Background: RAC1 is a small G-protein of the Rho family that activates the transcription factor NF-κB to elicit an inflammatory response.
Results: We have found that RAC1 also induces the NRF2/ARE pathway, which in term blocks RAC1-dependent NF-κB activation.
Conclusion: RAC1 modulates inflammation by coordinating the activity of pro-inflammatory NF-κB and anti-oxidant NRF2 transcription factors.
Significance: Therapeutic intervention on RAC1 could help modulate pro- and anti-inflammatory processes.
Keywords: Inflammation, Lipopolysaccharide (LPS), Microglia, NF-κ B (NF-KB), Nuclear Factor 2 (Erythroid-derived 2-like Factor) (NFE2L2) (Nrf2), Oxidative Stress, Ras-related C3 Botulinum Toxin Substrate 1 (Rac1), Rho GTPases
Abstract
The small GTPase protein RAC1 participates in innate immunity by activating a complex program that includes cytoskeleton remodeling, chemotaxis, activation of NADPH oxidase, and modulation of gene expression. However, its role in regulating the transcriptional signatures that in term control the cellular inflammatory profiles are not well defined. Here we investigated the functional and mechanistic connection between RAC1 and the transcription factor NRF2 (nuclear factor erythroid 2-related factor 2), master regulator of the anti-oxidant response. Lipopolysaccharide and constitutively active RAC1Q61L mutant induced the anti-oxidant enzyme heme-oxygenase-1 (HO-1) through activation of NRF2. The use of KEAP1-insensitive NRF2 mutants indicated that RAC1 regulation of NRF2 is KEAP1-independent. Interestingly, NRF2 overexpression inhibited, whereas a dominant-negative mutant of NRF2 exacerbated RAC1-dependent activation of nuclear factor-κB (NF-κB), suggesting that NRF2 has an antagonistic effect on the NF-κB pathway. Moreover, we found that RAC1 acts through NF-κB to induce NRF2 because either expression of a dominant negative mutant of IκBα that leads to NF-κB degradation or the use of p65-NF-κB-deficient cells demonstrated lower NRF2 protein levels and basally impaired NRF2 signature compared with control cells. In contrast, NRF2-deficient cells showed increased p65-NF-κB protein levels, although the mRNA levels remain unchanged, indicating post-translational alterations. Our results demonstrate a new mechanism of modulation of RAC1 inflammatory pathway through a cross-talk between NF-κB and NRF2.
Introduction
The Rho GTPase RAC1 is a pleiotropic regulator of many cellular processes. It is a well established that RAC1 is a mediator in execution of the inflammatory program of the innate immune system (1–3). Among several roles assigned to RAC1 in inflammation, its participation in cytoskeleton remodeling and chemotaxis as well as in NADPH oxidase-dependent production of superoxide have been well established (4, 5). Moreover, RAC1 leads to activation of the transcription factor NF-κB and its target genes, including several proinflammatory cytokines such as TNF (4, 6). However, it is not known how cells activated by RAC1 ultimately change their gene expression back to a “resting” state or toward a resolution profile.
One of the pathways implicated in the control of inflammation is the transcription factor nuclear factor (erythroid-derived 2)-like 2 (NRF2),2 which is a master regulator of redox homeostasis (7). NRF2 regulates the expression of a battery of cytoprotective genes that share in common a cis-acting enhancer sequence termed antioxidant response element (ARE) (8, 9). These genes include those coding the antioxidant enzymes heme oxygenase-1 (HO-1) and NADP(H) quinone oxidoreductase (NQO1), enzymes of glutathione metabolism and protein degradation, through the proteasome and autophagy routes (10, 11). Previous data from our group indicated that NRF2 knock-out mice are hypersensitive to the inflammation induced by LPS (12) and exhibit exacerbated activation of the brain resident macrophage, microglia, in comparison to their wild type littermates. Interestingly, the treatment with sulforaphane, a compound that induces NRF2-dependent gene expression, attenuated the LPS-induced microglial infiltration to the hippocampus, reinforcing the idea of NRF2 anti-inflammatory properties. This evidence has been corroborated in other recent reports by analyzing other cells of the monocyte-macrophage linage (8, 13–15). On the other hand, the transcription factor nuclear factor κB (NF-κB) plays a crucial role in immune responses through the regulation of genes encoding proinflammatory cytokines, adhesion molecules, chemokines, growth factors, and inducible enzymes (16). It is remarkable that NRF2 and NF-κB influence each other and coordinate the final fate of innate immune cells (17), but it is not known how this interconnection takes place.
In this study we report that RAC1 induces the anti-inflammatory NRF2/HO-1 pathway. We also provide evidence that NF-κB activity is induced by active RAC1 and that NRF2 can modulate this effect. These results uncover a new mechanism of regulation of inflammatory events trough a RAC1/NRF2/HO-1 axis.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Human embryonic kidney (HEK) 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 80 μg/ml gentamycin. Transient transfections were performed with calcium phosphate using reagents from Sigma. BV-2 microglial cells were cultured in RPMI 1640 medium supplemented with 10% FCS and 80 μg/ml gentamicin. Cells were changed to serum-free RPMI without antibiotics 16 h before the addition of LPS (Sigma). HEK-TLR4-MD2/CD14 cells were kindly provided by Dr. Manuel Fresno (Centro de Biología Molecular Severo Ochoa (CBMSO), Madrid, Spain). These cells were grown in DMEM containing 4.5 g/liter glucose, 4 mm l-glutamine, 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/ml streptomycin, 10 μg/ml blasticidin and 50 μg/ml Hygrogold. The p65−/− mouse embryo fibroblasts (MEFs) and their corresponding wild type p65+/+ MEFs were kindly provided by Dr. Jianping Ye (Pennington Biomedical Research Center, Louisiana State University, Baton Rouge, LA) (18). The NRF2−/− and NRF2+/+ MEFs were isolated around embryonal day 17. Skin pieces were minced in 3-cm Petri dishes and underwent enzymatic digestion at 37 °C for 30 min in Falcon tubes with 1 ml of HyQTase (Hyclone). The digested tissue was passed through a 160-μm Nitex filter into 3-cm Petri dishes, and the enzymes were neutralized with DMEM plus 15% FCS (Sigma). The resulting suspension was centrifuged at 450 × g for 5 min, and the cells were plated at a density of 1 × 106 cells/cm2 in culture flasks. The fibroblasts were placed in an incubator at 37 °C supplemented with 5% CO2, and medium was changed daily for the first 3 days. After near confluence, cells were detached with HyQTase and replated at a density of 1 × 106 cells/cm2 in 6-cm plates every third day. MEFs were grown in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2 mm l-glutamine. Sulforaphane (SFN) (LKT Laboratories, St. Paul, MN) was used at 14 μm for 6 h.
GST Pulldown Assays
The RAC1 binding domain from GST fusion of the PAK1 p21 binding domain (19) was expressed in Escherichia coli as a fusion protein with glutathione S-transferase. BV-2 were treated for different times with LPS (500 ng/ml) and then lysed in a buffer composed of 50 mm Tris-HCl, pH 7.5, 1% Triton X-100, 100 mm NaCl, 10 mm MgCl2, 5% glycerol, 1 mm Na3VO4, and protease inhibitors (20 μg/ml aprotinin; pepstatin and leupeptin (1 μg/ml each). Lysates were centrifuged at 15,000 × g for 10 min, and the Triton X-100-soluble fraction was recovered. A bacterial lysate containing the GST-PAK1 fusion protein was added to BV-2 lysates together with glutathione-Sepharose beads. After 1 h the beads were collected by centrifugation and washed 3 times in 25 mm Tris-HCl, pH 7.5, 1% Triton X-100, 1 mm dithiothreitol, 100 mm NaCl, and 30 mm MgCl2. The beads were then resuspended in Laemmli sample buffer and boiled under reducing conditions for 5 min. The precipitated proteins were subjected to 12% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked in TBS supplemented with 0.2% Tween 20 and 5% milk and then incubated for 1 h with a primary antibody (anti-RAC1) and thereafter washed 3 times for 5 min in TBS supplemented with 0.2% Tween 20. The membranes were subsequently incubated for 1 h with peroxidase-conjugated anti-mouse IgGs (1:10,000) in TBS supplemented with 0.2% Tween 20. The blots were extensively washed, and antibody binding was visualized by enhanced chemiluminescence. In another set of experiments we confirmed RAC1 activation with anti-active GTP-RAC1 antibody (#26903, NewEast Biosciences).
Plasmids
Expression vectors pcDNA3.1/V5HisB-mNRF2 and pcDNA3.1/V5HisB-mNRF2ΔETGE are described in McMahon et al. (20). For luciferase assays, transient transfections of HEK293T cells were performed with the expression vectors pHO1–15-LUC, ARE-LUC, pEF-ΔNRF2(DN), and ARE-mut-LUC (Dr. J. Alam, Dept. of Molecular Genetics, Ochsner Clinic Foundation, New Orleans, LA). pcDNA3-IκBS32A/S36A was described in Traenckner et al. (21), and pCEFL-AU5-RAC1Q61L was kindly provided by Dr. Silvio Gutkind, NIDCR, National Institutes of Health, Bethesda, MD. pCCMV-p65, pCCMV-p50, and NF-κB-dependent (−453/+80)-HIV-LUC are described in Rojo et al. (22). pcDNA3-Gal4-NRF2 was generated in our laboratory and described in Espada et al. (23).
Luciferase Assays
Transient transfections of HEK293T cells were performed with the expression vectors for TK-Renilla (Promega, Madison, CA) and the corresponding firefly luciferase reporters. Cells were seeded on 24-well plates (100,000 cells per well), cultured for 16 h, and transfected using calcium phosphate. After 24 h of recovery from transfection, the cells were lysed and assayed with a dual-luciferase assay system (Promega) according to the manufacturer's instructions. Relative light units were measured in a GloMax 96 microplate luminometer with dual injectors (Promega).
Immunoblotting
Cells were washed once with cold PBS and lysed on ice with radioimmune precipitation assay lysis buffer (150 mm NaCl, 25 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride, 1 mm NaF, 1 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 μg/ml leupeptin, 1 mm EGTA). Precleared cell lysates were resolved in SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Billerica, MA). The primary antibodies used were anti-V5 (Invitrogen), anti-GAPDH (Merck), anti-NF-κB (p65, RelA) (Calbiochem, Merck), anti-β-actin (sc-1616) and anti-lamin B (sc-6217) (Santa Cruz Biotechnology), anti-HO-1 (AB1284, Millipore), anti-AU5 (Covance, Berkeley, CA), anti-IBA1 (ab5076, Abcam, Cambridge, UK), anti-NRF2 (1:1000, Abyntek, Spain), anti-MnSOD (Stressgen), and anti-XIAP (Sigma). These membranes were analyzed using the primary antibodies indicated above and the appropriate peroxidase-conjugated secondary antibodies. Proteins were detected by enhanced chemiluminescence (GE Healthcare).
Preparation of Nuclear and Cytosolic Extracts
HEK293T and HEK-TLR4-MD2/CD14 cells were seeded in p100 plates (2 × 106 cells/plate) and transfected with calcium phosphate or treated with 1 mg/ml LPS, respectively. Cytosolic and nuclear fractions were prepared as described previously (22). Briefly, cells were washed with cold PBS and harvested by centrifugation at 1100 rpm for 10 min. The cell pellet was resuspended in 3 pellet volumes of cold buffer A (20 mm HEPES, pH 7.0, 0.15 mm EDTA, 0.015 mm EGTA, 10 mm KCl, 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 20 mm NaF, 1 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 μg/ml leupeptin) and incubated in ice for 30 min. Then the homogenate was centrifuged at 500 × g for 5 min. The supernatants were taken as the cytosolic fraction. The nuclear pellet was resuspended in 5 volumes of cold buffer B (10 mm HEPES, pH 8.0, 0.1 mm EDTA, 0.1 mm NaCl, 25% glycerol, 1 mm phenylmethylsulfonyl fluoride, 20 mm NaF, 1 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 μg/ml leupeptin). After centrifugation in the same conditions indicated above, the nuclei were resuspended in loading buffer containing 0.5% SDS. The cytosolic and nuclear fractions were resolved in SDS-PAGE and immunoblotted with the indicated antibodies.
Analysis of mRNA Levels by Quantitative Real-time PCR
Total RNA from microglial primary cultures and from BV-2 cells was extracted using TRIzol reagent according to the manufacturer's instructions (Invitrogen). One microgram of RNA from each experimental condition was treated with DNase (Invitrogen) and reverse-transcribed using 4 μl of High capacity RNA-to-cDNA Master Mix (Applied Biosystems, Foster City, CA). For real-time PCR analysis, we performed the method previously described by Rojo et al. (13). Primer sequences are shown in Table 1. To ensure that equal amounts of cDNA were added to the PCR, the β-actin housekeeping gene was amplified. Data analysis is based on the ΔΔCt method with normalization of the raw data to housekeeping genes as described in the manufacturer's manual (Applied Biosystems, Invitrogen). All PCRs were performed in triplicate.
TABLE 1.
Genes and primers for quantitative PCR amplification
Genes and primers used for quantitative RT-PCR amplification. NQO1, NADP(H) quinone oxidoreductase; Gstm3, glutathione S-transferase M3; Gpx, glutathione peroxidase.
| Gene product | Forward primer | Reverse primer |
|---|---|---|
| Gpx | 5′-GGACTACACCGAGATGAACG-3′ | 5′-GATGTACTTGGGGTCGGTCA-3′ |
| Gstm3 | 5′-GAGAAGCAGAAGCCAGAGT-3′ | 5′-ATACGATACTGGTCAAGAAT-3′ |
| Ho-1 | 5′-CACAGATGGCGTCACTTCGTC-3′ | 5′-GTGAGGACCCACTGGAGGAG-3′ |
| IL-1β | 5′-CTGGTGTGTGACGTTCCCATTA-3′ | 5′-CCGACAGCACGAGGCTTT-3′ |
| Mn-SOD | 5′-AACAATCTCAACGCCACCGA-3′ | 5′-GATTAGAGCAGGCAGCAATC-3′ |
| NRF2 | 5′-CCCGAAGCACGCTGAAGGCA-3′ | 5′-CCAGGCGGTGGGTCTCCGTA-3′ |
| NQO1 | 5′-GGTAGCGGCTCCATGTACTC-3′ | 5′-CATCCTTCCAGGATCTGCAT-3′ |
| p65 | 5′-GGCCTCATCCACATGAACTT-3′ | 5′-CACTGTCACCTGGAAGCAGA-3′ |
| TNF | 5′-CATCTTCTCAAAATTCGAGTGACAA-3′ | 5′-TGGGAGTAGACAAGGTACAACCC-3′ |
| β-Actin | 5′-TCCTTCCTGGGCATGGAG-3′ | 5′-AGGAGGAGCAATGATCTTGATCTT-3′ |
Reactive Oxygen Species (ROS) Production
The fluorescent probe 2′-7′-dihydrodichlorofluorescein diacetate was used to assess ROS production (24). Cells were transfected, and after 24 h of recovery from transfection, the cells were incubated with 2′-7′-dihydrodichlorofluorescein diacetate 100 mm for 1 h. Fluorescence intensity was measured in a Synergy fluorescence microplate reader (Bio-Tek Instruments, Inc., Winooski, VT).
Electrophoretic Mobility Shift Assays
The double-stranded oligonucleotide used for the NF-κB probe was 5′-AGTTGAGGGACTTTCCCAGGC-3′ (25) and for Nrf2 probe was 5′-TTTTATGCTGTGTCATGGTT-3′ (26). Then 250 ng/μl concentrations of each oligonucleotide was annealed by incubation in STE buffer (100 mm NaCl, 10 mm Tris-HCl, pH 8.0, and 1 mm EDTA) at 80 °C for 2 min. The mixture was cooled down slowly to 4 °C, with a thermal profile of 1 °C/min. Annealed oligonucleotides were diluted to 25 ng/μl in STE buffer. 5′-End labeling was performed with T4 polynucleotide kinase (Promega) using 25 ng of double-stranded oligonucleotide and 25 μCi of [γ-32P]ATP (3000 Ci/mmol; Amersham Biosciences). The labeled probes were purified in a G-25 spin column (Amersham Biosciences). The binding reaction mixture contained 5 μg of nuclear protein extract diluted in a buffer containing, at final concentration, 40 mm HEPES, pH 8.0, 50 mm KCl, 0.05% Nonidet P-40, 1% dithiothreitol, and 10 μg/ml poly(dI-dC) in a total volume of 20 μl. Unlabeled competitor probe (25 ng) was added, and the reaction was incubated for 1 h at 25 °C. Labeled DNA (0.25 ng) was added to the mixture and was submitted to an additional 20 min incubation at 25 °C. Samples were resolved at 4 °C in a 6% non-denaturing polyacrylamide gel in 0.5× Tris borate/EDTA buffer. After electrophoresis the gel was dried and autoradiographed.
Statistical Analyses
Data are presented as the mean ± S.E. One and two-way analysis of variance with post hoc Newman-Keuls test and Bonferroni's test were used as appropriate. Student's t test was used to assess differences between groups. The Prism Version 5.03 software for Windows (GraphPad, San Diego, CA) was used as described in each figure legend.
RESULTS
LPS Activates RAC1 and Induces HO-1 in BV-2 Microglial Cells
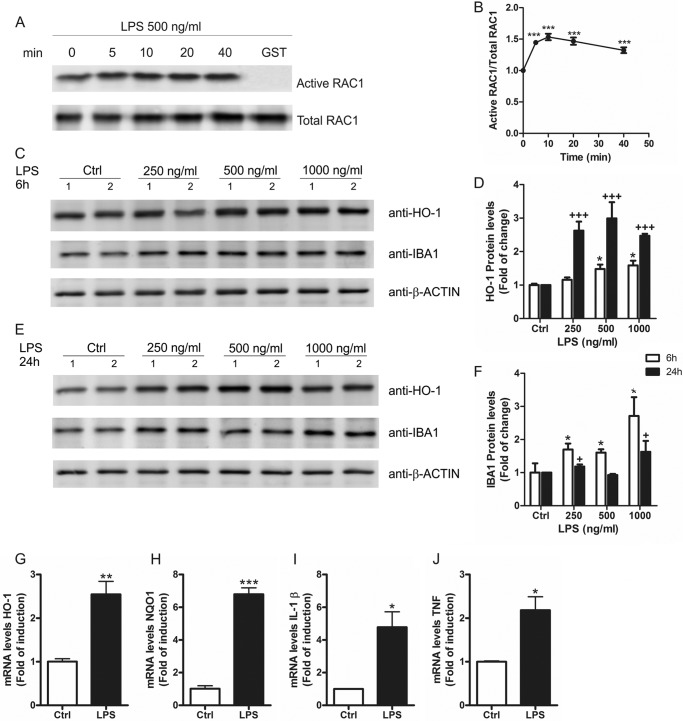
RAC1 activity was analyzed in cell lysates of BV-2 cells treated with LPS in a GST pulldown assay. This assay was based on the use of a chimeric protein made of glutathione S-transferase fused to the RAC1 interactive binding domain of p21-activated kinase 1 (Fig. 1A). We found that LPS (500 ng/ml) increased the levels of active RAC1 within 5 min and lasted for at least 40 min (Fig. 1B), indicating that RAC1 is implicated in LPS signaling. Moreover, LPS induced the expression of the antioxidant enzyme HO-1 in a dose-dependent fashion after 6 h (Fig. 1, C and D) and more markedly after 24 h (Fig. 1, E and D). To control that LPS was inducing an inflammatory response in BV-2 microglial cells, we determined the protein levels of IBA1, which is expressed in microglia and is up-regulated during the activation of these cells. LPS induced a dose-dependent and time-dependent increase of IBA1 protein levels (Fig. 1, C, E, and F), which correlated with the induction of HO-1. We also measured messenger RNA (mRNA) levels by qRT-PCR of two NRF2-regulated genes coding HO-1 and NAD(P)H dehydrogenase, quinone 1(NQO1) and two proinflammatory cytokines, IL-1β and TNF (Fig. 1, G–J, respectively) and found that treatment with LPS for 24 h increased the mRNA expression of NRF2-regulated genes and proinflammatory cytokines. These results indicate that RAC1 is activated very shortly after LPS stimulation leading to long term transcriptional modifications that include increased expression of HO-1.
FIGURE 1.
LPS activates RAC1 and increases HO-1 protein levels in microglia. A, BV-2 cells were treated with LPS (500 ng/ml) for 5, 10, 20, and 40 min, and cell lysates were used to perform a GST-PAK1-PBD pulldown assay as described under “Experimental Procedures.” As a negative control we used the same cell lysate from 40 min of treatment but with a GST empty vector. Immunoblots with anti-RAC1 antibody: upper panel, active RAC1; lower panel, total RAC1. B, quantification of immunoblots from two independent experiments for active RAC1 and total RAC1. One-way ANOVA followed by Newman-Keuls test was used to assess differences among groups. Asterisks denote significant differences. ***, p < 0.001, comparing the basal group to the indicated groups. C, BV-2 cells were treated with different doses of LPS for 6 h. Upper panel, immunoblots with anti-HO-1 antibody; middle panel, immunoblots with anti-IBA1 antibody as a control for microglial activation after LPS exposure; lower panel, anti-β-actin as protein load control. D, quantification of immunoblots for HO-1 at 6 and 24 h after LPS treatment. One-way ANOVA followed by Newman-Keuls test was used to assess differences among groups. Asterisks and plus symbols denote significant differences. *, p < 0.05; **, p < 0.01; ***, p < 0.001 comparing the indicated groups. E, BV-2 cells were treated with different doses of LPS for 24 h. Upper panel, immunoblots with anti-HO-1 antibody; middle panel, immunoblots with anti-IBA1 antibody as a control for microglial activation after LPS exposure; lower panel, anti-β-actin as protein load control. F, quantification of immunoblots for IBA1 at 6 and 24 h after LPS treatment. The statistical analysis was performed as in D. G–J, qRT-PCR determination of mRNA levels of Ho-1 (G), Nqo1 (H), Il-1β (I), and Tnf (J) normalized by β-actin levels. Student's t test was used to assess differences between groups. Asterisks denote statistically significant differences with: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RAC1 Induces the ARE/HO-1 Axis
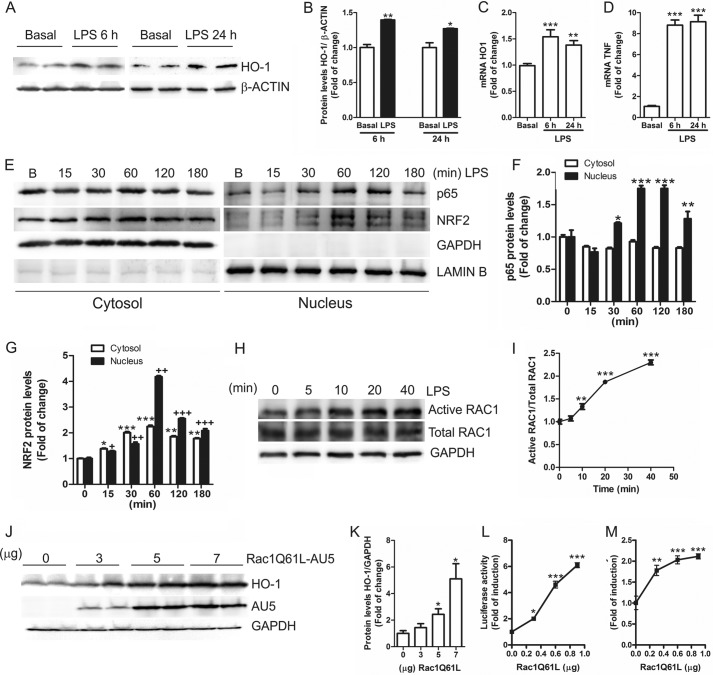
LPS stimulation of HEK-TLR4-MD2/CD14 cells (human HEK 293T cells are completely unresponsive to LPS but acquire high LPS sensitivity after transient transfection with CD14, TLR4, and MD-2) induced the expression of protein and mRNA levels of HO-1 (Fig. 2, A–C) after 6 and 24 h. Increased TNF mRNA levels confirmed the induction of inflammation due to LPS treatment (Fig. 2D). Besides, LPS induced nuclear translocation of p65 (Fig. 2, E and F) and NRF2 (Fig. 2, E and G) in a time-dependent manner and reached a maximum after 60 min. As observed in BV2 cells, LPS increased the levels of endogenous active RAC1 (Fig. 2, H and I), indicating that RAC1 activation is an essential effector of LPS inflammatory process. To analyze in depth the relevance of RAC1 activation on the NRF2 and NF-κB pathways, we transfected HEK293T cells with a constitutively active mutant of RAC1 (AU5-tagged RAC1Q61L) and analyzed the endogenous protein levels of HO-1 as a reporter of NRF2 transcriptional activity. Our results showed that RAC1Q61L increased HO-1 levels in a dose-response manner (Fig. 2, J and K), suggesting a direct connection between RAC1 activation and expression of the gene coding HO-1 (HMOX1). To further confirm these data, we analyzed the induction of two firefly luciferase reporters containing either 15 kb of the mouse Hmox1 promoter or three tandem sequences of the NRF2-dependent antioxidant response element (ARE-LUC) derived from the Hmox1 promoter. We found a dose-dependent activation of both reporters (Fig. 2, L and M). These results indicate that NRF2 mediates the RAC1-dependent up-regulation of HO-1 expression.
FIGURE 2.
RAC1 activates the ARE/HO-1 pathway. A, HEK-TLR4-MD2/CD14 cells were treated with 1 μg/ml LPS for 6 and 24 h. Upper panel, immunoblots with anti-HO-1 antibody; lower panel, anti-β-actin as protein load control. B, quantification of immunoblots for HO-1 at 6 and 24 h after LPS treatment. Student's t test was used to assess differences between groups. Asterisks denote statistically significant differences with: *, p < 0.05; **, p < 0.01. C and D, qRT-PCR determination of mRNA levels of Ho-1 (C) and Tnf (D) normalized by β-actin levels. One-way ANOVA followed by Newman-Keuls test was used to assess differences among groups. Asterisks denote significant differences (**, p < 0.01; ***, p < 0.001) comparing the basal group to the indicated groups. E, LPS induces p65 and NRF2 nuclear translocation. HEK-TLR4-MD2/CD14 cells were treated with 1 μg/ml for 15, 30, 60, 120, and 180 min, and subcellular fractionation was performed. Upper panels, immunoblots with anti-p65 or NRF2 antibodies; middle panel, immunoblots with GAPDH antibody as a control for cytosolic protein load; lower panel, anti-lamin B (B) as a control for nuclear protein load. F and G, quantification of immunoblots for p65 (F) or NRF2 (G) after LPS treatment. One-way ANOVA followed by Newman-Keuls test was used to assess differences among groups. Asterisks and plus denote significant differences: *, p < 0.05; **, p < 0.01; ***, p < 0.001 comparing the indicated groups. H, HEK-TLR4-MD2/CD14 cells were treated with LPS (1 μg/ml) for 5, 10, 20, and 40 min, and cell lysates were used to analyze active RAC1 by an active-RAC1-specific antibody (see “Experimental Procedures”). Immunoblots: upper panel, active RAC1; middle panel, total RAC1; lower panel, GAPDH as protein load control. I, quantification of immunoblots from two independent experiments for active RAC1 and total RAC1. One-way ANOVA followed by Newman-Keuls test was used to assess differences among groups. Asterisks denote significant differences (**p < 0.01; ***, p < 0.001) comparing the basal group to the indicated groups. J, HEK293T cells were transfected with different concentrations of the expression vector for AU5-tagged RAC1Q61L. Upper panel, HO-1; middle panel, AU5; lower panel, anti-GAPDH as protein loading control. K, quantification of immunoblots for HO-1. The statistical analysis was performed as in I. L and M, HEK293T cells were transfected with the HO1–15-LUC or ARE-LUC and TK-Renilla control vectors, with different amounts of RAC1Q61L vector. One-way ANOVA followed by Newman-Keuls test was used to assess differences among groups. Asterisks denote significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RAC1Q61L Induces the NRF2/HO-1 Pathway in a Dose-dependent Manner
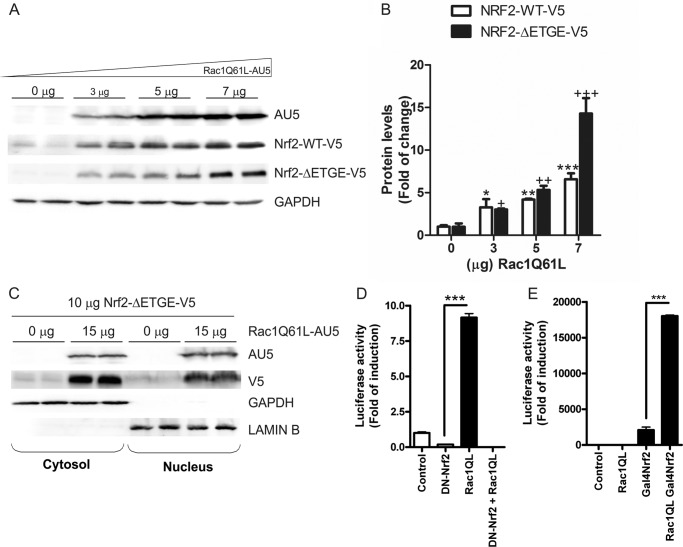
To determine if RAC1Q61L leads to an increase in NRF2 protein levels, we co-transfected HEK293T cells with different concentrations of RAC1Q61L and a constant concentration of a V5-tagged wild type form of NRF2 (NRF2-V5) or a stable mutant of NRF2, NRF2ΔETGE-V5, that lacks four residues (ETGE) essential for recognition by the E3 ligase complex Cul3/Keap1. The results indicated that RAC1Q61L produced a dose-dependent accumulation of both wild type NRF2-V5 and mutant NRF2ΔETGE-V5 (Fig. 3, A and B). Moreover, we observed by subcellular fractionation that the overexpression of RAC1Q61L induced a very strong increase in NRF2 protein levels in both cytosol and nucleus (Fig. 3C).
FIGURE 3.
RAC1 induces NRF2 signaling pathway. A, RAC1 induces NRF2 expression in a dose-dependent manner. Cells were transfected with different doses of RAC1Q61L and a constant amount of V5-tagged NRF2 or NRF2ΔETGE-mutant plasmids. Upper panels, immunoblots with anti-AU5 antibody; middle panel, immunoblots with anti-V5; lower panel, anti-GAPDH as protein load control. B, quantification of immunoblots for NRF2-WT-V5 and NRF2-ΔETGE-V5. One-way ANOVA followed by Newman-Keuls test was used to assess differences among groups as previously. C, RAC1 induces NRF2 nuclear translocation. Cells were co-transfected with or without RAC1Q61L and a constant dose of NRF2ΔETGE-V5 plasmid, and subcellular fractionation was performed. Upper panels, immunoblots with anti-AU5 or V5 antibodies; middle panel, immunoblots with GAPDH antibody as a control for cytosolic protein load; lower panel, anti-lamin B as a control for nuclear protein load. D, RAC1 requires NRF2 to activate the HO-1 promoter. HEK293T cells were co-transfected with RAC1Q61L expression vector, pHO1–15-LUC, Renilla control vectors, and either empty vector or dominant negative NRF2, (DN)-NRF2, expression vector. E, RAC1 uses the transactivating activity of NRF2 to induce AREs. Cells were co-transfected with Gal4-LUC (or pGL3-basic as a control) and either empty vector of expression vectors for Gal4NRF2 and RAC1 as indicated. Luciferase experiments were performed at least three times using three-four samples per group. The values in graphs correspond to the mean ± S.E. Student's t test was used to assess differences between groups or one-way ANOVA followed by a Newman-Keuls post-test (C). Asterisks denote statistically significant differences with: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Moreover, the implication of NRF2 in RAC1-induced HO-1 expression was analyzed with two different strategies. First, HEK293T cells were co-transfected with the expression vector for RAC1Q61L, a dominant-negative mutant of NRF2 (DN-NRF2) lacking the transcriptional activation domain and the luciferase reporter HO-1-LUC. We observed that DN-NRF2 abolished the RAC1Q61L-mediated induction of HO-1-LUC reporter (Fig. 3D). Second, we used a one-hybrid assay to study the transactivating activity of NRF2 in the presence of RAC1Q61L. The assay consisted of the use of an expression vector for a fusion protein containing the DNA binding domain of the yeast transcription factor Gal4 and NRF2 (Gal4-NRF2). HEK293T cells were co-transfected with RAC1Q61L, Gal4-NRF2, and the Gal4-LUC reporter plasmid as indicated in Fig. 3E. After 16 h, cells co-transfected with RAC1Q61L and Gal4-NRF2 exhibited a very strong increase in luciferase activity compared with that of cells co-transfected with Gal4-LUC and empty vector. Interestingly, RAC1Q61L highly increased the transactivating activity of NRF2 compared with NRF2 alone (Fig. 3E), indicating that RAC1Q61L activates NRF2 by targeting its transactivation domain. Taken together, all these data showed that active RAC1 led to accumulation of NRF2, resulting in increased ARE gene expression.
NRF2 Modulates RAC1-dependent Activation of the NF-κB Pathway
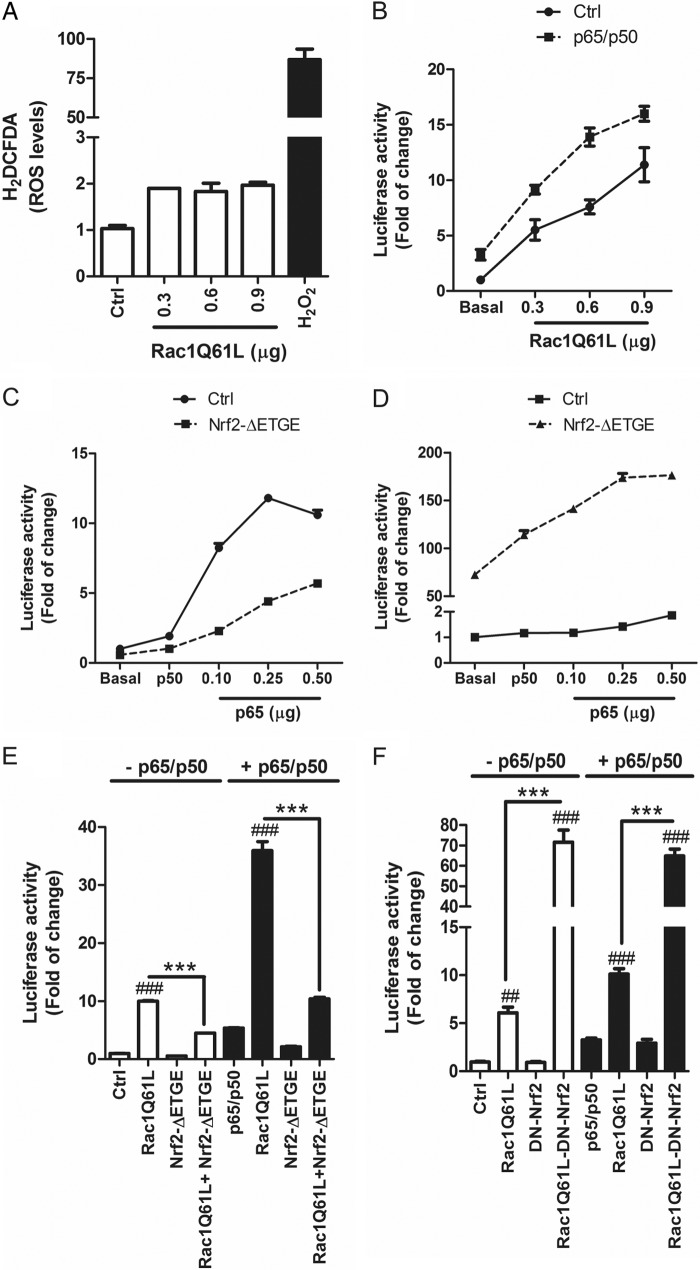
Considering that RAC1 may participate in production of NADPH oxidase-dependent ROS, we further analyzed the relative production of ROS by RAC1Q61L overexpression in HEK293T cells by using a dichlorofluorescein probe that emits green fluorescence in the oxidized form. Although we used different amounts of transfected RAC1Q61L, we only observed a slight increase in ROS levels in comparison with the effect of H2O2, used as positive control (Fig. 4A). These results were in concordance with results obtained with the antioxidant tripeptide GSH, which did not have any significant influence in the activation of the NRF2/HO-1 axis (data not shown), indicating that the effect of RAC1 on the NRF2 axis may not be mediated by ROS formation.
FIGURE 4.
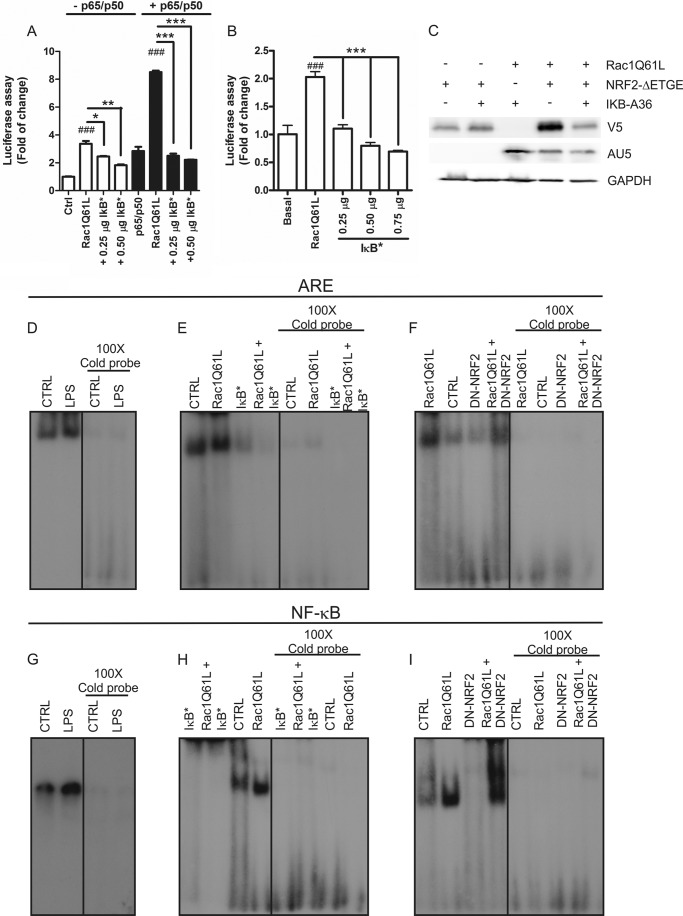
RAC1 activates NF-κB pathways and NRF2 acts as modulator. A, RAC1 overexpression induces generation of intracellular ROS. HEK293T cells were transfected with different doses of RAC1Q61L or empty (Ctrl) vectors, and after 16 h cells were incubated with 10 μm 2′-7′-dihydrodichlorofluorescein diacetate (H2DCFDA) for 1 h before fluorescence measurement. 2 mm H2O2 was used as positive control. B, RAC1 induces NF-κB pathway. Cells were co-transfected with NF-κB-dependent (−453/+80) HIV-LUC reporter and different concentrations of RAC1Q61L vector in absence or presence of p65/p50 plasmids. C, NRF2 inhibits NF-κB pathway. Cells were co-transfected with HIV-LUC reporter, p50, and increasing amounts of p65 in the absence or presence of NRF2ΔETGE-V5. D, p65/p50 increased the ARE activity mediated by NRF2. Cells were co-transfected with 3XARE-LUC, p50, and different concentrations of p65 in absence or presence of NRF2ΔETGE-V5. E, cells were co-transfected with HIV-LUC reporter, RAC1Q61L, NRF2ΔETGE-V5, or both in the absence (white bars) or presence (black bars) of p65/p50. F, cells were co-transfected with HIV-LUC reporter, RAC1Q61L, (DN)-NRF2, or both in the absence (white bars) or presence (black bars) of p65/p50. One-way ANOVA followed by a Newman-Keuls post-test was used to assess significant differences among groups. Asterisks denote statistically significant differences with: ***, p < 0.001; ##, p < 0.01; ###, p < 0.001 (respect to their control).
We further examined if RAC1Q61L could activate NF-κB by using a luciferase reporter carrying a promoter sequence (−453/+80) (HIV-LUC) (27), and we found that RAC1Q61L activated NF-κB in a dose-dependent fashion (Fig. 4B). Additionally, when we co-transfected different doses of RAC1Q61L with a constant concentration of p65 and p50 isoforms of NF-κB, we observed an exacerbated effect of RAC1 on the NF-κB transcriptional activity (Fig. 4B). These data indicate that RAC1 can induce the NF-κB responsive element and that this effect is more potent when both p65 and p50 are increased.
Our data indicate that RAC1 induces the anti-oxidant NRF2/HO-1 pathway and, on the other hand, that it increases NF-κB activity. To gain further insight into this paradox, we analyzed the cross-talk between NF-κB and NRF2 and the implication of RAC1 in this process. First, we determined if NRF2 could modulate NF-κB activity by using the HIV-LUC reporter. As expected, in the presence of a constant dose of p50, we observed a dose-dependent activation of the reporter by p65 (Fig. 4C). But, interestingly, co-transfection with the stable form of NRF2 (NRF2ΔETGE) reduced >50% of this effect (Fig. 4C), indicating that NRF2 inhibits NF-κB activity. Conversely, when we co-transfected p50 with increasing doses of p65, we could not observe an induction of the ARE reporter (Fig. 4D). Additionally, co-transfection of NRF2ΔETGE with p50/p65 exacerbated the ARE activity in comparison with that produced by NRF2ΔETGE alone (Fig. 4D). Then, we determined the cross-talk between NRF2 and NF-κB by using the HIV-LUC reporter. RAC1Q61L induced NF-κB activity (Fig. 4E) as mentioned before, but when we co-transfected RAC1Q61L with NRF2ΔETGE, the activity was reduced >50%. When p65 and p50 were present, the effects were more pronounced (Fig. 4E). Furthermore, when we co-transfected RAC1Q61L with DN-NRF2, NF-κB transcriptional activity was dramatically increased (Fig. 4F). A similar effect was observed when p65 and p50 were present. Taken together, our results suggest that RAC1Q61L induces inflammation by activating NF-κB activity, and p65/p50 cooperate with this effect. Moreover, the transcription factor NRF2 modulates this pathway and the absence of NRF2 exacerbates RAC1-dependent activation of NF-κB.
IκB Is Implicated in NRF2 and NF-κB-p65/p50 Activation by RAC1Q61L
To investigate the implication of the canonical pathway of NF-κB in the effects mediated by RAC1Q61L, we used a dominant-negative mutant of IκBα (IκBαS32A/S36A) in which Ser32 and Ser36 residues had been changed to Ala to block their phosphorylation and, consequently, their degradation. IκBαS32A/S36A produced a dose-dependent inhibition of NF-κB by RAC1Q61L (Fig. 5A). This effect was more pronounced in the presence of p65/p50. Then we analyzed the implication of IκBαS32A/S36A on ARE activity, and we observed that the increase of ARE activity induced by RAC1Q61L was inhibited by IκBS32A/S36A in a dose-dependent manner (Fig. 5B). These data were confirmed by immunoblot analysis. RAC1Q61L expression increased the protein levels of NRF2ΔETGE, and co-transfection of IκBαS32A/S36A abolished this effect (Fig. 5C), indicating that RAC1Q61L induced the activation of the NF-κB pathway and consequently the activation of the NRF2/HO-1 axis.
FIGURE 5.
IκBα mediates the activation of NRF2 and NF-κB induced by RAC1. A, a dominant-negative mutant of IκBα (IκBαS32A/S36A) inhibits RAC1-dependent induction of the NF-κB pathway. HEK293T cells were co-transfected with HIV-LUC reporter, RAC1Q61L, IκBαS32A/S36A, or both in the absence (white bars) or presence (black bars) of p65/p50. B, IκBα is implicated in the induction of NRF2 by RAC1. Cells were co-transfected with ARE-LUC promoter, RAC1Q61L, IκBαS32A/S36A, or both in the absence (white bars) or presence (black bars) of p65/p50. One-way ANOVA followed by a Newman-Keuls post-test was used to assess significant differences among groups. Asterisks denote statistically significant differences with: *, p < 0.05; **, p < 0.005; ***, p < 0.001; ###, p < 0.001 (with respect to their control). C, Western blot analysis of cells co-transfected with RAC1Q61L, IκBαS32A/S36A, and NRF2ΔETGE-V5 and their respective controls. Upper panel, immunoblot with anti-V5 antibody; middle panel, immunoblot with anti-AU5; lower panel, anti-GAPDH as protein load control. D, EMSA, using a double-stranded oligonucleotide containing the core ARE sequence and nuclear extracts from HEK-TLR4-MD2/CD14 cells treated with 1 μg/ml LPS for 1 h. E, EMSA, using a double-stranded oligonucleotide containing the core ARE sequence and nuclear extracts from HEK293T cells expressing RAC1Q61L ± IκBαS32A/S36A. F, EMSA, using a double-stranded oligonucleotide containing the core ARE sequence and nuclear extracts from HEK293T cells expressing RAC1Q61L ± DN-NRF2. In all cases the concentration of unlabeled oligonucleotide was 100× in the competition binding between labeled and unlabeled ARE probe. G, EMSA, using a double-stranded oligonucleotide containing the NF-κB binding element and nuclear extracts from HEK-TLR4-MD2/CD14 cells treated with 1 μg/ml LPS for 1 h. H, EMSA, using a double-stranded oligonucleotide containing the NF-κB promoter sequence and nuclear extracts from HEK293T cells expressing RAC1Q61L ± IκBS32A/S36A. I, EMSA, using a double-stranded oligonucleotide containing the NF-κB promoter sequence and nuclear extracts from HEK293T cells expressing RAC1Q61L ± DN-NRF2. In all cases the concentration of unlabeled oligonucleotide was 100× in the competition binding between labeled and unlabeled NF-κB probe. IkB* stands for IkB S32A/S36A mutations (a mutated plasmid). + indicates that the complete sample is Rac1Q61L + DN-NRF2, for example.
Taken together, those results indicated that IκBα is necessary for signaling to NF-κB and NRF2 through RAC1 activation. The mechanisms implicated in the cross-talk between NRF2 and NF-κB were analyzed in electrophoretic mobility shift assays (EMSAs). This method allowed us to establish whether LPS or RAC1Q61L could promote binding of NRF2 and p65-NF-κB to their respective ARE and NF-κB sequences and to analyze functional interference between them. Nuclear extracts from HEK-TLR4-MD2/CD14 cells treated for 1 h with 1 μg/ml LPS were incubated with a 32P-labeled, double-stranded oligonucleotide comprising the ARE or NF-κB sequence (see “Experimental Procedures”). As shown in Fig. 5D, the ARE complex was stronger in the presence of LPS, in agreement with the accumulation of NRF2 in the nucleus (Fig. 2E). A similar result was observed for the NF-κB binding (Fig. 5G). In addition, nuclear extracts from HEK293T cells transfected with RAC1Q61L showed increased complex formation with the ARE and NF-κB sequence (Fig. 5, panels E and F and panels H and I, respectively). Interestingly, co-transfection with IκBαS32A/S36A abolished this complex formation for both probes (Fig. 5, E and H, respectively), confirming the relevance of IκBα in signaling to NF-κB and NRF2 through RAC1 activation. Moreover, when RAC1Q61L was co-transfected with DN-NRF2, the intensity of the ARE complex was decreased in comparison to RAC1Q61L alone (Fig. 5F), indicating the loss of NRF2 in the complex. On the other hand, when RAC1Q61L was co-transfected with DN-NRF2 and NF-κB was analyzed, we observed several retarded complexes formed with the NF-κB probe, and the intensity of this retarded band was increased significantly (Fig. 5I), indicating that the absence of NRF2 increased the NF-κB complex formation after RAC1 activation.
NF-κB-p65 Regulates the NRF2/ARE Pathway
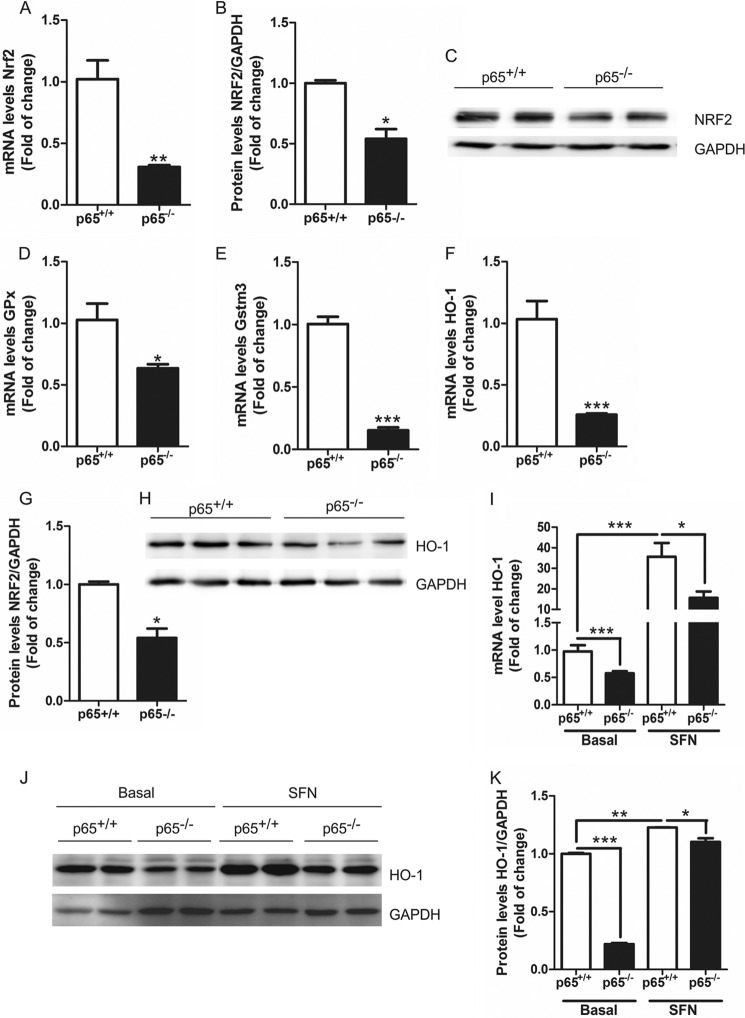
To establish the relevance of NF-κB-p65 in the cross-talk with NRF2, we used MEFs from p65-knock-out mice (28). Interestingly, the absence of p65 significantly reduced the mRNA and protein levels of NRF2 (Fig. 6, A–C). In addition, the mRNA levels of NRF2-regulated enzymes glutathione peroxidase (Gpx, Fig. 6D), glutathione S-transferase M3 (Gstm3, Fig. 6E), and HO-1 (Fig. 6F) were significantly decreased as well as the protein levels of HO-1 (Fig. 6, G and H).
FIGURE 6.
NRF2/ARE pathway is modulated by p65-NF-κB. A and B, NRF2 expression is decreased in p65−/− MEFs. A, qRT-PCR determination of mRNA levels of Nrf2 normalized by β-actin levels. B, immunoblot analysis of NRF2 protein levels, GAPDH was used as load control. C, quantification of immunoblots for NRF2 protein levels. Decreased levels of phase two enzymes in p65−/− MEFs. qRT-PCR determination of mRNA levels of Gstm3 (D), Gpx (E), and Ho-1 (F) normalized by β-actin levels. Dpx, glutathione peroxidase. G, immunoblot analysis of HO-1 protein levels, GAPDH was used as load control. H, quantification of immunoblots for HO-1 protein levels. Student's t test was used to assess differences between groups. Asterisks denote statistically significant differences with: *, p < 0.05; **, p < 0.01; ***, p < 0.001. p65+/+ and p65−/− MEF were incubated in a serum-free medium with 14 μm of SFN for 6 h. I, qRT-PCR determination of mRNA levels of Ho-1 normalized by β-actin levels. J, immunoblot analysis of HO-1 protein levels; GAPDH was used as load control. K, quantification of immunoblots for HO-1 protein levels. Two-way ANOVA followed by a Bonferroni post-test was used to assess significant differences among groups. Asterisks denote statistically significant differences with: *, p < 0.05; ***, p < 0.001.
Finally, we treated these MEFs with SFN, an isothiocyanate that induces the NRF2 transcriptional signature (29). SFN activated the transcriptional activity of NRF2 in MEFs from wild type mice as determined by increased mRNA and protein levels of HO-1 (Fig. 6, I–K). In the case of MEFs from p65-knock-out mice, although SFN was able to induce HO-1 expression, this effect was greatly attenuated in comparison with wild type MEFs. These results indicate that the regulation of NRF2 by NF-κB is not related elimination of the KEAP1 constrain and suggest that the changes in NRF2 levels are due to regulation of gene expression by NF-κB.
NRF2 Modulates the NF-κB Pathway at the Post-translational Level
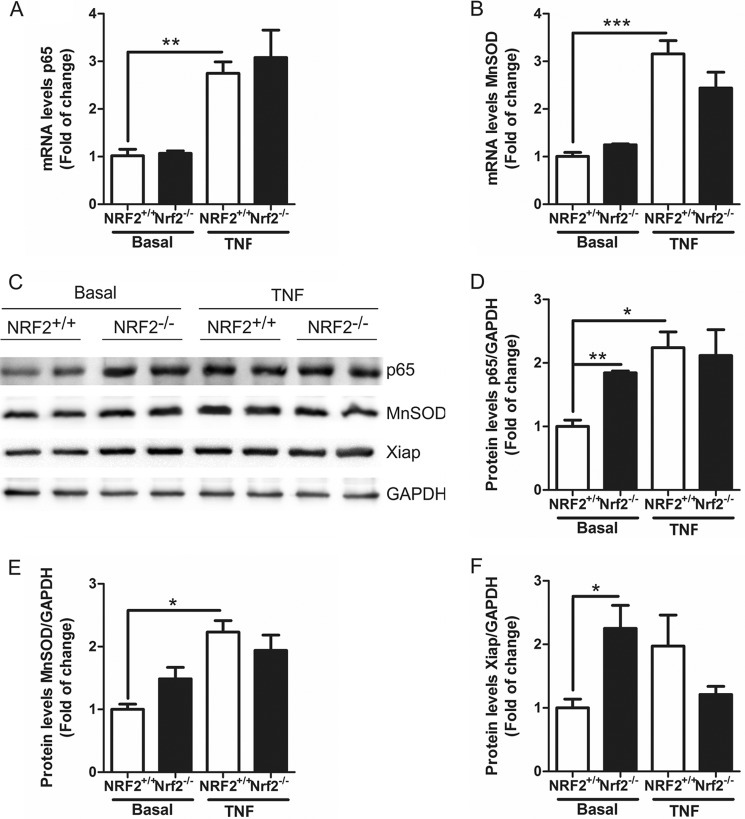
We used MEFs from NRF2-knock-out mice to determine the implication of NRF2 on NF-κB pathway. The absence of NRF2 did not modify the mRNA levels of either p65-NF-κB (Fig. 7A) or a p65-related gene Mn-SOD (Fig. 7B). After 6 h of treatment with 20 ng/ml TNF there was an increase in both p65-NF-κB and MnSOD mRNAs, although there were not significant changes between genotypes (Fig. 7, A and B), indicating that the NRF2 does not modify p65-NF-κB gene expression. However, the absence of NRF2 increased the amount of p65-NF-κB protein (Fig. 7, C and D) and p65-NF-κB-dependent proteins like MnSOD (Fig. 7, C and E) and XIAP (Fig. 7, C and F). TNF treatment for 6 h induced the protein expression of p65-NF-κB, MnSOD, and XIAP (Fig. 7, C–F) without changes between genotypes except for XIAP. We found that NRF2-deficient MEFs expressed decreased XIAP levels in comparison to NRF2+/+-MEFs treated with TNF. Taken together, these results indicate that there is cross-talk between NRF2 and NF-κB pathways at protein and transactivation levels.
FIGURE 7.
NRF2 deficiency increased p65-NF-κB protein levels. A and B, qRT-PCR determination of mRNA levels of p65-NF-κB (A) or MnSOD (B) normalized by β-actin levels in NRF2+/+ and NRF2−/− MEFs at basal conditions and after 6 h of TNF-α (20 ng/ml) treatment. C, immunoblots analysis of p65, MnSOD, and XIAP protein levels GAPDH was used as load control in NRF2+/+ and NRF2−/− MEFs at basal conditions and after 6 h TNF-α (20 ng/ml) treatment. D--F, quantification of immunoblots for p65-NF-κB (D), MnSOD (E), and XIAP (F) protein levels. Two-way ANOVA followed by a Bonferroni post-test was used to assess significant differences among groups. Asterisks denote statistically significant differences with: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
Low grade inflammation is present in several degenerative diseases, and RAC1 is a mediator of inflammatory signals. For instance, in amyotrophic lateral sclerosis (ALS), SOD1 mutations disrupt redox-sensitive RAC1 regulation of NADPH oxidase (30). In Fragile X syndrome, the most common hereditary form of mental retardation, lack of Fragile X mental retardation protein (FMRP) induced an overactivation of RAC1 in the mouse brain, indicating that regulation of RAC1 may provide a functional link among deficient neuronal morphology, aberrant synaptic plasticity, and cognition impairment in this disease (31). Interestingly, in Alzheimer disease it has been found that RAC1 inhibition targets amyloid precursor protein processing by γ-secretase and decreases Aβ production in vitro and in vivo (32). But RAC1 is not only involved in neurodegenerative disorders. Mutations in the RAC1 gene are associated with higher risk of inflammatory bowel disease (33). Intriguingly, loss of RAC1 expression results in protection from colitis (34). We hypothesize that RAC1 may be an essential mediator in several inflammation-related disorders through the activation of NRF2 and NF-κB pathways.
Taking RAC1 as a crucial mediator in inflammatory processes, our study focused on the modulation by this protein of two transcription factors, NRF2 and NF-κB, which are the master regulators of anti-oxidant (HO-1, NADP(H) quinone oxidoreductase (NQO1), and others) and proinflammatory (IL-1β, TNF, and others) responses, respectively. We describe that RAC1 induces the expression of the anti-oxidant enzyme HO-1 through the NRF2/ARE axis. Interestingly, RAC1 induces the nuclear translocation of NRF2 and increases its transactivating activity, suggesting that RAC1 could modulate the NRF2/ARE pathway under pathophysiological conditions. In concordance with these results, it has been described that auranofin, a glod (I)-containing anti-rheumatic drug, possesses anti-inflammatory properties due to HO-1 induction and NF-κB inhibition, and this correlates with the activation of RAC1 (35). Also, 15-deoxy-D12,14-prostaglandin J2, which is an immunoregulatory lipid metabolite derived from prostaglandin D2 dehydration, exerts anti-inflammatory activity through the RAC1-NADPH oxidase-ROS-p38 signaling and up-regulation of HO-1 in murine macrophages treated with LPS (36). This pathway differs from ours because, although RAC1Q61L induced a slight increase in ROS production (Fig. 4A), treatment with the antioxidant GSH did not reverse HO-1 induction, indicating that other pathways, independent of NADPH oxidase, could be involved as well.
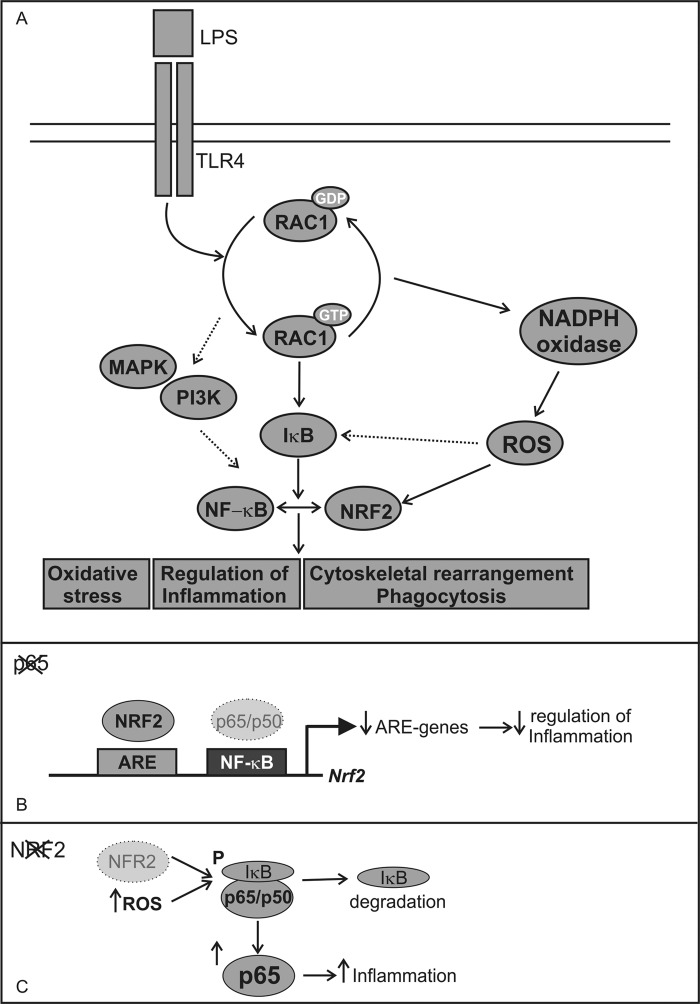
Constitutively active RAC1 induces the accumulation and nuclear translocation of NRF2 (Fig. 3, A and C, respectively) in a KEAP1-independent manner because RAC1 was able to induce NRF2ΔETGE-V5, which lacks four residues (ETGE) essential for recognition by this E3 ligase adapter. It seems that NRF2 induction through RAC1 is KEAP1-independent in our model even though KEAP1 overexpression inhibits RAC1 on the formation of E-cadherin-mediated cell-cell adhesion (37). It has been widely reported that RAC1 participates in several signaling kinase pathways, including MAPKs and PI3K (38, 39). Our group has reported a KEAP-independent regulation of NRF2 that relies on signaling kinases PI3K/AKT (40). Therefore, as shown in Fig. 8A, we suggest that through activation of the PI3K/AKT axis, RAC1 leads to NRF2 transcriptional activity.
FIGURE 8.
Implication of RAC1 in NF-κB and NRF2 transcriptional activities mediated by IκBα. A, RAC1 activation induces NF-κB and NRF2 transcription factors through IκBα implicated in different processes like oxidative stress, inflammation, or phagocytosis. But there are other pathways that could be implicated in these activation like PI3K and MAPK or the generation of ROS generated by NAPDH oxidase. B, in the absence of p65 there is a suboptimal expression of the Nrf2 and ARE-regulated genes, which are unable to regulate properly inflammation. The Nrf2 gene has a NF-κB binding site at the +270 bp from the TSS to which p65/p50 heterodimer is recruited (11). C, in the absence of NRF2 there is no proper modulation of redox homeostasis, which could lead to IκBα degradation, and therefore, we observed increased p65 protein levels, which could induce inflammation.
The emerging roles of RAC1 in inflammation led us to analyze the cross-talk between RAC1, NRF2, and NF-κB. Our results showed that activated RAC1 can induce the NF-κB pathway and cooperate with p65 and p50 in a dose-response manner (Fig. 4B) and that NRF2 acts as a modulator in this scenario. NRF2 overexpression inhibits RAC1-dependent activation of the NF-κB pathway and the absence of NRF2 due to overexpression of the DN-NRF2, exacerbated the activation of NF-κB-dependent inflammatory markers (Fig. 4, E and F, respectively). Therefore, our data indicate that RAC1 through IκBα (Fig. 5, A–C) induces NF-κB, which in turn modulates NRF2. Then NRF2 inhibits NF-κB as a regulatory feedback loop (Fig. 8A). Regarding this, we had previously observed that α-synuclein induced both the NF-κB proinflammatory and NRF2 anti-inflammatory responses but with different kinetics in BV2 microglial cells. We suggested that NF-κB activation is an early and direct event of α-synuclein signaling, whereas Nrf2 is a secondary event that participates in a later negative loop of NF-κB regulation (14). Moreover, we propose that these new results may explain our previous observation of exacerbated inflammatory response to LPS (12) or overexpression of α-synuclein (14) or TAU (41) in NRF2-deficient mice. In the absence of NRF2, NF-κB lacks a controller to switch off the inflammatory signal. Consistently, in animals treated with sulforaphane, an NRF2 inducer, the production of inflammatory markers in response to LPS was attenuated. These results are sustained by the presence of a NF-κB binding site in the Nrf2 gene (42) (Fig. 8B) and by the fact that IκK-β (IκBα kinase) contains an ETGE motif that enables it to bind to KEAP1 (43, 44). Also, NRF2 deficiency results in increased ROS levels, which induce IκBα phosphorylation and subsequent degradation, increasing p65-NF-κB levels and NF-κB proinflammatory processes (Fig. 8C). Therefore, RAC1 has an essential role modulating NRF2 and NF-κB pathways.
One of the hallmarks of RAC1 is its pleiotropic effects, regulating different cellular pathways. RAC1 participates in β-actin reorganization, and thus it has been implicated in inflammatory phagocytosis (45, 46). On the other hand, recent evidences showed that NRF2 is also implicated in phagocytosis. We and others have reported lower phagocytic index in microglial cells and macrophages from NRF2-deficient mice (14, 47), although the mechanism is unknown. Interestingly, sulforaphane is able to stimulate phagocytosis, indicating again a relevant role of NRF2 in this process (47). We hypothesize that the activation of RAC1 promotes β-actin-dependent cytoskeletal changes as well as oxidative processes, which lead to the induction of NRF2 and finally to phagocytosis. This mechanism could be highly relevant in neurodegenerative diseases like Alzheimer disease, where RAC1 participates in microglial phagocytosis of β-amyloid (48) and regulates the transcriptional activity of the amyloid precursor protein gene (49). Together, our results provide a new cross-talk mechanism of regulation of inflammatory processes involving RAC1 and the balance between NRF2 and NF-κB activities, which could be relevant in several pathophysiological events related to inflammation.
This work was supported by Spanish Ministerio de Ciencia e Innovacion Grant SAF2010-17822.
- NRF2
- nuclear factor erythroid 2-related factor 2
- ARE
- antioxidant response element
- HO-1
- heme oxygenase-1
- SFN
- sulforaphane
- SOD
- superoxide dismutase
- DN
- dominant-negative
- ROS
- reactive oxygen species
- MEF
- mouse embryo fibroblast
- XIAP
- X-chromosome-linked inhibitor of apoptosis
- qRT
- quantitative real-time
- Keap1
- Kelch-like ECH-associated protein 1.
REFERENCES
- 1. Arbibe L., Mira J. P., Teusch N., Kline L., Guha M., Mackman N., Godowski P. J., Ulevitch R. J., Knaus U. G. (2000) Toll-like receptor 2-mediated NF-κB activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533–540 [DOI] [PubMed] [Google Scholar]
- 2. Bokoch G. M. (2005) Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 15, 163–171 [DOI] [PubMed] [Google Scholar]
- 3. Ohsawa K., Imai Y., Kanazawa H., Sasaki Y., Kohsaka S. (2000) Involvement of Iba1 in membrane ruffling and phagocytosis of macrophages/microglia. J. Cell Sci. 113, 3073–3084 [DOI] [PubMed] [Google Scholar]
- 4. Sanlioglu S., Williams C. M., Samavati L., Butler N. S., Wang G., McCray P. B., Jr., Ritchie T. C., Hunninghake G. W., Zandi E., Engelhardt J. F. (2001) Lipopolysaccharide induces Rac1-dependent reactive oxygen species formation and coordinates tumor necrosis factor-α secretion through IKK regulation of NF-κB. J. Biol. Chem. 276, 30188–30198 [DOI] [PubMed] [Google Scholar]
- 5. Bosco E. E., Mulloy J. C., Zheng Y. (2009) Rac1 GTPase: a “Rac” of all trades. Cell. Mol. Life Sci. 66, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Utsugi M., Dobashi K., Ishizuka T., Kawata T., Hisada T., Shimizu Y., Ono A., Mori M. (2006) Rac1 negatively regulates lipopolysaccharide-induced IL-23 p19 expression in human macrophages and dendritic cells and NF-κB p65 transactivation plays a novel role. J. Immunol. 177, 4550–4557 [DOI] [PubMed] [Google Scholar]
- 7. Bryan H. K., Olayanju A., Goldring C. E., Park B. K. (2013) The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 85, 705–717 [DOI] [PubMed] [Google Scholar]
- 8. Innamorato N. G., Lastres-Becker I., Cuadrado A. (2009) Role of microglial redox balance in modulation of neuroinflammation. Curr. Opin. Neurol. 22, 308–314 [DOI] [PubMed] [Google Scholar]
- 9. Jazwa A., Cuadrado A. (2010) Targeting heme oxygenase-1 for neuroprotection and neuroinflammation in neurodegenerative diseases. Curr. Drug Targets 11, 1517–1531 [DOI] [PubMed] [Google Scholar]
- 10. Joshi G., Johnson J. A. (2012) The Nrf2-ARE pathway: a valuable therapeutic target for the treatment of neurodegenerative diseases. Recent Pat. CNS Drug Discov. 7, 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayes J. D., Dinkova-Kostova A. T. (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 39, 199–218 [DOI] [PubMed] [Google Scholar]
- 12. Innamorato N. G., Rojo A. I., García-Yagüe A. J., Yamamoto M., de Ceballos M. L., Cuadrado A. (2008) The transcription factor Nrf2 is a therapeutic target against brain inflammation. J. Immunol. 181, 680–689 [DOI] [PubMed] [Google Scholar]
- 13. Rojo A. I., Innamorato N. G., Martín-Moreno A. M., De Ceballos M. L., Yamamoto M., Cuadrado A. (2010) Nrf2 regulates microglial dynamics and neuroinflammation in experimental Parkinson's disease. Glia 58, 588–598 [DOI] [PubMed] [Google Scholar]
- 14. Lastres-Becker I., Ulusoy A., Innamorato N. G., Sahin G., Rábano A., Kirik D., Cuadrado A. (2012) α-Synuclein expression and Nrf2 deficiency cooperate to aggravate protein aggregation, neuronal death, and inflammation in early-stage Parkinson's disease. Hum. Mol. Genet. 21, 3173–3192 [DOI] [PubMed] [Google Scholar]
- 15. Brandenburg L. O., Kipp M., Lucius R., Pufe T., Wruck C. J. (2010) Sulforaphane suppresses LPS-induced inflammation in primary rat microglia. Inflamm. Res. 59, 443–450 [DOI] [PubMed] [Google Scholar]
- 16. Li Q., Verma I. M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734 [DOI] [PubMed] [Google Scholar]
- 17. Wakabayashi N., Slocum S. L., Skoko J. J., Shin S., Kensler T. W. (2010) When NRF2 talks, who's listening? Antioxid. Redox Signal. 13, 1649–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao Z., Yin J., Zhang J., He Q., McGuinness O. P., Ye J. (2009) Inactivation of NF-κB p50 leads to insulin sensitization in liver through post-translational inhibition of p70S6K. J. Biol. Chem. 284, 18368–18376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teramoto H., Coso O. A., Miyata H., Igishi T., Miki T., Gutkind J. S. (1996) Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c-Jun N-terminal kinase/stress-activated protein kinase pathway. A role for mixed lineage kinase 3/protein-tyrosine kinase 1, a novel member of the mixed lineage kinase family. J. Biol. Chem. 271, 27225–27228 [DOI] [PubMed] [Google Scholar]
- 20. McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J. D. (2004) Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 279, 31556–31567 [DOI] [PubMed] [Google Scholar]
- 21. Traenckner E. B., Pahl H. L., Henkel T., Schmidt K. N., Wilk S., Baeuerle P. A. (1995) Phosphorylation of human IκB-α on serines 32 and 36 controls I κ B-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 14, 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rojo A. I., Salinas M., Martín D., Perona R., Cuadrado A. (2004) Regulation of Cu/Zn-superoxide dismutase expression via the phosphatidylinositol 3 kinase/Akt pathway and nuclear factor-κB. J. Neurosci. 24, 7324–7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Espada S., Rojo A. I., Salinas M., Cuadrado A. (2009) The muscarinic M1 receptor activates Nrf2 through a signaling cascade that involves protein kinase C and inhibition of GSK-3β: connecting neurotransmission with neuroprotection. J. Neurochem. 110, 1107–1119 [DOI] [PubMed] [Google Scholar]
- 24. Hernández-Fonseca K., Cárdenas-Rodríguez N., Pedraza-Chaverri J., Massieu L. (2008) Calcium-dependent production of reactive oxygen species is involved in neuronal damage induced during glycolysis inhibition in cultured hippocampal neurons. J. Neurosci Res. 86, 1768–1780 [DOI] [PubMed] [Google Scholar]
- 25. Pan Q., Bao L. W., Merajver S. D. (2003) Tetrathiomolybdate inhibits angiogenesis and metastasis through suppression of the NFκB signaling cascade. Mol. Cancer Res. 1, 701–706 [PubMed] [Google Scholar]
- 26. Stewart D., Killeen E., Naquin R., Alam S., Alam J. (2003) Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 278, 2396–2402 [DOI] [PubMed] [Google Scholar]
- 27. Devary Y., Rosette C., DiDonato J. A., Karin M. (1993) NF-κB activation by ultraviolet light not dependent on a nuclear signal. Science 261, 1442–1445 [DOI] [PubMed] [Google Scholar]
- 28. Song L., Li J., Zhang D., Liu Z. G., Ye J., Zhan Q., Shen H. M., Whiteman M., Huang C. (2006) IKKβ programs to turn on the GADD45α-MKK4-JNK apoptotic cascade specifically via p50 NF-κB in arsenite response. J. Cell Biol. 175, 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jazwa A., Rojo A. I., Innamorato N. G., Hesse M., Fernández-Ruiz J., Cuadrado A. (2011) Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxid. Redox Signal. 14, 2347–2360 [DOI] [PubMed] [Google Scholar]
- 30. Harraz M. M., Marden J. J., Zhou W., Zhang Y., Williams A., Sharov V. S., Nelson K., Luo M., Paulson H., Schöneich C., Engelhardt J. F. (2008) SOD1 mutations disrupt redox-sensitive Rac regulation of NADPH oxidase in a familial ALS model. J. Clin. Invest. 118, 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bongmba O. Y., Martinez L. A., Elhardt M. E., Butler K., Tejada-Simon M. V. (2011) Modulation of dendritic spines and synaptic function by Rac1: a possible link to Fragile X syndrome pathology. Brain Res. 1399, 79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Désiré L., Bourdin J., Loiseau N., Peillon H., Picard V., De Oliveira C., Bachelot F., Leblond B., Taverne T., Beausoleil E., Lacombe S., Drouin D., Schweighoffer F. (2005) RAC1 inhibition targets amyloid precursor protein processing by γ-secretase and decreases Aβ production in vitro and in vivo. J. Biol. Chem. 280, 37516–37525 [DOI] [PubMed] [Google Scholar]
- 33. Atreya R., Atreya I., Neurath M. F. (2006) Novel signal transduction pathways: analysis of STAT-3 and Rac-1 signaling in inflammatory bowel disease. Ann. N.Y. Acad. Sci. 1072, 98–113 [DOI] [PubMed] [Google Scholar]
- 34. Muise A. M., Walters T., Xu W., Shen-Tu G., Guo C. H., Fattouh R., Lam G. Y., Wolters V. M., Bennitz J., van Limbergen J., Renbaum P., Kasirer Y., Ngan B. Y., Turner D., Denson L. A., Sherman P. M., Duerr R. H., Cho J., Lees C. W., Satsangi J., Wilson D. C., Paterson A. D., Griffiths A. M., Glogauer M., Silverberg M. S., Brumell J. H. (2011) Single nucleotide polymorphisms that increase expression of the guanosine triphosphatase RAC1 are associated with ulcerative colitis. Gastroenterology 141, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim N. H., Oh M. K., Park H. J., Kim I. S. (2010) Auranofin, a gold(I)-containing antirheumatic compound, activates Keap1/Nrf2 signaling via Rac1/iNOS signal and mitogen-activated protein kinase activation. J. Pharmacol. Sci. 113, 246–254 [DOI] [PubMed] [Google Scholar]
- 36. Hong H. Y., Jeon W. K., Kim B. C. (2008) Up-regulation of heme oxygenase-1 expression through the Rac1/NADPH oxidase/ROS/p38 signaling cascade mediates the anti-inflammatory effect of 15-deoxy-Δ12,14-prostaglandin J2 in murine macrophages. FEBS Lett. 582, 861–868 [DOI] [PubMed] [Google Scholar]
- 37. Kusano Y., Horie S., Shibata T., Satsu H., Shimizu M., Hitomi E., Nishida M., Kurose H., Itoh K., Kobayashi A., Yamamoto M., Uchida K. (2008) Keap1 regulates the constitutive expression of GST A1 during differentiation of Caco-2 cells. Biochemistry 47, 6169–6177 [DOI] [PubMed] [Google Scholar]
- 38. Tanaka T., Terada M., Ariyoshi K., Morimoto K. (2010) Monocyte chemoattractant protein-1/CC chemokine ligand 2 enhances apoptotic cell removal by macrophages through Rac1 activation. Biochem. Biophys. Res. Commun. 399, 677–682 [DOI] [PubMed] [Google Scholar]
- 39. Wang X., Zhang F., Chen F., Liu D., Zheng Y., Zhang Y., Dong C., Su B. (2011) MEKK3 regulates IFN-γ production in T cells through the Rac1/2-dependent MAPK cascades. J. Immunol. 186, 5791–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salazar M., Rojo A. I., Velasco D., de Sagarra R. M., Cuadrado A. (2006) Glycogen synthase kinase-3β inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J. Biol. Chem. 281, 14841–14851 [DOI] [PubMed] [Google Scholar]
- 41. Lastres-Becker I., Innamorato N. G., Jaworski T., Rábano A., Kügler S., Van Leuven F., Cuadrado A. (2014) Fractalkine activates NRF2/NFE2L2 and heme oxygenase 1 to restrain tauopathy-induced microgliosis. Brain 137, 78–91 [DOI] [PubMed] [Google Scholar]
- 42. Rushworth S. A., Zaitseva L., Murray M. Y., Shah N. M., Bowles K. M., MacEwan D. J. (2012) The high Nrf2 expression in human acute myeloid leukemia is driven by NF-κB and underlies its chemo-resistance. Blood 120, 5188–5198 [DOI] [PubMed] [Google Scholar]
- 43. Lee D. F., Kuo H. P., Liu M., Chou C. K., Xia W., Du Y., Shen J., Chen C. T., Huo L., Hsu M. C., Li C. W., Ding Q., Liao T. L., Lai C. C., Lin A. C., Chang Y. H., Tsai S. F., Li L. Y., Hung M. C. (2009) KEAP1 E3 ligase-mediated downregulation of NF-κB signaling by targeting IKKβ. Mol. Cell 36, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang Z. Y., Chu H. X., Xi M. Y., Yang T. T., Jia J. M., Huang J. J., Guo X. K., Zhang X. J., You Q. D., Sun H. P. (2013) Insight into the intermolecular recognition mechanism between Keap1 and IKKβ combining homology modelling, protein-protein docking, molecular dynamics simulations, and virtual alanine mutation. PLoS ONE 8, e75076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Masters T. A., Pontes B., Viasnoff V., Li Y., Gauthier N. C. (2013) Plasma membrane tension orchestrates membrane trafficking, cytoskeletal remodeling, and biochemical signaling during phagocytosis. Proc. Natl. Acad. Sci. U.S.A. 110, 11875–11880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beemiller P., Zhang Y., Mohan S., Levinsohn E., Gaeta I., Hoppe A. D., Swanson J. A. (2010) A Cdc42 activation cycle coordinated by PI 3-kinase during Fc receptor-mediated phagocytosis. Mol. Biol. Cell 21, 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suganuma H., Fahey J. W., Bryan K. E., Healy Z. R., Talalay P. (2011) Stimulation of phagocytosis by sulforaphane. Biochem. Biophys. Res. Commun. 405, 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kitamura Y., Shibagaki K., Takata K., Tsuchiya D., Taniguchi T., Gebicke-Haerter P. J., Miki H., Takenawa T., Shimohama S. (2003) Involvement of Wiskott-Aldrich syndrome protein family verprolin-homologous protein (WAVE) and Rac1 in the phagocytosis of amyloid-β(1–42) in rat microglia. J. Pharmacol. Sci. 92, 115–123 [DOI] [PubMed] [Google Scholar]
- 49. Wang P. L., Niidome T., Akaike A., Kihara T., Sugimoto H. (2009) Rac1 inhibition negatively regulates transcriptional activity of the amyloid precursor protein gene. J. Neurosci. Res. 87, 2105–2114 [DOI] [PubMed] [Google Scholar]