heparanase in the lysosome (30). Heparanase is reported to be synthesized as a 65-kDa non-active precursor that subsequently undergoes proteolytic cleavage in the lysosome, yielding 8- and 50-kDa protein subunits that heterodimerize to form an active enzyme (31). In addition, the amount of HS was increased by the treatment of cells with TD4-143-1 (Fig. 2), whereas the amount of CS was not significantly altered (data not shown). This is consistent with the finding that TD4-143-1 inhibits degradation of HS by heparanase.
So far, HS-related heparanase inhibitors have been reported (32, 33). Examples of such inhibitors are heparin, heparin-mimicking compounds, unfractionated heparin (average Mr ∼15,000), low molecular weight heparin (average Mr ∼6,000), and ultra-low molecular weight heparin (average Mr ∼2,000). Although unfractionated heparin prepared from animal tissues has been used as a century-old drug, it shows side effects, such as thrombocytopenia and bleeding. Low molecular weight and ultra-low molecular weight heparin also have the risk of bleeding due to overdosing. In contrast, chemically synthesized HS tetrasaccharides, TD4-143-1 (Mr 1,008), are supposed to exhibit no anticoagulant activity, because they do not possess critical structures for complex formation of antithrombin III (34). Previously, contaminated batches of heparin and low molecular weight heparin caused severe side effects (i.e. a rapid drop in blood pressure) (33). Because TD4-143-1 is a small molecule and is chemically synthesized, its quality can be tightly controlled. In addition, PI-88 (phosphomannopentaose sulfate) is a recently developed heparanase inhibitor currently undergoing phase II/III clinical trials in cancer patients with metastatic melanoma. Compared with PI-88 derived from the extracellular phosphomannan of the yeast Pichia, TD4-143-1 might take effect specifically and inflict little side effect because TD4-143-1 was chemically synthesized based on HS structures closely associated with malignant human breast cancer.
N-Unsubstituted glucosamine residues have previously been detected by amine-reactive fluorescence labeling and deaminative cleavage at pH 3.9 (1) and were recently observed in various rat organ tissues using a combination of liquid chromatography and mass spectrometry (35). In addition, a monoclonal antibody that recognized GlcNH3+ residues also revealed the presence of N-unsubstituted glucosamine in native HS (20). Although the content of GlcNH3+ residues is low, they are widely expressed in mammalian organ tissues and implicated in important biological and pathological phenomena. Previous reports have shown that the GlcNH3+ residues expressed in bovine and human endothelial cells are involved in the binding to L-selectin (2). Moreover, it has been indicated that 3-O-sulfated GlcNH3+ generates a binding site in cell surface HS for the herpes simplex virus 1 glycoprotein gD, a key player in viral invasion (36). In addition, cyclophillin B, which triggers chemotaxis and integrin-mediated adhesion of T lymphocytes, binds to cell surface HS, depending on the presence of a 3-O-sulfated GlcNH3+ residue (37). Furthermore, another report showed that HS chains containing GlcNH3+ residues were present in the deposition area of abnormal prion protein, PrPSc (25). Here, we found that HS oligosaccharides containing GlcNH3+ residues inhibit heparanase activity and tumor cell invasion in vitro. This observation suggests the possibility that tumor metastasis is suppressed by virtue of GlcNH3+-containing HS oligosaccharides.
Although the mechanism of GlcNH3+ formation during HS biosynthesis remains largely unclear, several possibilities have been discussed. During the formation of HS chains, the nascent polymer composed of alternating GlcA and GlcNAc units is first subject to partial N-deacetylation and N-sulfation of GlcNAc residues. Sometimes, the N-deacetylation/N-sulfation reaction might be incomplete, giving rise to a fraction of N-unsubstituted glucosamine residues. Conceivably, residual N-acetyl groups could be selectively removed by N-acetylase, or N-sulfate groups could be selectively eliminated by an unidentified endo-sulfamidase at a later stage of HS biosynthesis. Moreover, Carlsson et al. showed that variations in the N-substitution pattern could be affected by varying the concentration of the sulfate donor, PAPS (38). N-Deacetylation of a fully N-acetylated HS produced in the absence of PAPS might generate a limited proportion of GlcNH3+ residues. However, it is thought that another mechanism regulates the generation of GlcNH3+ residues, because depletion of PAPS affects every sulfotransferase. NDST-3, one of four distinct NDST isoforms, has much higher N-deacetylase compared with N-sulfotransferase activity than any of the other NDST enzymes. Thus, a role for NDST-3 in the generation of GlcNH3+ residues would seem mechanistically attractive; however, previous study on NDST-3 knock-out mice indicated little contribution of NDST-3 to the production of GlcNH3+ residues (11). In addition, overexpression in HEK293 cells of mutant NDST-1 lacking N-sulfotransferase activity resulted in no increase in GlcNH3+ residues (39). Furthermore, Gesteira et al. (40) have screened the peptide NMQALSMPVT, which inhibits N-sulfotransferase activity of mNDST-1, using a phage display library. This result suggests the possibility that endogenous proteins containing similar sequences inhibit N-sulfotransferase activity of NDST-1. In this study, we demonstrated that NDST-1 sulfotransferase activity is controlled by formation of a complex between NDST-1 and EXTL3 (Fig. 5, C and E) and that generation of GlcNH3+ residues can be regulated (Fig. 4B). Currently, a model in which the enzymes of HS biosynthesis form a complex (GAGosome) has been proposed. A previous report showed that the interaction of NDST-1 with EXT2 could enhance NDST-1 protein stability and that it results in elevated HS sulfation (15). It is probable that the GAGosome also regulates the generation of GlcNH3+ residues.
We thought that GlcNH3+ residues generated by the interaction of EXTL3 with NDST-1 might control the metabolism of HS chains. Thus, it is suggested that heparanase-resistant HS chains are produced by up-regulation of EXTL3. Although early and ongoing work with heparanase has focused on cancer, emerging evidence shows that heparanase plays important roles in other diseases states, including diabetes (41). Recent reviews describe exciting findings that HS within β-cells in the pancreatic islet acts to protect these cells from free radical damage and death (42). This protective antiapoptotic effect is neutralized when nearby autoreactive T cells secrete heparanase that subsequently degrade HS, leading to onset of Type I diabetes. On the other hand, it has been reported (43) that a lack of EXTL3 in β-cells decreased the β-cell number in the islet and caused glucose intolerance due to defective insulin secretion. Therefore, it is conceivable that dysregulation of HS biosynthesis by the loss of EXTL3 may be a potential factor in the pathogenesis of diabetes mellitus (43). Taken together, it is speculated that HS chains synthesized in the presence of EXTL3 are resistant to cleavage by heparanase and thus prevent the onset of diabetes. In contrast, GlcNH3+ residues are reported to be a nitric oxide (NO) cleavage site (28, 30). A few GlcNH3+ structures are reportedly located close to the linkage region to the core protein (28). When NO is released, it catalyzes deaminative cleavage of HS at the GlcNH3+ structures. In the case of Gpc-1 (glypican-1), an HS-PG, Cys residues in Gpc-1 can become S-nitrosylated by NO. After internalization of S-nitrosylated Gpc-1, the HS chains of Gpc-1 are degraded either enzymatically by heparanase or nonenzymatically by NO, derived from S-nitrosylation of Gpc-1. These degradations take place while Gpc-1 recycles via the late endosomes to lysosomes (28). Unless NO is not released, HS chains are degraded by heparanase except at GlcNH3+ residues. Thus, HS oligosaccharides containing GlcNH3+ residues remain and accumulate in the lysosome and might have an inhibitory effect on HS degradation by heparanase.
Why do endogenous GlcNH3+ structures have no inhibitory effect on breast cancer cells themselves? It has been reported that cell surface HS-PGs, especially Gpc-1, control uptake of polyamines involved in cell proliferation (44, 45). Tumor cells synthesizing heparanase-resistant HS chains with high GlcNH3+ contents might uptake polyamines during invasion, even if they are exposed to a high concentration of heparanase. Furthermore, after polyamines tightly bound to HS chains of Gpc-1 are internalized into the cells, deaminative cleavage of HS chains at the GlcNH3+ structures is required for release of polyamines from HS chains (44). Free polyamine molecules can function for cell proliferation (44, 45). Therefore, GlcNH3+ structures are considered to have little inhibitory effect on breast cancer growth, although GlcNH3+ structures inhibit heparanase.
This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas 23110003 (to H. K.) and for Scientific Research (B) 25293014 (to H. K.) and (C) 25460080 (to S. N.) and the Supported Program for the Strategic Research Foundation at Private Universities, 2012–2016 (to H. K.) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
- HS-PG
- heparan sulfate proteoglycan
- HS
- heparan sulfate
- CS
- chondroitin sulfate
- GlcA
- glucuronic acid
- GlcN
- glucosamine
- NDST
- N-deacetylase/N-sulfotransferase
- PAPS
- 3′-phosphoadenosine 5′-phosphosulfate
- GlcNH3+
- N-unsubstituted glucosamine
- Ni-NTA
- nickel-nitrilotriacetic acid
- NST
- N-sulfotransferase
- NDA
- N-deacetylase.
REFERENCES
- 1. Rees M. D., Pattison D. I., Davies M. J. (2005) Oxidation of heparan sulphate by hypochlorite: role of N-chloro derivatives and dichloramine-dependent fragmentation. Biochem. J. 391, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Norgard-Sumnicht K., Varki A. (1995) Endothelial heparan sulfate proteoglycans that bind to L-selectin have glucosamine residues with unsubstituted amino groups. J. Biol. Chem. 270, 12012–12024 [DOI] [PubMed] [Google Scholar]
- 3. van den Born J., Gunnarsson K., Bakker M. A., Kjellén L., Kusche-Gullberg M., Maccarana M., Berden J. H., Lindahl U. (1995) Presence of N-unsubstituted glucosamine units in native heparan sulfate revealed by a monoclonal antibody. J. Biol. Chem. 270, 31303–31309 [DOI] [PubMed] [Google Scholar]
- 4. Westling C., Lindahl U. (2002) Location of N-unsubstituted glucosamine residues in heparan sulfate. J. Biol. Chem. 277, 49247–49255 [DOI] [PubMed] [Google Scholar]
- 5. Cheng F., Mani K., van den Born J., Ding K., Belting M., Fransson L. A. (2002) Nitric oxide-dependent processing of heparan sulfate in recycling S-nitrosylated glypican-1 takes place in caveolin-1-containing endosomes. J. Biol. Chem. 277, 44431–44439 [DOI] [PubMed] [Google Scholar]
- 6. Murali S., Leong D. F., Lee J. J., Cool S. M., Nurcombe V. (2011) Comparative assessment of the effects of gender-specific heparan sulfates on mesenchymal stem cells. J. Biol. Chem. 286, 17755–17765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fujii M., Yusa A., Yokoyama Y., Kokuryo T., Tsunoda N., Oda K., Nagino M., Ishimaru T., Shimoyama Y., Utsunomiya H., Iwata H., Itoh Y., Itoh J., Kannagi R., Kyogashima M. (2010) Cytoplasmic expression of the JM403 antigen GlcA-GlcNH3+ on heparan sulfate glycosaminoglycan in mammary carcinomas: a novel proliferative biomarker for breast cancers with high malignancy. Glycoconj. J. 27, 661–672 [DOI] [PubMed] [Google Scholar]
- 8. Fan G., Xiao L., Cheng L., Wang X., Sun B., Hu G. (2000) Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Lett. 467, 7–11 [DOI] [PubMed] [Google Scholar]
- 9. Forsberg E., Pejler G., Ringvall M., Lunderius C., Tomasini-Johansson B., Kusche-Gullberg M., Eriksson I., Ledin J., Hellman L., Kjellén L. (1999) Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature 400, 773–776 [DOI] [PubMed] [Google Scholar]
- 10. Aikawa J., Grobe K., Tsujimoto M., Esko J. D. (2001) Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. Structure and activity of the fourth member, NDST4. J. Biol. Chem. 276, 5876–5882 [DOI] [PubMed] [Google Scholar]
- 11. Pallerla S. R., Lawrence R., Lewejohann L., Pan Y., Fischer T., Schlomann U., Zhang X., Esko J. D., Grobe K. (2008) Altered heparan sulfate structure in mice with deleted NDST3 gene function. J. Biol. Chem. 283, 16885–16894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Okada M., Nadanaka S., Shoji N., Tamura J., Kitagawa H. (2010) Biosynthesis of heparan sulfate in EXT1-deficient cells. Biochem. J. 428, 463–471 [DOI] [PubMed] [Google Scholar]
- 13. Mizushima S., Nagata S. (1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saito T., Seki N., Yamauchi M., Tsuji S., Hayashi A., Kozuma S., Hori T. (1998) Structure, chromosomal location, and expression profile of EXTR1 and EXTR2, new members of the multiple exostoses gene family. Biochem. Biophys. Res. Commun. 243, 61–66 [DOI] [PubMed] [Google Scholar]
- 15. Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., Kjellén L. (2008) Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takeda N., Ikeda-Matsumi R., Ebara-Nagahara K., Otaki-Nanjo M., Taniguchi-Morita K., Nanjo M., Tamura J. (2012) Synthesis of heparan sulfate tetrasaccharide as a substrate for human heparanase. Carbohydr. Res. 353, 13–21 [DOI] [PubMed] [Google Scholar]
- 17. Kim B. T., Kitagawa H., Tanaka J., Tamura J., Sugahara K. (2003) In vitro heparan sulfate polymerization: crucial roles of core protein moieties of primer substrates in addition to the EXT1-EXT2 interaction. J. Biol. Chem. 278, 41618–41623 [DOI] [PubMed] [Google Scholar]
- 18. Tsutsumi K., Shimakawa H., Kitagawa H., Sugahara K. (1998) Functional expression and genomic structure of human chondroitin 6-sulfotransferase. FEBS Lett. 441, 235–241 [DOI] [PubMed] [Google Scholar]
- 19. van den Born J., Pikas D. S., Pisa B. J., Eriksson I., Kjellen L., Berden J. H. (2003) Antibody-based assay for N-deacetylase activity of heparan sulfate/heparin N-deacetylase/N-sulfotransferase (NDST): novel characteristics of NDST-1 and -2. Glycobiology 13, 1–10 [DOI] [PubMed] [Google Scholar]
- 20. van den Born J., Salmivirta K., Henttinen T., Ostman N., Ishimaru T., Miyaura S., Yoshida K., Salmivirta M. (2005) Novel heparan sulfate structures revealed by monoclonal antibodies. J. Biol. Chem. 280, 20516–20523 [DOI] [PubMed] [Google Scholar]
- 21. Nadanaka S., Zhou S., Kagiyama S., Shoji N., Sugahara K., Sugihara K., Asano M., Kitagawa H. (2013) EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. J. Biol. Chem. 288, 9321–9333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim B. T., Kitagawa H., Tamura J., Saito T., Kusche-Gullberg M., Lindahl U., Sugahara K. (2001) Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode α1,4-N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/heparin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 98, 7176–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitagawa H., Shimakawa H., Sugahara K. (1999) The tumor suppressor EXT-like gene EXTL2 encodes an α1,4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region: the key enzyme for the chain initiation of heparan sulfate. J. Biol. Chem. 274, 13933–13937 [DOI] [PubMed] [Google Scholar]
- 24. Kitagawa H., Egusa N., Tamura J. I., Kusche-Gullberg M., Lindahl U., Sugahara K. (2001) rib-2, a Caenorhabditis elegans homolog of the human tumor suppressor EXT genes encodes a novel α1,4-N-acetylglucosaminyltransferase involved in the biosynthetic initiation and elongation of heparan sulfate. J. Biol. Chem. 276, 4834–4838 [DOI] [PubMed] [Google Scholar]
- 25. Grobe K., Ledin J., Ringvall M., Holmborn K., Forsberg E., Esko J. D., Kjellén L. (2002) Heparan sulfate and development: differential roles of the N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes. Biochim. Biophys. Acta 1573, 209–215 [DOI] [PubMed] [Google Scholar]
- 26. Sueyoshi T., Kakuta Y., Pedersen L. C., Wall F. E., Pedersen L. G., Negishi M. (1998) A role of Lys614 in the sulfotransferase activity of human heparan sulfate N-deacetylase/N-sulfotransferase. FEBS Lett. 433, 211–214 [DOI] [PubMed] [Google Scholar]
- 27. Weyers A., Yang B., Yoon D. S., Park J. H., Zhang F., Lee K. B., Linhardt R. J. (2012) A structural analysis of glycosaminoglycans from lethal and nonlethal breast cancer tissues: toward a novel class of theragnostics for personalized medicine in oncology? OMICS 16, 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ding K., Sandgren S., Mani K., Belting M., Fransson L. A. (2001) Modulations of glypican-1 heparan sulfate structure by inhibition of endogenous polyamine synthesis. Mapping of spermine-binding sites and heparanase, heparin lyase, and nitric oxide/nitrite cleavage sites. J. Biol. Chem. 276, 46779–46791 [DOI] [PubMed] [Google Scholar]
- 29. Vlodavsky I., Mohsen M., Lider O., Svahn C. M., Ekre H. P., Vigoda M., Ishai-Michaeli R., Peretz T. (1994) Inhibition of tumor metastasis by heparanase inhibiting species of heparin. Invasion Metastasis 14, 290–302 [PubMed] [Google Scholar]
- 30. Mani K., Cheng F., Sandgren S., Van Den Born J., Havsmark B., Ding K., Fransson L. A. (2004) The heparan sulfate-specific epitope 10E4 is NO-sensitive and partly inaccessible in glypican-1. Glycobiology 14, 599–607 [DOI] [PubMed] [Google Scholar]
- 31. Zetser A., Levy-Adam F., Kaplan V., Gingis-Velitski S., Bashenko Y., Schubert S., Flugelman M. Y., Vlodavsky I., Ilan N. (2004) Processing and activation of latent heparanase occurs in lysosomes. J. Cell Sci. 117, 2249–2258 [DOI] [PubMed] [Google Scholar]
- 32. Afratis N., Gialeli C., Nikitovic D., Tsegenidis T., Karousou E., Theocharis A. D., Pavão M. S., Tzanakakis G. N., Karamanos N. K. (2012) Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J. 279, 1177–1197 [DOI] [PubMed] [Google Scholar]
- 33. Linhardt R. J., Liu J. (2012) Synthetic heparin. Curr. Opin. Pharmacol. 12, 217–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Desai U. R., Petitou M., Björk I., Olson S. T. (1998) Mechanism of heparin activation of antithrombin: evidence for an induced-fit model of allosteric activation involving two interaction subsites. Biochemistry 37, 13033–13041 [DOI] [PubMed] [Google Scholar]
- 35. Shi X., Zaia J. (2009) Organ-specific heparan sulfate structural phenotypes. J. Biol. Chem. 284, 11806–11814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tiwari V., Clement C., Xu D., Valyi-Nagy T., Yue B. Y., Liu J., Shukla D. (2006) Role for 3-O-sulfated heparan sulfate as the receptor for herpes simplex virus type 1 entry into primary human corneal fibroblasts. J. Virol. 80, 8970–8980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vanpouille C., Deligny A., Delehedde M., Denys A., Melchior A., Liénard X., Lyon M., Mazurier J., Fernig D. G., Allain F. (2007) The heparin/heparan sulfate sequence that interacts with cyclophilin B contains a 3-O-sulfated N-unsubstituted glucosamine residue. J. Biol. Chem. 282, 24416–24429 [DOI] [PubMed] [Google Scholar]
- 38. Carlsson P., Presto J., Spillmann D., Lindahl U., Kjellén L. (2008) Heparin/heparan sulfate biosynthesis: processive formation of N-sulfated domains. J. Biol. Chem. 283, 20008–20014 [DOI] [PubMed] [Google Scholar]
- 39. Bengtsson J., Eriksson I., Kjellén L. (2003) Distinct effects on heparan sulfate structure by different active site mutations in NDST-1. Biochemistry 42, 2110–2115 [DOI] [PubMed] [Google Scholar]
- 40. Gesteira T. F., Coulson-Thomas V. J., Taunay-Rodrigues A., Oliveira V., Thacker B. E., Juliano M. A., Pasqualini R., Arap W., Tersariol I. L., Nader H. B., Esko J. D., Pinhal M. A. (2011) Inhibitory peptides of the sulfotransferase domain of the heparan sulfate enzyme, N-deacetylase-N-sulfotransferase-1. J. Biol. Chem. 286, 5338–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vlodavsky I., Iozzo R. V., Sanderson R. D. (2013) Heparanase: multiple functions in inflammation, diabetes and atherosclerosis. Matrix Biol. 32, 220–222 [DOI] [PubMed] [Google Scholar]
- 42. Ziolkowski A. F., Popp S. K., Freeman C., Parish C. R., Simeonovic C. J. (2012) Heparan sulfate and heparanase play key roles in mouse β cell survival and autoimmune diabetes. J. Clin. Invest. 122, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takahashi I., Noguchi N., Nata K., Yamada S., Kaneiwa T., Mizumoto S., Ikeda T., Sugihara K., Asano M., Yoshikawa T., Yamauchi A., Shervani N. J., Uruno A., Kato I., Unno M., Sugahara K., Takasawa S., Okamoto H., Sugawara A. (2009) Important role of heparan sulfate in postnatal islet growth and insulin secretion. Biochem. Biophys. Res. Commun. 383, 113–118 [DOI] [PubMed] [Google Scholar]
- 44. Fransson L. A., Belting M., Cheng F., Jönsson M., Mani K., Sandgren S. (2004) Novel aspects of glypican glycobiology. Cell Mol. Life Sci. 61, 1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Belting M., Borsig L., Fuster M. M., Brown J. R., Persson L., Fransson L. A., Esko J. D. (2002) Tumor attenuation by combined heparan sulfate and polyamine depletion. Proc. Natl. Acad. Sci. U.S.A. 99, 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tang P., Skinner K. A., Hicks D. G. (2009) Molecular classification of breast carcinomas by immunohistochemical analysis: are we ready? Diagn. Mol. Pathol. 18, 125–132 [DOI] [PubMed] [Google Scholar]