Background: Heme insertion into souble guanylate cyclase (sGC) enables it to bind nitric oxide (NO) for cell signaling.
Results: NO triggered a rapid, reversible, and hsp90-dependent heme insertion into sGC-β1 and an association with sGC-α1 subunit. sGC activator BAY 60-2770 did the same.
Conclusion: NO dynamically impacts the maturation and stability of active sGC heterodimer.
Significance: The data uncover new mechanisms that regulate cellular NO signaling cascades.
Keywords: Guanylate Cyclase (Guanylyl Cyclase), Heme, Hsp90, Nitric Oxide, Nitrosative Stress, Signal Transduction
Abstract
The chaperone heat shock protein 90 (hsp90) associates with signaling proteins in cells including soluble guanylate cyclase (sGC). hsp90 associates with the heme-free (apo) sGC-β1 subunit and helps to drive heme insertion during maturation of sGC to its NO-responsive active form. Here, we found that NO caused apo-sGC-β1 to rapidly and transiently dissociate from hsp90 and associate with sGC-α1 in cells. This NO response (i) required that hsp90 be active and that cellular heme be available and be capable of inserting into apo-sGC-β1; (ii) was associated with an increase in sGC-β1 heme content; (iii) could be mimicked by the heme-independent sGC activator BAY 60-2770; and (iv) was followed by desensitization of sGC toward NO, sGC-α1 disassociation, and reassociation with hsp90. Thus, NO promoted a rapid, transient, and hsp90-dependent heme insertion into the apo-sGC-β1 subpopulation in cells, which enabled it to combine with the sGC-α1 subunit to form the mature enzyme. The driving mechanism likely involves conformational changes near the heme site in sGC-β1 that can be mimicked by the pharmacologic sGC activator. Such dynamic interplay between hsp90, apo-sGC-β1, and sGC-α1 in response to NO is unprecedented and represent new steps by which cells can modulate the heme content and activity of sGC for signaling cascades.
Introduction
Soluble guanylyl cyclase (sGC)2 is an intracellular enzyme that plays a primary role in sensing NO and transducing its multiple signaling effects in mammals (1, 2). The active mammalian sGC is a heterodimer made up of slightly dissimilar α and β subunits that each contain a N-terminal regulatory, middle dimerization, and C-terminal catalytic domains (3–6). A metal cofactor (iron protoporphyrin IX, heme) binds only in the regulatory domain of the β subunit and is critical for sGC function because it enables NO to bind and activate the enzyme (3–6). Although the sGC heme normally functions in its reduced (ferrous) oxidation state, an increase in cell oxidant stress that can occur under many inflammatory conditions (7) can cause oxidation and loss of the sGC heme, thus creating a population of heme-free (apo) sGC that is insensitive to NO (8, 9). This led to development of novel drug candidates that can activate sGC independent of NO or its heme (10, 11) and has piqued interest in the cellular mechanisms that control the heme content, protein associations, and activity of sGC.
hsp90 is a ubiquitously expressed, ATP-dependent chaperone that helps to fold, stabilize, or modify the functions of select client proteins (12, 13). We recently found that hsp90 drives heme insertion into sGC during its maturation in cells (14). Hsp90 is bound primarily to the heme-free sGC-β1 subunit in cells, drives heme insertion into the apo-sGC-β1 in an ATP-dependent process, and then dissociates afterward. In the same study, we saw that the hsp90 association with apo-sGC-β1 fell off rapidly when we added an NO donor to cells to activate their sGC. This was surprising because it suggested that NO might play additional roles in addition to simply activating the heme-replete, mature sGC. Our current study explores the basis for the NO effect and revealed that NO triggers dynamic and transient rearrangements between hsp90, sGC-β1, and sGC-α1 in conjunction with a rapid heme insertion into apo-sGC-β1 in the cells.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were purchased from Sigma or Thermo Fisher Scientific. Succinyl acetone (SA), ascorbic acid, hemoglobin, radicicol, and phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine were purchased from Sigma. BAY 60-2770 and BAY 41-2272 were obtained from Bayer, NO donors, S-nitroso-N-acetyl-d,l-penicillamine (SNAP), 3-ethyl-3-(ethylaminoethyl)-1-hydroxy-2-oxo-1-triazene (NOC-12) and sodium nitroprusside were purchased from Sigma, and Lipofectamine was purchased from Invitrogen. cDNAs for sGC α1, β1, and sGC-β1H105F mutant were gifts from Dr. Andreas Papapetropoulos (University of Patras, Patras, Greece). Green African monkey kidney cells COS-7, rat fetal lung fibroblast (RFL-6) cells, and bovine aortic endothelial cells were purchased from ATCC. cGMP ELISA assay kit was obtained from Cell Signaling Technology.
Antibodies
Rabbit polyclonal sGC-β1 and sGC-α1 antibodies were obtained from Cayman Chemicals and Novus Biologicals, respectively, whereas mouse monoclonal and goat polyclonal sGC-β1 antibodies were purchased from Santa Cruz Biotechnology. Rabbit polyclonal hsp90 and monoclonal Myc tag antibodies were purchased from Cell Signaling Technology, epitope-tagged anti-V5 antibody was purchased from Invitrogen, and biotin antibody was obtained from Sigma. Goat polyclonal GAPDH antibody was purchased from Genscript.
Cell Culture and Transient Transfection of Cells
All cell lines were grown and harvested as described previously (14). Cultures (50–60% confluent) of COS-7 cells were transfected with expression constructs of sGC subunits (α1 and β1 or β1 alone) or sGC-β1H105F. After 42 h of transient transfection the COS-7 cells or cells (RFL-6 and bovine aortic endothelial cells) expressing endogenous levels of sGC were treated with 0.5 mm phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine for 10 min followed by NO donors (SNAP or sodium nitroprusside (50 μm) or NOC-12 (35 μm)) or either a heme-dependent (BAY 41-2272, 10 μm) or heme-independent (BAY 60-2270, 10 μm) sGC activator from varying time points between 0–45 min before being harvested. For NO scavenging experiments, hemoglobin (3 μm) and ascorbic acid (1 mm) were added to RFL-6 cultures after 3 min of SNAP activation, and cells were harvested at indicated time points. In all cases, the cells were treated with cycloheximide (10 μg/ml) for 30 min before sGC activation. To inhibit heme biosynthesis and deplete stores of intracellular heme, 400 μm SA was added to the cells 48 h prior to transfection or to activation of sGC (14). In such cases, the heme depletion was followed either by transient transfection or sGC activation. In other cases, to study the effect of hsp90 inhibition or study the effects of heme-repletion, RFL-6 cells were pretreated with radicicol (20 μm) or hemin (5 μm) for 2 h prior to sGC activation. The transfection experiments were carried out in duplicate plates, and the experiments were repeated at least three times.
Western Blots and Immunoprecipitations
Standard protocols were followed as mentioned previously (14). For immunoprecipitations, 500 μg of the total cell supernatant was precleared with 20 μl of protein G-Sepharose beads (Amersham Biosciences) for 1 h at 4 °C, beads were pelleted, and the supernatants incubated overnight at 4 °C with 3 μg of anti-V5 or anti-sGC-β1 antibody. Protein G-Sepharose beads (20 μl) were then added and incubated for 1 h at 4 °C. The beads were microcentrifuged (6000 rpm), washed three times with wash buffer (50 mm HEPES pH 7.6, 100 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40), and then boiled with SDS buffer and centrifuged. The supernatants were then loaded on SDS-PAGE gels and Western blotted using respective antibodies. For immunoprecipitations involving sGC-β1 antibody, mouse monoclonal sGC-β1 antibody was used for pulldowns, and bead-bound β1 protein was probed with goat polyclonal sGC-β1.
Gel Filtration Chromatography
Size exclusion chromatography was performed on RFL-6 cell supernatants at 4 °C as described previously for RAW cells (15). The column was equilibrated at 0.5 ml/min with 40 mm EPPS buffer, pH 7.6, containing 3 mm DTT, 5% glycerol, and 150 mm NaCl. Equal amounts (2.5 mg) in 100 μl were injected onto the column. The molecular weights of the protein fractions were estimated relative to gel filtration protein molecular weight standards. The relative distribution of sGC-β1 and hsp90 in the fractions was determined from band intensities from Western blots using ImageJ quantification software.
cGMP Enzyme-linked Immunosorbent Assay
The cGMP concentration in various cell supernatants made from intact cells that were given sGC activators was estimated using the cGMP ELISA assay kit (Cell Signaling Technology). sGC enzymatic activity (16) in reactions containing aliquots of the sizing column fractions was determined by adding 500 μm GTP and 20 μm of sGC activators BAY 41-2272 or BAY 60-2770 and incubating for 10 min at 37 °C. Reactions were quenched by addition of 10 mm Na2CO3 and Zn (CH3CO3)2. In certain other cases, sGC activity in RFL-6 supernatants was determined by passing the supernatants through the desalting PD-spin trap G-25 columns (GE Healthcare) prior to addition of the above constituents. The cGMP concentration was then determined by ELISA.
Measurement of NO Release Rate
The NO-mediated conversion of oxy-hemoglobin to methemoglobin was used to determine the rate of NO release from SNAP or NOC-12 at 25 °C, following procedures as described previously (17).
Biotin Switch Assay
The biotin switch assay was performed to determine S-nitrosated proteins as described previously (18), and the presence of the S-nitrosated target protein was assayed by immunoblotting with specific antibodies.
RESULTS
NO Transiently Alters the hsp90-sGC Association
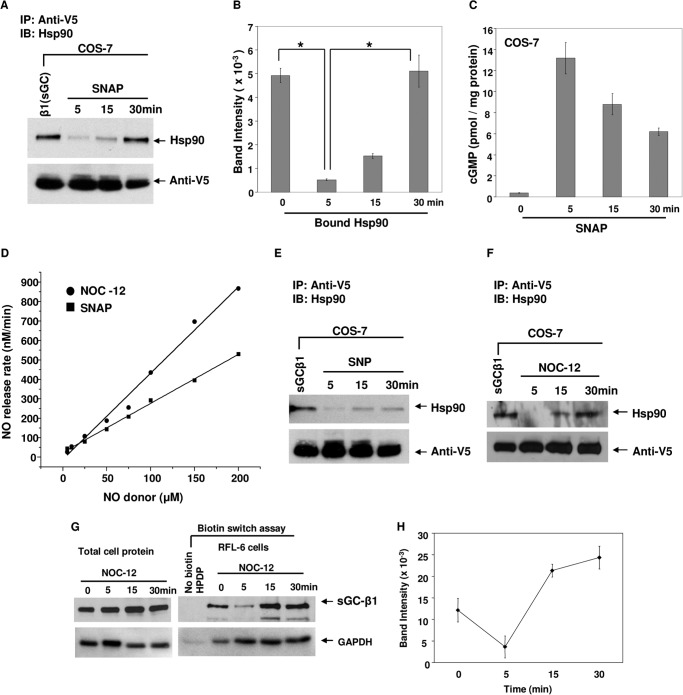
We studied how a brief NO exposure influenced the hsp90-sGC protein association by transiently expressing a V5-tagged sGC-β1 in COS-7 cells, adding the NO donor SNAP (in the presence of a phosphodiesterase inhibitor) and assessing subsequent hsp90 protein association and the cellular cGMP content at three time points within the first 30 min. As shown in Fig. 1, A and B, there was detectable hsp90-sGC-β1 association in the resting cells that fell with SNAP treatment by the 5th minute and then gradually recovered by the 30th minute. Measures of cell cGMP accumulation imply that the sGC was active only during the first 5 min of SNAP treatment (Fig. 1C), despite continuous NO release from SNAP occurring over the 30-min period, which we measured to be 144 nm NO per min from 50 μm SNAP (Fig. 1D). Similar or identical changes in the hsp90-sGC-β1 association dynamics were observed using the alternative NO donors sodium nitroprusside or NOC-12 (Fig. 1, E and F), which released NO at a similar rate to SNAP (Fig. 1D).
FIGURE 1.
Changes in hsp90-sGC interaction during and after NO-dependent activation. COS-7 cells that were transfected with a V5-tagged sGC-β1 construct for 42 h, or RFL-6 cells expressing endogenous sGC, were treated with SNAP (50 μm), sodium nitroprusside (SNP) (50 μm) or NOC-12 (35 μm) and cell supernatants generated between 0–30 min as indicated. Supernatant aliquots (equal protein) were analyzed for cGMP level by ELISA and were immunoprecipitated with an anti-V5 antibody followed by SDS-PAGE and Western analysis with anti-hsp90 and V5 antibodies. Likewise, supernatants harvested from RFL-6 cultures were subjected to biotin switch assays. A, E, and F, immunoprecipitation showing bound hsp90 and sGC-β1 (input 20%) retained on the beads. B, densitometric quantification of bound hsp90 with sGC-β1 in SNAP-treated COS-7 cells (n = 3). C, cell supernatant cGMP concentrations. Values depicted are mean ± S.D. of three independent experiments (*, p < 0.05, by one-way ANOVA). D, rates of NO release from SNAP and NOC-12 under culture conditions as determined by the oxyhemoglobin assay. G (left and right), supernatants and eluates from NOC-12-treated cells that underwent biotin switch assays were Western blotted with sGC-β1 and GAPDH antibodies. H, densitometric quantification of S-nitroso-sGC-β1 levels as revealed by corresponding panel in G (upper right). Values depicted are mean ± S.D. of three independent experiments. IB, immunoblot.
We studied the NO effect on the hsp90-sGC-β1 association in two other cell types that constitutively express the sGC enzyme (bovine aortic endothelial and RFL-6 cells). The NO donor again caused a quick loss and gradual return in hsp90 association with sGC-β1 in both cell types (Fig. 2, A and C), and the endogenous sGC was only active during the first 5 min of the SNAP treatment (Fig. 2, B and D). When we performed a follow-up study with RFL-6 cells in a smaller time window, we found that the hsp90-sGC-β1 association was entirely lost even within the first 2 min after SNAP addition, whereas steady sGC activity continued through the 6th min (Fig. 2, E and F).
FIGURE 2.
Hsp90 interaction with endogenous sGC is dynamic following activation by NO and further NO exposure. Confluent cultures of RFL-6 or bovine aortic endothelial cells (BAEC) had their endogenous sGC activated by SNAP (50 μm), and supernatants were prepared at indicated time points. Parallel experiments added hemoglobin (Hb, 3 μm) and ascorbic acid (AA, 1 mm) to RFL-6 cultures after 3 min of SNAP addition to scavenge the excess NO, and cells were harvested at indicated time points. Aliquots (equal protein) of cell supernatants were subjected to immunoprecipitation and assayed for cGMP concentration by ELISA. A, C, E, and G, immunoprecipitation showing bound hsp90 and sGC-β1 (input 20%) retained on the beads. B, D, and F, cGMP concentrations in the corresponding supernatants made at the indicated time points. Values are mean ± S.D. of three independent experiments. IB, immunoblot.
We saw that the reassociation of sGC-β1 with hsp90 during longer NO exposure was associated with an increase in sGC-β1 S-nitrosation levels (Fig. 1, G and H). The sGC-β1 reassociation was significantly diminished if hemoglobin and ascorbic acid were added to RFL-6 cultures after 3 min of SNAP activation to scavenge NO (Fig. 2G), suggesting that the continuous and/or excess exposure to NO may be driving the process. Together, our results show that the hsp90-sGC-β1 association is dynamic and quickly falls off during NO-dependent sGC activation, but under continued NO exposure returns with time and after the sGC becomes insensitive toward NO activation.
NO Transiently Alters sGC-β1 Distribution in Cells
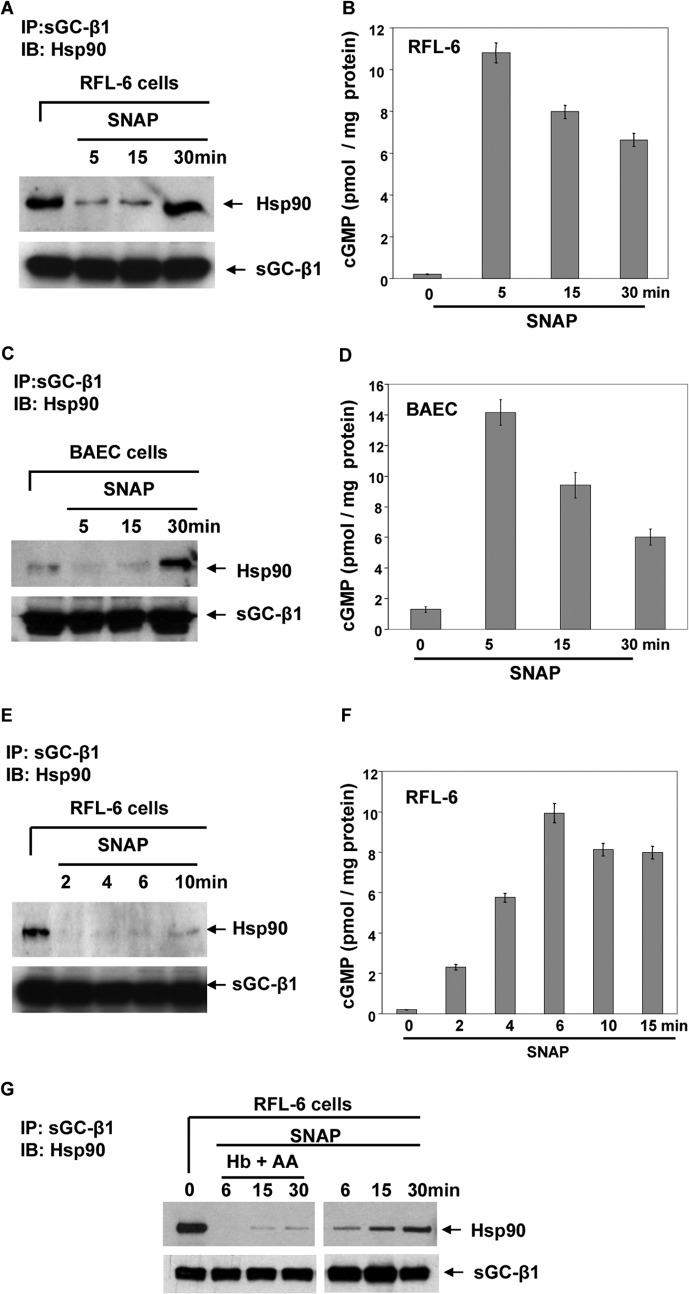
We went on to examine the apparent mass distributions of sGC-β1, sGC-α1, and hsp90 in the RFL-6 cell supernatants, to see whether their distributions became altered during the SNAP treatment. Cultures of RFL-6 cells were treated with vehicle or SNAP for 5 or 30 min, supernatants were prepared at 4 °C and then run on a high performance size exclusion column at 4 °C, with column fractions undergoing Western analysis using anti-sGC-β1, sGC-α1, or hsp90 antibodies, and also undergoing sGC activity measurements. Figs. 3, A and D, show that resting cells (-SNAP) contained two main molecular mass populations of sGC-β1, whose apparent molecular weight (Mr) ranges (see Mr scale in B) suggest that the sGC-β1 was primarily in complex with other cell proteins, possibly including hsp90 and sGC-α1, which co-eluted in fractions that contained sGC-β1 (Fig. 4, A and B). Interestingly, the predominant form of sGC-α1 expressed in the RFL-6 cells was a truncated splice variant of 52 kDa, probably similar to the Δ1–239 splice variant that is expressed in BE2 neuroblastoma cells and in lung tissue, which is missing a portion of the α subunit N-terminal regulatory domain (19, 20). The mass distribution of Δ1–239 sGC-α1 in the cell supernatant was broad and similar to that of the sGC-β1 subunit, consistent with the truncated sGC-α1 subunit and the sGC-β1 subunit still being able to combine and form an active heterodimer in cells (19, 20).
FIGURE 3.
NO alters Mr distributions of sGC-β1 and hsp90 in RFL-6 cells. RFL-6 cells were given vehicle or SNAP for 5 or 30 min, and then supernatants were prepared. Equal protein amounts of supernatants (2.5 mg in 100 μl) were fractionated on a Superdex 200 gel filtration column. The fractions were analyzed by Western blotting with sGC-α1/β1 or hsp90 antibodies and for sGC activity. A–C, sGC-β1 and hsp90 protein levels in the column fractions under various conditions as indicated. B also has an Mr scale for the eluted proteins based on injected protein Mr standards. D and E, densitometric quantification of sGC-β1 and hsp90 levels in column fractions as shown in A and B, respectively. F and G, cGMP concentrations achieved in reactions containing column fractions from A and B in response to sGC activators BAY 41-2272 or BAY 60-2770 (n = 3). H, supernatants generated from control or 5 or 30 min SNAP-treated RFL-6 cells were buffer-exchanged (PD-25, spin trap), and sGC activity in the eluants was assayed in response to BAY 41-2272 or BAY 60-2770 (n = 3). Values are mean ± S.D. of three independent experiments (*, p < 0.05, by one-way ANOVA; ns, not statistically significant).
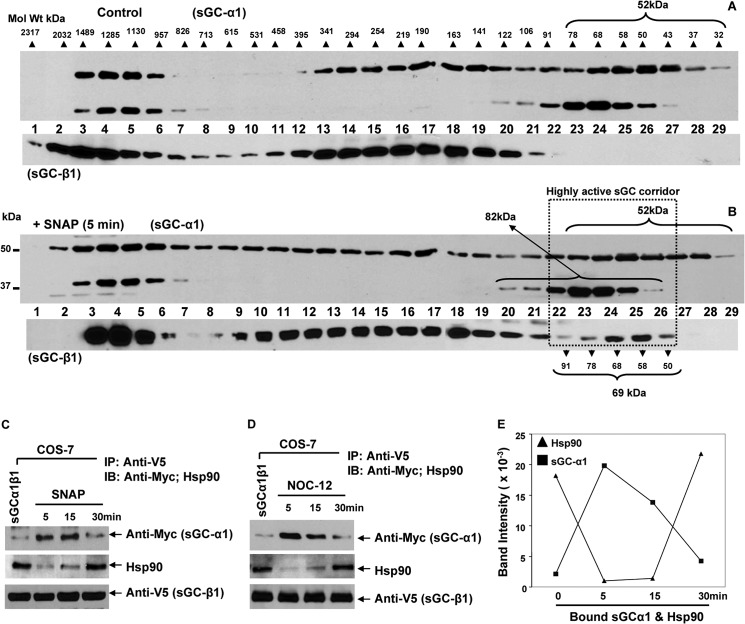
FIGURE 4.
NO does not alter the sGC-α1 Mr distribution profile but causes sGCβ1 to shift its interaction from hsp90 to sGCα1 and back. COS-7 cells that transiently expressed V5-tagged sGC-β1 and Myc-tagged sGCα1 constructs were treated with SNAP (50 μm) or NOC-12 (35 μm), and cell supernatants were generated at indicated times. A and B, Western analysis of gel filtration column fractions showing the sGC-α1 Mr distribution profile in RFL-6 cell supernatants samples. Experimental details are outlined in the legend to Fig. 3, and the same sGC-β1 blots are included here for orientation. C and D, Western analysis of V5-based immunoprecipitations showing bound hsp90, sGC-α1, and sGC-β1 (input 20%) retained on the beads. E, band intensities versus time of NO treatment for the hsp90 and sGC-α1 bound to sGC-β1 as determined from D. Data are representative of three replica experiments.
Activity measurements on the supernatant column fractions prepared from resting cells showed that the two Mr populations of sGC-β1 both displayed a blend of heme-dependent and heme-independent enzyme activity, as judged from the nearly equivalent cGMP production measured in reactions that contained either a heme-dependent or heme-independent sGC activator (BAY 41-2272 and BAY 60-2770, respectively) (Fig. 3, F and H, left three bars). These activity data confirm that both heme-free and heme-replete forms of sGC-β1 were present in the resting RFL-6 cells and show that these two forms were distributed fairly similarly among the higher and lower Mr populations in the cell supernatant.
In supernatant prepared from RFL-6 cells that had been given SNAP for 5 min, the Mr distribution patterns shifted for sGC-β1 and hsp90 proteins (+SNAP, 5 min, Fig. 3, B and E). In particular, there emerged a lower Mr subpopulation of sGC-β1 that clearly had no hsp90 associated with it (lanes 23–26). This subpopulation existed only transiently in the cells because it was no longer present in cells that underwent a longer SNAP exposure for 30 min (Fig. 3C). This transient low Mr sGC-β1 subpopulation could be activated by the heme-dependent activator BAY 41-2272 (Fig. 3G), but the heme-independent activator BAY 60-2770 no longer activated, thus identifying this low Mr sGC-β1 subpopulation as exclusively heme-replete. These fractions also contained sGC-α1 (Fig. 4, A and B), consistent with their having enzymatic activity.
Because most of the column fractions from the cells that were given 5-min SNAP treatment appeared to have greater BAY 41-2272 activities and lower BAY 60-2770 activities relative to the replica column fractions that were created from the resting cell supernatant (compare Fig. 3, F and G), this suggested that the 5-min SNAP treatment may have increased the heme content of the sGC-β1. We examined this in a semi-quantitative way by summing the total heme-dependent (BAY 41-2272) activities measured for the two sets of column fractions that were derived from equal amounts of supernatant protein from RFL-6 cells that either had or had not received the 5-min SNAP treatment (i.e. the data from the graphs depicted in Fig. 3, F and G). The total cGMP production in response to BAY 41-2772 for the control cells was 1049 ± 19 nm, compared with 1752 ± 29 nm for the cells treated with SNAP for 5 min. This 67% increase is consistent with transformation of apo-sGC-β1 into heme-containing sGC-β1 in response to the 5-min NO treatment. This was consistent with our finding that the unfractionated cell supernatant prepared from the 5 min SNAP-treated cells had an increased BAY 41-2272 activity and a decreased BAY 60-2770 activity relative to the resting cell supernatant (Fig. 3H). This change appeared to be time-sensitive because the supernatant prepared from cells given 30-min SNAP treatment had lower sGC activity toward both BAY 41-2272 and BAY 60-2770 compared with resting cells.
Together, our findings suggest a dynamic change in the hsp90-apo-sGCβ1 association occurs upon NO exposure, irrespective of cell type or whether the sGC is endogenously or transiently expressed. The process creates a larger population of heme-containing, active sGC that within a short time becomes inactive and ultimately reassociates with hsp90 and possibly other proteins into higher Mr complexes.
NO Causes an Increase in sGCα1-β1 Heterodimer Association
To investigate whether NO may alter interaction of sGC-α1 with the apo-sGCβ1 in our system, we transiently transfected sGC-α1 (Myc-tagged) and β1 (v5-tagged) constructs into COS-7 cells, and 42 h post transfection, the cells were either given SNAP or the NO donor NOC-12 for various times. Because the NO release rate of NOC-12 (210 nm/min) was faster than SNAP (144 nm/min, Fig. 1D) we adjusted the concentration of NOC-12 to 35 μm to give an equivalent NO flux in the cultures. Cell supernatants prepared at various time points were subjected to immunoprecipitation with anti-v5 antibody and immunoblotted with anti-Myc, hsp90, and anti-v5 antibodies. As shown in Fig. 4, C and D, both NO donors caused a transient decay in the sGC-β1 association with hsp90, followed by a recovery by the 30th min, as we saw earlier. In the same cells, the sGC-β1 association with sGCα1 was initially low, was greater at the 5- and 15-min time points, and then decayed by the 30th min, the exact inverse of what we observed for the hsp90 association. This implied that interactions of sGC-β1 with hsp90 and sGC-α1 may be mutually exclusive and can change quickly and reversibly during the NO exposure (Fig. 4E). Taken together, our data suggest that NO induced apo-sGC-β1 to incorporate heme and shift its association from hsp90 to sGC-α1 to form an active sGC heterodimer, but more prolonged NO exposure reversed this process.
Heme Must Be Available and Be Insertable into Apo-sGC-β1 for NO to Alter the hsp90 Association
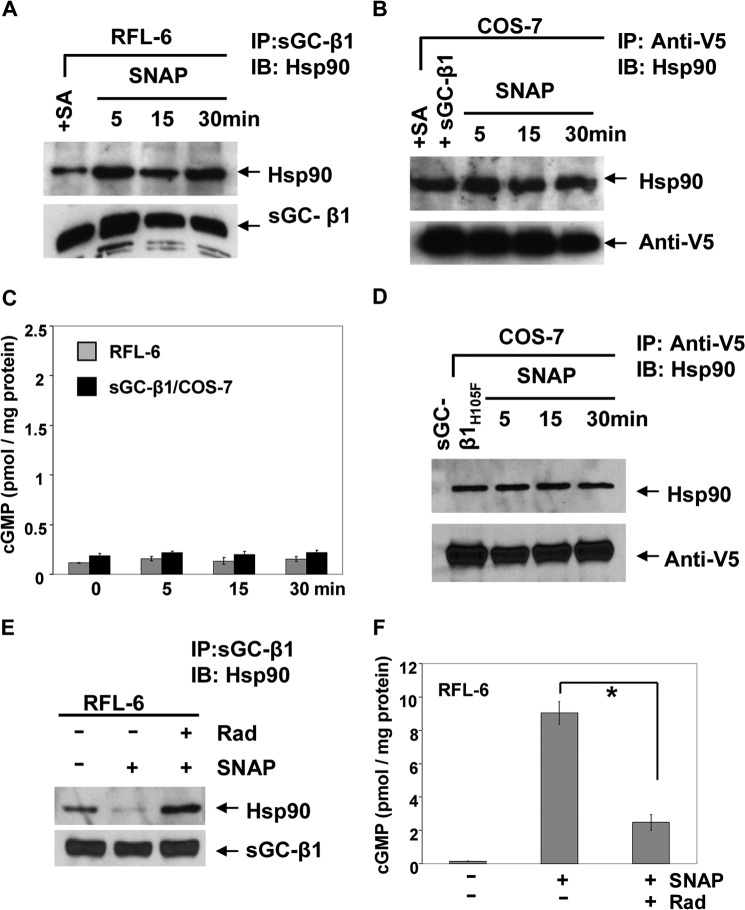
We sought to define how NO diminishes the hsp90-apo-sGC-β1 association. To test a role for cell heme, we used RFL-6 and COS-7 cells that had been made heme-deficient and thus could constitutively or transiently express only the apo-sGC-β1, respectively (14). Fig. 5, A and B, shows that a 5-min SNAP treatment did not alter the hsp90-apo-sGC-β1 association in this circumstance and did not activate sGC catalysis in either cell type (Fig. 5C). This implied the NO effect requires heme to be generally available in cells for insertion into apo-sGC-β1. We also found that the 5-min SNAP treatment did not diminish hsp90 association with a V5-tagged sGC-β1 mutant that is defective in heme binding (sGC-β1H105F), even when the cells that expressed the mutant were heme-replete (Fig. 5D). This suggested that actual heme insertion into sGC-β1 must take place to diminish the hsp90 binding in response to NO. Heme insertion into apo-sGC-β1 also requires that hsp90 have an intact ATPase activity (14). Here, we found that NO did not diminish the hsp90-apo-sGC-β1 association if the hsp90 ATPase activity was inhibited with radicicol, and under this circumstance, less cGMP accumulated in the cells in response to the 5-min SNAP treatment (Fig. 5, E and F). Thus, the NO effect on hsp90 dissociation required two features that enable heme insertion into apo-sGC-β1, namely, an active hsp90 ATPase activity, and a functional heme binding site within the sGC-β1. This further supports the concept that NO caused its effects by stimulating heme insertion into sGC-β1.
FIGURE 5.
Factors that are required for heme insertion into apo-sGC also drive the NO effect on hsp90 binding. A, B, and D, Western analyses of immunoprecipitations of sGC-β1 showing how 50 μm SNAP treatment for indicated times impacts level of hsp90 associated with apo-sGC-β1 expressed in heme-deficient (+SA) RFL-6 or COS-7 cells or with the sGC-β1H105F heme-free mutant expressed in heme-replete COS-7 cells (input 20%). C, cGMP levels in supernatants of cells in A and B. E and F, impact of an hsp90 inhibitor (radicicol; Rad) on hsp90-sGC-β1 association levels and cGMP production in cells given a 5-min SNAP treatment. Values are mean ± S.D. of three independent experiments (*, p < 0.05, by one-way ANOVA). IB, immunoblot.
Dissecting the Importance of Heme Site Occupancy
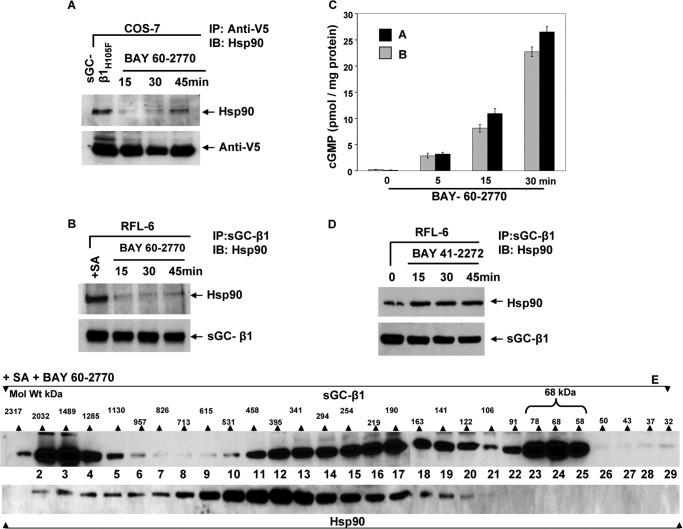
To further probe the mechanism, we utilized the apo-sGC activator BAY 60-2770. This molecule binds in the heme pocket of the sGC-β1 homolog Nostoc H-NOX domain (21) and is thought to activate sGC by triggering protein conformational changes in the sGC-β1 subunit that mimic those caused by NO binding to the sGC-β1 heme (21). When BAY 60-2770 was given to cells that transiently expressed the heme-free mutant sGC-β1H105F, or to heme-deficient RFL-6 cells that expressed endogenous apo-sGC-β1, it caused rapid dissociation of the hsp90·apo-sGC-β1 complex in both cases (Fig. 6, A and B, respectively), followed by a much weaker and more gradual hsp90 reassociation than what was observed in our previous experiments using NO donors (see Fig. 1). This behavior correlated with BAY 60-2770 stimulating a much longer-lived activation of sGC in the cells (Fig. 6C) compared with the NO donors. Gel filtration analysis of the heme-deficient RFL-6 cell supernatant (Fig. 6E) revealed that the BAY 60-2770 treatment shifted the apparent Mr distribution of sGC-β1 to cause significant buildup of the low Mr, hsp90-free sGC-β1 subpopulation in the cells (fractions 22–25). Similar results were obtained with RFL-6 cells that had not been made heme-deficient prior to the BAY 60-2770 treatment (data not shown), consistent with their containing some apo-sGC-β1 under normal culture conditions. Thus, pharmacore occupancy of the sGC-β1 heme binding site was also able to mimic NO in causing hsp90 dissociation and the Mr redistribution of apo-sGC-β1. Importantly, BAY 60-2770 did so even in heme-deficient cells. This distinguishes it from NO, which only diminished the hsp90 interaction and altered the apparent Mr distribution of sGC-β1 when cellular heme was available for insertion into apo-sGC-β1.
FIGURE 6.
Hsp90-sGC interaction dynamics in response to heme-dependent versus heme-independent sGC activators. COS-7 cells expressing a V5-tagged heme-free mutant sGC-β1H105F or heme-deficient (SA-pretreated) RFL-6 cells expressing endogenous apo-sGC were given the heme-independent activator BAY 60-2770 (10 μm), and supernatants were made at 0, 15, 30, and 45 min. Parallel experiments utilized the heme-dependent activator BAY 41-2272 (10 μm). A and B, gel and Western analysis of immunoprecipitations with anti-V5 and sGC-β1 antibodies showing hsp90 associated with sGC-β1H105F or apo-sGC-β1, respectively (input 20%). C, cGMP concentrations in supernatants as indicated. D, hsp90 associated with sGC-β1 (input 20%) after cell treatment with BAY 41-2272. E, gel filtration fractions of supernatant from SA-pretreated RFL-6 cultures given BAY 60-2770 for 30 min, analyzed by Western blotting using sGC-β1 or hsp90 antibodies. Scale indicates Mr range of column fractions determined with protein Mr standards. Values in the bar graph are mean ± S.D. of three independent experiments. IB, immunoblot.
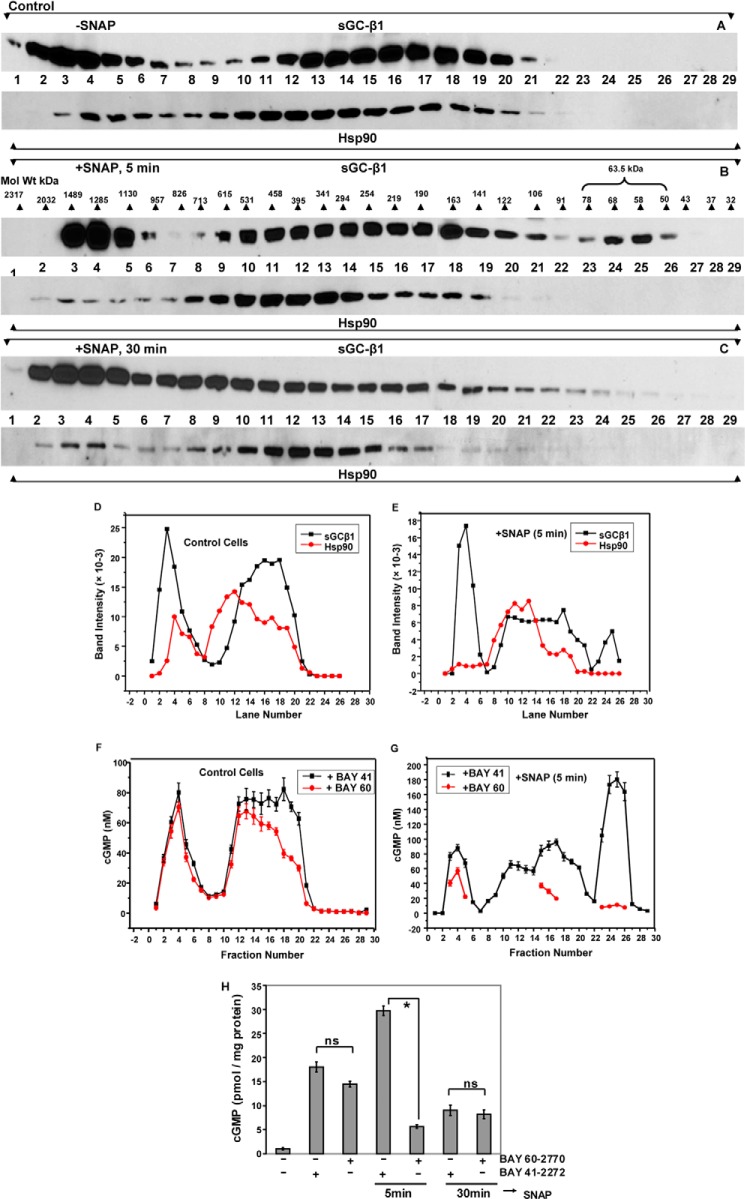
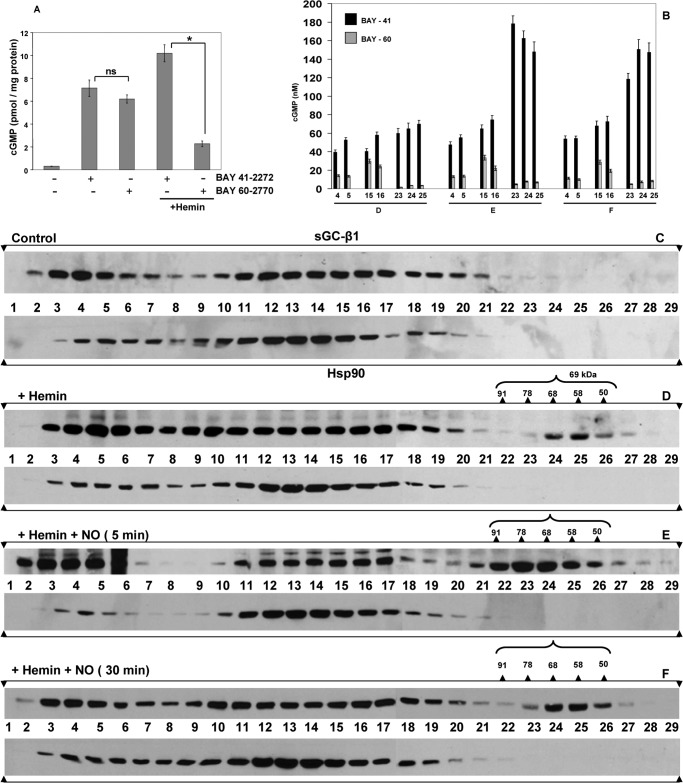
To further examine the role of heme site occupancy, we added hemin to RFL-6 cultures to promote heme insertion into the subpopulation of apo-sGC-β1. We previously reported that adding hemin to cells enabled heme insertion into apo-sGC-β1 and resulted in its dissociation from hsp90 (14). Here, we assessed how hemin treatment with or without a subsequent exposure to SNAP would impact the apparent Mr distribution of the sGC-β1. As expected, giving cells hemin for 2 h increased the proportion of heme-replete sGC-β1, as judged by the increased sGC activation in response to the heme-dependent drug (BAY 41-2272) and the decreased response to the heme-independent drug (BAY 60-2770) (Fig. 7A). However, the hemin treatment only caused a relatively modest shift in the apparent Mr distribution of sGC-β1 and created a relatively small subpopulation of the hsp90-free, low Mr, heme-replete sGC-β1, as judged by the elution profile and drug response profile of the column fractions to BAY 41-2272 versus BAY 60-2770 (Fig. 7, B, fraction D, and D). This low Mr sGC-β1 subpopulation formed to a much greater extent if the cells that received hemin also received a subsequent dose of SNAP for 5 min (Fig. 7, B, fraction E, and E), and in cells given hemin the low Mr sGC-β1 population partly persisted even after the 30-min SNAP exposure (Fig. 7, B, fractions F, and F). This persistence suggested that the added hemin stabilized the heme-replete sGC-β1 species in the continued presence of NO. Taken together, our data suggest that incorporating heme into apo-sGC-β1, despite its causing hsp90 dissociation (14), did not promote extensive Mr redistribution of sGC-β1 in the cells, unless NO was subsequently added.
FIGURE 7.
Hemin treatment of RFL-6 cells causes heme insertion into apo-sGCβ1 but only marginally alters its Mr distribution. RFL-6 cells were pretreated with vehicle or hemin (5 μm) for 2 h and then given sGC activators BAY 41-2272, BAY 60-2770 (30 min), or SNAP (5 min), and cell supernatants were prepared. A, cGMP generated in supernatant reactions. B, cGMP generated in reactions that contained an equal volume of each column fraction (from experiments in D–F) and given BAY 41-2272 or BAY 60-2770 to stimulate sGC activity. C–F, representative Western analyses of sGC-β1 and hsp90 in column fractions after gel filtration of supernatants. Activity values are mean ± S.D. of three independent experiments (*, p < 0.05, by one-way ANOVA; ns, not statistically significant).
Dissecting the Importance of sGC Activation
Both NO and BAY 60-2770 activate sGC by directly interacting with its β subunit. To better understand the role of sGC activation, we investigated the response to BAY 41-2272, which activates heme-containing sGC by binding to its α subunit (22). Adding BAY 41-2272 to the RFL-6 cells did not diminish association of apo-sGC-β1 with hsp90 (Fig. 6D), despite its activating sGC catalysis in the cells (Fig. 7A). This poor response toward BAY 41-2272 contrasted with the response toward BAY 60-2770, which caused hsp90 to dissociate from apo-sGC-β1 and altered its apparent Mr distribution. We conclude that sGC activation per se does not impact these parameters unless it occurs through a mechanism that directly involves the sGC-β1 subunit.
DISCUSSION
We found that NO triggers a dynamic change in association among hsp90, apo-sGC-β1, and sGC-α1 in cells. NO quickly diminished apo-sGC-β1 association with hsp90 and caused a concomitant increase in its association with sGC-α1 that was independent of cell type or whether the sGC was transiently or naturally expressed. These NO effects were transient and reversed with further NO exposure and after sGC became desensitized toward NO and its catalysis had stopped.
Possible Mechanism of Action
One reason that hsp90 associates with apo-sGC-β1 in cells is to drive heme insertion into the enzyme, and hsp90 dissociates from sGC-β1 after heme insertion takes place (14). Our observing an hsp90·sGC-β1 complex in all the cell types used in our study implies that cells contain a mixture of apo-sGC-β1 and holo-sGC-β1 under normal culture conditions. This concept is supported by our observing a strong sGC activation to the heme-independent sGC activator BAY 60-2770 in the various cell types, and by the BAY 60-2770 response becoming muted (and the corresponding response to BAY 41-2272 increasing) when the cells were incubated with hemin to increase the sGC-β1 heme content. Thus, we can surmise that NO caused hsp90 to quickly dissociate from the apo-sGC-β1 subpopulation that was present in cells. But how might this occur?
In principle, NO could weaken the hsp90 association with apo-sGC-β1 by several ways. We saw that the heme-independent sGC activator BAY 60-2770 could mimic the effect of NO in promoting hsp90 dissociation, whereas the heme-dependent sGC activator BAY 41-2272 could not. The ability of BAY 60-2770 to do so is perhaps the best indicator that the mechanism of NO action does not necessarily require any NO-based protein modifications such as protein S-nitrosation or tyrosine nitration, which can otherwise occur in hsp90 and sGC proteins when cells are exposed to NO (23–25). Rather, our results suggest a mechanism of action that involves fundamental changes in the apo-sGC-β1 subunit.
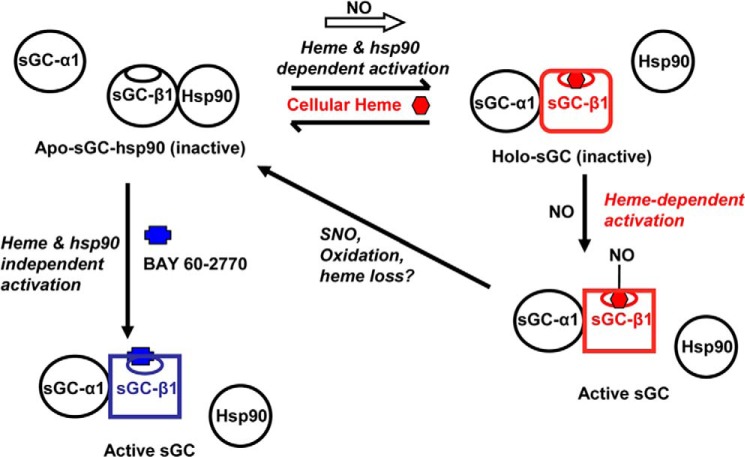
A model that is consistent with the data is illustrated in Fig. 8. It has NO-stimulating heme insertion into the subpopulation of apo-sGC-β1, leading to dissociation of hsp90 and to association of sGC-α1 to form the active heterodimeric enzyme. This model is consistent with the following: (i) NO boosting sGC activation by the heme-dependent activator BAY 41-2272 and causing a concurrent loss in sGC activation by the heme-independent activator BAY-60-2770, as should occur when heme incorporates into apo-sGC-β1; (ii) NO being unable to diminish the hsp90 association if the cells are heme-deficient and thus lacking available heme for insertion, or if the sGC-β1 contains a mutation that impairs its ability to incorporate heme, because heme incorporation causes hsp90 dissociation (14); (iii) NO having no effect if the cell hsp90 ATPase activity is inhibited because the ATPase activity of hsp90 is needed for heme insertion to occur (14), (iv) BAY 60-2770 triggering hsp90 dissociation on its own because the drug is thought to bind in sGC-β1 as a structural cognate of the heme·NO complex itself (21). Thus, hsp90 dissociation likely indicates that NO-driven heme insertion (or insertion of BAY 60-2770 in place of heme) has occurred in the apo-sGC-β1 subunit. Because the NO effect occurred within the first 2 min, it would seem that a pool of cellular heme exists that can rapidly insert into the apo-sGC-β1. Alternatively, NO might speed up the normal heme insertion process, which otherwise occurs over tens of minutes in the absence of added NO (14). That NO can quickly shift the equilibrium between apo- and holo-sGC-β1 in cells is remarkable and should be further investigated.
FIGURE 8.
Model that connects sGC-β1 protein interactions, heme content, and activity and shows the influence of heme-dependent (NO) or heme-independent (BAY 60-2770) sGC activators. An equilibrium exists in cells between a hsp90-bound apo-sGC-β1 (top left, black β subunit) and a heme-replete sGC-β1 that is instead associated with sGC-α1 (top right, red β subunit). NO can rapidly shift this equilibrium to the right when cell heme levels are sufficient and hsp90 is active. NO can then bind to the heme in the sGC heterodimer and activate catalysis (bottom right). The distinct structural changes in the sGC-β1 subunit caused by the heme insertion and NO binding steps are indicated by changes in the β subunit shape. Further NO exposure may cause S-nitrosation (SNO) of sGC-β1 and heme oxidation/loss and thereby desensitize sGC toward NO and promote its hsp90 reassociation. Binding of the heme-independent activator BAY 60-2770 (blue) to the apo-sGC-hsp90 species can occur independently of active hsp90 and cellular heme, and this triggers the same changes in sGC-β1 structure and protein interactions that are needed to activate its catalysis (lower left, blue β subunit).
Structural Insights
The crystal structures of the Nostoc H-NOX domain are regarded to be good models of the mammalian sGC-β1 regulatory domain structure (21, 26), whose structure remains to be solved. In comparing the structures of a H-NOX domain containing bound BAY 58-2667 or BAY 60-2770 with that of the drug free, heme-containing form, the authors (21, 26) identified some specific structural changes that occur with drug binding, that mainly involve the α-F helix and flanking residues that are located proximal to the bound heme. Although these structural changes help to show how sGC-β1 may achieve a catalytically-active state in response to NO binding to the sGC-β1 heme, these particular structural changes are unlikely to be the ones that weaken the apo-sGC-β1 interaction with hsp90 because we know that heme insertion into apo-sGC-β1 alone, without any NO or sGC activation, is sufficient to weaken its hsp90 association (14). However, the same subset of structural changes (21, 26) is possibly associated with the process that led to a Mr redistribution of sGC-β1 in the cells. Thus, BAY 60-2770 may promote extensive protein conformational changes within apo-sGC-β1 that lead to hsp90 dissociation, sGC-β1 redistribution in cells, and activation of sGC enzymatic activity. Although it is remarkable that BAY 60-2770 binding can mimic the effects of heme incorporation plus heme-NO binding in sGC-β1, this behavior is completely consistent with BAY 60-2770 binding in the heme site in Nostoc H-NOX to adopt a porphyrin-like structure (21).
In comparison, BAY 41-2272 showed no ability to diminish hsp90 interaction with sGC-β1, despite its activating sGC to a similar extent as did BAY 60-2770 in the RFL-6 cells. BAY 41-2272 is thought to bind to the sGC-α1 subunit near amino acid residues 236–290 (22, 27). This region would still be present in the N-terminally truncated sGC-α1 form that we found is predominantly expressed in RFL-6 cells and is consistent with BAY 41-2272 being able to activate sGC catalysis in the RFL-6 cells. Several studies show that BAY 41-2272 binding to the sGC-α1 subunit can measurably alter the structure and properties of the heme site in the partner sGC-β1 subunit of a heterodimer (28–30). However, our study suggests that sGC-α1 does not bind to apo-sGC-β1 and so cannot transduce any effects under this circumstance. Alternatively, if it did bind to apo-sGC-β1, perhaps the structural changes induced by BAY 41-2272 binding to sGC-α1 are unable to alter hsp90 binding or may not occur at all if heme is absent in the sGC-β1 subunit. Exploring these possibilities may help improve our understanding of the fundamental mechanisms of sGC activation.
Redistribution of sGC-β1 in the Cell
We found that NO and BAY 60-2770 drove a reorganization of the sGC-β1 protein in cells that was manifested by the appearance of a lower Mr sGC-β1 subpopulation. In contrast, we found that the Mr distribution profile of sGC-α1 was largely unaltered by NO (Fig. 4, A and B). Such changes in sGC enzyme distribution have not been noted previously or appreciated. We surmise that the intracellular redistribution of sGC-β1 involves distinct structural changes that occur when NO stimulates heme incorporation into apo-sGC-β1 and binds to its heme, or alternatively when BAY 60-2770 binds in the apo-sGC-β1, that are related to the specific structural changes that were identified in the Nostoc H-NOX protein when BAY 60-2770 binds (21), as discussed above. In any case, it is interesting that activating sGC through its β subunit (either by NO or BAY 60-2770) can cause a temporary Mr redistribution of sGC-β1 within the cell cytosol, which likely reflects changes in sGC-β1 protein-protein interactions and/or intracellular compartmentalization. The mechanisms involved and the relationship to cellular sGC activity and biological function deserve further study.
Hemeprotein Maturation Shows a Complex Response to NO
Our current study suggests that NO can have a more nuanced impact on heme protein maturation and function than was previously appreciated. NO appears to impact sGC maturation at three levels. (i) It can promote rapid heme insertion into apo-sGC-β1, as described in the present study. (ii) Prolonged NO exposure generally blocks cellular heme insertion into apo-hemeproteins (17) through a mechanism involving buildup of S-nitrosated proteins in cells (31). (iii) Prolonged NO exposure can also promote oxidation and loss of heme from sGC-β1, due to increased oxidative stress (8, 9, 16, 32). There are likely to be important and fascinating distinctions among these three types of NO responses with regard to timing, concentration response, mechanism, and when they come into play in biology.
sGC Reassociation with hsp90
The transient nature of the NO effects and their relationship to sGC activation were striking. Specifically, we found that sGC-β1 reassociated with hsp90 in cells during a longer (10–30 min) exposure to the NO donor. The reassociation depended on NO, was affiliated with desensitization of sGC toward NO activation and its consequent loss of activity, and correlated with sGC-β1 dissociation from sGC-α1 and its cytosol Mr redistribution back toward the pattern seen in resting cells. The fact that sGC-β1 reassociation with hsp90 was greatly reduced when BAY 60-2770 was used in place of NO donor suggests that the hsp90 reassociation may be a consequence of an NO-based event like sGC desensitization. Indeed, biotin switch assays showed that sGC-β1 became S-nitrosated in the cells over time with exposure to NOC-12 (Fig. 1, G and H). Previous studies showed there are two different time frames for sGC desensitization. Cell sGC activity can deflect very quickly (within seconds) after being activated by NO (33). NO and its derived oxidants can also desensitize sGC over a longer time frame of minutes, either through S-nitrosation of specific Cys residues in sGC-β1 (16, 34) or by causing oxidation or loss of heme from sGC-β1 (32). It will be important to examine whether these events help to drive the sGC-β1 reassociation with hsp90. Whether hsp90 reassociation is a natural process that helps to protect sGC-β1 from degradation (35) and whether there are accompanied changes in sGC-β1 heme site occupancy (36, 37) are interesting possibilities to be addressed.
Implications
That NO triggers dynamic and reversible change in sGC-β1 heme content, protein interactions, and apparent Mr distribution is unexpected and provides new insight on the cellular mechanisms that may activate or desensitize sGC in response to NO. The key role of hsp90 in determining sGC outcomes has important implications for cancer drug development programs that target hsp90 (38, 39). It is important to emphasize that the activation pathway we describe involves the heme-free sGC-β1 subpopulation that is present to various extents in healthy cells but is likely present to a greater extent under inflammatory or diseased states due to greater oxidant stress (8, 14, 40, 41). The ability of BAY 60-2770 to trigger all of the same changes in apo-sGC-β1 that we saw with NO, while bypassing the requirements for active hsp90 and cellular heme, suggests that BAY 60-2770 could activate cGMP-based signal cascades in disease states that may compromise hsp90 activity or cellular heme or that lead to accumulation of desensitized sGC-β1.
This work was supported, in whole or in part, by National Institutes of Health Grants GM51491, HL076491, and GM 097041 (to D. J. S.).
- sGC
- soluble guanylyl cyclase
- NO
- nitric oxide
- EPPS
- 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- SA
- succinyl acetone
- SNAP
- S-nitroso-N-acetyl-d,l-penicillamine
- NOC-12
- 3-ethyl-3-(ethylaminoethyl)-1-hydroxy-2-oxo-1-triazene
- RFL
- fetal lung fibroblast
- ANOVA
- analysis of variance.
REFERENCES
- 1. Lucas K. A., Pitari G. M., Kazerounian S., Ruiz-Stewart I., Park J., Schulz S., Chepenik K. P., Waldman S. A. (2000) Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 52, 375–414 [PubMed] [Google Scholar]
- 2. Garthwaite J., Boulton C. L. (1995) Nitric oxide signaling in the central nervous system. Annu. Rev. Physiol. 57, 683–706 [DOI] [PubMed] [Google Scholar]
- 3. Gilles-Gonzalez M. A., Gonzalez G. J. (2005) Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 99, 1–22 [DOI] [PubMed] [Google Scholar]
- 4. Fritz B. G., Roberts S. A., Ahmed A., Breci L., Li W., Weichsel A., Brailey J. L., Wysocki V. H., Tama F., Montfort W. R. (2013) Molecular model of a soluble guanylyl cyclase fragment determined by small-angle X-ray scattering and chemical cross-linking. Biochemistry 52, 1568–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Derbyshire E. R., Marletta M. A. (2012) Structure and regulation of soluble guanylate cyclase. Annu. Rev. Biochem. 81, 533–559 [DOI] [PubMed] [Google Scholar]
- 6. Underbakke E. S., Iavarone A. T., Marletta M. A. (2013) Higher-order interactions bridge the nitric oxide receptor and catalytic domains of soluble guanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 110, 6777–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tabima D. M., Frizzell S., Gladwin M. T. (2012) Reactive oxygen and nitrogen species in pulmonary hypertension. Free Radic. Biol. Med. 52, 1970–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stasch J. P., Schmidt P. M., Nedvetsky P. I., Nedvetskaya T. Y., H S A. K., Meurer S., Deile M., Taye A., Knorr A., Lapp H., Müller H., Turgay Y., Rothkegel C., Tersteegen A., Kemp-Harper B., Müller-Esterl W., Schmidt H. H. (2006) Targeting the heme-oxidized nitric oxide receptor for selective vasodilatation of diseased blood vessels. J. Clin. Invest. 116, 2552–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fritz B. G., Hu X., Brailey J. L., Berry R. E., Walker F. A., Montfort W. R. (2011) Oxidation and loss of heme in soluble guanylyl cyclase from Manduca sexta. Biochemistry 50, 5813–5815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stasch J. P., Pacher P., Evgenov O. V. (2011) Soluble guanylate cyclase as an emerging therapeutic target in cardiopulmonary disease. Circulation 123, 2263–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kots A. Y., Bian K., Murad F. (2011) Nitric oxide and cyclic GMP signaling pathway as a focus for drug development. Curr. Med. Chem. 18, 3299–3305 [DOI] [PubMed] [Google Scholar]
- 12. Li J., Buchner J. (2013) Structure, function and regulation of the hsp90 machinery. Biomed. J. 36, 106–117 [DOI] [PubMed] [Google Scholar]
- 13. Makhnevych T., Houry W. A. (2012) The role of Hsp90 in protein complex assembly. Biochim. Biophys. Acta 1823, 674–682 [DOI] [PubMed] [Google Scholar]
- 14. Ghosh A., Stuehr D. J. (2012) Soluble guanylyl cyclase requires heat shock protein 90 for heme insertion during maturation of the NO-active enzyme. Proc. Natl. Acad. Sci. U.S.A. 109, 12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghosh A., Chawla-Sarkar M., Stuehr D. J. (2011) Hsp90 interacts with inducible NO synthase client protein in its heme-free state and then drives heme insertion by an ATP-dependent process. FASEB J. 25, 2049–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fernhoff N. B., Derbyshire E. R., Underbakke E. S., Marletta M. A. (2012) Heme-assisted S-nitrosation desensitizes ferric soluble guanylate cyclase to nitric oxide. J. Biol. Chem. 287, 43053–43062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Waheed S. M., Ghosh A., Chakravarti R., Biswas A., Haque M. M., Panda K., Stuehr D. J. (2010) Nitric oxide blocks cellular heme insertion into a broad range of heme proteins. Free Radic. Biol. Med. 48, 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jaffrey S. R., Snyder S. H. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001, pl1. [DOI] [PubMed] [Google Scholar]
- 19. Sharina I. G., Cote G. J., Martin E., Doursout M. F., Murad F. (2011) RNA splicing in regulation of nitric oxide receptor soluble guanylyl cyclase. Nitric Oxide 25, 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharina I. G., Jelen F., Bogatenkova E. P., Thomas A., Martin E., Murad F. (2008) Alpha1 soluble guanylyl cyclase (sGC) splice forms as potential regulators of human sGC activity. J. Biol. Chem. 283, 15104–15113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar V., Martin F., Hahn M. G., Schaefer M., Stamler J. S., Stasch J. P., van den Akker F. (2013) Insights into BAY 60-2770 activation and S-nitrosylation-dependent desensitization of soluble guanylyl cyclase via crystal structures of homologous nostoc H-NOX domain complexes. Biochemistry 52, 3601–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pal B., Kitagawa T. (2010) Binding of YC-1/BAY 41-2272 to soluble guanylate cyclase: A new perspective to the mechanism of activation. Biochem. Biophys. Res. Commun. 397, 375–379 [DOI] [PubMed] [Google Scholar]
- 23. Franco M. C., Ye Y., Refakis C. A., Feldman J. L., Stokes A. L., Basso M., Melero Fernández de Mera R. M., Sparrow N. A., Calingasan N. Y., Kiaei M., Rhoads T. W., Ma T. C., Grumet M., Barnes S., Beal M. F., Beckman J. S., Mehl R., Estévez A. G. (2013) Nitration of Hsp90 induces cell death. Proc. Natl. Acad. Sci. U.S.A. 110, E1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martínez-Ruiz A., Villanueva L., González de Orduña C., López-Ferrer D., Higueras M. A., Tarín C., Rodríguez-Crespo I., Vázquez J., Lamas S. (2005) S-Nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc. Natl. Acad. Sci. U.S.A. 102, 8525–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Retzlaff M., Stahl M., Eberl H. C., Lagleder S., Beck J., Kessler H., Buchner J. (2009) Hsp90 is regulated by a switch point in the C-terminal domain. EMBO Rep. 10, 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin F., Baskaran P., Ma X., Dunten P. W., Schaefer M., Stasch J. P., Beuve A., van den Akker F. (2010) Structure of cinaciguat (BAY 58-2667) bound to Nostoc H-NOX domain reveals insights into heme-mimetic activation of the soluble guanylyl cyclase. J. Biol. Chem. 285, 22651–22657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Becker E. M., Alonso-Alija C., Apeler H., Gerzer R., Minuth T., Pleiss U., Schmidt P., Schramm M., Schröder H., Schroeder W., Steinke W., Straub A., Stasch J. P. (2001) NO-independent regulatory site of direct sGC stimulators like YC-1 and BAY 41-2272. BMC Pharmacol. 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu X., Feng C., Hazzard J. T., Tollin G., Montfort W. R. (2008) Binding of YC-1 or BAY 41-2272 to soluble guanylyl cyclase induces a geminate phase in CO photolysis. J. Am. Chem. Soc. 130, 15748–15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoo B. K., Lamarre I., Rappaport F., Nioche P., Raman C. S., Martin J. L., Negrerie M. (2012) Picosecond to second dynamics reveals a structural transition in Clostridium botulinum NO-sensor triggered by the activator BAY-41-2272. ACS Chem. Biol. 7, 2046–2054 [DOI] [PubMed] [Google Scholar]
- 30. Ibrahim M., Derbyshire E. R., Soldatova A. V., Marletta M. A., Spiro T. G. (2010) Soluble guanylate cyclase is activated differently by excess NO and by YC-1: resonance Raman spectroscopic evidence. Biochemistry 49, 4864–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chakravarti R., Aulak K. S., Fox P. L., Stuehr D. J. (2010) GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 107, 18004–18009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gladwin M. T. (2006) Deconstructing endothelial dysfunction: soluble guanylyl cyclase oxidation and the NO resistance syndrome. J. Clin. Invest. 116, 2330–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bellamy T. C., Garthwaite J. (2001) Sub-second kinetics of the nitric oxide receptor, soluble guanylyl cyclase, in intact cerebellar cells. J. Biol. Chem. 276, 4287–4292 [DOI] [PubMed] [Google Scholar]
- 34. Sayed N., Baskaran P., Ma X., van den Akker F., Beuve A. (2007) Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc. Natl. Acad. Sci. U.S.A. 104, 12312–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nedvetsky P. I., Meurer S., Opitz N., Nedvetskaya T. Y., Müller H., Schmidt H. H. (2008) Heat shock protein 90 regulates stabilization rather than activation of soluble guanylate cyclase. FEBS Lett. 582, 327–331 [DOI] [PubMed] [Google Scholar]
- 36. Hoffmann L. S., Schmidt P. M., Keim Y., Schaefer S., Schmidt H. H., Stasch J. P. (2009) Distinct molecular requirements for activation or stabilization of soluble guanylyl cyclase upon haem oxidation-induced degradation. Br. J. Pharmacol. 157, 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meurer S., Pioch S., Pabst T., Opitz N., Schmidt P. M., Beckhaus T., Wagner K., Matt S., Gegenbauer K., Geschka S., Karas M., Stasch J. P., Schmidt H. H., Müller-Esterl W. (2009) Nitric oxide-independent vasodilator rescues heme-oxidized soluble guanylate cyclase from proteasomal degradation. Circ. Res. 105, 33–41 [DOI] [PubMed] [Google Scholar]
- 38. Trepel J., Mollapour M., Giaccone G., Neckers L. (2010) Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 10, 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miyata Y., Nakamoto H., Neckers L. (2013) The therapeutic target Hsp90 and cancer hallmarks. Curr. Pharm. Des. 19, 347–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torregrossa A. C., Aranke M., Bryan N. S. (2011) Nitric oxide and geriatrics: Implications in diagnostics and treatment of the elderly. J. Geriatr. Cardiol. 8, 230–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller T. W., Isenberg J. S., Roberts D. D. (2010) Thrombospondin-1 is an inhibitor of pharmacological activation of soluble guanylate cyclase. Br. J. Pharmacol. 159, 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]